The idea of using nucleic acid molecules as therapeutic agents was conceived in the 1970s with the development of antisense strategies. Antisense compounds are single-stranded nucleic acids that, in principle, disrupt the synthesis of a targeted protein by hybridizing in a sequence-dependent manner to the mRNAs that encode it. The mechanism of inhibition by nucleic acid aptamers is fundamentally different. Aptamers are single-stranded nucleic acids that directly inhibit a protein’s function by folding into a specific three-dimensional structure that dictates high-affinity binding to the targeted protein.
Through iterative in vitro selection techniques, aptamers can be generated that bind essentially any protein (or small molecule) target. A high-affinity, specific inhibitor can theoretically be made to order, provided that a small quantity of pure target is available. Because they inhibit the activity of existing proteins directly, aptamers are more similar to monoclonal antibody or small molecule drugs than to antisense compounds, and this property greatly increases the number of clinical indications that are potentially treatable by nucleic acid–based compounds. Aptamers have been generated against a wide variety of targets, a complete discussion of which is beyond the scope of this article (for review, see ref. 1).
As with any molecular therapeutic approach, the inhibitor is only as effective as the target is important. Through their ability to specifically inhibit a molecule of interest, aptamers have already proven useful as reagents for target validation in a variety of disease models. The next step, therapeutic utility, will depend on the efficacy of these novel compounds in humans. In this Perspective, we will focus on the development and status of aptamers as therapeutic molecules.
Nucleic acids as direct antagonists of protein function
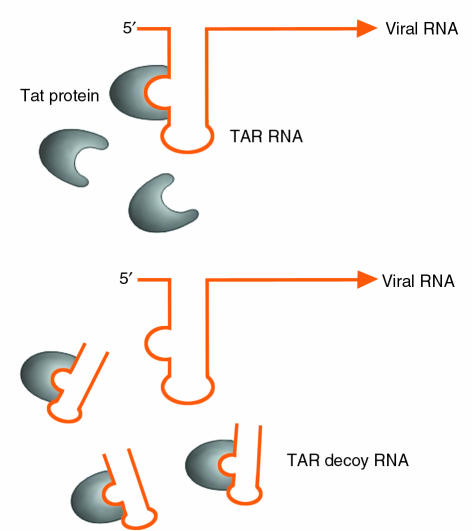
Many viruses, including HIV, have evolved small, structured RNA sequences to recruit viral and host cell proteins to perform essential functions in viral replication. The HIV virus, for example, employs the TAR and RRE RNA sequences to recruit the essential viral regulatory proteins tat and rev, which aid in induction and regulation of viral gene expression. The use of small, structured RNAs to inhibit the activity of viral proteins was first explored employing a short transcript corresponding to the TAR RNA sequence (2). This “decoy” RNA competes with the virally encoded TAR sequence for binding to tat (Figure 1). By sequestering the tat protein from the real TAR RNA sequence present in the HIV viral RNA, the decoy RNA can inhibit HIV RNA transcription and thus reduce viral replication. Expression of these TAR decoys in CD4+ T-cell lines and in bulk populations of transduced immortalized T lymphocytes renders these cells highly resistant to HIV replication. These initial studies not only demonstrated that such RNA decoys could be used as antiviral agents, but in a more general sense, suggested that short RNA molecules could bind to specific proteins and inhibit their activity. Recently, the use of the RRE decoy to render cells resistant to HIV infection has been translated to the clinic, where its safety, feasibility, and efficacy are being evaluated in pilot clinical trials following retroviral-mediated gene transfer into CD34+ cells from the bone marrow of HIV-1 infected pediatric patients (3).
Figure 1.
Decoy RNA corresponding to the TAR RNA sequence competes with the viral encoded sequence for binding of tat protein. By sequestering the tat protein from the real TAR RNA sequence present in the HIV viral RNA, the decoy RNA can inhibit HIV RNA transcription and thus reduce viral replication.
