Abstract
Oxygen (O2) is an essential nutrient that serves as a key substrate in cellular metabolism and bioenergetics. In a variety of physiological and pathological states, organisms encounter insufficient O2 availability, or hypoxia. In order to cope with this stress, evolutionarily conserved responses are engaged. In mammals, the primary transcriptional response to hypoxic stress is mediated by the Hypoxia-inducible factors (HIFs). While canonically regulated by prolyl hydroxylase domain-containing enzymes (PHDs), the HIFα subunits are intricately responsive to numerous other factors including Factor Inhibiting HIF-1α (FIH1), sirtuins, and metabolites. These transcription factors function in normal tissue homeostasis and impinge on critical aspects of disease progression and recovery. Insights from basic HIF biology are being translated into pharmaceuticals targeting the HIF pathway.
Introduction
Aerobic organisms require oxygen (O2) to produce energy. For this reason, O2 deprivation creates significant stress in living cells. O2 deprivation is also paradoxically linked to the inappropriate accumulation of free radicals, which cause additional stress on proteins and DNA in the cell. During low O2 (hypoxic) conditions, therefore, cells activate a number of adaptive responses to match O2 supply with metabolic, bioenergetic, and redox demands. Cells temporarily arrest in the cell cycle, reduce energy consumption, and secrete survival and proangiogenic factors. These events are coordinated by various cellular pathways, including the unfolded protein response, mTOR signaling, and gene regulation by hypoxia inducible factors (HIFs). Initially identified as a regulator of erythropoietin (EPO) production, HIF is recognized as a key modulator of the transcriptional response to hypoxic stress. Besides its adaptive function in cellular stress responses, recent work has also revealed important roles for HIF in both physiological and pathological processes.
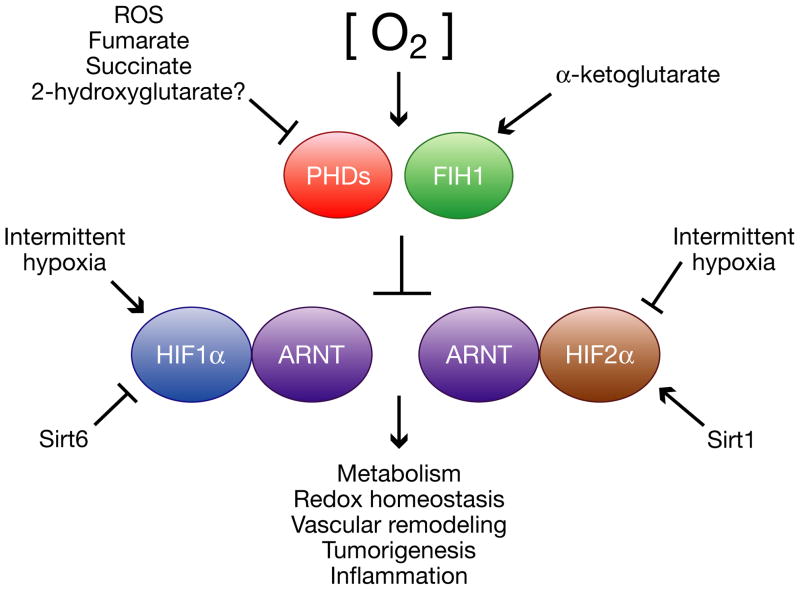
HIFs are obligate heterodimers consisting of an O2-labile α subunit and a stable β subunit. Mammals possess three isoforms of HIFα, of which HIF1α and HIF2α (also known as EPAS1) are the most structurally similar and best characterized. HIF3α (or IPAS) exists as multiple splice variants, some of which inhibit HIF1α and HIF2α activity in a dominant-negative fashion (reviewed in Kaelin and Ratcliffe, 2008). HIF1α is expressed ubiquitously in all cells, whereas HIF2α and HIF3α are selectively expressed in certain tissues, including vascular endothelial cells, type II pneumocytes, renal interstitial cells, liver parenchymal cells, and cells of the myeloid lineage (reviewed in Bertout et al., 2008). In Figure 1, we highlight some of the novel mechanisms of HIFα regulation and HIFα transcriptional activity.
Figure 1. Regulation and function of hypoxia inducible factor-α (HIFα) subunits.
In the presence of oxygen and α-ketoglutarate, HIF prolyl hydroxylase domain-containing enzymes (PHDs) and Factor Inhibiting HIF (FIH1) hydroxylate and inactivate HIFα. PHD and FIH1 activity are inhibited by hypoxia and certain intracellular metabolites, including reactive oxygen species, fumarate, succinate, and potentially 2-hydroxyglutarate, resulting in HIFα stabilization. HIFα expression and/or activity is also regulated post-translationally by sirtuins and intermittent hypoxia. HIFα stabilization results in the activation of a transcriptional program with effects on metabolism, redox homoeostasis, vascular remodeling, tumorigenesis, inflammation, and other processes.
HIFα subunits heterodimerize with the stable HIF1β, or ARNT, subunit through their HLH and PAS domains. HIF heterodimers recognize and bind to hypoxia response elements (HREs) in the genome, which are similar to Enhancer box (E-box) motifs and have the consensus sequence G/ACGTG. Genome-wide chromatin immunoprecipitation experiments indicate that the correlation between HRE occupancy and hypoxic gene induction ranges from high for HIF1α upregulated genes, to low for both HIF2α induced genes and HIF repressed genes (Mole et al., 2009; Xia et al., 2009). In these latter cases, flanking sequences and additional regulatory elements appear to further specify HIF binding and target gene regulation. Recent examples of additional modifiers of HIF dependent gene regulation include the forkhead transcription factor FOXA2 and the chromatin modifier Reptin (Qi et al., 2010; Lee et al., 2010).
In this review, we will focus on recent insights into HIF expression, activation, and function in various cellular and tissue stress responses. We will also summarize emerging data on the crosstalk between HIFs and other metabolic regulators and provide a survey of recent pharmacologic strategies to modulate HIF activity in the treatment of diseases.
HIFα Regulation by O2 Availability
In well oxygenated environments, HIFα subunits are hydroxylated at conserved proline residues. These modifications are mediated by PHDs, whose activities are regulated by O2 availability (reviewed in Kaelin and Ratcliffe, 2008). Hydroxylated HIFα is, in turn, recognized and marked for proteosomal destruction by an E3 ubiquitin ligase, the von Hippel-Lindau protein (pVHL) complex. In the setting of hypoxic stress, PHD activity is diminished, and stabilized HIFα proteins can induce transcription of genes with adaptive functions.
PHD regulation by O2 and metabolites
Because of their dependence on O2 as a direct substrate, PHDs have been proposed to be ‘oxygen sensors’ linking cellular O2 concentration to HIF molecular responses. This notion is supported by measurements of the apparent KM of PHD enzymes in vitro (reviewed in Kaelin and Ratcliffe, 2008). The KM values for oxygen were considerably higher than intracellular pO2, suggesting that PHDs may function below saturation kinetics in vivo. While the enzymatic properties of PHDs in vitro do not necessarily reflect their properties in vivo, these measurements imply that O2 availability can influence PHD activity across the entire physiological range.
Cellular metabolites can also influence PHD activity. PHDs utilize the tricarboxylic acid (TCA) cycle intermediate 2-oxoglutarate (α-ketoglutarate) as a substrate and can be inhibited by other TCA intermediates (Kaelin and Ratcliffe, 2008; Klimova and Chandel, 2008; Sudarshan et al., 2009). As we will describe later, several types of cancer have been shown to exploit this metabolic regulation and bear mutations that are likely to stimulate HIF activity.
O2-dependent Regulation of HIFα by FIH1
HIFα subunits are also substrates for an asparaginyl hydroxylase: Factor Inhibiting HIF-1α (FIH1). This enzyme is O2-dependent and, thus, represents another component of the oxygen-sensing machinery. Hydroxylation by FIH1 disrupts a critical interaction between HIFα and co-activators p300/CBP, impairing HIF transcriptional activity (reviewed in Webb et al., 2009a; Mahon et al., 2001). However, FIH1 has other targets (Webb et al., 2009a, 2009b), indicating it may have HIF-independent functions as well.
Fih1−/− mice were recently generated (Zhang et al., 2010) and found to be viable, unlike Vhl−/− and Phd2−/− animals (Gnarra et al., 1997; Takeda et al., 2006). Mutant adult mice displayed a range of metabolic phenotypes including decreased weight, decreased adiposity, hyperventilation and increased insulin sensitivity. Remarkably, several of these phenotypes were recapitulated by neuronal-specific deletion of Fih1. Moreover, when placed on a high fat diet, Fih1−/− mice were less likely to develop insulin resistance, weight gain, and hepatic steatosis (or fatty liver). In correlation with these phenotypes, the authors observed that Fih1 deletion affected the activity of AMPK, PGC1α and PPARγ—factors which have been implicated in metabolism and metabolic diseases (Muoio and Koves, 2007; Towler and Hardie, 2007; Picard and Auwerx, 2002). Collectively, these data indicate that FIH1 regulates fat storage, insulin sensitivity, and organism growth. Furthermore, FIH1 deficiency can prevent features of obesity, diabetes and non-alcoholic fatty liver disease (Adams and Lindor, 2007) in animals with increased caloric intake. Thus, FIH1 represents a novel drug target for treating these diseases.
Although several in vitro phenotypes of Fih1−/− fibroblasts were HIF1α-dependent or synergized with Vhl deficiency, it is unclear if FIH1 negatively regulates the HIF pathway in vivo. HIF1α activation can inhibit adipocyte differentiation in vitro (Yun et al., 2002), consistent with the reduced adipocyte density observed in Fih1−/− mice. On the other hand, HIFα activation in Vhl-deficient livers actually promotes hepatic steatosis (Rankin et al., 2009), in contrast to Fih1−/− mice. Therefore, further investigation is required to determine whether, as a general rule, FIH1 deficiency promotes metabolic phenotypes in vivo through HIFα activity.
Mitochondria as oxygen sensors and PHD regulators
Considerable evidence indicates that mitochondria also participate in O2 sensing. Genetic and pharmacological approaches have been employed to inhibit components of the electron transport chain (ETC) in mitochondria. These studies have shown that in moderate hypoxia (1.5% O2), mitochondria stimulate the production of cellular reactive oxygen species (ROS), which inhibit PHD activity and HIFα degradation (reviewed in Kaelin, 2005; Klimova and Chandel, 2008). These oxygen radicals emanate specifically from complex III of the ETC (Klimova and Chandel, 2008). Moreover, Waypa and colleagues were able to visualize redox changes within specific cellular compartments using a novel redox-sensitive fluorescent protein (RoGFP) (Waypa et al., 2010). Hypoxia-induced oxidants were observed in the inner-membrane space of mitochondria as well as in the cytosol, where ROS could influence PHD activity (Waypa et al., 2010). These findings support a model in which mitochondria sense O2 deprivation and produce ROS to regulate PHD activity.
The role of mitochondria in O2 sensing may be restricted, however, to moderate hypoxia (1.5%). As O2 levels decline further to anoxia (0% O2), HIFα can be stabilized in the absence of functional mitochondria, suggesting that factors in addition to mitochondrial ROS antagonize PHD activity in more severe O2 deprivation (Kaelin, 2005; Klimova and Chandel, 2008). For example, this may represent a setting in which PHDs directly sense O2 through its availability as a substrate.
In spite of the available data, the notion of mitochondria as O2 sensors is controversial, and many questions remain unanswered. For instance, it is unclear what triggers mitochondria to release ROS in response to low intracellular pO2 and whether ROS modulate PHD function directly or indirectly. Furthermore, it has been suggested that mitochondria signal to PHDs indirectly through their consumption of O2 and not through ROS production (Klimova and Chandel, 2008). This model could also explain the observation that mitochondrial inhibitors impair hypoxic stabilization of HIF1α: specifically, decreased mitochondrial O2 consumption due to ETC inhibition could help maintain cytosolic pO2 and, consequently, PHD activity. However, studies using cytochrome b mutant cells suggest this is not the case (Klimova and Chandel, 2008). In these cells, which generate mitochondrial ROS but fail to consume O2, HIF1α can be stabilized by hypoxia in an oxidant-dependent manner. This indicates that mitochondrial ROS production, but not O2 consumption, is important for HIFα stabilization.
Therefore, there may be multiple O2 sensors—PHDs, FIH1, mitochondria, others—which collectively promote HIF molecular responses in hypoxia.
The Growing Complexity of HIF Regulation
O2-sensing via hydroxylases and mitochondria define a core feature of HIF regulation. However, the list of additional cues, which modulate the HIF pathway, is growing. These factors range from microRNAs to oncogenic signals, and some prominent examples are listed in Table 1. Two of the more novel aspects of HIF regulation will be described below.
Table 1.
