Abstract
Objectives:
Brucellosis is the most common zoonotic disease that has been diagnosed mainly by serological tests and blood culture to some extent. This study was designed to establish a PCR technique for rapid diagnosis to be used in surveillance activities.
Methods:
The purpose of this study was firstly explained to the study population and verbal consent was obtained before sample collection. Peripheral blood was collected from 116 occupationally exposed groups with and without pyrexia of unknown origin from various districts of Punjab. Samples were subjected to blood culture, serological tests and DNA extraction was done using conventional laboratory extraction procedure. A primer pair B4/B5 that amplifies a gene encoding a 31 kDa immunogenic outer membrane protein (bcsp31) of Brucella species was used for PCR amplification.
Results:
The results showed that 8 (7%) of the cases had positive PCR and the detection threshold of primers used in this study were 715 cfu/ml. PCR results were 51.3% accurate for sensitivity of 12.6% and specificity of 100% using STAT as gold standard.
Conclusions:
Early-case reporting is possible by rapid tests like PCR. Thus, PCR is a promising diagnostic tool for routine investigation and surveillance of brucellosis which is the key element for management of prevention and control programmes. But patient condition before testing, optimal clinical specimen, sample volume used, simple and efficient DNA extraction protocol are the points of concern for PCR to be used as a routine test in clinical laboratory practice.
Keywords: Brucella, PCR, human brucellosis, blood, DNA extraction, India
INTRODUCTION
Brucellosis is the most common zoonotic disease that leads considerable economic losses in livestock industry and serious public health consequences in many parts of the world.1,2 The diagnosis of human brucellosis remains a clinical challenge especially to those unaware in view of the fact that its presentation can affect any organ or system.3 Even then the clinical picture of brucellosis alone cannot always lead to diagnosis since the symptoms are nonspecific and often atypical; therefore, diagnosis needs to be supported by laboratory tests. Although many serological tests and new automated blood culture techniques have been developed to diagnose brucellosis, there are still many difficulties in the diagnosis of the disease.4
Numerous PCR-based assays for Brucella have been developed and published since 1987 across the globe. The earliest assays were designed to exploit a single unique genetic locus that was highly conserved in Brucella like the BCSP31 or the 16S rRNA genes.5 The first published PCR-based diagnostic assay was reported by Fekete et al.6 This assay was based on the amplification of a 635-bp sequence from a gene encoding a 43-kDa outer membrane protein of B. abortus S19. However, the sensitivity and specificity of PCR for Brucella vary between laboratories and no standardization of sample preparation, target genes and detection methods have been established yet.7
In India, a lot of studies have been done on diagnosis of human brucellosis using conventional serological tests.8,9,10,11 But, there is scanty information on application of this molecular method for diagnosis, prevention and control of human brucellosis.12 Therefore, the purpose of this study was to apply PCR assay for rapid diagnosis of human brucellosis that helps in the management of prevention and control programmes.
METHODS
Clinical sample
The purpose of this study was explained to the study population and verbal consent was obtained from them before sample collection. About 10 ml of peripheral blood was collected from 116 occupationally exposed groups with and without pyrexia of unknown origin from various districts of Punjab over a period of 10 months. For serology, 5 ml venous blood was transferred to plain tubes and serum was separated from clotted blood by centrifuging at 1200 rpm for 10 min. Separated serum was collected in a screw caped sterilized plastic vial and stored at -20°C until use. For blood culture and PCR 5 ml of whole blood was asceptically transferred to screw-caped sterilized vials containing anticoagulant sodium citrate and stored at -20°C until use.
Bacteriological method
Conventional culture method was done for isolation and identification from 68 blood samples.13,14 A medium consisting of both a solid and a liquid phase in the same bottle, first described by Castaneda, was used to avoid the necessity for making repeated subcultures from liquid on to solid medium. Brucella agar and Brucella broth from Difco laboratories (BD India Pvt. Ltd., 204, Tolstoy House 15, Tolstoy Rd, New Delhi-110 00l) were used as solid and liquid phase, respectively.
Serological methods
Sera from 116 individuals were screened by RBPT and diagnosis was established in 64 (55.2%) cases using STAT with titre range between 80-1280 IU per ml.13,15 Rose Bengal and plain Brucella antigen required for this test was procured from Punjab Veterinary Vaccine Institute, Ludhiana, Punjab and stored at 4°C until use. PCR was applied on 64 serologically positive and 52 serologically negative cases.
DNA extraction from blood
A modification of the method described by Miller et al. was used for extraction of DNA from whole blood.16 Briefly, 0.5 ml of blood collected in sodium citrate was suspended in 1 ml of erythrocyte lysis solution (320 mM saccharose, 5 mM MgCl2, 1% Triton X-100, 10 mM Tris HCl [pH 7.5]), mixed, and centrifuged at 15,000 g for 3 min. The supernatant was discarded and the pellet was washed with 1 ml of Milli-Q water to remove the heme. Treatment with water was repeated until the leukocyte pellet lost all reddish colouring.