Designer nucleic acid antagonists
Iterative in vitro selection
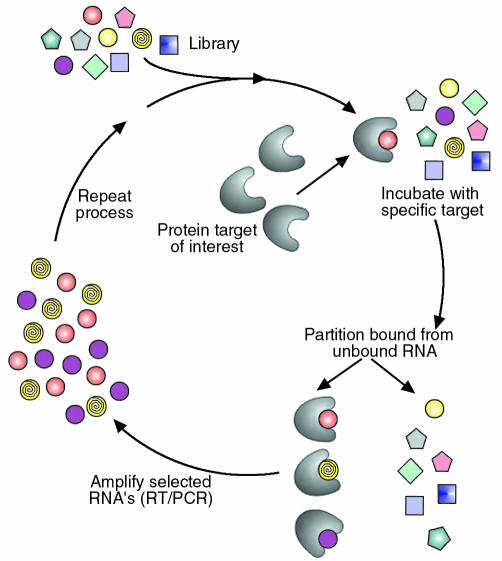
Concurrent with the advent of RNA decoys was the demonstration by several laboratories that iterative in vitro selection techniques could be employed to isolate, from randomized RNA libraries, short RNA ligands to a wide variety of proteins and small molecules (4, 5). The basic approach to such in vitro selection is outlined in Figure 2 and was named SELEX (systematic evolution of ligands by exponential enrichment) by Tuerk and Gold. In principle, SELEX can be considered an extremely powerful purification method in which very rare binding activities (with frequencies of 1 in 1011 to 1 in 1013) are isolated by affinity purification from a large combinatorial library. The starting point for the in vitro selection process is a combinatorial library composed of single-stranded nucleic acids (RNA, DNA, or modified RNA) usually containing 20–40 randomized positions. Randomization creates an enormous diversity of possible sequences (e.g., four different nucleotides at 40 randomized positions gives a theoretical possibility of 440 or ∼1024 different sequences). Because short single-stranded nucleic acids adopt fairly rigid structures that are dictated by their sequences, such a library contains a vast number of molecular shapes or conformations. To isolate high affinity nucleic acid ligands to a given target protein, the starting library of nucleic acids (in practice ∼1014 to 1015 different sequences) is incubated with the protein of interest. Nucleic acid molecules that adopt conformations that allow them to bind to a specific protein are then partitioned from other sequences in the library that are unable to bind to the protein under the conditions employed. The bound sequences are then removed from the protein and amplified by reverse transcription and PCR (for RNA-based libraries) or just PCR (for DNA-based libraries) to generate a reduced complexity library enriched in sequences that bind to the target protein. This library is then transcribed in vitro (for RNA-based libraries), or its strands are separated (for DNA libraries) to generate molecules for use in the next round of selection. After several rounds (usually 8–12), which are typically performed with increasing stringency, the selected ligands are sequenced and evaluated for their affinity for the targeted protein and their ability to inhibit the activity of the targeted protein in vitro.
Figure 2.
An overview of the SELEX process. A starting library of nucleic acids is incubated with the protein target of interest. Molecules that bind to the target protein are then partitioned from other sequences in the library. The bound sequences are then amplified to generate a library enriched in sequences that bind to the target protein.
General properties of aptamer compounds
Affinities of aptamers for the targeted proteins tend to be very high, with typical dissociation constants ranging from low picomolar (1 × 10–12 M) to low nanomolar (1 × 10–9 M). As in vitro selection techniques have improved, the generation of aptamers with subnanomolar affinities for the target has become increasingly common. These affinities are similar to those measured for interactions between monoclonal antibodies and antigens. However, since the dissociation constants measured for aptamer-target proteins are true affinities, reflecting a bimolecular interaction in solution, they are more accurately compared to the affinities of Fab fragments for their target antigens. On average, the affinities of aptamers for a targeted protein are stronger than is typical for interactions between Fab fragments and their target antigens (1).
High-affinity nucleic acid–protein interactions require specific complementary contacts between functional groups on both the nucleic acid and the protein. Because the specific three-dimensional arrangement of complementary contact sites that mediate the protein-aptamer interaction are unlikely to be recapitulated in other proteins, aptamers are generally specific for their targets. For example, aptamers have been generated that are capable of discriminating between isoforms of protein kinase C that share a high degree of identity (6). Modified-RNA aptamers to coagulation factor VIIa (FVIIa) exhibit a greater than 500-fold specificity for FVIIa relative to coagulation factor Xa and greater than 1000-fold relative to coagulation factor IXa, although these proteins share a common set of structural domains (7). Employing subtractive selection strategies can yield aptamers with even greater ability to discriminate between the target and related nontargeted proteins (8, 9). Furthermore, because the selection process is performed in vitro, schemes to improve the specificity of a given aptamer for the target, or to direct the binding of the aptamer to a particular site on the target, are only limited by knowledge of the target and the investigator’s imagination.