Regulators of HIF activity
| HIF regulators | |
| VHL | ↓ HIFα stability (see text) |
| PHD1/2/3 | ↓ HIFα stability (see text) |
| FIH1 | ↓ HIFα transcriptional activity (see text) |
| Siah1a/2 | ↑ HIF1α stability; promotes degradation of PHD1/3 in hypoxia (Simon, 2004) |
| RSUME | ↑ HIF1α stability; Enhances SUMOylation (Carbia-Nagashima et al., 2007) |
| SENP1 | ↑ HIF1α stability; Removes SUMO moieties (Cheng et al., 2007) |
| HSP90 | ↑ HIF1α stability (Isaacs et al., 2002) |
| COMMD1 | ↓ HIF1α stability and disrupts HIFα/β dimerization (van de Sluis et al., 2010, 2009) |
| HSP70/CHIP | ↓ HIF1α stability but not HIF2α stability (Luo et al., 2009) |
| CITED2 | ↓ HIF1α activity (Bakker et al., 2007) |
| Metabolites/Related | |
| 2-oxoglutarate | ↓ HIFα stability as PHD co-factor (see text) |
| Ascorbate | ↓ HIFα stability as PHD co-factor (Kaelin and Ratcliffe, 2008) |
| Iron (Fe2+) | ↓ HIFα stability as PHD co-factor (Kaelin and Ratcliffe, 2008) |
| IRP | ↓ HIF2α mRNA translation in response to high intracellular iron (see text) |
| NO | Modulates HIFα expression (Kaelin and Ratcliffe, 2008) |
| Intermittent Hypoxia | ↑ HIF1α stability but ↓ HIF2α stability (see text) |
| Redox | |
| ROS | ↑ HIFα stability (see text); observed in inflammatory cells (Shatrov et al., 2003) |
| Sirt1 | ↑ HIF2α, but not HIF1α, transcriptional activity (see text) |
| Sirt6 | Binds to and ↓ HIF1α stability/activity (see text) |
| microRNAs | |
| miR-107 | microRNA leads to ↓ ARNT expression (Yamakuchi et al., 2010) |
| miR-17–92 | miRNA cluster; microRNAs lead to ↓ HIF1α expression (Taguchi et al., 2008) |
| Oncogenes/Tumor Suppressors | |
| PI3K/Akt | ↑ HIF1α expression (Brugarolas and Kaelin Jr., 2004; Mottet et al., 2003) |
| mTORC1 | ↑ HIF1α mRNA translation (Brugarolas and Kaelin Jr., 2004; Bernardi et al., 2006) |
| GSK3β | ↓ HIF1α stability (Mottet et al., 2003) |
| p53 | ↓ HIF1α/ARNT expression (Blagosklonny et al., 1998; Yamakuchi et al., 2010; Sano et al., 2007; Schmid et al., 2004; Ravi et al., 2000) |
| β-catenin | Binds to HIF1α; ↑ HIF1α transcriptional activity (Kaidi et al., 2007) |
| Ras | ↑ HIF1α expression by ROS generation (Gerald et al., 2004) |
| ERβ | ↓ HIF1α stability (see text) |
| SDH/FH | Mutations in these genes lead to ↑ HIF1α stability (see text) |
| Inflammation | |
| NF↓ κB | ↑ HIF1α transcription (see text) |
| p44/42 MAPK | ↑ HIF1α expression downstream of LPS (Frede et al., 2006) |
| IFN-γ | ↑ HIF1α and HIF2α(?) expression (see text) |
| IL-4 | ↑ HIF1α and HIF2α(?) expression (see text) |
Sirtuins
Sirtuins are a stress-responsive family of nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases that influence gene transcription, metabolism, DNA repair and organism lifespan (Haigis and Sinclair, 2010). These enzymes respond to perturbations in the ratio of oxidized NAD+/reduced NADH and, therefore, represent sensors of the cellular redox state (Denu, 2003). Sirtuins have recently been shown to modulate HIF activity, strengthening the link between cellular stress and HIF responses (Dioum et al., 2009; Zhong et al., 2010).
Garcia and colleagues showed that Sirt1 forms a complex with HIF2α, but not HIF1α, and deacetylates lysine residues in the HIF2α protein (Dioum et al., 2009). Sirt1 also co-occupies the Epo promoter with HIF2α and enhances HIF2α transcriptional activity in vitro. Finally, the Sirt1/HIF2α axis functions in vivo to regulate renal and hepatic erythropoietin expression. These data suggest that HIF2α-dependent EPO production is responsive to perturbations in the cellular redox state in addition to changes in systemic O2 availability (see HIF in a Systemic Response to Hypoxia).
Sirt6 was also recently linked to HIF (Zhong et al., 2010). Sirt6−/− cells and mice display increased rates of glucose consumption and elevated expression of glycolytic genes, many of which are HIF1α targets. Sirt6 occupies the promoters of these genes and appears to enhance the deacetylation of associated histones, consistent with transcriptional repression. Furthermore, Sirt6 deficiency stimulates HIF1α protein expression. Using chemical inhibitors and RNA interference, the authors showed that HIF1α expression contributes, in part, to the metabolic phenotypes observed in Sirt6−/− cells and mice. Because the chemical inhibitors employed may lack target specificity, it will be important to determine if genetic loss of HIF1α also rescues phenotypes in Sirt6−/− animals. Lastly, Sirt6 and HIF1α appear to form a weak interaction, although the structure and functional significance of this complex requires further characterization. Based on these findings, Sirt6 appears to regulate glucose homeostasis by inhibiting HIF1α and glycolytic gene expression.
In sum, sirtuin activity can modulate HIF-dependent regulation of homeostatic processes such as erythropoiesis and glucose metabolism. The interplay between HIFs and sirtuins may also extend to stress settings such as hypoxic tumors, in which cellular redox balance is perturbed. However, the nature of the HIF response will depend on which sirtuin is engaged, for Sirt1 activates HIF2α, while Sirt6 represses HIF1α.
Intermittent hypoxia
Intermittent hypoxia (IH) occurs when tissue O2 tension cycles between normal and hypoxic levels. This pattern of O2 deprivation is particularly relevant during recurrent sleep apnea, in which transient pauses in breathing lead to chronic IH during sleep (Basner, 2007). Patients with this disease have a higher likelihood of developing hypertension, atherosclerosis, myocardial infarction and stroke (Basner, 2007).
Prabhakar and colleagues have detailed how HIFα subunits are differentially regulated by IH. HIF1α protein expression is induced during IH, secondary to many factors including NADPH oxidase-dependent ROS generation (Peng et al., 2006; Yuan et al., 2008). Of note, this source of ROS is distinct from the mitochondrial ROS which inhibits PHD activity during continuous hypoxia. Importantly, studies in Hif1α+/− mice demonstrated that HIF1α promotes the acute respiratory and cardiovascular consequences of IH. In contrast, HIF2α expression is repressed by IH through calpain-dependent mechanisms (Nanduri et al., 2009). In turn, HIF2α inhibition appears to contribute to IH-induced oxidative stress and cardiovascular responses in vivo. These data indicate that HIF1α and HIF2α may play opposing roles in IH-associated disease.
Differential regulation of HIFα subunits
As the cases of IH and Sirt1 indicate, HIF1α and HIF2α can be regulated through distinct mechanisms. Additional examples from the literature support this concept. HIF1α, but not HIF2α, appears to be degraded in hypoxia in an Hsp70/CHIP-dependent fashion (Luo et al., 2009). On the other hand, HIF2α mRNA translation is uniquely responsive to iron content through an iron-response element in its 5′ UTR (Sanchez et al., 2007). These observations underscore the complexity of the HIF response, and suggest that HIFα subunits require distinct forms of regulation because they mediate non-overlapping biological effects, as was observed in mouse models of IH.
The Role of HIF in Metabolism and Redox Homeostasis
In artherosclerotic diseases, tissues such as the heart, brain, and limb muscles are susceptible to ischemic insults (Beckman et al., 2002). Deprivation of O2, nutrients, and growth factors causes stress in these pathologies, and cell death can ensue. The HIF pathway is activated and applies a critical adaptive response (Ratan et al., 2007; Shohet and Garcia, 2007). For instance, HIF1α expression correlates with increased preservation of brain and heart tissue in many (Ratan et al., 2007; Shohet and Garcia, 2007; Baranova et al., 2007) but not all (Helton et al., 2005) models of strokes and heart attacks, respectively.
HIF1α and glucose catabolism
HIF1α is thought to mediate cardio- and neuro-protection, in part, by reprogramming cellular metabolism. Because molecular O2 serves as an electron acceptor in oxidative phosphorylation, a central adaptation to hypoxia is a shift towards non-oxidative forms of carbon metabolism and ATP production, such as anaerobic glycolysis (reviewed in Gordan et al., 2007b). HIF1α guides this shift by promoting the expression of glucose transporters, glycolytic enzymes and LDHA, which replenishes NAD+ for further glycolysis (Gordan et al., 2007b).
Moreover, studies from the Semenza and Denko laboratories showed that pyruvate dehydrogenase kinase 1—encoded by the HIF1α target gene PDK1—represses the flux of pyruvate into acetyl-CoA, diverting carbon away from mitochondria and suppressing O2 consumption (reviewed in Simon, 2006). In HIF1α deficient cells, O2 deprivation leads to reduced ATP levels, elevated ROS, and apoptosis (Simon, 2006; Gordan et al., 2007b). The oxidant stress observed in hypoxic HIF1α mutant cells appears to be secondary to inappropriate acetyl-CoA generation and TCA cycle activity, such that forced PDK1 expression reduces ROS and promotes survival in hypoxia. These findings clearly demonstrate that a HIF1α-dependent metabolic shift promotes viability during hypoxic stress.
HIF1α activity was more recently shown to influence the pentose phosphate pathway or PPP (Zhao et al., 2010). The PPP converts glycolytic intermediates into ribose-5-phosphate (R5P), a substrate for nucleotide biosynthesis (Tong et al., 2009). In drug resistant leukemia cells, HIF1α promotes the flux of glucose carbon through a non-oxidative arm of PPP relative to the oxidative arm (Zhao et al., 2010). These effects are critical for leukemia cell growth and survival. HIF1α, therefore, redirects the metabolism of glucose for use both as an energy source and as a building block for RNA and DNA synthesis, and these adaptations are likely important for facilitating cell growth and survival in hypoxic tumors.
Overall, HIF1α can directly reprogram the metabolic state in cells, and this response is important in hypoxic settings such as vascular disease and cancer.
HIF2α functions in metabolism
Many of the metabolic genes described above are directly regulated by HIF1α but not HIF2α (Hu et al., 2003; Raval et al., 2005). Nevertheless, HIF2α also plays a critical role in metabolism. Semenza and colleagues demonstrated that both HIF1α and HIF2α can modulate the expression of cytochrome c oxidase isoforms so as to maximize efficiency of the electron transport chain (reviewed in Gordan et al., 2007b). Defects in this response lead to impaired ATP production and elevated oxidant production in hypoxia.
HIF2α also has some unique targets in cellular redox homeostasis. Garcia and colleagues showed that HIF2α stimulates the expression of genes encoding anti-oxidant enzymes, such as SOD2, in mice (reviewed in Gordan et al., 2007b). This transcriptional program appears to suppress aberrant ROS accumulation, such that HIF2α deficiency leads to severe striated muscle damage. More recently, HIF2α has been demonstrated to play a similar function in renal cancer: HIF2α depletion leads to reduced expression of genes with anti-oxidant functions, such as heme oxygenase 1 (HMOX1) and others (Bertout et al., 2009). In correlation, HIF2α loss induces increased cellular ROS, activation of p53 and tumor cell death. These effects are all enhanced with ionizing radiation treatment, which robustly elevates cellular ROS levels. Therefore, HIF2α promotes redox homeostasis and cellular viability in multiple settings. Moreover, HIF2α can mediate tumor cell resistance to ionizing radiation.
Studies in Phd1−/− mice provide further evidence that HIF2α can buffer tissues from hypoxic stress (Aragones et al., 2008). When placed under ischemic stress, Phd1−/− limb skeletal muscle is protected from oxidative damage and cell death. Interestingly, similar effects of PHD1 inhibition have been observed in liver ischemia (Schneider et al., 2010). The ischemic tolerance in Phd1−/− skeletal muscle is predominantly dependent on HIF2α, which is negatively regulated by PHD1. HIF2α may promote this tolerance by modulating glucose metabolism, for Phd1−/− muscle displays a metabolic shift towards anaerobic glycolysis and elevated expression of Pdk4. Like Pdk1, Pdk4 inhibits the mitochondrial consumption of glucose-derived carbon (Huang et al., 2002). Although HIF2α does not alter glucose utilization directly (Hu et al., 2003; Raval et al., 2005), it could function indirectly through regulation of PPARα, which is essential for ischemic tolerance in Phd1−/− skeletal muscle (Aragones et al., 2008) and may regulate Pdk4 expression (Huang et al., 2002). Alternatively, HIF2α’s more established role in redox homeostasis (reviewed in Gordan et al., 2007b; Bertout et al., 2009) could contribute to the ischemic phenotypes in Phd1−/− skeletal muscle.
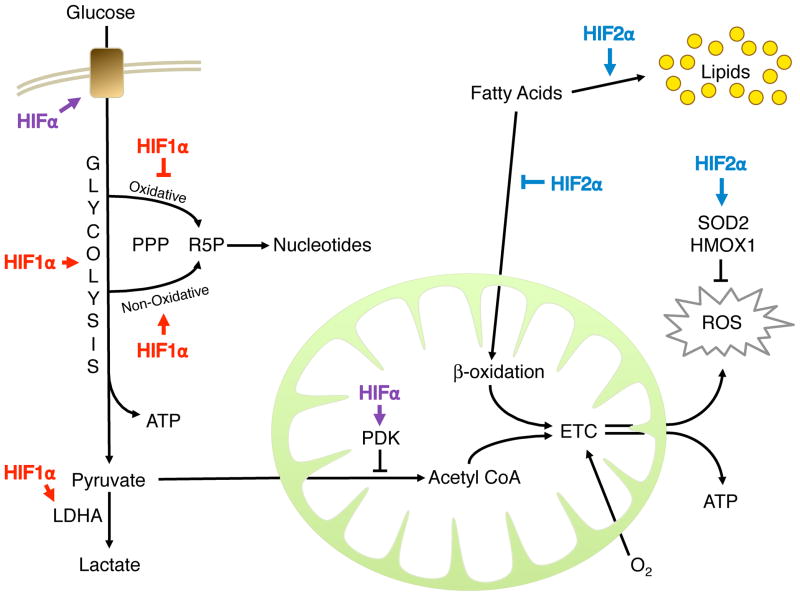
Collectively, these observations indicate that both HIF1α and HIF2α control cell metabolism and redox homeostasis through non-overlapping transcriptional programs (Figure 2).
