Abstract
XIST RNA paints and induces silencing of one X chromosome in mammalian female cells, providing a powerful model to investigate long-range chromosomal regulation. This review focuses on events downstream of the spread of XIST RNA across the interphase chromosome, to consider how this large non-coding RNA interacts with and silences a whole chromosome. Several lines of evidence are summarized which point to the involvement of repeat sequences in different aspects of the X-inactivation process. Although the “repeat genome” comprises close to half the human genome, the potential of abundant repeats to contribute to genome regulation has been largely overlooked and may be underestimated. X-inactivation has the potential to reveal roles of interspersed and other repeats in the genome. For example, evidence indicates XIST RNA acts at the architectural level of the whole chromosome to induce formation of a silent core enriched for non-genic and repetitive (Cot-1) DNA, which corresponds to the DAPI-dense Barr Body. Expression of repeat RNAs may contribute to chromosome remodeling, and evidence suggests that other types of repeat elements may be involved in escape from X-inactivation. Despite great progress in decoding the rest of the genome, we suggest that the repeat genome may contain meaningful but complex language that remains to be better studied and understood.
The inactivation of one X chromosome in mammalian females is a powerful model for epigenetic regulation in early development, involving formation of facultative heterochromatin that sweeps across an entire chromosome. Remarkably, a large non-coding RNA from the X-linked human XIST/mouse Xist gene (Brockdorff et al., 1992; Brown et al., 1992) “paints” its parent chromosome (Brockdorff et al., 1992; Brown et al., 1992; Chow et al., 2007; Clemson et al., 1996) and induces a silencing cascade throughout the chromosome territory (reviewed in Hall and Lawrence, 2003 and Heard and Disteche, 2006). A central question has now become: how does XIST RNA localize across its chromosome and impact its architecture and expression? Importantly, XIST RNA is strictly localized to its parent chromosome in cis, yet how it binds and what confers susceptibility (or resistance) of particular genes to silencing remains poorly understood.
In recent years substantial progress has been made in defining the series of heterochromatic chromatin modifications that spread across the chromosome shortly after XIST RNA first paints the chromosome (reviewed in (Heard, 2005). Evidence suggests that XIST RNA can “recruit” polycomb group proteins that induce heterochromatin modifications, such as EZH2 which methylates histone H3K27 (Schoeftner et al., 2006) or RING-1 which ubiquinates H2A (Fang et al., 2004). However, it is increasingly appreciated that genome regulation involves not only “local” changes, at the level of chromatin modifications or transcription factor/repressor binding, but also higher-order changes to nuclear and chromosome/chromatin architecture. In fact, the inactive X-chromosome exemplifies a now well-established principle of nuclear structure, that the nuclear interior is highly “compartmentalized” into non-membrane bound compartments devoted to particular functions (Moen et al., 1995); (Chubb and Bickmore, 2003); (Cremer and Cremer, 2001). The inactive X-chromosome (Xi) consistently resides in the heterochromatic compartment at the nuclear or nucleolar periphery (FIGURE 1), which electron microscopic studies have long shown is enriched for more densely packaged chromatin less active in transcription. Thus the inactive X is largely excluded from the internal euchromatic compartment that is punctuated by a number of smaller domains enriched in RNA metabolic factors, forming “hubs” of higher activity (Reviewed in: (Hall et al., 2006; Lamond and Spector, 2003; Meaburn and Misteli, 2007).
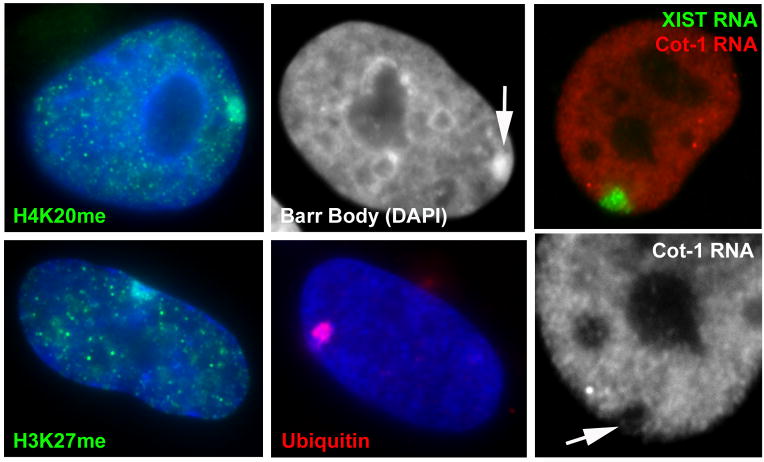
FIGURE 1. XIST RNA paints the inactive X-chromosome (Xi) altering its chromatin composition, structure, and nuclear organization.
Note that most images show the Xi in the peripheral heterochromatic compartment of the nucleus in these human somatic cells. A) Histone H4K20 methylation, and B) histone H3K27 methylation are enriched on the Xi. C) DAPI DNA stain reveals the condensed Barr Body (arrow) at the periphery of the nucleus. D) Ubiquitinated H2A K119 is also enriched on the Xi. E) XIST RNA paints the Xi, and repeat RNA (Cot-1 RNA) is present throughout the non-nucleolar nucleoplasm, but absent from the Xi (F: arrow) and other regions of heterochromatin.
The organization of the Xi within overall nuclear structure, as well as most biochemical modifications across the chromosome, can be readily visualized by light microscopy (FIGURE 1). However a greater challenge is to investigate what organizational changes may be occurring to particular sequence elements within the chromosome itself. Since some genes are known to escape X-inactivation, particularly in humans, a key question is whether those sequences are organized differently at a cytological level within the interphase chromosome territory, or whether escape from silencing is controlled at a more local level. Irrespective of how some genes escape silencing, the XIST-RNA coated chromosome undergoes large-scale structural changes to form the DNA-dense Barr Body, which is readily seen in human cells by light microscopy with DAPI staining (FIGURE 1), and corresponds to a condensed chromatin mass by electron microscopy (Rego et al., 2008). Our lab recently took two different approaches to investigate the relationship between human Xi regulation and sequence elements, one which employed molecular cytology and another using a bioinformatic word-count analysis. In both cases the findings led us to the potential roles of repeat elements, of different types, in chromosome structure and regulation.
The human genome is not a linear entity, but a complex three dimensional structure with several levels of higher-order packaging, likely influenced by what we refer to as the “fabric” of sequences underlying it. In addition, the overall perspective of the human genome is changing dramatically as it becomes evident that the meaningful information occurs at least as much in the “non-coding” DNA as in protein coding “genes”. While the importance of non-coding DNA as a source of small and large non-coding RNAs (microRNAs and LincRNAs) is now being intensely investigated (reviewed in (Koziol and Rinn, 2010), what we would term the “repeat genome” is still largely unexplored. Highly repetitive sequences comprise close to half of the human genome; in addition to the satellite sequences at centromeres, the bulk of the repeat genome is comprised of SINE elements (Alu in human, B1 or B2 in mouse), LINE elements and simple sequence repeats (reviewed in Lander et al., 2001; Smit, 1996). We share the perspective stated by Britten and Kohne (1968) over forty years ago, “A concept that is repugnant to us is that about half of the DNA of higher organisms is trivial or permanently inert.” Several years ago we began using the Cot-1 (rapidly reannealing) fraction of the genome not as a means to mask repeats from consideration, but rather as a labeled probe to broadly survey the organization and/or expression of repeats throughout nuclei and chromosomes (Hall et al., 2002a); (Clemson et al., 2006). As we consider how the non-coding XIST RNA paints a chromosome, or how some genes escape the silencing cascade induced by XIST, we will keep in mind the possible roles for the presumptive junk of the genome in chromosome structure and regulation.