Translation of aptamers from binding agents into potential therapeutic compounds
Postselection compound optimization
The completion of the aptamer selection process typically yields a high affinity and specific antagonist of the targeted protein. Several postselection optimization steps generally must be performed to translate a molecule from an in vitro antagonist to a molecule that can be tested for pharmacologic effect in animals or used in vivo in target-validation studies. Chemical synthesis is required to produce the quantities of aptamer needed for most if not all in vivo experiments. Currently, for efficient and cost-effective chemical synthesis, the size of an aptamer must be reduced to fewer than 40 nucleotides, from a starting molecule of approximately 80 to 100 nucleotides. A number of strategies can be used to determine the minimum-sized aptamer that retains high affinity for the target. Fortunately, unlike peptides isolated from structurally constrained peptide-based combinatorial libraries, most aptamers can be readily reduced in size with minimal loss of affinity for the target protein. Depending on the indication, once an aptamer suitable for chemical synthesis has been generated, additional modifications may need to be made to the aptamer to enhance its bioavailability or delivery. Ultimately, the postselection modification of an aptamer is an empirical process, with each alteration requiring retesting to determine the effects of the modification on the affinity and inhibitory activity of the aptamer.
The bioavailability of an aptamer is determined by two key properties of the molecule — stability in biologic fluids and systemic clearance. The stability of a nucleic acid in plasma is determined by its backbone composition. The in vitro half-life of a typical RNA oligonucleotide in plasma is a few seconds, while that of a typical DNA oligonucleotide is 30 to 60 minutes. The plasma stability of an RNA oligonucleotide can be increased by substitution of ribonucleotides with 2′-amino, 2′-fluoro, or 2′-O-alkyl nucleotides (10, 11). Modified-RNA oligonucleotides containing these substitutions are quite stable in plasma, with in vitro half-lives in the 5 to 15 hour range. Furthermore, because 2′-amino or 2′-fluoro CTP and UTP can be readily incorporated into RNA by in vitro transcription, these backbone modifications can be introduced into the combinatorial library at the outset of the selection process (12, 13). Even when the composition of the starting library has been chosen judiciously, the plasma stability of an aptamer may need to be further enhanced after selection. An aptamer can be protected from exonuclease degradation by capping its 3′ end (11). Resistance to endonuclease degradation can be further increased by additional substitution of ribose and deoxyribose nucleotides with modified nucleotides or nonnucleotide linkers where possible. Most aptamers can be heavily modified after selection to yield molecules that are very stable in plasma; however, determining the positions within an aptamer that are amenable to such modification is not a trivial task.
A minimized aptamer 25 to 40 nucleotides in length corresponds to a molecular weight of about 8,250–13,200 kDa. Due to this small size, aptamer compounds are cleared from animals via the kidneys in minutes. Therefore, for molecules with even modest plasma stability, the in vivo plasma half-life is governed by the rate of clearance from the animal. The clearance rate of an aptamer can be rationally altered by increasing its effective molecular size, such as by the site-specific addition of various molecular weight polyethylene glycol (PEG) moieties or other hydrophobic groups, or by attachment of the aptamer to the surface of a liposome (14, 15). Thus, depending on the indication, a given aptamer can be formulated in such a way as to have a half-life in vivo of a few minutes or several hours.
Challenges
The challenges facing aptamer technology are not entirely unique. Some valuable lessons about oligonucleotide pharmacology have been learned from antisense, ribozyme (see Usman and Blatt, this Perspective series [ref. 16]), and DNAzyme molecules (see Khachigian, this Perspective series [ref. 17]). Sequence-independent, nonspecific effects of the phosphorothioate backbones commonly used for antisense molecules have been observed at the micromolar levels required for efficacy (18). In contrast, most aptamers currently feature phosphodiester backbones with modified sugars to enhance nuclease resistance. Although comprehensive toxicity profiles for these molecules have not been published, no untoward effects have been reported in the several animal studies in the literature. Furthermore, the high affinity of most aptamers should allow for submicromolar therapeutic levels with less potential for nonspecific effects.