Figure 2. HIFα Control of Cell Metabolism.
HIFs modulate cellular metabolism to faciliate cellular adaptation to low oxygen environments. Glucose consumption and glycolysis are promoted primarily by HIF1α, while fatty acid storage is promoted by HIF2α. Both factors inhibit mitochondrial consumption and oxidation of carbon, leading to a decreased production of ATP through oxidative phosphorylation and less reactive oxygen species (ROS) as a by-product. Instead, glycolysis makes a larger contribution to ATP synthesis in the cell. HIF2α, furthermore, inhibits ROS production through SOD2 and other targets.
HIF regulation of lipid metabolism
In tissues such as the heart and liver, lipids provide a rich source of energy via oxidative phosphorylation (Jungermann, 1988; Shohet and Garcia, 2007). In the setting of hypoxic stress, lipid metabolism is reprogrammed to suppress mitochondrial oxidation of lipid-derived carbon. Specifically, hypoxia stimulates lipid storage and inhibits lipid catabolism through β-oxidation (Huss et al., 2001; Whitmer et al., 1978; Bostrom et al., 2006).
It was previously unclear if HIFs control these adaptations. However, a recent study implicates HIF2α in the regulation of lipid metabolism (Rankin et al., 2009). Mice with liver-specific deletion of Vhl exhibit fatty livers (or hepatic steatosis), marked by increased lipid droplet deposition and decreased fatty acid consumption. In mutant livers, genes involved in β-oxidation are reduced, while those important in lipid storage are enhanced. These phenotypes are dependent on HIF2α more so than HIF1α. Therefore, in the pseudo-hypoxic context of Vhl-deficient livers, HIF2α represses lipid catabolism and oxidation (Figure 2).
The Role of HIF in Vascular Responses to Hypoxia
It is well established that, in response to hypoxia, HIF1α and HIF2α regulate angiogenic genes such as vascular endothelial growth factor (Manalo et al., 2005; Hu et al., 2003; Kelly et al., 2003). In correlation, HIFs are known to play essential roles in embryonic vascular development (Ramirez-Bergeron et al., 2006; Covello and Simon, 2004). Recent studies have demonstrated how HIFs can influence the adult vascular system, specifically during angiogenesis in pathologic settings.
HIF1α in ischemia-induced angiogenesis
Growing evidence supports a role for HIF1α activity in the neoangiogenic response to tissue ischemia. In a femoral artery ligation model of hindlimb ischemia, ischemic limb muscle in Hif1α+/− mice exhibits impaired HIF1α induction, defective activation of angiogenic factors, and decreased reperfusion (Bosch-Marce et al., 2007). Conversely, adenoviral delivery of constituitively active HIF1α to the site of ligation leads to enhanced reperfusion. Therefore, global HIF1α expression is essential and sufficient to promote reperfusion in ischemic skeletal muscle.
The authors also demonstrated that as mice age, the ischemic induction of HIF1α diminishes, and, in association, reperfusion is compromised. This is more than a correlation, for ectopic HIF1α expression partially rescues limb perfusion in old mice. Interestingly, impaired HIF1α activation is also observed in the hypoxic skin wounds of aged diabetic mice (Liu et al., 2008). These findings underscore the importance of aging as a modulator of ischemic responses. This is especially crucial because peripheral arterial disease, the vascular disease which femoral artery ligation models, is associated with age (Beckman et al., 2002).
A pro-angiogenic role for HIF1α has also been described in other injury models such as hypertrophic cardiomyopathy, myocardial infarction, skin wound healing, and retinal neo-vascularization (Sano et al., 2007; Jiang et al., 2009; Yoshida et al., 2010; Heinl-Green et al., 2005; Mace et al., 2007; Liu et al., 2008; Botusan et al., 2008). Relative to HIF1α, the angiogenic functions of HIF2α have not been as extensively tested in these disease models (Dioum et al., 2008). In sum, these data suggest that HIFα expression, and in particular HIF1α expression, in ischemic tissues promotes angiogenesis. This has strong clinical implications for angiogenic therapies in patients with ischemic disease (see HIF-targeted Therapies).
These findings also raise questions about the critical cell types in which HIFs operate during angiogenesis. While HIF activation in tissue parenchyma can influence endothelial cell behavior indirectly through expression of angiogenic factors, HIFs have also been shown to play essential roles within the vascular endothelium during vessel formation (see next section).
HIF in endothelial cells
In ischemic tissues, endothelial cells (ECs) respond to cues provided by numerous distinct cell populations—including parenchyma, pericytes, and inflammatory cells—to form new blood vessels (Carmeliet, 2005). They also alter their function in direct response to changes in O2 availability. For instance, hypoxia and HIF1α can direct ECs to form tube-like structures in vitro, mimicking morphologic changes which arise during angiogenesis (Yamakawa et al., 2003; Manalo et al., 2005).
Several studies have evaluated how HIFs regulate endothelial cell functions in hypoxic settings in vivo. This began with studies of mice with EC-specific deletion of Hif1α (Tang et al., 2004). Mutant mice exhibit defective blood vessel growth in hypoxic settings such as skin wounds and xenograft tumors. In correlation, isolated mutant endothelial cells exhibit defective hypoxic activation of VEGF and its receptor VEGFR2 as well as impaired cell proliferation and migration. Therefore, the authors proposed that HIF1α promotes an autocrine VEGF/VEGFR2 loop in endothelial cells that promote their functions in tissue angiogenesis.
HIF2α is highly expressed in endothelial cells during development, suggesting it also may play a cell-autonomous role in this cell type (Ema et al., 1997). Mice with EC-specific deletion of Hif2α exhibit homeostatic defects in vessel integrity as well as impaired tumor angiogenesis (Skuli et al., 2009). Xenograft tumors in mutant mice are smaller, more hypoxic and possess fewer luminized (functional) vessels relative to tumors in control mice. Mutant ECs from these animals exhibit defective adherence and impaired hypoxic induction of genes with functions in cell adhesion. These findings indicate that HIF2α instructs endothelial cells to form more functional blood vessels, and this role is critical for tumor development.
Of note, VEGF expression is unaffected in HIF2α mutant ECs, implying that it is a HIF1α-specific target in endothelial cells (Tang et al., 2004; Skuli et al., 2009; Manalo et al., 2005). This is consistent with prior studies indicating that HIF1α and HIF2α can promote the expression of distinct genes in endothelial cells and, therefore, may carry out unique functions in this compartment (Elvert et al., 2003).
Overall, these studies highlight how endothelial cells respond to local hypoxia during vessel growth and how HIFs mediate this response, particularly in tumor settings (Figure 4C). Therefore, inhibiting HIF function in endothelial cells could have utility in the treatment of cancer. This could be problematic, however, in situations where HIFs play tumor suppressive roles (see The Role of HIF in Cancer).
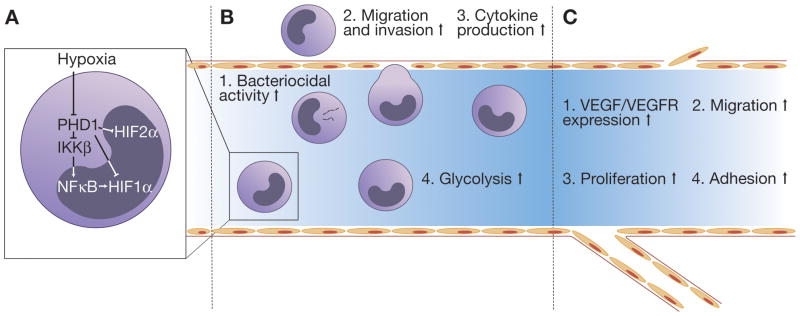
Figure 4. Macrophage and vascular responses to HIF.
A, NFκB dependent regulation of HIF in macrophages. In addition to direct HIF stabilization, hypoxic inhibition of PHDs result in IKK-mediated degradation of the NFκB inhibitor IκB. Activated NFκB directly transactivates HIF1α. B, HIF activity is involved in multiple aspects of macrophage behavior via the induction of genes involved in 1. bacterial killing (NOS2, CRAMP), 2. migration and invasion (CXCR4, FN1, MCSFR), 3. cytokine production (IL1β, IL6, IL12, TNFα), and 4. metabolism (GLUT1, PGK1). C, HIF1α stabilization in endothelial cells increase 1. VEGF expression, 2. migration, and 3. proliferation, whereas HIF2α stabilization promotes 4. endothelial cell adhesion to the extracellular matrix.
PHD2 in endothelial cells
On a side note, studies in Phd2+/− mice indicate that PHD inhibition in endothelial cells may have clinical implications (Mazzone et al., 2009). Xenograft tumors grown in Phd2 heterozygotes are less hypoxic and have more functional vessels than those in control mice. ECs from these animals are more quiescent and exhibit an altered transcriptional program dependent on HIF2α. The authors propose that this transcriptional response directs ECs to form more organized vessels in tumors, and that this “normalization” plays a causal role in reducing the number of tumor metastases in mutant mice. It has also been suggested that normalization of tumor vessels can enhance tumor perfusion and potentially drug delivery (Jain et al., 2005). Therefore, PHD inhibition could be effective in several aspects of cancer therapy. However, the authors caution that in their models, PHD activity was impaired in the microenvironment but not in the tumor cells. This is an important distinction given the oncogenic role of HIFs (see next section).
The Role of HIF in Cancer
There is ample evidence that solid tumors frequently encounter hypoxic stress. Rapidly proliferating cancer cells may outgrow their vascular network, limiting O2 diffusion within the tumor. Hypoxic stress can also be caused by perfusion defects as a result of abnormal tumor blood vessel structure and function. Not surprisingly, therefore, solid tumors often exhibit high levels of HIFα accumulation (reviewed in Bertout et al., 2008). It should be noted that HIFα expression in cancer cells is also increased via hypoxia-independent mechanisms (see Table 1 and relevant sections). Genetic alterations such as VHL mutation in renal cell carcinoma, mutations in the Wnt/β-catenin signaling pathway in colon carcinoma, and other oncogenic events have been reported to result in HIFα stabilization (reviewed in Kaelin, 2008). Collectively, these findings indicate that HIFα expression and the downstream activation of the hypoxic stress response are widespread in many cancers.
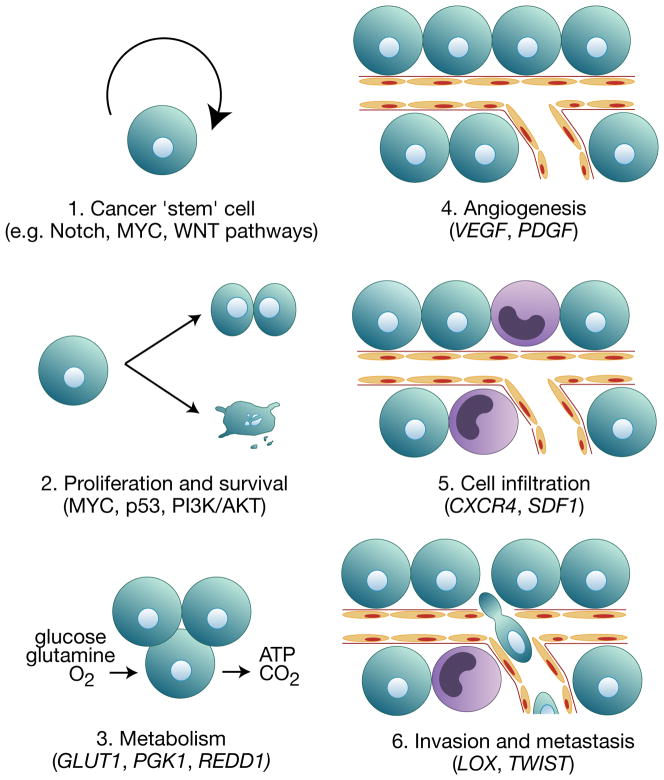
Work from many laboratories has revealed that HIF-regulated gene responses play key roles in various aspects of cancer development, including proliferation (MYC), angiogenesis (VEGF, PDGF), apoptosis/autophagy (NDRG2, BNIP3), metabolism (PDK1, LDHA), DNA damage response (GADD45A), microRNAs (MIR210), extracellular matrix remodeling (LOX, MMP1), cell migration and invasion (CXCR4, SDF1) (Huang et al., 2009; reviewed in Bertout et al., 2008; reviewed in Kaelin, 2008) (Figure 3). The importance of HIF activity in cancer is demonstrated by the fact that increased HIFα expression correlates with poor clinical prognosis in many cancer types (reviewed in Semenza, 2007).
Figure 3. Effects of HIF on multiple steps of cancer development.
HIF is stabilized by hypoxia and other nonhypoxic stimuli in many cancers. HIF activity in cancer has been associated with 1. putative cancer ‘stem’ cell maintenance and increased expression of genes involved in 2. proliferation and survival, 3. metabolism, 4. angiogenesis, 5. recruitment of infiltrating cells such as tumor-associated macrophages and bone marrow-derived cells, and 6. tumor cell metastasis. Some examples of HIF regulated genes and oncogenic pathways are given in parentheses.