XIST RNA binds and induces silencing of one X-chromosome in mammalian female cells
To balance gene dosage between male and females, one X-chromosome in female cells is almost entirely silenced, providing the preeminent model of facultative heterochromatin, with biochemical and structural changes manifest across the entire chromosome. While the mouse trophectoderm shows imprinted inactivation of the paternal chromosome, human extraembryonic tissues do not, and in both species X-chromosome inactivation (XCI) within the inner cell mass and embryo proper is random (reviewed in (Payer and Lee, 2008). Although the timing may differ slightly between human and mouse, this random XCI initially occurs as cells of the inner cell mass begin to differentiate to specific lineages, and involves mechanisms that specify counting and “choice” of which X-chromosome will be silenced in each cell, although these are only partially understood. Recent work suggested that the X-linked gene Rlm/Rnf12 has a role in regulating Xist/X-inactivation (Jonkers et al., 2009), and this transcription factor was further shown to be essential for imprinted paternal X-inactivation and thus viability of the embryo (Shin et al., 2010). Whatever the mechanism whereby XIST is initially expressed from just one homolog, the biology of how this novel chromosomal RNA interacts with and impacts a whole chromosome is fascinating, and will likely reveal fundamental insights into the structure and regulation of chromosomes in general.
Human XIST RNA is poly-adenylated and spliced to generate a ~19kb RNA expressed exclusively from the Xi (Brown et al., 1992), and silenced by methylation on Xa. XIST RNA forms a large accumulation tightly restricted to the Xi chromosome territory in interphase (Clemson et al., 1996), and in mouse an antisense termed Tsix regulates Xist expression itself (Payer and Lee, 2008). Most studies of the initiation of chromosome silencing have been done in mouse ES cells, where it was shown that shortly following Xist RNA accumulation across the chromosome, a series of chromatin modifications occur, including increased H3K27me3, H3K9me, and H2AK119ub and decreased H3K4me and H3K4ace. XIST RNA is maintained on the Xi in all adult somatic cells, where it is associated with the late replicating DAPI-dense Barr Body and contributes to faithful maintenance of the silent state. Although Xist/XIST RNA is essential to initiate X-inactivation in the embryo (Penny et al., 1996), silencing is maintained in somatic cells by the synergistic effects of multiple heterochromatic changes to the chromosome (Csankovszki et al., 2001). How this long non-coding RNA binds to and interacts with the chromosome is not clear; but at some level this must involve the sequence/structure of XIST RNA itself and its affinity for components of chromatin. However, a major question is whether XIST RNA is indifferent to the sequence of the underlying DNA. We briefly consider below current knowledge regarding RNA and chromatin protein factors, but focus primarily on evolving evidence which suggests interplay between XIST RNA and underlying chromosomal DNA, leading to the potential role of repeat sequences in chromosome regulation.
Affinity of a long non-coding RNA for its parent chromosome and potential roles for tandem repeats within Xist RNA
XIST RNA has now been studied for almost 20 years, yet only recently has there been movement towards understanding how or what regulates binding of this large RNA to the chromosome. Since XIST RNA binds across the chromosome, the underlying DNA logically plays some role in XIST RNA localization, either directly or indirectly. We have previously shown that the RNA remains surprisingly tightly bound and well localized to the chromosome territory in nuclei after histone extraction and digestion of the bulk (~95%) of chromosomal DNA (Clemson et al., 1996; Hall and Lawrence, 2003). This indicates the RNA is not localized via hybridization to chromosomal DNA, but suggests the RNA likely bridges chromatin with the residual non-soluble structural elements (scaffold) of the nucleus (model, FIGURE 2). In addition, as the RNA spreads from its site of transcription on the X and coats the rest of the chromosome, it must somehow recognize some architectural boundary of the interphase chromosome territory, since it does not promiscuously associate with the neighboring chromatin despite the intimate packaging of chromosomes within nuclei (Hall et al., 2002b). Thus, the chromosome territory likely has an underlying scaffold that somehow limits spread of Xist RNA.
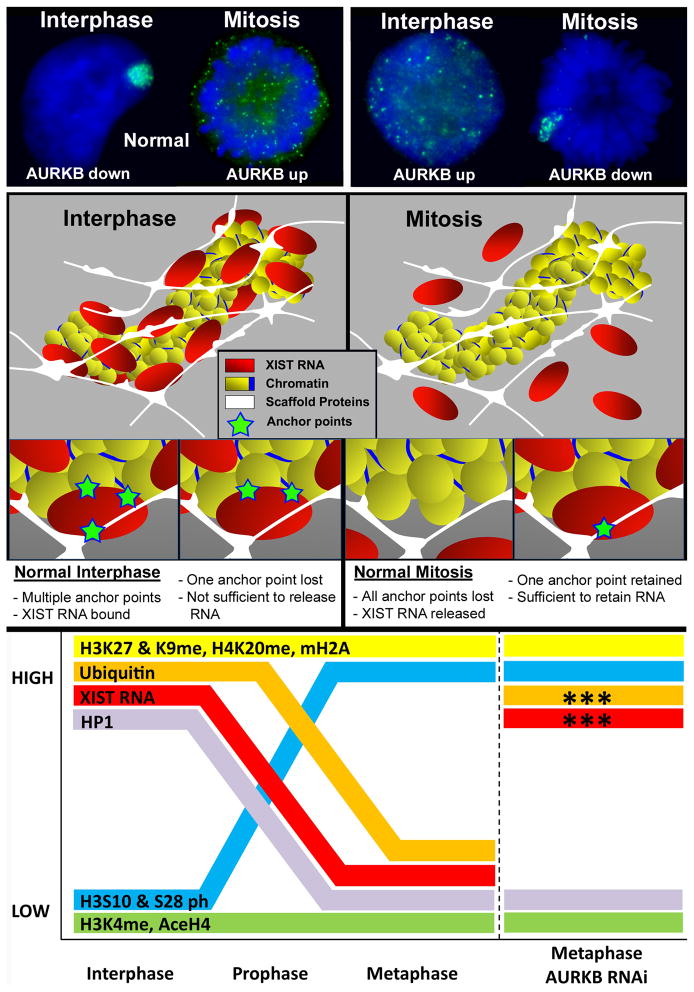
FIGURE 2. AURKB chromatin phosphorylation affects the binding of XIST RNA to multiple anchor points which we suggest bridge chromatin and insoluble nuclear structure.
A) In normal cells (left) XIST RNA paints the chromosomes territory at interphase and releases from the chromosome early in mitosis. In cells in which AURKB is manipulated (right), use of an inhibitor of PP1 (which releases repression of AURKB) causes XIST RNA to release from the interphase chromosome, whereas RNAi or inhibition of AURKB causes XIST RNA to be retained on metaphase chromosomes. B) Model of XIST RNA interaction with the interphase chromosome proposes that XIST RNA is anchored at multiple points and bridges chromatin with insoluble scaffold (matrix) proteins of the nucleus. To release XIST RNA in interphase all anchor points must be abrogated, but retention of one anchor point may be sufficient to force chromosomal retention of XIST RNA at mitosis. C) Manipulation of XIST RNA binding in live cells provides a strategy to determine which chromatin proteins or modifications most closely parallel XIST behavior. In this analysis, only histone ubiquitination followed the same pattern as XIST RNA under all conditions. Figures adapted from Hall et al., 2009.
Because the RNA has significant capacity to silence autosomal chromatin, as detailed below, it is essential that its binding be strictly cis limited. We interpret this to likely require multiple redundant anchor points and cooperative players to assure strict and stable XIST RNA binding. Both human XIST and mouse Xist RNAs contain multiple regions of tandem repeats (identified as repeat A, B, C, D). Studies of Xist RNA transgenes in mouse ES cells suggest that the A-repeat region is required for Xist RNA to silence chromatin (Wutz et al., 2002) and is important for PRC2 (EZH2) recruitment to enact H3K27 methylation (Zhao et al., 2008). With respect to RNA localization, however, (Wutz et al., 2002) reported that a number of different regions along the length of the transcript contribute to its proper chromosomal localization. These sites do not exhibit a common motif and are functionally redundant, suggesting multiple low affinity cooperative binding sites for XIST RNA binding. This finding is consistent with other evidence from our lab (below) that there are likely multiple anchor points involving distinct modifications to chromosomal proteins (Hall et al., 2009).