While the plasma pharmacokinetics of aptamers are gradually being elucidated, the efficient delivery of aptamers remains a consideration for the inhibition of intracellular targets. Methods adopted from other molecular therapeutic strategies may ultimately prove useful for the delivery of aptamers into cells, such as incorporation into liposomes or generation of high effective concentrations using various means of local delivery.
As one advantage over peptide-based inhibitors, aptamers are purported to be nonimmunogenic. Their small size and similarity to endogenous molecules theoretically makes them poor antigens. Experiments designed with the intent to elicit a humoral response to modified-RNA aptamers have failed to generate a specific response, despite the generation of responses to a variety of adjuvants employed (B.A. Sullenger et al., unpublished results). However, the presence of antibodies against nucleic acids in autoimmune diseases makes the possibility of antiaptamer antibodies one that is plausible and worthy of investigation.
The great specificity that aptamers can achieve represents both a strength and a potential weakness of the technology. Being too specific may be problematic if species crossreactivity is desired, as is often the case during the preclinical evaluation of a molecule in animal models. Obviously, the more conserved the protein, the less likely this situation is to occur. With knowledge of the target, subtractive selection schemes may direct the aptamer to a conserved portion of the molecule. Alternatively, this problem may be obviated by driving the selection with targets from different species either in parallel or alternating in series.
Finally, the expense of synthesis is not always considered in the early developmental stages of a novel therapeutic agent. While small-scale synthesis is feasible for most in vitro applications, the scale of synthesis necessary for transition of a candidate aptamer to animal or even human models may be cost-prohibitive for even a well-funded academic laboratory.
Indications
Given the current state of aptamer technology, the ideal indication for a therapeutic aptamer is an acute condition for which an important, spatially confined, extracellular target has been identified and for which no good alternative therapies exist. In such a situation, local delivery of even a small dose of aptamer may be effective, both in terms of clinical response and cost. Aptamers targeting conditions that meet some or all of these criteria have made the most progress towards clinical utility. We will discuss three indications for which aptamers have progressed to animal and/or human models. By no coincidence, all three fall under the umbrella of vascular biology. For these common clinical conditions, several plasma- or endothelium-based targets have been characterized. Drug delivery can thus be accomplished either intravascularly or locally in the setting of surgery or percutaneous vascular intervention.
Coagulation and thrombosis
The delicate balance between hemostasis and hemorrhage is maintained by a complex system of plasma, cellular, and endothelial factors. Coagulation, the normal process by which a fibrin clot is generated in response to a vascular injury, is to be distinguished from thrombosis, the pathological formation of clot in response to injury, stasis, or hypercoagulability. The latter is widespread in conditions such as acute coronary syndrome, stroke, peripheral vascular disease, and deep vein thrombosis, and can also occur in response to iatrogenic vascular injury. The clinical demand for more and better antithrombotics, which inhibit the initial formation of the platelet plug, and anticoagulants, which inhibit the cascade of reactions leading to the crosslinking of fibrin, is testimony to the importance of this process. In addition, the variety of patients and scenarios in which such agents are utilized requires an array of inhibitors with different mechanisms, properties, and toxicity profiles.
Thrombin is the most obvious target for the generation of both anticoagulant and antithrombotic compounds. Bock et al. (19) generated a 15 nucleotide DNA-based thrombin aptamer that binds thrombin with moderate affinity (apparent Kd ∼10–7 M) and can prolong the clotting time of human plasma. To take advantage of its rapid clearance (in vivo half-life of approximately 1–2 minutes), the thrombin DNA aptamer was developed largely as an anticoagulant for use in surgical indications requiring regional anticoagulation of an extracorporeal circuit. When administered by constant infusion, this molecule was successfully used to maintain the patency of an extracorporeal circuit in sheep and was substituted for heparin in a canine cardiopulmonary bypass model (20, 21). Furthermore, because of its rapid clearance, once infusion of the aptamer was stopped, no reversal of the anticoagulant activity of this molecule was required. Based on its ability to inhibit clot-bound thrombin and platelet thrombus formation in an ex vivo whole artery angioplasty model, this aptamer also exhibited potential as a novel antithrombotic (22). However, for this thrombin aptamer to be successfully tested in animal models of arterial thrombosis, it would probably be necessary to modify it to improve its circulating half-life.