HIFα subunits and tumorigenesis
Surprisingly, HIF1α expression correlates with lower cancer stage or decreased patient mortality in certain cancers; examples include non-small cell lung cancer, head and neck squamous cell carcinoma, and neuroblastoma. HIF2α expression in these malignancies, on the other hand, is a negative prognostic factor (reviewed in Bertout et al., 2008). This difference between HIF1α and HIF2α expression suggests that HIFα subunits may contribute differently towards tumorigenesis in certain cancers.
The distinct roles of HIF1α and HIF2α in tumorigenesis have been studied most thoroughly in VHL-deficient clear cell renal cell carcinoma (ccRCC). VHL-deficient ccRCC cluster into tumors which express either both HIF1α and HIF2α or HIF2α only. Overexpression and knockdown studies of HIF1α and HIF2α in VHL-deficient ccRCC cell lines indicate that HIF2α, but not HIF1α, is necessary for tumor growth (reviewed in Kaelin, 2008). One possible explanation for this effect is that HIF1α antagonizes MYC function, whereas HIF2α promotes MYC activity (Gordan et al., 2007a). Microarray profiling of ccRCC specimens revealed that compared to tumors expressing both HIFα isoforms, tumors exclusively expressing HIF2α up-regulate MYC target genes, proliferate faster, and are relatively resistant to replication stress (Gordan et al., 2008). Another mechanism by which HIFs exert opposing effects on tumor behavior lies in the hypoxic regulation of the tumor suppressor protein p53. HIF1α binds to p53, resulting in p53 stabilization and hypoxia induced cell death (Moeller et al., 2005; An et al., 1998). This interaction between HIF1α and p53 is probably a late evolutionary development specific to higher organisms, as HIF1α indirectly inhibits the p53 homolog CEP-1 in C. elegans after radiation induced DNA damage (Sendoel et al., 2010). In contrast, recent experiments have shown that HIF2α indirectly suppresses p53 activity and thereby promotes radioresistance and chemoresistance in tumor cells (Bertout et al., 2009; Roberts et al., 2009). These findings indicate that certain cancers, including but not limited to renal cell carcinoma, may differ in their tumor behavior and drug response according to the expression of HIF1α and/or HIF2α. Whether selective pressures exist in renal cell carcinomas for the loss of HIF1α and gain of HIF2α expression and whether HIFα expression patterns influence renal cancer progression remain subjects for further study.
Does HIF2α have greater oncogenic capacity than HIF1α in non-VHL malignancies? A recent report demonstrated that shRNA-mediated inhibition of HIF2α, but not HIF1α, in multiple human cancer cell lines reduced cell proliferation in vitro and subcutaneous xenograft growth in mice (Franovic et al., 2009). However, functional rescue experiments using exogenous HIF2α were lacking. In vivo models of lung tumorigenesis suggest a more complex role for HIF2α in cancer. Constitutively stabilized HIF2α increases lung tumour burden, tumour vascularity, and local invasion in Kras mutant mice (Kim et al., 2009). Intriguingly, a lung-specific deletion of HIF2α in the same Kras mutant model similarly enhances lung tumorigenesis (Mazumdar et al., 2010). A clue to this paradox may lie in the observation that HIF2α gain of function and HIF2α loss of function promote tumorigenesis via two unrelated mechanisms (Mazumdar et al., 2010; Kim et al., 2009). If we assume that different HIF2α targets have varying activation thresholds—due to HRE sequence conservation, co-regulation by other transcription factors, composition of the transcription machinery, etc. —then according to the extent of HIF2α stabilization, either tumor suppressive (e.g. AKT inhibition) or tumor promoting effects (e.g. angiogenesis, epithelial to mesenchymal transition) may ensue.
The biology of HIF3α in relation to tumorigenesis remains largely unstudied. HIF3α is down-regulated in renal cell carcinoma specimens, consistent with its known function as a dominant negative inhibitor of HIF1α and HIF2α (Maynard et al., 2007). In summary, HIF1α, HIF2α, and HIF3α have varying effects on cancer development because of their context-dependent functions and distinct modes of action.
HIF and metastasis
Epithelial to mesenchymal transition (EMT) is a key feature of invasive cells and can be characterized by the loss of epithelial cell-cell contact and the acquisition of mesenchymal features and motility. Hypoxia and HIF influence the expression of many EMT regulators to promote metastasis. Studies by Maxwell and colleagues revealed that HIF1α expression in renal cell carcinoma is sufficient to induce the loss of E-cadherin and an increase in invasion (reviewed in Kaelin, 2008). HIF1α directly regulates TWIST1 transcription and increases tumor cell invasiveness and metastasis in head and neck squamous cell cancer (Yang et al., 2008). In prostate cancer, HIF1α promotes SNAIL1 nuclear localization in a VEGF dependent manner (Mak et al., 2010). This finding is clinically relevant and implicates HIF1α expression in prostate cancer progression, because low grade prostate tumors repress HIF1α via estrogen receptor β (ERβ) activity, while high grade tumors downregulate ERβ, resulting in increased HIF1α expression, SNAIL1 nuclear localization, and metastasis (Mak et al., 2010). HIF1α also induces lysyl oxidase (LOX), which is an extracellular matrix remodeling enzyme as well as an upstream regulator of SNAIL1. As Giaccia and colleagues demonstrated, inhibition of LOX reduces tumor cell invasion, adhesion, and metastasis in an orthotopic breast cancer model (reviewed in Bertout et al., 2008). Recent work from the same group indicates that LOX secreted by the primary tumor remodels distant premetastatic sites to recruit tumor and stromal cells (Erler et al., 2009). The hypoxic tumor microenvironment therefore promotes metastasis via the activation of multiple HIF-responsive genes that together regulate all stages of cancer spread, including invasion, intravasation, and distant extravasation.
HIF and tumor angiogenesis
HIF exerts similar effects on endothelial cells in both tumor and nonmalignant tissues to mediate angiogenesis (see relevant section). However, unlike ‘normal’ blood vessels, tumor-associated vasculature is leaky, tortuous, and noncontiguous (reviewed in Jain, 2005). Tumor-associated endothelium interact with tumor cells as well as nonmalignant stromal cells, such as fibroblasts and infiltrating bone marrow derived cells. These cell types differ widely in their responses to hypoxic stress and therefore may contribute differently to tumor angiogenesis. For example, HIF activity in glioblastoma promotes tumor angiogenesis, as HIF1α inhibition in glioblastoma cells reduces vascular remodeling and normalizes tumor vasculature (Du et al., 2008). Paradoxically, HIF1α depletion in these cells also increases perivascular invasion, because of the direct effect of decreased VEGF levels on glioblastoma cell migration (Du et al., 2008). The development of tumor vasculature also appears to require myeloid-derived VEGF specifically. Deletion of the HIF target gene Vegf in myeloid cells increases murine mammary tumor growth, tumor oxygenation, and tumor sensitivity to chemotherapy, most likely due to “normalization” of tumor vessels (Stockmann et al., 2008). In contrast, haploinsufficiency of the HIF regulator Phd2 in nonmalignant tissues allowed the ‘normalization’ of xenograft tumor vasculature, improved oxygenation, and reduced metastasis (Mazzone et al., 2009). However, the dependence of these effects on HIF stabilization remains uncertain. These studies illustrate that the tumor vasculature responds to distinct and perhaps opposing HIF activities in different cell types. Therefore, selective manipulation of the hypoxic stress response in distinct tumor subcompartments may be more effective than systemic HIF inhibition as an antitumor strategy.
HIF and cancer stem cells
Hypoxia can promote an undifferentiated state in certain populations of stem and progenitor cells (Yoshida et al., 2009; Keith and Simon, 2007). Similarly, hypoxia and HIFs may contribute to the maintenance of putative cancer ‘stem’ cells. HIF depletion in CD133+ glioblastoma cells, which are enriched for cancer stem cells, reduces their tumorigenic and angiogenic potential in vitro and in vivo (Li et al., 2009). Furthermore, HIF2α is selectively expressed in the CD133+ subpopulation of glioblastoma cells whereas HIF1α expression is widespread among both tumorigenic and nontumorigenic cells, suggesting that HIF2α may fulfill a specific function in glioblastoma stem cells (Li et al., 2009). In a separate study, a small subset of immature cells in human neuroblastoma specimens was found to express neural crest markers and HIF2α; upon HIF2α knockdown, these cells underwent early sympathetic differentiation (Pietras et al., 2009). However, the precise identity and function of these cells remain unclear. It is interesting to note that both CD133+ glioblastoma cells and putative neuroblastoma progenitor cells express high levels of HIF2α while residing in periendothelial niches (Pietras et al., 2009; Calabrese et al., 2007). Although the extent of O2 saturation within these capillaries is unknown, these findings suggest that HIFα expression in certain cancer cell subpopulations may be controlled by both hypoxia and nonhypoxic stimuli, including metabolic aberrations in cancer.
HIF regulation by cancer metabolism
Inactivating homozygous mutations in the TCA cycle genes fumarate hydratase (FH) and succinate dehyrogenase (SDH) lead to elevated HIF1α expression in cells and human tumors (reviewed in King et al., 2006). Mice bearing kidney specific inactivation of Fh1 were generated and develop renal cysts marked by HIF activation (reviewed in Kaelin and Ratcliffe, 2008). HIF1α appears to be induced because of increased cellular levels of fumarate and succinate, which can inhibit PHD activity directly or indirectly through promotion of cellular ROS (Sudarshan et al., 2009; reviewed in Kaelin and Ratcliffe, 2008; reviewed in Klimova and Chandel, 2008). These findings clearly demonstrate the extent to which metabolism influences HIFα expression in cancer. While it is assumed that HIF1α activation is promoting oncogenesis in these tumor settings, HIFs can be tumor suppressive in some contexts (see below). The Fh1 mutant mice will be a useful tool for testing this premise directly.
More recently, heterozygous mutations in isocitrate dehydrogenase 1/2 (IDH1/2) have been linked to cancer. Mutant IDH1 proteins are defective in their ability to oxidize isocitrate into α-ketoglutarate (αKG) (Dang et al., 2009; Zhao et al., 2009). Zhao and colleagues suggested that through a dominant negative effect, cells expressing mutant IDH1 have reduced αKG levels, a substrate for PHDs. Therefore, HIF1α is indirectly stabilized. HIF1α expression is also elevated in human gliomas with mutant IDH1. Hence, they proposed that mutations in IDH1 promote cancer by indirectly activating HIF1α, in the manner of FH and SDH mutations.
On the other hand, multiple studies have since shown that lysates from IDH1/2 wild-type and mutant cancers have comparable levels of αKG, indicating that one wild-type allele of IDH1/2 may be sufficient to generate αKG in vivo (Dang et al., 2009; Ward et al., 2010; Gross et al., 2010). As a consequence, comparable levels of substrate should be available for PHD activity in wild-type and mutant cancers. This indicates the initial IDH1/HIF1α model may be inaccurate: IDH1 mutations may stimulate oncogenic HIF responses, but not necessarily through decreased αKG levels.
Moreover, mutant IDH1 has an unexpected reverse activity which reduces αKG and generates 2-hydroxyglutarate (2HG), a metabolite associated with brain tumors (Aghili et al., 2009). Elevated 2HG is also observed in IDH1/2 mutant glioblastoma and acute myeloid leukemia (Dang et al., 2009; Ward et al., 2010; Gross et al., 2010). Therefore, it has been suggested that this metabolite, synthesized by mutant IDH1/2, promotes cancers of the brain and blood. The molecular and cellular functions of 2HG, however, are poorly understood. One possibility is that 2HG inhibits PHD activity and, in turn, promotes oncogenic HIF responses in IDH1/2 mutant tumors. 2HG may promote cancer through HIF-independent mechanisms as well. Further investigation is required to test the role of 2HG in the HIF pathway and, more generally, in tumor cell functions.
Overall, the latter studies on IDH1/2 mutations underscore the notion that metabolic enzymes can be oncogenic as well as tumor suppressive (e.g. FH and SDH). They also challenge the concept that mutations in metabolic genes induce cancer solely through aberrant HIF responses.
The Role of HIF in Inflammation
In response to inflammatory stimuli, local vascular permeability increases, resulting in the increased delivery of effector cells and nutrients to the inflamed tissue. The combination of reduced circulation at the site of inflammation and increased metabolic demand from infiltrating immune cells and pathogens eventually leads to the local depletion of O2, resulting in hypoxia. Hypoxia and associated HIF activation have been observed in tissue specimens from patients with inflammatory conditions such as arthritis, artherosclerosis, and autoimmune diseases (reviewed in Nizet and Johnson, 2009).
HIF regulation in inflammatory cells
The response to hypoxic stress is tightly coupled to the immune response via NF-κB signaling (Figure 4A). Work from Nizet, Chilvers, Taylor and their colleagues has demonstrated that HIF promotes NF-κB activity in macrophages, neutrophils, and non-immune cells (reviewed in Nizet and Johnson, 2009). Hypoxia inhibits PHD1 activity and thereby IKK hydroxylation, resulting in IKK activation and phosphorylation of IκB. Subsequent IκB degradation liberates NF-κB from the cytoplasm, resulting in the transcription of downstream target genes, including inflammatory cytokines (Cummins et al., 2006). Interestingly, NF-κB signaling in activated macrophages directly regulates Hif1α transcription (Rius et al., 2008). Macrophages lacking IKKβ cannot stabilize HIF1α after hypoxic or microbial challenge and exhibit decreased HIF target gene expression. However, activation of NF-κB alone is insufficient for HIF1α stabilization, indicating that maximal HIF1α accumulation depends on both transcriptional regulation by NF-κB and post-translational regulation by hypoxia (Rius et al., 2008). This coordinated response suggests a mechanism for graded HIF1α expression in immune cells, in which maximal HIF1α activity is induced in cells located at the site of most severe inflammation, where hypoxic and inflammatory stresses are simultaneously present.