Studies from the Strauss and Lee labs have found that introduction of PNA or LNA oligos respectively, to Xist in living cells can dislodge Xist RNA from the chromosome (Memili et al., 2001) and (Sarma et al., 2010). Interestingly, both studies report that oligos which target the C repeat of Xist are more effective in disrupting its localization, although this happens most rapidly when the LNA oligos are used. This suggests that the C repeat is important in Xist RNA binding, however an involvement of other sequences cannot be ruled out because some parts of Xist RNA may be inaccessible to oligos in live cells. Nonetheless, this is an intriguing approach for future studies.
AURKB regulates XIST RNA localization independent of mitosis, allowing manipulation of XIST RNA interphase binding or mitotic release
Although many labs have attempted to identify proteins that specifically interact with XIST RNA using standard biochemical approaches, identification of proteins that bind and localize XIST RNA has long frustrated researchers in this field. Evidence indicates that Xist RNA can recruit certain chromatin modifiers to Xi, as shown for EZH2/PRC2 (Schoeftner et al., 2006; Fang et al., 2004), but identification of proteins that localize or regulate Xist RNA has been a challenge. One study implicated the tumor suppressor, BRCA1, as localizing across the Xi to support XIST RNA binding (Ganesan et al., 2002); however, our group found that BRCA1 associates with replicating satellite heterochromatin on many chromosomes but does not “paint” the Xi or play a direct role in XIST localization (Pageau et al., 2007b; Pageau and Lawrence, 2006). Interestingly, loss of the Barr Body and XIST RNA is common in several types of aggressive breast and other tumors, and there is now consensus that this primarily reflects a broader effect of BRCA1 or other tumor suppressor loss on genetic and epigenetic instability of cells (Pageau et al., 2007a). Very recently, Hasegawa et al (2010) appear to have had success in identifying an Xist RNA binding factor, implicating the matrix protein hnRNPu/SAF-A in Xist RNA binding (Hasegawa et al., 2010). This finding followed on an earlier report showing that SAF-A localized to Xi with XIST RNA (Helbig and Fackelmayer, 2003).
Since biochemical analysis of such a large, tightly chromatin bound RNA has been difficult by standard extraction-based approaches, we pursued a different approach to manipulate the binding or release of XIST RNA directly within living cells. This strategy built upon a significant clue from our earlier demonstration that XIST RNA releases from the mitotic Xi during early prophase in human cells, and slightly later in mouse (Clemson et al., 1996; Hall et al., 2009). Upon mitotic release, the RNA is visible as bright punctate dots throughout the cytoplasm (FIGURE 2), and it is re-synthesized in early G1 daughter cells (Clemson et al., 1996). Tests of inhibitors that mimic mitotic chromatin modifications implicated an indirect role of protein phosphatase 1 (PP1), where only PP1 inhibitors released XIST RNA to disperse throughout the nucleoplasm at interphase, similar to its punctuate distribution at mitosis (FIGURE 2). PP1 normally inhibits AURKB at interphase, and AURKB normally localizes across the chromosome arms at prophase (when XIST RNA detaches) where it phophorylates H3 and other chromatin proteins and releases HP1. Our results indicated that the improper activation of AURKB in interphase caused H3Ser10 phosphorylation of interphase chromatin concomitant with release of the RNA, whereas RNAi depletion of AURKB prevented the normal release of XIST RNA from condensed mitotic chromosomes (FIGURE 2). Thus, the release of XIST RNA at mitosis is independent of chromosome condensation but appears to involve chromatin modifications controlled directly or indirectly by AURKB mediated phosphorylation. AURKB, which is highly over-expressed in many cancers, is known for its role at the mitotic spindle and centromeres, but Hall et al. (2009) surprisingly identified AURKB as a key player in regulation of RNA binding to heterochromatin. Although H3Ser10 and HP1 (regulated by AURKB phosphorylation) may be involved, phosphorylation of H3Ser10 alone at interphase was not sufficient to release XIST RNA. However, as shown in the model in FIGURE 2, these and above results suggest XIST RNA binding likely involves multiple distinct anchor points. As summarized in FIGURE 2, the ability to manipulate XIST RNA in vivo (to remain bound at metaphase or to release at interphase) provides a new approach to further advance the thorny question of what regulates XIST RNA binding and identify the biochemical changes most closely linked to XIST RNA behavior (see FIGURE 2).
XIST RNA can repress most autosomal chromatin but is not indifferent to DNA sequence content
A remarkable aspect of Xist/XIST chromosome biology is that this novel RNA can silence autosomal chromatin, as indicated by silencing of autosomal chromatin in X;autosome translocations (White et al., 1998) which is associated with XIST RNA (Hall et al., 2002a), as well as silencing by Xist/XIST transgenes inserted into autosomes (Lee and Jaenisch, 1997); (Hall et al., 2002a). While it was suggested that mouse ES cells which had begun to differentiate were no longer competent to enact chromosome silencing in response to Xist transgenes (Wutz et al., 2002), we and our collaborators found that human XIST transgenes could inactivate autosomal chromosomes in somatic cell lines (Hall et al., 2002a); (Chow et al., 2003); (Chow et al., 2007). In fact, the most “robust” example of a condensed Barr Body currently in our lab is actually a Chromosome 4 Barr Body, carrying a cosmid XIST transgene (FIGURE 1) in HT1080 fibrosarcoma cells. However, since the HT1080 and 293 cell lines used have neoplastic origins, we suggested that such cells may have greater epigenetic plasticity (Hall et al., 2002a), which the Wutz lab (Agrelo et al., 2009) recently reported involves a requirement for the SATB1 protein.
Despite the finding that XIST RNA is retained in nuclear structure after digestion of chromosomal DNA, numerous observations indicate that XIST/Xist RNA is not indifferent to the chromosomal DNA sequence. Foremost among them is that in humans many genes escape X-inactivation, to varying degrees (Carrel and Willard, 2005). Additionally, in mouse mitotic cells, before Xist RNA detaches, it appears that the RNA does not coat the centromeric region, and the RNA selectively detaches to form a discrete banding pattern (Smith et al., 2004), similar to observations on vole chromosomes (Nesterova et al., 2002). Although transgene studies generally have not thoroughly analyzed the extent of chromosomal silencing, studies of human X;autosome translocations demonstrated that XIST RNA shows a compromised affinity for autosomal chromatin, as it is not maintained long-term across the full autosomal material (Hall et al., 2002a; Hall et al., 2002b). This is consistent with earlier clinical studies of patient translocations showing variations in the spread and fidelity of autosomal-inactivation, leading Mary Lyon to suggest that LINE repeats may play a role in propagation and binding of silencing (Lyon Repeat Hypothesis) (reviewed in (Lyon, 2003). Recently the Brockdorff lab has found further evidence that domains that are low in gene density and high in L1 density are more efficiently silenced (Tang et al., 2010). This impact of sequence content is exemplified by the presence of two XIST transgenes integrated in the same cells, where XIST RNA localizes to the chromosome at one site and not the other (Hall et al., 2002a); (Hall and Lawrence, 2003). This impact of chromosomal sequence context could be either direct or indirect, as the DNA sequence must impact chromatin proteins in the region as well as potentially higher order chromatin folding elements that may be recognized by XIST RNA.
It is notable that Migeon et al., 1999 and others have reported that human XIST RNA is capable of anchoring to mouse chromosomes, even though most of the primary sequence of human and mouse XIST RNA is not well conserved (Migeon et al., 1999); (Brockdorff et al., 1992; Brown et al., 1992). As the above studies on AURKB strongly indicate that protein modifications regulate XIST RNA binding, this suggests that human XIST RNA can recognize mouse chromatin proteins on a mouse chromosome. However, two studies (Clemson et al., 1998); (Hansen et al., 1998) showed that, in mouse human hybrid cells carrying just one human chromosome, human XIST RNA cannot localize to the human X chromosome in the otherwise mouse nucleus, indicating that mouse chromosomal proteins do not properly interact with the human chromosomal DNA to stabilize XIST RNA binding. As recently reviewed elsewhere (Minks and Brown, 2009), Xist/XIST transgene studies to date are limited by differences in random insertion sites, copy numbers, and epigenetic factors of different cells. However, use of targeted, single-copy XIST transgenes in the future has great potential to further elucidate fundamental aspects of XIST RNA and chromosome biology.