In addition to thrombin, most of the serine proteases and their cofactors in the coagulation pathway are suitable targets for generation of experimental anticoagulants or antithrombotics. Novel inhibitors of these proteins may also prove useful for the experimental dissection of thrombus formation in different pathologic states. Physiologic coagulation is initiated by enzymatic activity of the protease FVIIa in complex with its requisite cofactor, tissue factor. Recently, a 2′-amino modified-RNA aptamer has been generated to coagulation FVIIa (7). This aptamer binds FVIIa with high affinity and specificity and has been shown to block the activity of FVIIa by preventing formation of an active FVIIa–tissue factor complex. This aptamer is stable in plasma, with a half-life of approximately 15 hours, and can prolong the clot time of human plasma. While this aptamer has not been studied in animals, it highlights the potential use of this technology to generate a panel of antagonists to the factors that compose the coagulation pathway.
Intimal hyperplasia
The two inner layers — the intima and the media — of a normal vessel wall exist in a nearly quiescent state. When a vessel is injured (for example, during angioplasty or endarterectomy) or subjected to hemodynamic stress (for example, during the arterialization of a venous graft), a complex series of mechanical, cellular, and molecular factors activate key intracellular pathways within the smooth muscle cells (SMCs) of the media. Intimal hyperplasia is the abnormal proliferation and migration of vascular SMCs in response to injury. This process underlies the accelerated restenosis of venous bypass grafts and of arteries following angioplasty or endarterectomy. Intimal hyperplasia occurs to varying degrees in all such patients over the months following revascularization.
The roles of several cell cycle regulatory proteins in this process have been investigated. In particular, blockade of the transcription factor E2F — the common effector of numerous signaling pathways — has been shown to inhibit SMC proliferation both in vitro and in vivo. Double-stranded DNA featuring a highly-conserved promoter sequence functions as an E2F decoy; intraoperative administration markedly inhibits intimal hyperplasia in balloon-injured rat carotid arteries for up to 6 months and significantly decreases failure rates in human lower extremity bypass grafts at 12 months (see Perspective by Mann and Dzau [ref. 23]; refs. 24, 25). These findings corroborate those of other studies in which acute suppression of cellular proliferation and migration following insult may contribute to long-term prevention of intimal hyperplasia and stenosis.
The affinity of decoy DNA for E2F can at best approximate that of native DNA (∼10–7 M, several orders of magnitude looser than that of a typical aptamer for its target protein), requiring high pharmacologic doses of oligonucleotide for inhibition. A “first-generation,” unmodified-RNA aptamer against E2F binds the DNA-binding site of E2F with high affinity and inhibits fibroblast proliferation when microinjected into cells (26). The transition of such an intracellular inhibitor to in vivo models requires one of two approaches. Gene therapeutic methods utilizing viral constructs may be used to “express” small, therapeutic RNAs following gene transfer into cells (27). Alternatively, an aptamer may be selected de novo with nuclease-resistant nucleotides and delivered locally or systemically.
To avoid the need for intracellular delivery altogether, Leppanen et al. (28) are studying the effects of NX1975 (see ref. 29), a highly modified, PEG-conjugated aptamer against PDGF. The importance of PDGF in intimal hyperplasia has been demonstrated in animal models utilizing neutralizing antibodies against PDGF or the PDGF receptor (30). When applied to a restenosis model in rats, intraperitoneal administration of the PDGF aptamer several hours before and after injury led to a 50% reduction in the ratio of intimal to medial thickness, measured one week after the injury.
Angiogenesis
Angiogenesis, the growth of new blood vessels from pre-existing vessels, has been implicated in a variety of human diseases. VEGF is a central positive regulator of physiological angiogenesis, and growing evidence supports its importance in several pathological conditions. Inhibition of VEGF function by means of VEGF-neutralizing antibodies has been shown to reduce tumor growth and vascularization in animal models (31). In addition, its expression is elevated in most solid tumors, as well as in mesangioproliferative glomerulonephritis, rheumatoid arthritis, psoriasis, and several retinopathies. Although many other proangiogenic factors have been identified, VEGF has attracted the most interest as a therapeutic target due to its unique ability to potently and specifically stimulate endothelial cell proliferation and increase blood vessel permeability.