In contrast, hypoxic induction of HIF2α does not require IKKβ (Rius et al., 2008). A recent report proposes that HIF1α and HIF2α are differentially stabilized in macrophages exposed respectively to TH1 cytokines such as IFN-γ and to TH2 cytokines such as IL-4 (Takeda et al., 2010). Because IFN-γ but notIL-4 activates the NF-κB pathway in macrophages (Thieu et al., 2007), this finding appears to support the notion that HIF1α expression, but not that of HIF2α, is dependent on NF-κB. However, a subsequent study showed significant HIF2α induction with TH1 cytokines IFN-γ and LPS (Imtiyaz et al., 2010). Available data on this aspect of HIF2α regulation in macrophages are therefore conflicting. Given the role of HIF-2α in myeloid mediated inflammatory responses (see below), it will be important to fully assess the contribution of NF-κB signaling towards HIF2α activity in immune settings, as well as other mechanistic differences between HIF1α and HIF2α stabilization in hypoxic and inflammatory stress.
HIF and myeloid cell function
Myeloid cells neutrophils, macrophages, and dentritic cells—play key effector roles in acute and chronic inflammation. Conditional inactivation of HIF1α or HIF2α in these cells has revealed crucial roles for both subunits in mediating inflammatory responses (Figure 4B). The absence of HIF1α in myeloid cells significantly weakens the inflammatory response in mouse models of arthritis and chronic dermatitis, and confers increased protection against LPS induced sepsis (Peyssonnaux et al., 2007; Cramer et al., 2003). Macrophages lacking HIF1α exhibit decreased motility, invasiveness, and bacteriocidal activity (Peyssonnaux et al., 2005; Cramer et al., 2003). Similarly, conditional HIF2α loss in myeloid cells decreases macrophage motility and invasion and reduces the severity of cutaneous inflammation and LPS-induced endotoxemia (Imtiyaz et al., 2010). The cytokine response to LPS-induced sepsis, characterized by the production of TNF-α, IL-1β, IL-6, and IL-12, is reduced in the absence of either HIF1α or HIF2α (Imtiyaz et al., 2010; Peyssonnaux et al., 2007). Interestingly, HIF1α and HIF2α coordinate similar inflammatory responses via distinct mechanisms. HIF1α controls the metabolic shift from oxidative phosphorylation to glycolysis in activated macrophages (Cramer et al., 2003). In contrast, HIF2α regulates the transcription of cytokines, such as IL1β, IL12, and TNFα, as well as genes involved in macrophage migration and chemotaxis, such as FN1 and CXCR4 (Imtiyaz et al., 2010). A gene expression profile analysis of primary human macrophages in which HIF1α or HIF2α were depleted confirms that HIF1α and HIF2α induce overlapping but distinct sets of genes (Fang et al., 2009).
Hypoxia and HIFs regulate other myeloid lineages also. Neutrophils express HIF1α, but not HIF2α, under hypoxic and inflammatory stress (Imtiyaz et al., 2010). HIF1α expression in these cells is required for host pathogen defence and neutrophil survival (Walmsley et al., 2006; Peyssonnaux et al., 2005). HIF1α expression in dentritic cells is required for the expression of co-stimulatory molecules after LPS exposure and the induction of allogenic T cell proliferation (Jantsch et al., 2008). In mast cells, HIF1α expression regulates the production of pro-inflammatory cytokines, vasodilators such as VEGF, and the histamine synthesizer histidine decarboxylase (reviewed in Nizet and Johnson, 2009). Therefore, HIFs coordinate the behavior of different immune cells in a hypoxic inflammatory milieu to produce a unified immune response.
HIF and tumor associated macrophages
Studies in many cancer types have shown that macrophage infiltration correlates with unfavorable clinical prognosis (reviewed in Lewis and Pollard, 2006). Macrophages are recruited to tumor areas primarily by the production of chemoattractants by hypoxic tumor and stromal cells, such as the HIF target genes CSF1 and VEGF (reviewed in Murdoch et al., 2004). Recent work has also shown that apoptotic cells, such as those in hypoxic regions of a tumor, produce soluble factors such as TGF-β to attract monocytes and macrophages (Herr et al., 2009). Once recruited, tumor associated macrophages (TAMs) exhibit a highly dynamic immune phenotype which promotes tumor growth, angiogenesis, metastasis, and tumor immunosuppression. Furthermore, as Harris and colleagues noted, TAMs exhibit elevated HIF1α and HIF2α expression due to the hypoxic tumor microenvironment (reviewed in Murdoch et al., 2004).
HIF2α expression in breast and cervical cancer TAMs is correlated with unfavorable prognoses, suggesting a functional relevance for HIF2α in this setting (Kawanaka et al., 2008; Leek et al., 2002). Conditional HIF2α deletion in the myeloid lineage in mice has revealed key roles for HIF2α and TAMs in tumorigenesis. In mouse models of hepatocellular carcinoma and colitis-associated colon carcinoma, mice lacking HIF2α in their myeloid cells exhibit decreased recruitment of TAMs into tumor areas (Imtiyaz et al., 2010). This finding correlates with reduced tumor mitotic index, lower tumor grade, and a downward trend in the number and size of colitis-induced colon carcinomas (Imtiyaz et al., 2010). It will be of interest to explore whether HIF1α expression in TAMs is functionally relevant to tumor progression in similar in vivo models, and if so, whether HIF1α and HIF2α complement each other in this context. Recent work suggests that the absence of HIF1α in macrophages has no effect on tumor spheroid infiltration, tumor cell proliferation, or tumor invasiveness in vitro, but reduces cell death in tumor spheroid cultures (Werno et al., 2010).
HIF in a Systemic Response to Hypoxia
Organs involved in erythropoiesis (red blood cell production) can respond to systemic hypoxia to increase red blood cell numbers and the O2-carrying capacity of blood (reviewed in Fandrey, 2004). EPO, a preferential HIF2α target, stimulates this process (reviewed in Lee, 2008; reviewed in Fandrey, 2004). Abundant data suggest that the PHD2/pVHL/HIF2α axis controls EPO levels and, therefore, adult erythropoiesis in humans and mice. Human genetic studies of familial polycythemia (abnormally elevated hemoglobin or red blood cell count) identified mutations in VHL, which impaired HIF1α degradation (reviewed in Lee, 2008). Subsequent genetic and biochemical analyses have identified inactivating mutations in PHD2 and activating lesions in EPAS1/HIF2α (reviewed in Lee, 2008; Furlow et al., 2009). Mouse models bearing mutations in these genes have also been generated and exhibit abnormal erythropoiesis (reviewed in Lee, 2008). More recent studies indicate that while the kidney and liver are the main producers of EPO, HIFα expression in other distant tissues—skin and glial cells—also seems to influence EPO production in response to hypoxia (Boutin et al., 2008; Weidemann et al., 2009).
Furthermore, studies comparing Tibetan highlanders and the closely related lowland Han Chinese have provided an evolutionary link between the PHD2/pVHL/HIF2α axis and erythropoiesis (Yi et al., 2010; Beall et al., 2010; Simonson et al., 2010). The authors compared the frequencies of single-nucleotide polymorphism (SNP) alleles between these groups, and noted significant divergence in allelic frequency of SNPs located in or near the PHD2/EGLN1 and EPAS1/HIF2α genes. These findings correlated with lower hemoglobin and erythrocyte levels in the blood of Tibetan subjects, suggesting that the PHD2 and HIF2α alleles common to Tibetan highlanders cause relatively decreased erythropoiesis. It has been proposed that the divergence in HIF2α and PHD2 occurred through natural selection, whereby Tibetan HIF2α and PHD2 alleles facilitate survival in the high altitude of the Tibetan plateau. For instance, Tibetans are resistant to developing chronic mountain sickness, which is marked by elevated erythropoiesis. These reports highlight the evolutionary implications of PHD2 and HIF2α function in red blood cell production.
In sum, studies in humans and rodents have contributed to a body of knowledge linking the PHD/pVHL/HIF pathway to erythropoiesis. This information is now being applied to the treatment of anemia (see below).
HIF-independent Responses to Hypoxic Stress
While this review emphasizes the role of HIFs in response to O2 deprivation, the hypoxic response is an integration of multiple O2 sensing pathways. Substantial evidence indicates that mTOR signaling and the unfolded protein response (UPR) play critical roles in hypoxic adaptations by modulating protein translation, cell metabolism, and cell fate (Wouters and Koritzinsky, 2008). We must also consider that other transcriptional regulators, such as PGC1α, can complement the HIF response in ischemic settings: PGC1α, independent of HIF, promotes VEGF expression and neo-angiogenesis in a model of hindlimb ischemia (Arany et al., 2008).
In addition, PHDs and FIH1 have non-HIFα substrates which may underlie some of their biological functions (Webb et al., 2009a). For example, recent reports indicate that prolyl hydroxylases play significant HIF-independent roles in cancer. PHD2 suppresses growth of xenograft tumors in a HIF-and, surprisingly, hydroxylase-independent fashion (Chan et al., 2009). PHD1/EglN2, on the other hand, promotes tumor growth through HIF-independent regulation of Cyclin D1 (Zhang et al., 2009). It is also important to note that all 2-oxoglutarate-dependent dioxygenases, including but not limited to PHDs, require O2 for their enzymatic activity and therefore could potentially mediate HIF-independent responses to hypoxia.
These observations emphasize that hypoxic adaptations are mediated by more than the HIF response. Hypoxia can activate many distinct pathways and influence less established branches of the canonical PHD/pVHL/HIFα pathway.
HIF-targeted Therapies
Many insights from HIF biology are being translated into clinical applications and, in particular, drug discovery. The most advanced HIF-targeted pharmaceuticals in terms of clinical development, to date, are PHD inhibitors. These compounds, FG-2216 and FG-4592, are being evaluated for treatment of anemia and are currently in Phase I and II clinical trials (ClinicalTrials. gov Identifiers NCT00456053, NCT00761657, NCT00978198, and NCT00978198).
HIF activators
In addition to PHD inhibition, several strategies to promote HIF1α activity and angiogenesis are in development for use in ischemic disease. In models of hindlimb ischemia, adenoviral delivery of constituitively active HIF1α has shown promise when administered alone or in combination with bone-marrow derived angiogenic cells (Bosch-Marce et al., 2007; Rey et al., 2009). HIF1α adenoviral therapy has also shown benefit in limb ischemia models in aged and diabetic mice (Bosch-Marce et al., 2007; Sarkar et al., 2009). These findings are significant if one considers that two large patient populations afflicted by atherosclerosis and associated ischemic diseases are diabetics and the elderly (Beckman et al., 2002).
Similar approaches with hybrid HIF1α/VP16 have been used in rabbit and diabetic rat models of limb ischemia and have even progressed through Phase I and II clinical studies in patients with severe peripheral arterial disease (Vincent et al., 2000; Rajagopalan et al., 2007; Kajiwara et al., 2009). These interventions have also been applied to other ischemic injuries such as wound healing and myocardial infarction (Liu et al., 2008; Mace et al., 2007; Heinl-Green et al., 2005). In addition to gene therapy, PHD inhibitors have also shown utility in wound healing in diabetic animals (Botusan et al., 2008).
Overall, these findings indicate that HIF activating therapies are effective in pre-clinical studies of ischemia and merit investigation in patients suffering from ischemic disease.
HIF inhibitors
Transcription factors have historically been considered undruggable targets. However, interest in HIF inhibition as a therapeutic strategy remains high (Semenza, 2007). A high-throughput screen of FDA-approved drugs for anti-HIF activity revealed that digoxin and other cardiac glycosides inhibit HIFα translation and subcutaneous xenograft growth (Zhang et al., 2008). Importantly, HIFα inhibition is independent of digoxin’s known effect on the Na+/K+ ATPase, but necessary for its tumor suppressive effect (Zhang et al., 2008). Other HIF inhibitors identified in similar screens include the antiseptic dye acriflavine, and anthracyclines such as doxorubicin and daunorubicin (Lee et al., 2009a, 2009b). Screening for HIF inhibitors among approved agents means that identified compounds, although pharmacologically well studied and suitable for human use, can be presumed to exhibit HIF-independent effects stemming from their original therapeutic purposes. Therefore, for these drugs to be used in targeted HIF therapy, it will be crucial to demonstrate that HIF repression is sufficient for their intended biological effects.
Another challenge in HIF targeting involves the overlapping but distinct biological roles of HIFα subunits. Compounds that promote the binding of IRP1 to the 5′ UTR of HIF2α mRNA decrease HIF2α hypoxic induction, but also repress HIF1α synthesis via an independent mechanism (Zimmer et al., 2008). An RNA antagonist of HIF1α, EZN-2968, reduces HIF1α protein and target gene expression in vitro and in vivo, but not that of HIF2α, and is being evaluated in a Phase I clinical trial (Patnaik et al., 2009; Greenberger et al., 2008). We expect that RNAi will play a key role in targeted HIF therapy once effective and selective delivery methods become available.
With the emerging view of the importance of HIF1α and HIF2α in disease, there is a largely unmet need for specific HIF inhibitors. While combined inhibition of HIFα isoforms will be appropriate in certain disease situations, HIF1α or HIF2α specific therapies may be preferable in other scenarios.