Bioinformatic analysis implicates different types of repeat elements in chromosome silencing and escape
X-chromosome inactivation provides a singular opportunity to investigate the potential relationship between sequence elements and the structural and functional transformation of essentially a whole chromosome. Since at least hundreds or thousands of XIST transcripts bind across the chromosome, sequence motifs involved would likely be highly represented throughout the genome, potentially making it difficult to discriminate them from putative “junk”. It was long ago suggested that repetitive sequences may be involved in promoting chromosome inactivation (Gartler and Riggs, 1983), with LINE elements a suspect, as noted above (for review, see (Lyon, 2003). Using bioinformatic sequence analysis, Bailey et al. reported that the human X-chromosome has a ~1.7 fold higher level of L1 LINE elements than the autosomal average except in the region at Xp22 that escapes chromosome inactivation (Bailey et al., 2000). While this correlation is consistent with an involvement of L1 elements in promoting X-inactivation, L1 may also have accumulated on the X due to its lower meiotic recombination in all but the pseudoautosomal region (which also lacks L1 enrichment). Other studies of canonical repeats concluded that L1 elements are either not likely involved (Chureau et al., 2002); (Ke and Collins, 2003) or may not be solely responsible (Ross et al., 2005).
Rather than focus on candidate elements, we took an open, unbiased bioinformatic search for any motifs that are abundant, widely distributed, and specifically enriched on the X chromosome. Additionally, analyses were performed with and without masking for known interspersed repeat families (e.g., LINEs, SINEs, and LTRs), since these copious elements may well contribute, but would also likely obscure other repeated motifs. Using a linguistic approach, the number and distribution of all nine base-pair words in genomic sequence of all individual human chromosomes was examined. In addition, we focused on the X chromosome as comprising two distinct segments: XE, a ~10 Mb region at Xp22.3 (FIGURE 3 & 4) that includes the pseudoautosomal region and more fully escapes X inactivation (Carrel and Willard, 2005), and XS, the remainder of the chromosome which is largely silenced on Xi. While there are some genes scattered throughout XS that partially escape silencing in some cell types, XE is a large, unique chromosomal domain that is wholly resistant to X-inactivation, unlike autosomal chromatin which has substantial capacity for inactivation.
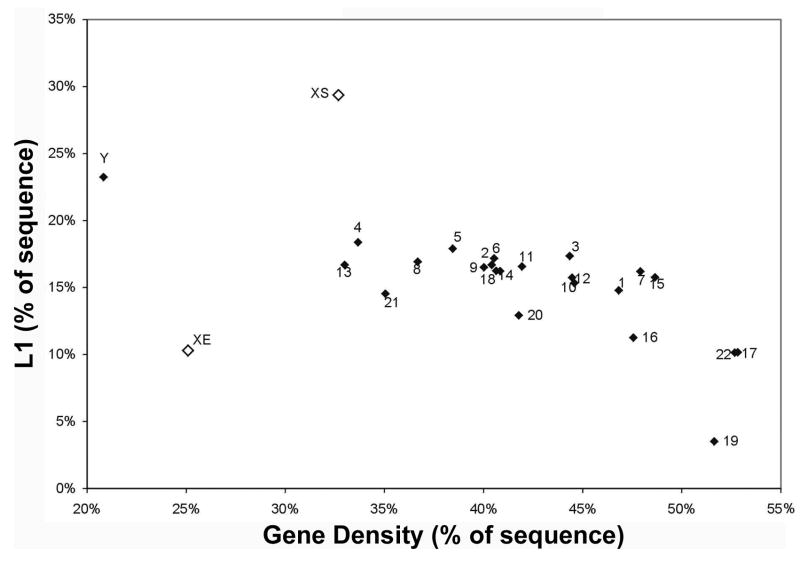
FIGURE 3. L1 LINE density versus gene density across all human chromosomes and the XE escape domain.
The silenced region of the X-chromosome (XS) has the highest LINE 1 (L1) density compared to each chromosome in the human genome as well as to the escape region of the X (XE). Note that autosomes show significant variations in the density of L1 elements, but this data shows the XS has significantly more than the most enriched autosome (Chr 4) which has similar gene density. Since LINE content is known to correlate with gene density, it was important to show that the difference is not simply explained by gene density. This data also shows that the Y chromosome is more enriched in L1s than autosomes, consistent with the possibility that some of the evolutionary accumulation of LINE elements on the sex chromosomes may have to do with their lower recombination frequency. Figure reproduced from McNeil et al., 2006.
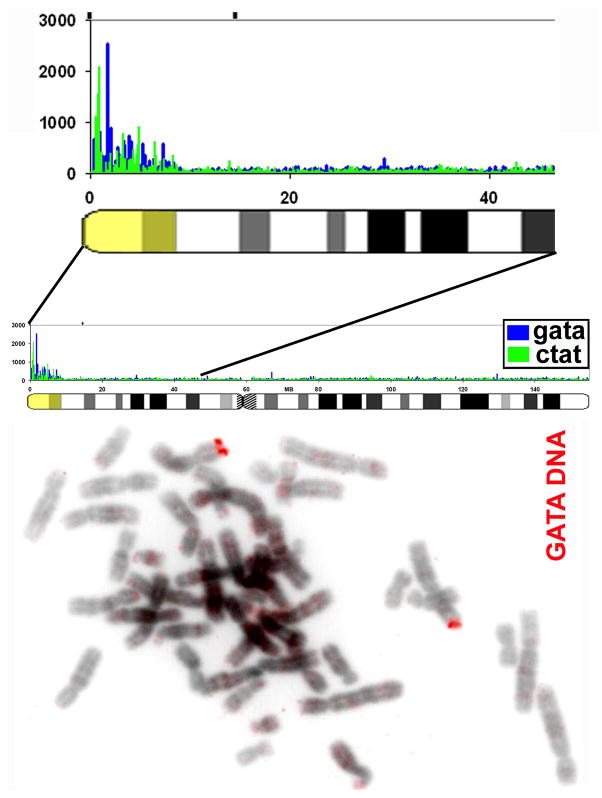
FIGURE 4. The pseudoautosomal region, which fully escapes inactivation on the Xi, exhibits a striking 11 fold enrichment in the GATA repeat sequence.
Right) Shows the distribution of GATA repeats (and CTAT) along the X chromosome determined by word count analysis, and a close-up view of the pseudoautosomal and XE region that escapes inactivation (yellow). Importantly, small repeats of (GATA)n were widely dispersed at very many sites throughout this ~10 Mb segment, and were not found in large blocks. Left) DNA FISH using a GATA DNA oligo probe labels the pseudoautosomal region of the X-chromosome strongly, confirming that it is unique in the genome for this striking sequence feature.
This analysis revealed several substantial new features of X chromosome sequence content. First, we confirmed that L1 is enriched on XS, but extended this to show that this enrichment on X is distinct from all individual autosomes (not just the autosomal average) (FIGURE 3). This was important since individual chromosomes can vary substantially; for example, gene rich Chr 19 is especially depleted in L1 elements in contrast to Chr 4. Notably, the rest of our findings identified differences in simple sequence repeats, which are typically excluded from such analyses. Results showed that the dinucleotide repeats [AT]n, [AC]n, and [AG]n are significantly enriched across the X chromosome compared to autosomes. Intriguingly, these repeats have the property of being able to form unusual DNA structures, which potentially could contribute to the regulation of facultative heterochromatin.