Ruckman et al. (32) characterized a family of high-affinity, nuclease-resistant aptamers against VEGF-165, the most abundant isoform. The association of these aptamers with large, inert entities such as liposomes and PEG has been shown to dramatically reduce aptamer clearance (14, 33, 34). For example, the biological half-life of the PEG-conjugated aptamer is improved to approximately 9 hours after intravenous administration and approximately 12 hours after subcutaneous injection (15). This PEG-conjugated 2′F aptamer, also known as NX1838, has demonstrated activity against both vascular cell proliferation and permeability. Intradermal delivery of NX1838 blocks the VEGF-induced increase in vascular permeability (32). Delivered intravenously, this aptamer can also significantly reduce the VEGF-induced angiogenic response in a rat corneal micropocket angiogenesis assay (33) or block glomerular endothelial cell proliferation and endothelial cell death in rats with mesangioproliferative nephritis. Renal function in normal rats does not appear to be compromised by this treatment (33). These in vivo results have paved the way for further animal and now human studies of NX1838 in the wet form of age-related macular degeneration and diabetic retinopathy, both of which are believed to involve VEGF-induced ocular neovascularization. Currently in phase I clinical trials, NX1838 is the first therapeutic aptamer to be administered to humans.
Future
Aptamers are a promising class of compounds, both for target validation and therapy. As designer drugs, they exhibit high specificity, high affinity, and modifiable bioavailability. The ability to generate inhibitors with such properties against a variety of target proteins will be invaluable as the human genome and proteome are deciphered.
Further animal and, in turn, human data are now necessary to facilitate the transition of these molecules from lab reagents to pharmaceuticals. Safety and efficacy in appropriate disease models must be established. Such development has been slowed by medium- and large-scale synthesis costs; however, these have been decreasing as a function of improved production capability. As has been witnessed with other new technologies, production capability will continue to improve if demand exists. Meanwhile, progress toward developing therapeutic aptamers will be expedited through collaborations between industry and academic researchers.
References
- 1.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 2.Sullenger BA, Gallardo HF, Ungers GE, Gilboa E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell. 1990;63:601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 3.Kohn DB, et al. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood. 1999;94:368–371. [PubMed] [Google Scholar]
- 4.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 5.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 6.Conrad R, Keranen LM, Ellington AD, Newton AC. Isozyme-specific inhibition of protein kinase C by RNA aptamers. J Biol Chem. 1994;269:32051–32054. [PubMed] [Google Scholar]
- 7.Rusconi, C., Yeh, A., Lyerly, H., Lawson, J., and Sullenger, B. 2000. Blocking the initiation of coagulation by RNA aptamers to Factor VIIa. Thromb. Haemost. In press. [PubMed]
- 8.Doudna JA, Cech TR, Sullenger BA. Selection of an RNA molecule that mimics a major autoantigenic epitope of human insulin receptor. Proc Natl Acad Sci USA. 1995;92:2355–2359. doi: 10.1073/pnas.92.6.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SW, Sullenger BA. Isolation of a nuclease-resistant decoy RNA that can protect human acetylcholine receptors from myasthenic antibodies. Nat Biotechnol. 1997;15:41–45. doi: 10.1038/nbt0197-41. [DOI] [PubMed] [Google Scholar]
- 10.Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2’-modified hammerhead ribozymes. Science. 1991;253:314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- 11.Beigelman L, et al. Chemical modification of hammerhead ribozymes. Catalytic activity and nuclease resistance. J Biol Chem. 1995;270:25702–25708. doi: 10.1074/jbc.270.43.25702. [DOI] [PubMed] [Google Scholar]
- 12.Aurup H, Williams DM, Eckstein F. 2’-Fluoro- and 2’-amino-2’-deoxynucleoside 5′-triphosphates as substrates for T7 RNA polymerase. Biochemistry. 1992;31:9636–9641. doi: 10.1021/bi00155a016. [DOI] [PubMed] [Google Scholar]