Synthetic lethality with VHL deficiency
In many cancers, re-activating tumor suppressors could provide a therapeutic benefit. However, it is challenging to design drugs for this purpose. Several groups, therefore, have adopted a synthetic lethal screening approach to overcome this hurdle. Two genes are synthetically lethal if inhibition of either gene is compatible with viability, but inhibition of both leads to cell death. In ccRCC, several groups have screened for genes which are synthetically lethal with VHL (Bommi-Reddy et al., 2008; Turcotte et al., 2008). One group screened a shRNA library targeting various kinases and identified several targets, such as CDK6, for which chemical inhibitors already exist (Bommi-Reddy et al., 2008). Another screen employed a drug library and identified a compound, STF-62247, which promoted autophagic cell death in VHL-deficient cells (Turcotte et al., 2008). Surprisingly, in several cases, VHL-replete cells with stable HIF2α expression were viable in the presence of drug or kinase-specific shRNA, suggesting the observed death in VHL-deficient cells is HIF-independent (Turcotte et al., 2008; Bommi-Reddy et al., 2008). In sum, these screens implicate new molecular targets and compounds in the treatment of VHL-deficient ccRCC.
Conclusion
Hypoxic stress is characteristic of many pathological settings, and the HIFs direct critical adaptations to enable cells, tissues, and organisms to survive and thrive in these conditions. Recent work has revealed new mechanisms of HIF induction, including PHD-dependent and -independent modes of regulation, as well as new effects of activating the HIF response in development and disease. In some cases, these responses promote disease progression, while in others, HIF responses are a part of disease recovery. Recent evidence has also highlighted the common and distinguishing features between HIF1α- and HIF2α-mediated responses in cancer, tissue ischemia, and inflammatory disease. A deeper understanding of how HIFα isoforms are uniquely regulated and how they can be selectively modulated will be essential for translating our current knowledge of the HIF pathway to clinical settings.
Acknowledgments
We thank Brian Keith and other members of our laboratory for helpful discussions, and Kelly Clark for assistance with figure preparation. We apologize to our colleagues whose work has not been directly cited because of space limitations.
Bibliography
- Adams L, Lindor K. Nonalcoholic Fatty Liver Disease. Ann Epidemiol. 2007;17:863–869. doi: 10.1016/j.annepidem.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Aghili M, Zahedi F, Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J Neurooncol. 2009;91:233–236. doi: 10.1007/s11060-008-9706-2. [DOI] [PubMed] [Google Scholar]
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1[alpha] Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- Aragones J, Schneider M, Van Geyte K, Fraisl P, Dresselaers T, Mazzone M, Dirkx R, Zacchigna S, Lemieux H, Jeoung NH, et al. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat Genet. 2008;40:170–180. doi: 10.1038/ng.2007.62. [DOI] [PubMed] [Google Scholar]
- Arany Z, Foo S, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1α. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- Bakker WJ, Harris IS, Mak TW. FOXO3a Is Activated in Response to Hypoxic Stress and Inhibits HIF1-Induced Apoptosis via Regulation of CITED2. Mol Cell. 2007;28:941–953. doi: 10.1016/j.molcel.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Baranova O, Miranda LF, Pichiule P, Dragatsis I, Johnson RS, Chavez JC. Neuron-Specific Inactivation of the Hypoxia Inducible Factor 1{alpha} Increases Brain Injury in a Mouse Model of Transient Focal Cerebral Ischemia. J Neurosci. 2007;27:6320–6332. doi: 10.1523/JNEUROSCI.0449-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basner RC. Continuous Positive Airway Pressure for Obstructive Sleep Apnea. N Engl J Med. 2007;356:1751–1758. doi: 10.1056/NEJMct066953. [DOI] [PubMed] [Google Scholar]
- Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, Li C, Li JC, Liang Y, McCormack M, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 2010;107:11459 –11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman JA, Creager MA, Libby P. Diabetes and Atherosclerosis: Epidemiology, Pathophysiology, and Management. JAMA. 2002;287:2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, Cordon-Cardo C, Celeste Simon M, Rafii S, Pandolfi PP. PML inhibits HIF-1[alpha] translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–785. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- Bertout JA, Majmundar AJ, Gordan JD, Lam JC, Ditsworth D, Keith B, Brown EJ, Nathanson KL, Simon MC. HIF2 inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc Natl Acad Sci U S A. 2009;106:14391–14396. doi: 10.1073/pnas.0907357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–975. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, Neckers L. p53 Inhibits Hypoxia-inducible Factor-stimulated Transcription. Journal of Biological Chemistry. 1998;273:11995 –11998. doi: 10.1074/jbc.273.20.11995. [DOI] [PubMed] [Google Scholar]
- Bommi-Reddy A, Almeciga I, Sawyer J, Geisen C, Li W, Harlow E, Kaelin WG, Grueneberg DA. Kinase requirements in human cells: III. Altered kinase requirements in VHL/cancer cells detected in a pilot synthetic lethal screen. Proc Natl Acad Sci U S A. 2008;105:16484–16489. doi: 10.1073/pnas.0806574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Marce M, Okuyama H, Wesley JB, Sarkar K, Kimura H, Liu YV, Zhang H, Strazza M, Rey S, Savino L, et al. Effects of Aging and Hypoxia-Inducible Factor-1 Activity on Angiogenic Cell Mobilization and Recovery of Perfusion After Limb Ischemia. Circ Res. 2007;101:1310–1318. doi: 10.1161/CIRCRESAHA.107.153346. [DOI] [PubMed] [Google Scholar]
- Bostrom P, Magnusson B, Svensson P, Wiklund O, Boren J, Carlsson LMS, Stahlman M, Olofsson S, Hulten LM. Hypoxia Converts Human Macrophages Into Triglyceride-Loaded Foam Cells. Arterioscler Thromb Vasc Biol. 2006;26:1871–1876. doi: 10.1161/01.ATV.0000229665.78997.0b. [DOI] [PubMed] [Google Scholar]
- Botusan IR, Sunkari VG, Savu O, Catrina AI, Grünler J, Lindberg S, Pereira T, Ylä-Herttuala S, Poellinger L, Brismar K, et al. Stabilization of HIF-1α is critical to improve wound healing in diabetic mice. Proc Natl Acad Sci U S A. 2008;105:19426–19431. doi: 10.1073/pnas.0805230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin AT, Weidemann A, Fu Z, Mesropian L, Gradin K, Jamora C, Wiesener M, Eckardt K, Koch CJ, Ellies LG, et al. Epidermal Sensing of Oxygen Is Essential for Systemic Hypoxic Response. Cell. 2008;133:223–234. doi: 10.1016/j.cell.2008.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Kaelin WG., Jr Dysregulation of HIF and VEGF is a unifying feature of the familial hamartoma syndromes. Cancer Cell. 2004;6:7–10. doi: 10.1016/j.ccr.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, et al. A Perivascular Niche for Brain Tumor Stem Cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, Holsboer F, Arzt E. RSUME, a Small RWD-Containing Protein, Enhances SUMO Conjugation and Stabilizes HIF-1[alpha] during Hypoxia. Cell. 2007;131:309–323. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- Chan DA, Kawahara TL, Sutphin PD, Chang HY, Chi J, Giaccia AJ. Tumor Vasculature Is Regulated by PHD2-Mediated Angiogenesis and Bone Marrow-Derived Cell Recruitment. Cancer Cell. 2009;15:527–538. doi: 10.1016/j.ccr.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kang X, Zhang S, Yeh ET. SUMO-Specific Protease 1 Is Essential for Stabilization of HIF1alpha during Hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello KL, Simon MC. HIFs, Hypoxia, and Vascular Development. Curr Top Dev Biol Volume. 2004;62:37–54. doi: 10.1016/S0070-2153(04)62002-3. [DOI] [PubMed] [Google Scholar]
- Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, et al. HIF-1[alpha] Is Essential for Myeloid Cell-Mediated Inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins EP, Berra E, Comerford KM, Ginouves A, Fitzgerald KT, Seeballuck F, Godson C, Nielsen JE, Moynagh P, Pouyssegur J, et al. Prolyl hydroxylase-1 negatively regulates IκB kinase-β, giving insight into hypoxia-induced NFκB activity. Proc Natl Acad Sci U S A. 2006;103:18154–18159. doi: 10.1073/pnas.0602235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD+-dependent deacetylases. Trends Biochem Sci. 2003;28:41–48. doi: 10.1016/s0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of Hypoxia-Inducible Factor 2 Signaling by the Stress-Responsive Deacetylase Sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Clarke SL, Ding K, Repa JJ, Garcia JA. HIF-2 -Haploinsufficient Mice Have Blunted Retinal Neovascularization Due to Impaired Expression of a Proangiogenic Gene Battery. Investigative Ophthalmology & Visual Science. 2008;49:2714–2720. doi: 10.1167/iovs.07-1469. [DOI] [PubMed] [Google Scholar]
- Du R, Lu KV, Petritsch C, Liu P, Ganss R, Passegué E, Song H, VandenBerg S, Johnson RS, Werb Z. HIF1α Induces the Recruitment of Bone Marrow-Derived Vascular Modulatory Cells to Regulate Tumor Angiogenesis and Invasion. Cancer Cell. 2008;13:206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvert G, Kappel A, Heidenreich R, Englmeier U, Lanz S, Acker T, Rauter M, Plate K, Sieweke M, Breier G, et al. Cooperative Interaction of Hypoxia-inducible Factor-2α (HIF-2α) and Ets-1 in the Transcriptional Activation of Vascular Endothelial Growth Factor Receptor-2 (Flk-1) J Biol Chem. 2003;278:7520–7530. doi: 10.1074/jbc.M211298200. [DOI] [PubMed] [Google Scholar]
- Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci U SA. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler J, Bennewith K, Cox T, Lang G, Bird D, Koong A, Le Q, Giaccia A. Hypoxia-Induced Lysyl Oxidase Is a Critical Mediator of Bone Marrow Cell Recruitment to Form the Premetastatic Niche. Cancer Cell. 2009;15:35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fandrey J. Oxygen-dependent and tissue-specific regulation of erythropoietin gene expression. Am J Physiol Regul Integr Comp Physiol. 2004;286:R977–988. doi: 10.1152/ajpregu.00577.2003. [DOI] [PubMed] [Google Scholar]
- Fang H, Hughes R, Murdoch C, Coffelt SB, Biswas SK, Harris AL, Johnson RS, Imityaz HZ, Simon MC, Fredlund E, et al. Hypoxia-inducible factors 1 and 2 are important transcriptional effectors in primary macrophages experiencing hypoxia. Blood. 2009;114:844–859. doi: 10.1182/blood-2008-12-195941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franovic A, Holterman CE, Payette J, Lee S. Human cancers converge at the HIF-2 oncogenic axis. Proc Natl Acad Sci U S A. 2009;106:21306–21311. doi: 10.1073/pnas.0906432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frede S, Stockmann C, Freitag P, Fandrey J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-κB. Biochem J. 2006;396:517–527. doi: 10.1042/BJ20051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlow PW, Percy MJ, Sutherland S, Bierl C, McMullin MF, Master SR, Lappin TRJ, Lee FS. Erythrocytosis-associated HIF-2α Mutations Demonstrate a Critical Role for Residues C-terminal to the Hydroxylacceptor Proline. J Biol Chem. 2009;284:9050–9058. doi: 10.1074/jbc.M808737200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, Pouyssegur J, Yaniv M, Mechta-Grigoriou F. JunD Reduces Tumor Angiogenesis by Protecting Cells from Oxidative Stress. Cell. 2004;118:781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Ward JM, Porter FD, Wagner JR, Devor DE, Grinberg A, Emmert-Buck MR, Westphal H, Klausner RD, Linehan WM. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc Natl Acad Sci U S A. 1997;94:9102–9107. doi: 10.1073/pnas.94.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan J, Lal P, Dondeti V, Letrero R, Parekh K, Oquendo C, Greenberg R, Flaherty K, Rathmell W, Keith B, et al. HIF-α Effects on c-Myc Distinguish Two Subtypes of Sporadic VHL-Deficient Clear Cell Renal Carcinoma. Cancer Cell. 2008;14:435–446. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Bertout JA, Hu C, Diehl JA, Simon MC. HIF-2[alpha] Promotes Hypoxic Cell Proliferation by Enhancing c-Myc Transcriptional Activity. Cancer Cell. 2007a;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: Sibling Rivals for Control of Cancer Cell Metabolism and Proliferation. Cancer Cell. 2007b;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberger LM, Horak ID, Filpula D, Sapra P, Westergaard M, Frydenlund HF, Albaek C, Schroder H, Orum H. A RNA antagonist of hypoxia-inducible factor-1, EZN-2968, inhibits tumor cell growth. Mol Cancer Ther. 2008;7:3598–3608. doi: 10.1158/1535-7163.MCT-08-0510. [DOI] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian Sirtuins: Biological Insights and Disease Relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinl-Green A, Radke PW, Munkonge FM, Frass O, Zhu J, Vincent K, Geddes DM, Alton EW. The efficacy of a ‘master switch gene’ HIF-1α in a porcine model of chronic myocardial ischaemia. Eur Heart J. 2005;26:1327–1332. doi: 10.1093/eurheartj/ehi223. [DOI] [PubMed] [Google Scholar]