Most importantly, this analysis uncovered a dramatic difference in the content of small simple repeats scattered throughout the whole region. As shown in FIGURE 4, a striking enrichment (> 10 fold) of (GATA)n repeats distinctly marks the 10 Mb segment at Xp22 that escapes inactivation, which is confirmed by FISH with an oligo GATA probe, and which, importantly, is also seen in other eutherians (McNeil et al., 2006). These findings suggested a new paradigm whereby a regional escape from X-inactivation in a large chromosomal domain may be due to the presence of elements that prevent heterochromatinization, rather than simply lack of elements that promote it. The GATA repeats are clearly a marked and conserved feature dispersed at many sites throughout the “fabric” of this large chromosomal segment; in fact, further analysis showed that no other 10 Mb chromosomal segment in the genome showed such a striking enrichment for any 9-mer word (McNeil and Lawrence, unpublished). Thus this strongly suggests that the GATA repeats are involved in the unique biology of this region, in either escape from silencing or potentially the obligatory meiotic recombination of this region in the XY body, or both (McNeil et al., 2006). Recent literature provides other examples in which gene regulation appears to be coordinate across a chromosomal domain, such as in hESC (Li et al., 2006), or silencing of a tumor suppressor gene in a band-sized chromatin domain (Frigola et al., 2006). We suggest that the broader sequence context of a chromosomal region may increasingly prove important in gene regulation, and the chromosomal domain may in part be defined by the repeated motifs or “words” that populate it.
While (GATA)n repeats clearly mark this unique chromosomal domain that more fully escapes silencing, GATA enrichment is not seen for the individual genes that partially or more variably escape inactivation throughout the rest of XS (McNeil et al., 2006). These may be regulated by a distinct mechanism, since in mouse the Jarid1C gene (and two other individual genes that escape silencing) were flanked by CTCF binding sites (Filippova et al., 2005), and Li and Carrel (2008) further showed this was an intrinsic property of the Jarid1C locus. Carrel et al. (2006) and Wang et al. (2006) each published that computer profiling could recognize motifs that predict genes that escape silencing even outside the XE region. While no discrete consensus motif was identified, this provides further evidence that XIST RNA is not indifferent to chromosomal sequence context.
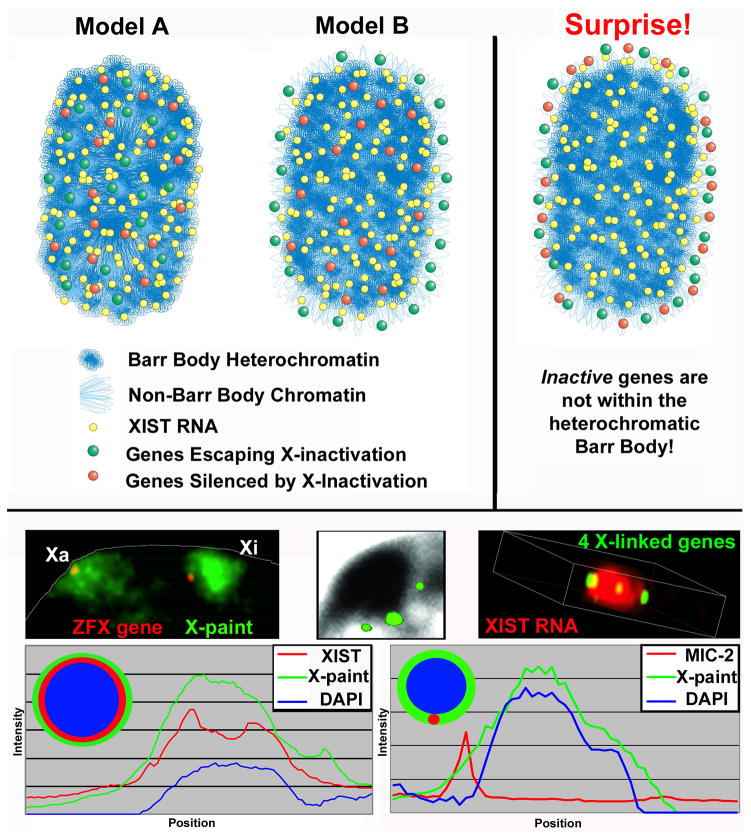
A Surprising Organization of the Xi: 14 genes position outside the Barr Body in the outer rim of the XIST RNA territory independent of activity
In addition to the bioinformatic studies above, our lab also took a molecular cytological approach to address whether genes that escape X-inactivation are organized differently at a cytological level. The Xi clearly exhibits a striking “architectural” change manifest by formation of the heterochromatic Barr Body, which is most obvious in human cells (FIGURE 1), but also visible in mouse cells. We hypothesized that genes which escape silencing might lie outside the XIST RNA territory and avoid being engulfed and silenced in the condensed Barr Body (Model B, FIGURE 5), or alternatively, be within the Barr Body but controlled more locally (Model A, FIGURE 5). The Barr Body has clear features of heterochromatin and, as further evidenced below is transcriptionally inactive, therefore it was logically and universally assumed that the Barr Body contained the silenced genes of the Xi. Surprisingly, however, through direct analysis of 14 genes (six that escape and eight that are silenced), we found that not only are the escape genes just outside the Barr Body, but the silenced genes are as well, a result we never anticipated (Last model, FIGURE 5). All 14 genes localized overwhelmingly on the rim of the XIST RNA territory (FIGURE 5) and, moreover, outside the DNA-dense Barr Body. Initially when we saw this result with the first few genes examined, we hesitated to publish these counter-intuitive results. However, we analyzed more loci in numerous experiments using different fixation methods and this ruled out artifacts due to hybridization efficiency, probe penetration, and 3-D imaging. This gene organization within the overall Xi architecture became clearer as we better defined three distinct ways to examine the Xi: the DNA-dense Barr Body, the XIST RNA territory, and the X-chromosome DNA territory (Clemson et al., 2006). While these are all used as ways to visualize Xi, they are not equivalent. As shown by the linescan analysis FIGURE 5, the DAPI dense Barr Body does not in fact encompass the entire X-chromosomal DNA territory, but occupies ~65% of its interior. Similarly, as previously indicated (Clemson et al., 1996), the XIST RNA territory is slightly smaller (80–85%) than the DNA territory, but larger than the Barr Body. Thus, the Barr Body is not the Xi, but a densely packaged core within Xi. Quantitative results showed that these gene-coding loci positioned predominantly, though not entirely, at the border or outer rim of the XIST RNA territory regardless of whether they escape or are silenced. While in a minority of cells the gene signal appears more internal within the territory, these frequently localized to an invagination or gap in the Xi DNA staining.
FIGURE 5. An unanticipated finding that inactive X-linked genes are predominantly not located within the heterochromatic Barr body (BB).
A) Two theoretical models of gene organization on Xi (left) and the model supported by empirical results (right). Initially, it was presumed that either (Model A) escape from silencing is controlled at the local level and all genes are located within the BB or (Model B) genes escape inactivation by positioning outside XIST RNA and the Barr Body with silenced genes well within the heterochromatic BB. However, the data supports the surprising result that all genes positioned with high probability on the outer border of the XIST RNA territory, outside the Barr Body. B) Examples of gene organization relative to the Xi and Xa DNA chromosome territory (left), Barr Body (middle), and XIST RNA (right). Left shows one gene at a peripheral position relative to the DNA territories of Xi and to a lesser extent Xa. Note: the densely packed interior region of the Xi is evident in contrast to a more extended conformation of the Xa. Middle shows three X-linked genes located just outside the DAPI dense BB. Right shows a 3D still-shot of a video showing four X-linked genes at the outer edge of the XIST RNA paint (from Clemson et al., 2006). C) The linescan on the left, through the Xi of one nucleus, shows relative sizes of the BB (DAPI), the XIST RNA painted region, and the X-chromosome DNA territory (X-paint). The BB comprises 62% of the X-territory and 87% of the XIST RNA paint, while the XIST RNA region covers 80% of the X-territory. The linescan on the right shows the position of an X-linked gene (MIC-2) just outside the BB and at the outer edge of the X DNA territory. Adapted from Clemson et al., 2006.