- 13.Jellinek D, et al. Potent 2’-amino-2’-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry. 1995;34:11363–11372. doi: 10.1021/bi00036a009. [DOI] [PubMed] [Google Scholar]
- 14.Willis MC, et al. Liposome-anchored vascular endothelial growth factor aptamers [erratum 1998, 9:633] Bioconjug Chem. 1998;9:573–582. doi: 10.1021/bc980002x. [DOI] [PubMed] [Google Scholar]
- 15.Tucker CE, et al. Detection and plasma pharmacokinetics of an anti-vascular endothelial growth factor oligonucleotide-aptamer (NX1838) in rhesus monkeys. J Chromatogr B Biomed Sci Appl. 1999;732:203–212. doi: 10.1016/s0378-4347(99)00285-6. [DOI] [PubMed] [Google Scholar]
- 16.Usman, N., and Blatt, L.M. 2000. Characterization and development of nuclease-resistant synthetic ribozymes as a new class of therapeutics. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 17.Khachigian, L.M. 2000. Catalytic DNA as potential therapeutic agents and sequence-specific molecular tools. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 18.Srinivasan SK, Iversen P. Review of in vivo pharmacokinetics and toxicology of phosphorothioate oligonucleotides. J Clin Lab Anal. 1995;9:129–137. doi: 10.1002/jcla.1860090210. [DOI] [PubMed] [Google Scholar]
- 19.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 20.Griffin LC, Tidmarsh GF, Bock LC, Toole JJ, Leung LL. In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood. 1993;81:3271–3276. [PubMed] [Google Scholar]
- 21.DeAnda A, Jr, et al. Pilot study of the efficacy of a thrombin inhibitor for use during cardiopulmonary bypass. Ann Thorac Surg. 1994;58:344–350. doi: 10.1016/0003-4975(94)92206-3. [DOI] [PubMed] [Google Scholar]
- 22.Li WX, Kaplan AV, Grant GW, Toole JJ, Leung LL. A novel nucleotide-based thrombin inhibitor inhibits clot-bound thrombin and reduces arterial platelet thrombus formation. Blood. 1994;83:677–682. [PubMed] [Google Scholar]
- 23.Mann, M.J., and Dzau, V.J. 2000. Therapeutic applications of transcription factor decoy oligonucleotides. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 24.Morishita R, et al. A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc Natl Acad Sci USA. 1995;92:5855–5859. doi: 10.1073/pnas.92.13.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann MJ, et al. Ex-vivo gene therapy of human vascular bypass grafts with E2F decoy: the PREVENT single-centre, randomised, controlled trial. Lancet. 1999;354:1493–1498. doi: 10.1016/S0140-6736(99)09405-2. [DOI] [PubMed] [Google Scholar]
- 26.Ishizaki J, Nevins JR, Sullenger BA. Inhibition of cell proliferation by an RNA ligand that selectively blocks E2F function. Nat Med. 1996;2:1386–1389. doi: 10.1038/nm1296-1386. [DOI] [PubMed] [Google Scholar]
- 27.Sullenger BA, Lee TC, Smith CA, Ungers GE, Gilboa E. Expression of chimeric tRNA-driven antisense transcripts renders NIH 3T3 cells highly resistant to Moloney murine leukemia virus replication. Mol Cell Biol. 1990;10:6512–6523. doi: 10.1128/mcb.10.12.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leppänen, O., et al. 2000. Intimal hyperplasia recurs after removal of PDGF-AB and -BB inhibition in a rat model of arterial restenosis. Atherosclerosis, Thrombosis and Vascular Biology. In press. [DOI] [PubMed]
- 29.Green LS, et al. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry. 1996;35:14413–14424. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 30.Ferns GA, et al. Inhibition of neointimal smooth muscle accumulation after angioplasty by an antibody to PDGF. Science. 1991;253:1129–1132. doi: 10.1126/science.1653454. [DOI] [PubMed] [Google Scholar]
- 31.Kim KJ, et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 32.Ruckman J, et al. 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 33.Ostendorf T, et al. VEGF(165) mediates glomerular endothelial repair. J Clin Invest. 1999;104:913–923. doi: 10.1172/JCI6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Floege J, et al. Novel approach to specific growth factor inhibition in vivo: antagonism of platelet-derived growth factor in glomerulonephritis by aptamers. Am J Pathol. 1999;154:169–179. doi: 10.1016/S0002-9440(10)65263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]