- Helton R, Cui J, Scheel JR, Ellison JA, Ames C, Gibson C, Blouw B, Ouyang L, Dragatsis I, Zeitlin S, et al. Brain-Specific Knock-Out of Hypoxia-Inducible Factor-1{alpha} Reduces Rather Than Increases Hypoxic-Ischemic Damage. J Neurosci. 2005;25:4099–4107. doi: 10.1523/JNEUROSCI.4555-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr B, Zhou J, Werno C, Menrad H, Namgaladze D, Weigert A, Dehne N, Brune B. The supernatant of apoptotic cells causes transcriptional activation of hypoxia-inducible factor-1{alpha} in macrophages via sphingosine-1-phosphate and transforming growth factor-{beta} Blood. 2009;114:2140–2148. doi: 10.1182/blood-2009-01-201889. [DOI] [PubMed] [Google Scholar]
- Hu C, Wang L, Chodosh LA, Keith B, Simon MC. Differential Roles of Hypoxia-Inducible Factor 1{alpha} (HIF-1{alpha}) and HIF-2{alpha} in Hypoxic Gene Regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wu P, Bowker-Kinley MM, Harris RA. Regulation of Pyruvate Dehydrogenase Kinase Expression by Peroxisome Proliferator–Activated Receptor-α Ligands, Glucocorticoids, and Insulin. Diabetes. 2002;51:276–283. doi: 10.2337/diabetes.51.2.276. [DOI] [PubMed] [Google Scholar]
- Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le Q, Giaccia AJ. Hypoxia-Inducible mir-210 Regulates Normoxic Gene Expression Involved in Tumor Initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Levy FH, Kelly DP. Hypoxia Inhibits the Peroxisome Proliferator-activated Receptor α/Retinoid X Receptor Gene Regulatory Pathway in Cardiac Myocytes. J Biol Chem. 2001;276:27605–27612. doi: 10.1074/jbc.M100277200. [DOI] [PubMed] [Google Scholar]
- Imtiyaz H, Williams E, Hickey M, Patel S, Durham A, Yuan L, Hammond R, Gimotty P, Keith B, Simon M. Hypoxia inducible factor 2alpha regulates macrophage function in mouse models of acute and tumor inflammation. J Clin Invest. 2010;120:2699–2714. doi: 10.1172/JCI39506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs JS, Jung Y, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 Regulates a von Hippel Lindau-independent Hypoxia-inducible Factor-1α-degradative Pathway. J Biol Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- Jantsch J, Chakravortty D, Turza N, Prechtel AT, Buchholz B, Gerlach RG, Volke M, Glasner J, Warnecke C, Wiesener MS, et al. Hypoxia and Hypoxia-Inducible Factor-1{alpha} Modulate Lipopolysaccharide-Induced Dendritic Cell Activation and Function. J Immunol. 2008;180:4697–4705. doi: 10.4049/jimmunol.180.7.4697. [DOI] [PubMed] [Google Scholar]
- Jiang J, Xia X, Xu H, Xiong Y, Song W, Xiong S, Li Y. Inhibition of retinal neovascularization by gene transfer of small interfering RNA targeting HIF-1alpha and VEGF. J Cell Physiol. 2009;218:66–74. doi: 10.1002/jcp.21566. [DOI] [PubMed] [Google Scholar]
- Jungermann K. Metabolic Zonation of Liver Parenchyma. Semin Liver Dis. 1988;8:329–341. doi: 10.1055/s-2008-1040554. [DOI] [PubMed] [Google Scholar]
- Kaelin WG. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- Kaelin WG. ROS: Really involved in Oxygen Sensing. Cell Metab. 2005;1:357–358. doi: 10.1016/j.cmet.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Ratcliffe PJ. Oxygen Sensing by Metazoans: The Central Role of the HIF Hydroxylase Pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Kaidi A, Williams AC, Paraskeva C. Interaction between β-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol. 2007;9:210–217. doi: 10.1038/ncb1534. [DOI] [PubMed] [Google Scholar]
- Kajiwara H, Luo Z, Belanger AJ, Urabe A, Vincent KA, Akita GY, Cheng SH, Mochizuki S, Gregory RJ, Jiang C. A hypoxic inducible factor-1alpha hybrid enhances collateral development and reduces vascular leakage in diabetic rats. J Gene Med. 2009;11:390–400. doi: 10.1002/jgm.1318. [DOI] [PubMed] [Google Scholar]
- Kawanaka T, Kubo A, Ikushima H, Sano T, Takegawa Y, Nishitani H. Prognostic significance of HIF-2alpha expression on tumor infiltrating macrophages in patients with uterine cervical cancer undergoing radiotherapy. J Med Invest. 2008;55:78–86. doi: 10.2152/jmi.55.78. [DOI] [PubMed] [Google Scholar]
- Keith B, Simon M. Hypoxia-Inducible Factors, Stem Cells, and Cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BD, Hackett SF, Hirota K, Oshima Y, Cai Z, Berg-Dixon S, Rowan A, Yan Z, Campochiaro PA, Semenza GL. Cell Type-Specific Regulation of Angiogenic Growth Factor Gene Expression and Induction of Angiogenesis in Nonischemic Tissue by a Constitutively Active Form of Hypoxia-Inducible Factor 1. Circ Res. 2003;93:1074–1081. doi: 10.1161/01.RES.0000102937.50486.1B. [DOI] [PubMed] [Google Scholar]
- Kim WY, Perera S, Zhou B, Carretero J, Yeh JJ, Heathcote SA, Jackson AL, Nikolinakos P, Ospina B, Naumov G, et al. HIF2α cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009;119:2160–2170. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- Klimova T, Chandel NS. Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ. 2008;15:660–666. doi: 10.1038/sj.cdd.4402307. [DOI] [PubMed] [Google Scholar]
- Lee FS. Genetic causes of erythrocytosis and the oxygen-sensing pathway. Blood Rev. 2008;22:321–332. doi: 10.1016/j.blre.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kim Y, Kim IS, Kim B, Choi HJ, Lee JM, Shin HR, Kim JH, Kim J, Seo S, et al. Negative Regulation of Hypoxic Responses via Induced Reptin Methylation. Molecular Cell. 2010;39:71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Qian DZ, Rey S, Wei H, Liu JO, Semenza GL. Anthracycline chemotherapy inhibits HIF-1 transcriptional activity and tumor-induced mobilization of circulating angiogenic cells. Proc Natl Acad Sci U S A. 2009a;106:2353–2358. doi: 10.1073/pnas.0812801106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci U S A. 2009b;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Leek RD, Talks KL, Pezzella F, Turley H, Campo L, Brown NS, Bicknell R, Taylor M, Gatter KC, Harris AL. Relation of Hypoxia-inducible Factor-2{alpha} (HIF-2{alpha}) Expression in Tumor-infiltrative Macrophages to Tumor Angiogenesis and the Oxidative Thymidine Phosphorylase Pathway in Human Breast Cancer. Cancer Res. 2002;62:1326–1329. [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE. Hypoxia-Inducible Factors Regulate Tumorigenic Capacity of Glioma Stem Cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Marti GP, Wei X, Zhang X, Zhang H, Liu YV, Nastai M, Semenza GL, Harmon JW. Age-dependent impairment of HIF-1alpha expression in diabetic mice: Correction with electroporation-facilitated gene therapy increases wound healing, angiogenesis, and circulating angiogenic cells. J Cell Physiol. 2008;217:319–327. doi: 10.1002/jcp.21503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Zhong J, Chang R, Hu H, Pandey A, Semenza GL. Hsp70 and CHIP Selectively Mediate Ubiquitination and Degradation of Hypoxia-inducible Factor (HIF)-1 but Not HIF-2. J Biol Chem. 2009;285:3651–3663. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace KA, Yu DH, Paydar KZ, Boudreau N, Young DM. Sustained expression of Hif-1alpha in the diabetic environment promotes angiogenesis and cutaneous wound repair. Wound Repair Regen. 2007;15:636–645. doi: 10.1111/j.1524-475X.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, Gouvin LM, Sharma VM, Mercurio AM. ERβ Impedes Prostate Cancer EMT by Destabilizing HIF-1α and Inhibiting VEGF-Mediated Snail Nuclear Localization: Implications for Gleason Grading. Cancer Cell. 2010;17:319–332. doi: 10.1016/j.ccr.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JGN, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- Maynard M, Evans A, Shi W, Kim W, Liu F, Ohh M. Dominant-Negative HIF-3α4 Suppresses VHL-Null Renal Cell Carcinoma Progression. Cell Cycle. 2007;6:2810–2816. doi: 10.4161/cc.6.22.4947. [DOI] [PubMed] [Google Scholar]
- Mazumdar J, Hickey M, Pant D, Durham A, Sweet-Cordero A, Jacks T, Chodosh L, Kissil J, Simon M, Keith B. HIF-2alpha deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1001296107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone M, Dettori D, Leite de Oliveira R, Loges S, Schmidt T, Jonckx B, Tian Y, Lanahan AA, Pollard P, Ruiz de Almodovar C. Heterozygous Deficiency of PHD2 Restores Tumor Oxygenation and Inhibits Metastasis via Endothelial Normalization. Cell. 2009;136:839–851. doi: 10.1016/j.cell.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY, Dewhirst MW. Pleiotropic effects of HIF-1 blockade on tumor radiosensitivity. Cancer Cell. 2005;8:99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ. Genome-wide Association of Hypoxia-inducible Factor (HIF)-1 and HIF-2 DNA Binding with Expression Profiling of Hypoxia-inducible Transcripts. J Biol Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottet D, Dumont V, Deccache Y, Demazy C, Ninane N, Raes M, Michiels C. Regulation of Hypoxia-inducible Factor-1α Protein Level during Hypoxic Conditions by the Phosphatidylinositol 3-Kinase/Akt/Glycogen Synthase Kinase 3β Pathway in HepG2 Cells. J Biol Chem. 2003;278:31277–31285. doi: 10.1074/jbc.M300763200. [DOI] [PubMed] [Google Scholar]
- Muoio DM, Koves TR. Skeletal muscle adaptation to fatty acid depends on coordinated actions of the PPARs and PGC1α: implications for metabolic disease. Appl Physiol Nutr Metab. 2007;32:874–883. doi: 10.1139/H07-083. [DOI] [PubMed] [Google Scholar]
- Nanduri J, Wang N, Yuan G, Khan SA, Souvannakitti D, Peng Y, Kumar GK, Garcia JA, Prabhakar NR. Intermittent hypoxia degrades HIF-2α via calpains resulting in oxidative stress: Implications for recurrent apnea-induced morbidities. Proc Natl Acad Sci U S A. 2009;106:1199–1204. doi: 10.1073/pnas.0811018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik A, Chiorean EG, Tolcher A, Papadopoulos K, Beeram M, Kee D, Waddell M, Gilles E, Buchbinder A. EZN-2968, a novel hypoxia-inducible factor-1{alpha} (HIF-1{alpha}) messenger ribonucleic acid (mRNA) antagonist: Results of a phase I, pharmacokinetic (PK), dose-escalation study of daily administration in patients (pts) with advanced malignancies. J Clin Oncol (Meeting Abstracts) 2009;27:2564. [Google Scholar]
- Peng Y, Yuan G, Ramakrishnan D, Sharma SD, Bosch-Marce M, Kumar GK, Semenza GL, Prabhakar NR. Heterozygous HIF-1α deficiency impairs carotid body-mediated systemic responses and reactive oxygen species generation in mice exposed to intermittent hypoxia. J Physiol. 2006;577:705–716. doi: 10.1113/jphysiol.2006.114033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyssonnaux C, Cejudo-Martin P, Doedens A, Zinkernagel AS, Johnson RS, Nizet V. Cutting Edge: Essential Role of Hypoxia Inducible Factor-1{alpha} in Development of Lipopolysaccharide-Induced Sepsis. J Immunol. 2007;178:7516–7519. doi: 10.4049/jimmunol.178.12.7516. [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. HIF-1α expression regulates the bactericidal capacity of phagocytes. J Clin Invest. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Auwerx J. PPARγ and glucose homeostasis. Annu Rev Nutr. 2002;22:167–197. doi: 10.1146/annurev.nutr.22.010402.102808. [DOI] [PubMed] [Google Scholar]
- Pietras A, Hansford LM, Johnsson AS, Bridges E, Sjolund J, Gisselsson D, Rehn M, Beckman S, Noguera R, Navarro S, et al. HIF-2 maintains an undifferentiated state in neural crest-like human neuroblastoma tumor-initiating cells. Proc Natl Acad Sci U S A. 2009;106:16805–16810. doi: 10.1073/pnas.0904606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Nakayama K, Cardiff RD, Borowsky AD, Kaul K, Williams R, Krajewski S, Mercola D, Carpenter PM, Bowtell D, et al. Siah2-Dependent Concerted Activity of HIF and FoxA2 Regulates Formation of Neuroendocrine Phenotype and Neuroendocrine Prostate Tumors. Cancer Cell. 2010;18:23–38. doi: 10.1016/j.ccr.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Olin J, Deitcher S, Pieczek A, Laird J, Grossman PM, Goldman CK, McEllin K, Kelly R, Chronos N. Use of a Constitutively Active Hypoxia-Inducible Factor-1{alpha} Transgene as a Therapeutic Strategy in No-Option Critical Limb Ischemia Patients: Phase I Dose-Escalation Experience. Circulation. 2007;115:1234–1243. doi: 10.1161/CIRCULATIONAHA.106.607994. [DOI] [PubMed] [Google Scholar]