A paper from the Heard lab included analysis of six X-linked genes in the mouse; while these findings do not disagree with those of Clemson et al. (2006), initially it may appear that they do (Chaumeil et al., 2006). Chaumeil et al. studied Xi genes during the initial formation of the Xi in mouse ES cells, and reported that the genes moved inward from a more distal location, relative to the Xist RNA territory, as X-inactivation proceeds. In this case, shift to a more “internal” location was apparently in comparison to the much looser chromosome packaging in ES cells (where genes can reside outside even the visible X DNA territory). In the discussion, Chaumeil et al. refer to the propensity of genes to be more peripheral in the Xi territory as shown by Clemson et al. (2006) and then state “It should be noted that all of the X-linked gene we have examined so far tend to be fairly peripherally located on the X-chromosome territory, whatever their status, as in the study by Clemson et al., (2006).” Thus in this study of the mouse, six genes (regardless of silencing) remained in the periphery of the territory and not in the center, consistent with our demonstration of gene organization on the human Xi in somatic cells.
In addition to this surprising organization of silenced genes, Clemson et al (2006) also concluded that, at the cytological levels examined, there was not a clear pattern of differential localization between silenced versus escape genes. Clemson et al. specifically state that a more subtle difference in packaging at the cytological level could not be ruled out, but the results indicate that escape from silencing appears to be controlled at a more “local” level of chromatin packaging or gene regulation, likely below the resolution of light microscopy. For example, it remains possible that a higher-resolution (ultrastructural) analysis of gene position relative to the dense heterochromatin of the Barr Body would show some difference in proximity of escape versus silenced genes. Chaumeil et al. reported that the Jarid1C gene which escapes inactivation had a slightly more external position in differentiating ES cells, however any difference was quite subtle and the consistency of this distinction remains to be established. Since many more genes escape inactivation in the human and the Barr Body is more readily seen, the human Xi provided an advantageous system to examine this for a larger sampling of genes, which in our hands showed for the “steady-state” of Xi of somatic cells no consistent difference in the organization of silent and escape genes within the chromosome territory.
Finally, as further evidenced below, although the silenced X-linked human genes were typically not within the core of the Barr Body, they were still positioned abutting this large block of repressed chromatin, which likely is key to their silencing. This is also reminiscent of the reported positioning of inactive loci adjacent to the large heterochromatic mouse chromocenters (Brown et al., 1997), suggested to contribute to their silencing. In addition, the localization of genes outside the Barr Body may place them in an environment more permissive to escape from inactivation, dependent upon whether local sequence context supports this, such as the possible abundance of GATA repeats (McNeil et al., 2006) or other elements discussed above.
XIST RNA impacts chromosome architecture and coats a “junk” rich core with a gene-rich outer rim
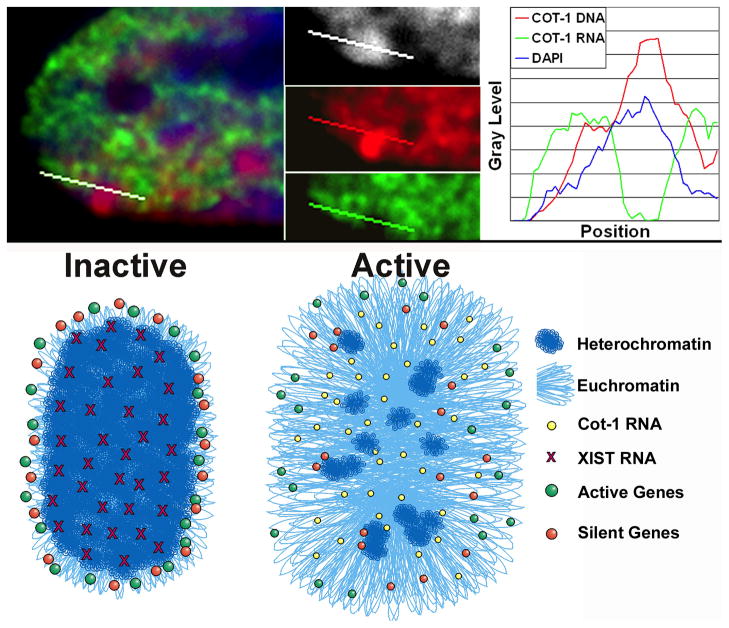
These unanticipated findings on gene organization on the human Xi led to two important related concepts, as forwarded by Clemson et al. (2006). First, since the numerous genes studied in human (with detection efficiencies of 90–99%) localized at the XIST RNA periphery and just outside the Barr Body, this begged the question: what sequences are within the Barr Body? Clemson et al. took a significant step towards addressing this by showing that hybridization to the “repeat genome”, using Cot-1 DNA as a probe; Cot-1 DNA was detected within the central regions of the XIST RNA territory and within the Barr Body (FIGURE 6), as was the X centromere in many cells. Since Cot-1 DNA is comprised mostly of interspersed LINE (L1) and SINE (Alu) sequences, this indicated that such abundant repeats may have a distinct role in Xi chromosome structure, facilitating the formation of a heterochromatic inner core that is disproportionately “non-coding” and repeat rich. Thus, Clemson et al. suggested that structural elements of the chromosome likely provide a framework that positions most protein coding genes at or near the surface of the chromosome DNA territory. A recent study by Chow et al., 2010, using a probe to LINE elements, corroborated that LINE repeats are found within the central regions of the mouse Xi DNA territory.
FIGURE 6. Results and model for the relationship of Cot-1 repeat RNA to active and inactive X chromosomes.
A) Cot-1 repeat RNA (green) is present throughout the nucleoplasm of active chromosomes but is essentially absent within the interior core of the Xi, even though Cot-1 repeat DNA (red) is detectable within DAPI dense Barr Body (white). Thus the Xi has a core of silent repeat elements. The linescans through the Xi in a normal female fibroblast quantifies the sharp distinction between Cot-1 DNA enrichment and lack of Cot-1 RNA. B) Model for the loosely peripheral organization of genes on Xa, which on Xi becomes more striking and surrounds a compacted and silent inner core. We suggest that Cot-1 RNA may be expressed throughout the central regions of euchromatic interphase chromosome territories even though evidence indicates the central regions have fewer genes.
Moreover, the surprising organization of genes at the peripheral rim provided an important new insight into how XIST RNA acts in chromosome silencing, in that these findings strongly indicated that XIST RNA does not act just at the local level on individual genes, as might have been anticipated, but has a more architectural relationship with the chromosome as a whole. For example, while a priori it would be plausible for XIST RNA to silence by binding near gene promoters; much of the interphase chromosome painted by XIST RNA is the DAPI-dense inner core. As XIST RNA detaches from mouse mitotic chromosomes, it produces a banding pattern because it is retained longest on gene-rich R bands, suggesting it could have a different affinity for gene-rich regions (Smith et al., 2004). However, it is the repeat rich inner core that is converted into a DAPI-dense heterochromatic ball on the Xi (similar to a mouse chromocenter) and this appears to be one of the early steps in initiation of inactivation. Thus, this indicates that as XIST RNA paints the Xi during initiation, its interaction with these copious repeats results in generation of a large (on a molecular scale) nuclear compartment in which the whole ~1–1.5 micron region is silent and devoid of RNA metabolic factors, as further discussed below.
Since most autosomal chromatin has substantial capacity to be silence by XIST RNA, it is important to note that XIST-silenced autosomal material also showed a similar organization of genes at the border of the XIST RNA territory (Clemson et al., 2006). Thus it is likely that the structural principles of chromosome organization that allow for formation of facultative heterochromatin may be shared between XIST-mediated facultative heterochromatin and other developmental mechanisms that control cell-type specific heterochromatin formation on autosomes.