- Ramirez-Bergeron DL, Runge A, Adelman DM, Gohil M, Simon MC. HIF-Dependent Hematopoietic Factors Regulate the Development of the Embryonic Vasculature. Dev Cell. 2006;11:81–92. doi: 10.1016/j.devcel.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin EB, Rha J, Selak MA, Unger TL, Keith B, Liu Q, Haase VH. Hypoxia-Inducible Factor 2 Regulates Hepatic Lipid Metabolism. Mol Cell Biol. 2009;29:4527–4538. doi: 10.1128/MCB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratan R, Siddiq A, Smirnova N, Karpisheva K, Haskew-Layton R, McConoughey S, Langley B, Estevez A, Huerta P, Volpe B, et al. Harnessing hypoxic adaptation to prevent, treat, and repair stroke. J Mol Med. 2007;85:1331–1338. doi: 10.1007/s00109-007-0283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval RR, Lau KW, Tran MGB, Sowter HM, Mandriota SJ, Li J, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Contrasting Properties of Hypoxia-Inducible Factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-Associated Renal Cell Carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1α. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- Rey S, Lee K, Wang CJ, Gupta K, Chen S, McMillan A, Bhise N, Levchenko A, Semenza GL. Synergistic effect of HIF-1α gene therapy and HIF-1-activated bone marrow-derived angiogenic cells in a mouse model of limb ischemia. Proc Natl Acad Sci U S A. 2009;106:20399–20404. doi: 10.1073/pnas.0911921106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AM, Watson IR, Evans AJ, Foster DA, Irwin MS, Ohh M. Suppression of Hypoxia-Inducible Factor 2{alpha} Restores p53 Activity via Hdm2 and Reverses Chemoresistance of Renal Carcinoma Cells. Cancer Res. 2009;69:9056–9064. doi: 10.1158/0008-5472.CAN-09-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2α expression in iron deficiency. Nat Struct Mol Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, Akazawa H, Tateno K, Kayama Y, Harada M, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- Sarkar K, Fox-Talbot K, Steenbergen C, Bosch-Marcé M, Semenza GL. Adenoviral transfer of HIF-1α enhances vascular responses to critical limb ischemia in diabetic mice. Proc Natl Acad Sci U S A. 2009;106:18769–18774. doi: 10.1073/pnas.0910561106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid T, Zhou J, Khl R, BrüNe B. p300 relieves p53-evoked transcriptional repression of hypoxia-inducible factor-1 (HIF-1) Biochem J. 2004;380:289. doi: 10.1042/BJ20031299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Van Geyte Katie, Fraisl Peter, Kiss Judit, Aragonés Julián, Mazzone Massimiliano, Mairbäurl Heimo, De Bock Katrien, Ho Jeoung Nam, Mollenhauer Martin, et al. Loss or Silencing of the PHD1 Prolyl Hydroxylase Protects Livers of Mice Against Ischemia/Reperfusion Injury. Gastroenterology. 2010;138:1143–1154.e2. doi: 10.1053/j.gastro.2009.09.057. [DOI] [PubMed] [Google Scholar]
- Semenza G. Evaluation of HIF-1 inhibitors as anticancer agents. Drug Discov Today. 2007;12:853–859. doi: 10.1016/j.drudis.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Sendoel A, Kohler I, Fellmann C, Lowe SW, Hengartner MO. HIF-1 antagonizes p53-mediated apoptosis through a secreted neuronal tyrosinase. Nature. 2010;465:577–583. doi: 10.1038/nature09141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatrov VA, Sumbayev VV, Zhou J, Brune B. Oxidized low-density lipoprotein (oxLDL) triggers hypoxia-inducible factor-1{alpha} (HIF-1{alpha}) accumulation via redox-dependent mechanisms. Blood. 2003;101:4847–4849. doi: 10.1182/blood-2002-09-2711. [DOI] [PubMed] [Google Scholar]
- Shohet R, Garcia J. Keeping the engine primed: HIF factors as key regulators of cardiac metabolism and angiogenesis during ischemia. J Mol Med. 2007;85:1309–1315. doi: 10.1007/s00109-007-0279-x. [DOI] [PubMed] [Google Scholar]
- Simon MC. Coming up for air: HIF-1 and mitochondrial oxygen consumption. Cell Metabolism. 2006;3:150–151. doi: 10.1016/j.cmet.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Simon M. Siah Proteins, HIF Prolyl Hydroxylases, and the Physiological Response to Hypoxia. Cell. 2004;117:851–853. doi: 10.1016/j.cell.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, et al. Genetic Evidence for High-Altitude Adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- Skuli N, Liu L, Runge A, Wang T, Yuan L, Patel S, Iruela-Arispe L, Simon MC, Keith B. Endothelial deletion of hypoxia-inducible factor-2{alpha} (HIF-2{alpha}) alters vascular function and tumor angiogenesis. Blood. 2009;114:469–477. doi: 10.1182/blood-2008-12-193581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sluis B, Groot AJ, Vermeulen J, van der Wall E, van Diest PJ, Wijmenga C, Klomp LW, Vooijs M. COMMD1 Promotes pVHL and O2-Independent Proteolysis of HIF-1α via HSP90/70. PLoS ONE. 2009;4:e7332. doi: 10.1371/journal.pone.0007332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sluis B, Mao X, Zhai Y, Groot AJ, Vermeulen JF, van der Wall E, van Diest PJ, Hofker MH, Wijmenga C, Klomp LW, et al. COMMD1 disrupts HIF-1α/β dimerization and inhibits human tumor cell invasion. J Clin Invest. 2010;120:2119–2130. doi: 10.1172/JCI40583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockmann C, Doedens A, Weidemann A, Zhang N, Takeda N, Greenberg JI, Cheresh DA, Johnson RS. Deletion of vascular endothelial growth factor in myeloid cells accelerates tumorigenesis. Nature. 2008;456:814–818. doi: 10.1038/nature07445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarshan S, Sourbier C, Kong H, Block K, Romero VAV, Yang Y, Galindo C, Mollapour M, Scroggins B, Goode N, et al. Fumarate Hydratase Deficiency in Renal Cancer Induces Glycolytic Addiction and Hypoxia-Inducible Transcription Factor 1{alpha} Stabilization by Glucose-Dependent Generation of Reactive Oxygen Species. Mol Cell Biol. 2009;29:4080–4090. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T. Identification of Hypoxia-Inducible Factor-1 as a Novel Target for miR-17–92 MicroRNA Cluster. Cancer Res. 2008;68:5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- Takeda K, Ho VC, Takeda H, Duan L, Nagy A, Fong G. Placental but Not Heart Defects Are Associated with Elevated Hypoxia-Inducible Factor {alpha} Levels in Mice Lacking Prolyl Hydroxylase Domain Protein 2. Mol Cell Biol. 2006;26:8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, O’Dea EL, Doedens A, Kim J, Weidemann A, Stockmann C, Asagiri M, Simon MC, Hoffmann A, Johnson RS. Differential activation and antagonistic function of HIF-α isoforms in macrophages are essential for NO homeostasis. Genes Dev. 2010;24:491–501. doi: 10.1101/gad.1881410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Wang L, Esko J, Giordano FJ, Huang Y, Gerber H, Ferrara N, Johnson RS. Loss of HIF-1alpha in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell. 2004;6:485–495. doi: 10.1016/j.ccr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Thieu VT, Nguyen ET, McCarthy BP, Bruns HA, Kapur R, Chang C, Kaplan MH. IL-4-stimulated NF-{kappa}B activity is required for Stat6 DNA binding. J Leukoc Biol. 2007;82:370–379. doi: 10.1189/jlb.1106707. [DOI] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler MC, Hardie DG. AMP-Activated Protein Kinase in Metabolic Control and Insulin Signaling. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A Molecule Targeting VHL-Deficient Renal Cell Carcinoma that Induces Autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent KA, Shyu K, Luo Y, Magner M, Tio RA, Jiang C, Goldberg MA, Akita GY, Gregory RJ, Isner JM. Angiogenesis Is Induced in a Rabbit Model of Hindlimb Ischemia by Naked DNA Encoding an HIF-1{alpha}/VP16 Hybrid Transcription Factor. Circulation. 2000;102:2255–2261. doi: 10.1161/01.cir.102.18.2255. [DOI] [PubMed] [Google Scholar]
- Walmsley SR, Cowburn AS, Clatworthy MR, Morrell NW, Roper EC, Singleton V, Maxwell P, Whyte MKB, Chilvers ER. Neutrophils from patients with heterozygous germline mutations in the von Hippel Lindau protein (pVHL) display delayed apoptosis and enhanced bacterial phagocytosis. Blood. 2006;108:3176–3178. doi: 10.1182/blood-2006-04-018796. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The Common Feature of Leukemia-Associated IDH1 and IDH2 Mutations Is a Neomorphic Enzyme Activity Converting [alpha]-Ketoglutarate to 2-Hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waypa GB, Marks JD, Guzy R, Mungai PT, Schriewer J, Dokic D, Schumacker PT. Hypoxia Triggers Subcellular Compartmental Redox Signaling in Vascular Smooth Muscle Cells. Circ Res. 2010;106:526–535. doi: 10.1161/CIRCRESAHA.109.206334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb J, Coleman M, Pugh C. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell Mol Life Sci. 2009a;66:3539–3554. doi: 10.1007/s00018-009-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JD, Murányi A, Pugh CW, Ratcliffe PJ, Coleman ML. MYPT1, the targeting subunit of smooth-muscle myosin phosphatase, is a substrate for the asparaginyl hydroxylase factor inhibiting hypoxia-inducible factor (FIH) Biochem J. 2009b;420:327–333. doi: 10.1042/BJ20081905. [DOI] [PubMed] [Google Scholar]
- Weidemann A, Kerdiles YM, Knaup KX, Rafie CA, Boutin AT, Stockmann C, Takeda N, Scadeng M, Shih AY, Haase VH, et al. The glial cell response is an essential component of hypoxia-induced erythropoiesis in mice. J Clin Invest. 2009;119:3373–3383. doi: 10.1172/JCI39378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werno C, Menrad H, Weigert A, Dehne N, Goerdt S, Schledzewski K, Kzhyshkowska J, Brune B. Knockout of Hif-1alpha in Tumor-Associated Macrophages Enhances M2 Polarization and Attenuates Their Pro-Angiogenic Responses. Carcinogenesis. 2010 doi: 10.1093/carcin/bgq088. [DOI] [PubMed] [Google Scholar]
- Whitmer JT, Idell-Wenger JA, Rovetto MJ, Neely JR. Control of fatty acid metabolism in ischemic and hypoxic hearts. J Biol Chem. 1978;253:4305–4309. [PubMed] [Google Scholar]
- Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proceedings of the National Academy of Sciences. 2009;106:4260 –4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa M, Liu LX, Date T, Belanger AJ, Vincent KA, Akita GY, Kuriyama T, Cheng SH, Gregory RJ, Jiang C. Hypoxia-Inducible Factor-1 Mediates Activation of Cultured Vascular Endothelial Cells by Inducing Multiple Angiogenic Factors. Circ Res. 2003;93:664–673. doi: 10.1161/01.RES.0000093984.48643.D7. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, Lowenstein CJ. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Wu M, Chiou S, Chen P, Chang S, Liu C, Teng S, Wu K. Direct regulation of TWIST by HIF-1[alpha] promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Zhang H, Iwase T, Shen J, Semenza GL, Campochiaro PA. Digoxin inhibits retinal ischemia-induced HIF-1{alpha} expression and ocular neovascularization. FASEB J. 2010;24:1759–1767. doi: 10.1096/fj.09-145664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia Enhances the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Yuan G, Nanduri J, Khan S, Semenza GL, Prabhakar NR. Induction of HIF-1alpha expression by intermittent hypoxia: Involvement of NADPH oxidase, Ca(2+) signaling, prolyl hydroxylases, and mTOR. J Cell Physiol. 2008;217:674–685. doi: 10.1002/jcp.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Z, Maecker HL, Johnson RS, Giaccia AJ. Inhibition of PPARgamma2 Gene Expression by the HIF-1-Regulated Gene DEC1/Stra13: A Mechanism for Regulation of Adipogenesis by Hypoxia. Dev Cell. 2002;2:331–341. doi: 10.1016/s1534-5807(02)00131-4. [DOI] [PubMed] [Google Scholar]
- Zhang H, Qian DZ, Tan YS, Lee K, Gao P, Ren YR, Rey S, Hammers H, Chang D, Pili R, et al. Digoxin and other cardiac glycosides inhibit HIF-1 synthesis and block tumor growth. Proc Natl Acad Sci U S A. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Fu Z, Linke S, Chicher J, Gorman JJ, Visk D, Haddad GG, Poellinger L, Peet DJ, Powell F. The Asparaginyl Hydroxylase Factor Inhibiting HIF-1α Is an Essential Regulator of Metabolism. Cell Metab. 2010;11:364–378. doi: 10.1016/j.cmet.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Gu J, Li L, Liu J, Luo B, Cheung H, Boehm JS, Ni M, Geisen C, Root DE. Control of Cyclin D1 and Breast Tumorigenesis by the EglN2 Prolyl Hydroxylase. Cancer Cell. 2009;16:413–424. doi: 10.1016/j.ccr.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Mancuso A, Bui TV, Tong X, Gruber JJ, Swider CR, Sanchez PV, Lum JJ, Sayed N, Melo JV, et al. Imatinib resistance associated with BCR-ABL upregulation is dependent on HIF-1α-induced metabolic reprograming. Oncogene. 2010;29:2962–2972. doi: 10.1038/onc.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. Glioma-Derived Mutations in IDH1 Dominantly Inhibit IDH1 Catalytic Activity and Induce HIF-1. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1[alpha] Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M, Ebert B, Neil C, Brenner K, Papaioannou I, Melas A, Tolliday N, Lamb J, Pantopoulos K, Golub T. Small-Molecule Inhibitors of HIF-2a Translation Link Its 5′UTR Iron-Responsive Element to Oxygen Sensing. Mol Cell. 2008;32:838–848. doi: 10.1016/j.molcel.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]