A potential role of Repeat RNA expression and silencing in chromosome regulation
In 2002 we demonstrated a new method to assay silencing across an XIST-transgenic chromosome by assessing what we referred to as hnRNA (Hall et al., 2002a), but which we long suspected suggests something more profound about repeat RNAs. Instead of using cold COT-1 DNA as a competitor to suppress repeat hybridization, we used labeled COT-1 DNA as a probe hybridized to non-denatured nuclei, thereby detecting single-stranded RNAs containing repeats, which would include introns in pre-mRNAs. Rather than laborious expression assays for individual genes along the chromosome, this new approach allowed a more rapid and convenient overview of long-rang transcriptional activity, which we found nicely distinguishes the Xa from Xi (Hall et al., 2002a) and delineates heterochromatin at centromeres or the nuclear or nucleolar periphery (Tam, 2004). We discussed above the evidence for involvement of repetitive DNA in chromosome structure and regulation, however analysis of “Cot-1 RNA’ led us to suggest that interspersed repeat RNAs may be involved as well (Clemson et al., 2006). Following the spread of XIST RNA across the chromosome, the repetitive DNA at the “core” of the territory is compacted into the Barr Body, which we showed lacked Cot-1 RNA despite containing copious Cot-1 DNA. In contrast, Cot-1 RNA is robustly detected throughout the nucleoplasm associated with the autosomal and Xa chromosome territories. Clearly, the repeats within the Barr Body are silent (FIGURE 1 & 6), and XIST RNA acts on the chromosome to generate this silent compartment. Since most LINE elements are truncated and generally believed silent, one possibility is that this already silent DNA is reorganized into a recognizable structure (the Barr Body). However, we suggest an alternative hypothesis may be true: that XIST RNA’s first function is not only to structurally re-organize this non-genic DNA in this core, but also to silence it. In either case, a nuclear compartment lacking in Cot-1 RNA, RNA pol II (Clemson et al., 2006); (Chaumeil et al., 2006) and other RNA metabolic factors (Clemson et al., 1996) is formed. Creating a large Barr Body to shut off at most a few hundred genes would seem “over-kill”, but this suggests that formation of this structural compartment is key to the subsequent genic silencing, which involves regulation at the level of the whole chromosome and its nuclear organization.
These observations likely have relevance to the structure and regulation of euchromatic chromosomes. Studies of several autosomal genes reported a tendency to be loosely localized within an outer zone of the chromosome territory (reviewed in Chubb and Bickmore, 2003 and Cremer and Cremer 2001). While the number of genes in most studies was small and the extent of this debated, Clemson et al., (2006) examined this for 15 loci on Xa as well as Xi, and showed a much less pronounced organization of Xa versus Xi genes, but nonetheless a loose propensity for genes to reside in the peripheral zone on Xa as well (Model FIGURE 6). While this indirectly suggests non-genic and repeat DNA may be preferentially enriched within the central regions, Cot-1 repeat RNA appears copiously expressed throughout euchromatic chromosome territories (Clemson et al. 2006; Gomez et al., in Preparation). Thus, we hypothesize that Cot-1 RNA may include substantial repeat RNA apart from that in introns of pre-mRNAs (Clemson et al., 2006), which we interpret to be most consistent with the results of Chaumeil et al. (2006) showing the Cot-1 RNA depleted compartment forms while gene transcription foci are still expressed. Currently it is widely believed that intergenic LINE elements are mostly inert, as only a small fraction is “active” and believed capable of transcription. However, years ago early evidence suggested expression of repeated sequences in cell type specific patterns (Britten and Kohne, 1968); (Davidson and Britten, 1979), as further evidenced recently by Faulkner et al. (2010), and by earlier evidence that murine B1 and B2 RNAs are involved in the stress response (Williams et al., 2004); (Allen et al., 2004).
More recently, Chow et al., (2010) reported that specific active full-length LINE elements are transiently upregulated as X-inactivation initiates in mouse ES cells. They interpret this to suggest that particular active L1 elements influence the regulation of a nearby gene which escapes silencing. These interesting findings suggest some role of particular L1 loci in regulation of specific neighboring genes, but this is distinct from the concept discussed above regarding the broader set of abundant repeats distributed throughout the chromosome contributing en mass to the regulation of the heterochromatic versus euchromatic status of a chromosome or chromosomal domain.
A word of caution about interpreting the COT-1 RNA assay as an indicator of chromosome silencing
The COT-1 RNA assay (Hall et al., 2002) has been widely adopted as an effective tool to delineate Xi or other heterochromatin, and in many cases this assay has been used to good effect. However, we believe it should be applied cautiously with at least two major caveats in mind: 1) As per the discussion above, silencing of Cot-1 RNA might be distinct from silencing of protein coding genes, or even specific non-coding RNAs. If so, as we suggest, this would be both interesting and important, but nonetheless should be considered in interpreting results. Cot-1 RNA does not necessarily equal genic transcription. 2) The smaller the Cot-1 RNA “hole” the more subject to interpretation and mis-interpretation the analysis is. We have noted in recent literature that quite small Cot-1 RNA “holes” are interpreted to support important points, but there are many small areas of the nucleoplasm that have diminished Co-t1 RNA expression, and neighboring structures that may lack Cot-1 RNA (e.g. clustered heterochromatin or centromeres) may be near an XIST RNA focus but be unrelated to XIST or chromosome silencing.
The Language of the Repeat Genome
As evident throughout the above review, the power of the X-chromosome inactivation model has begun to reveal new insights into the potential roles of repeat sequences in the genome, at multiple levels. Several different types of repeats are implicated in several distinct aspects of XIST RNA/chromosome regulation, and in addition to a structural role of DNA elements, their expression in RNA may undergo complex, possibly cell-type specific changes. While this area of research is still at a very early stage and the import and mechanisms of most of these findings remains to be established, the overall lesson in our view is that the “repeat genome” may contain substantial meaningful information well beyond the widely held view of most repeats as evolutionary debris. Repeats have physical properties that make them especially well suited for contributing to chromosomal regulation. For example, McNeil et al. (2006) discuss the potential for GATA and other simple sequence repeats to form unusual DNA structures (triplex and Z-DNA). Interspersed repeats, such as LINES and SINES, have the intriguing potential to form inter-strand molecular hybrids (with DNA or RNA) which could impact higher-order chromatin folding. In addition, due to their abundance and broad “interspersed” distribution, they have greater potential to regulate chromosomal domains, not just individual genes, by recruiting or sequestering substantial quantities of chromatin modifying RNAs or enzymes. Based on the various examples evidenced above and other theoretical considerations beyond the scope of this review, we speculate that there is a “language of the repeat genome” which we have yet to learn how to read. Ultimately, however, the drive to understand the regulation of protein coding genes will require greater attention to this very major part of the genome long masked from most genomic analyses.
Acknowledgments
Our research is supported by the NIH in grants to JBL from the Institute of General Medicine, GMGM053234 and GM085548. We thank Dawn Carone and Meg Byron for reading the manuscript and Charlene Baron for help with figure processing.
References
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, Wutz A. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell. 2009;16:507–516. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci U S A. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Kohne DE. Repeated sequences in DNA: Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The Product of the Mouse Xist Gene is a 15 kb Inactive X-Specific Transcript Containing No Conserved ORF and Located in the Nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence JB, Willard HF. The Human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Hall LL, Baldry SE, Thorogood NP, Lawrence JB, Brown CJ. Inducible XIST-dependent X-chromosome inactivation in human somatic cells is reversible. Proc Natl Acad Sci U S A. 2007;104:10104–10109. doi: 10.1073/pnas.0610946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Hall LL, Clemson CM, Lawrence JB, Brown CJ. Characterization of expression at the human XIST locus in somatic, embryonal carcinoma, and transgenic cell lines. Genomics. 2003;82:309–322. doi: 10.1016/s0888-7543(03)00170-8. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Bickmore WA. Considering nuclear compartmentalization in the light of nuclear dynamics. Cell. 2003;112:403–406. doi: 10.1016/s0092-8674(03)00078-3. [DOI] [PubMed] [Google Scholar]
- Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, Eggen A, Avner P, Duret L. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Chow JC, Brown CJ, Lawrence JB. Stabilization and localization of Xist RNA are controlled by separate mechanisms and are not sufficient for X inactivation. J Cell Biol. 1998;142:13–23. doi: 10.1083/jcb.142.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci U S A. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J of Cell Biol. 2001;153:773–783. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Britten RJ. Regulation of gene expression: possible role of repetitive sequences. Science. 1979;204:1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, Mok SC, Randrianarison V, Brodie S, Salstrom J, Rasmussen TP, Klimke A, Marrese C, Marahrens Y, Deng CX, Feunteun J, Livingston DM. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111:393–405. doi: 10.1016/s0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- Gartler SM, Riggs AD. Mammalian X-chromosome inactivation. Ann Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- Hall LL, Byron M, Pageau G, Lawrence JB. AURKB-mediated effects on chromatin regulate binding versus release of XIST RNA to the inactive chromosome. J Cell Biol. 2009;186:491–507. doi: 10.1083/jcb.200811143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci U S A. 2002a;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Clemson CM, Byron M, Wydner K, Lawrence JB. Unbalanced X;autosome translocations provide evidence for sequence specificity in the association of XIST RNA with chromatin. Hum Mol Genet. 2002b;11:3157–3165. doi: 10.1093/hmg/11.25.3157. [DOI] [PubMed] [Google Scholar]
- Hall LL, Lawrence JB. The cell biology of a novel chromosomal RNA: chromosome painting by XIST/Xist RNA initiates a remodeling cascade. Semin Cell Dev Biol. 2003;14:369–378. doi: 10.1016/j.semcdb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006 doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Canfield TK, Stanek AM, Keitges EA, Gartler SM. Reactivation of XIST in normal fibroblasts and a somatic cell hybrid: abnormal localization of XIST RNA in hybrid cells. Proc Natl Acad Sci U S A. 1998;95:5133–5138. doi: 10.1073/pnas.95.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Heard E. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr Opin Genet Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Helbig R, Fackelmayer FO. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma. 2003;112:173–182. doi: 10.1007/s00412-003-0258-0. [DOI] [PubMed] [Google Scholar]
- Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, Rentmeester E, Grosveld F, Grootegoed JA, Gribnau J. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- Ke X, Collins A. CpG islands in human X-inactivation. Ann Hum Genet. 2003;67:242–249. doi: 10.1046/j.1469-1809.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Li H, Liu Y, Shin S, Sun Y, Loring JF, Mattson MP, Rao MS, Zhan M. Transcriptome coexpression map of human embryonic stem cells. BMC Genomics. 2006;7:103. doi: 10.1186/1471-2164-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. The Lyon and the LINE hypothesis. Semin Cell Dev Biol. 2003;14:313–318. doi: 10.1016/j.semcdb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- McNeil JA, Smith KP, Hall LL, Lawrence JB. Word frequency analysis reveals enrichment of dinucleotide repeats on the human X chromosome and [GATA]n in the X escape region. Genome Res. 2006;16:477–484. doi: 10.1101/gr.4627606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- Memili E, Hong YK, Kim DH, Ontiveros SD, Strauss WM. Murine Xist RNA isoforms are different at their 3′ ends: a role for differential polyadenylation. Gene. 2001;266:131–137. doi: 10.1016/s0378-1119(01)00353-5. [DOI] [PubMed] [Google Scholar]
- Migeon BR, Kazi E, Haisley-Royster C, Hu J, Reeves R, Call L, Lawler A, Moore CS, Morrison H, Jeppesen P. Human X inactivation center induces random X chromosome inactivation in male transgenic mice. Genomics. 1999;59:113–121. doi: 10.1006/geno.1999.5861. [DOI] [PubMed] [Google Scholar]
- Minks J, Brown CJ. Getting to the center of X-chromosome inactivation: the role of transgenes. Biochem Cell Biol. 2009;87:759–766. doi: 10.1139/O09-040. [DOI] [PubMed] [Google Scholar]
- Moen PT, Jr, Smith KP, Lawrence JB. Compartmentalization of specific pre-mRNA metabolism: an emerging view. Human Molecular Genetics. 1995;4:1779–1789. doi: 10.1093/hmg/4.suppl_1.1779. [DOI] [PubMed] [Google Scholar]
- Nesterova TB, Mermoud JE, Hilton K, Pehrson J, Surani MA, McLaren A, Brockdorff N. Xist expression and macroH2A1.2 localisation in mouse primordial and pluripotent embryonic germ cells. Differentiation. 2002;69:216–225. doi: 10.1046/j.1432-0436.2002.690415.x. [DOI] [PubMed] [Google Scholar]
- Pageau GJ, Hall LL, Ganesan S, Livingston DM, Lawrence JB. The disappearing Barr body in breast and ovarian cancers. Nat Rev Cancer. 2007a;7:628–633. doi: 10.1038/nrc2172. [DOI] [PubMed] [Google Scholar]
- Pageau GJ, Hall LL, Lawrence JB. BRCA1 does not paint the inactive X to localize XIST RNA but may contribute to broad changes in cancer that impact XIST and Xi heterochromatin. J Cell Biochem. 2007b;100:835–850. doi: 10.1002/jcb.21188. [DOI] [PubMed] [Google Scholar]
- Pageau GJ, Lawrence JB. BRCA1 foci in normal S-phase nuclei are linked to interphase centromeres and replication of pericentric heterochromatin. J Cell Biol. 2006;175:693–701. doi: 10.1083/jcb.200602055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for XIST in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci. 2008;121:1119–1127. doi: 10.1242/jcs.026104. [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, Sudbrak R, Wen G, Jones MC, Hurles ME, Andrews TD, Scott CE, Searle S, Ramser J, Whittaker A, Deadman R, Carter NP, Hunt SE, Chen R, Cree A, Gunaratne P, Havlak P, Hodgson A, Metzker ML, Richards S, Scott G, Steffen D, Sodergren E, Wheeler DA, Worley KC, Ainscough R, Ambrose KD, Ansari-Lari MA, Aradhya S, Ashwell RI, Babbage AK, Bagguley CL, Ballabio A, Banerjee R, Barker GE, Barlow KF, Barrett IP, Bates KN, Beare DM, Beasley H, Beasley O, Beck A, Bethel G, Blechschmidt K, Brady N, Bray-Allen S, Bridgeman AM, Brown AJ, Brown MJ, Bonnin D, Bruford EA, Buhay C, Burch P, Burford D, Burgess J, Burrill W, Burton J, Bye JM, Carder C, Carrel L, Chako J, Chapman JC, Chavez D, Chen E, Chen G, Chen Y, Chen Z, Chinault C, Ciccodicola A, Clark SY, Clarke G, Clee CM, Clegg S, Clerc-Blankenburg K, Clifford K, Cobley V, Cole CG, Conquer JS, Corby N, Connor RE, David R, Davies J, Davis C, Davis J, Delgado O, Deshazo D, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO Journal. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Bossenz M, Chung Y, Ma H, Byron M, Taniguchi-Ishigaki N, Zhu X, Jiao B, Hall LL, Green MR, Jones SN, Hermans-Borgmeyer I, Lawrence JB, Bach I. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature. 2010;467:977–981. doi: 10.1038/nature09457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Smith KP, Byron M, Clemson CM, Lawrence JB. Ubiquitinated proteins including uH2A on the human and mouse inactive X chromosome: enrichment in gene rich bands. Chromosoma. 2004;113:324–335. doi: 10.1007/s00412-004-0325-1. [DOI] [PubMed] [Google Scholar]
- Tam R, Smith KP, Lawrence JB. The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatic unlike most human telomeres. Journal of Cell Biology. 2004;167:269–279. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YA, Huntley D, Montana G, Cerase A, Nesterova TB, Brockdorff N. Efficiency of Xist-mediated silencing on autosomes is linked to chromosomal domain organisation. Epigenetics Chromatin. 2010;3:10. doi: 10.1186/1756-8935-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White WM, Willard HF, Van Dyke DL, Wolff DJ. The Spreading of X Inactivation into Autosomal Material of an X;autosome Translocation: Evidence for a Difference between Autosomal and X-Chromosomal DNA. Am J Hum Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WP, Tamburic L, Astell CR. Increased levels of B1 and B2 SINE transcripts in mouse fibroblast cells due to minute virus of mice infection. Virology. 2004;327:233–241. doi: 10.1016/j.virol.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nature Genetics V. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]