Template DNA was obtained from the leukocytes by adding 400 μl of nucleic lysis buffer (60 mM NH4Cl, 24 mM Na2-EDTA [pH 8.0]) containing proteinase K (1 mg/ml) and sodium dodecyl sulfate (1%). The solution was mixed and incubated for 2 hrs at 55°C. After digestion, the samples were cooled at room temperature and 100 μl of ammonium acetate (7.5 M) was added, followed by centrifugation at 15,000 g for 15 min. The supernatant containing total DNA was transferred to a fresh tube. Two volumes of absolute ethanol at room temperature were added and the tubes were inverted several times until the DNA precipitated. DNA was recovered by centrifuging the samples at 15,000 g for 10 min; the pellets were rinsed with 1 ml of 70% ethanol, dried and re-suspended in 25 μl of Milli-Q water and stored at -20° C until use.
Positive control was genomic DNA isolated from Brucella abortus S99 by boiling and chilling method. For this bacterial lysate preparation, 1 ml of NSS was heated in boiling water bath for 10 min and then snap chilled. From this 5 μl was used as a template in PCR. DNA from Pasteurella multocida (P52) was used as negative control.
DNA amplification
A target sequence of 223 bp in a gene encoding a 31 kDa immunogenic outer membrane protein (bcsp 31) of Brucella species was used for PCR amplification. The sequences of the primers were: B4 5’- TGGCTCGGTTGCCAATATCAA -3’ B5 5’- CGCGCTTGCCTTTCAGGTCTG -3’. The primers were supplied by OPERON Bio-technologies, Nattermannallee 1, 50829 Cologne Germany. PCR amplification of DNA using primers B4 and B5 specific for genus Brucella was standardized in 25 μl volume by varying the concentration of the reaction mix and cycling conditions. The reaction mixture contained 20 pmol of each primer, 0.2 mM dNTPs (10mM), 1x PCR buffer (10x), 1.5 mM MgCl2, 0.5 U Taq DNA polymerase, and 10 μl of template DNA. The cycling conditions were optimized at: initial denaturation at 93°C for 5 min, 40 cycles of template denaturation at 90°C for 1 min, 30 sec of primer annealing at 580C and 60 sec of primer extension at 72°C with final extension at 72°C for 7 min. In each PCR run, positive and negative controls were included to monitor performance of the run and absence of cross contamination. All the reactions were performed in a Master cycle Gradient thermocycler (Hybaid) with a preheated lid.
Analysis of PCR product
Ten microliter of amplified products were analysed in 1.5% agarose gel containing ethidium bromide at a final concentration of 0.5 Āg/ml after electrophoresis as per the method described by Sambrook and Russel.17
Detection limit of PCR assay
To determine the diagnostic sensitivity of PCR from blood, known numbers of bacterial cells were prepared by spread plate method as per the procedure described by Quinn et al.14 Brucella abortus S99 culture was inoculated into Brucella broth and incubated at 37°C for 4 days in microaerophillic environment using anaerobic system. Tenfold serial dilutions (10-1 to 10-10) were prepared by transfer of 1ml Brucella broth in to 9 ml of NSS in first tube followed by thorough mixing and transfer of 1ml into 9 ml of NSS in the second test tube and so on up to 10-10 dilution. From the highest 5 dilutions, 200 ml suspensions were individually inoculated onto separate Brucella agar plates and incubated at 37°C for 5 days. Colonies on the plates were counted and cfu/ml was determined for each dilution. To know the detection limit, known numbers of bacterial cells were added to serologically as well as PCR negative blood samples and aliquots of 0.5ml were used for DNA extraction as described above. The concentration and purity of extracted DNA was determined by measuring OD at 260 and 280 nm spectrophotometrically. Hence, PCR was employed on the last five dilutions containing 715, 230, 85, 55 and 35 cfu/ml. The procedure was repeated to ascertain the repeatability of the results.
RESULTS
There were 113 (97.4%) males and 3 (2.6%) females enrolled in this study. Age range of this study population was 19-64 years with mean and SD of 38.63 ± 11.58 years. Information on medical history of the cases and seropositivity are shown in Table 1.
Table 1.
Medical history of cases at presentation and seropositivity

In the preset study, 8 (7%) of the cases had positive PCR and all blood cultures were negative (Table 2). The primer set B4/B5 used was able to amplify a target sequence of 223 bp in a gene encoding a 31 kDa immunogenic outer membrane protein of Brucella species (Fig. 1 and 2). None of the serologically negative cases were positive by PCR. The relationship of Brucella antibody titre and PCR positivity is shown in Table 3.
Table 2.
The results of various diagnostic tests

Figure 1.
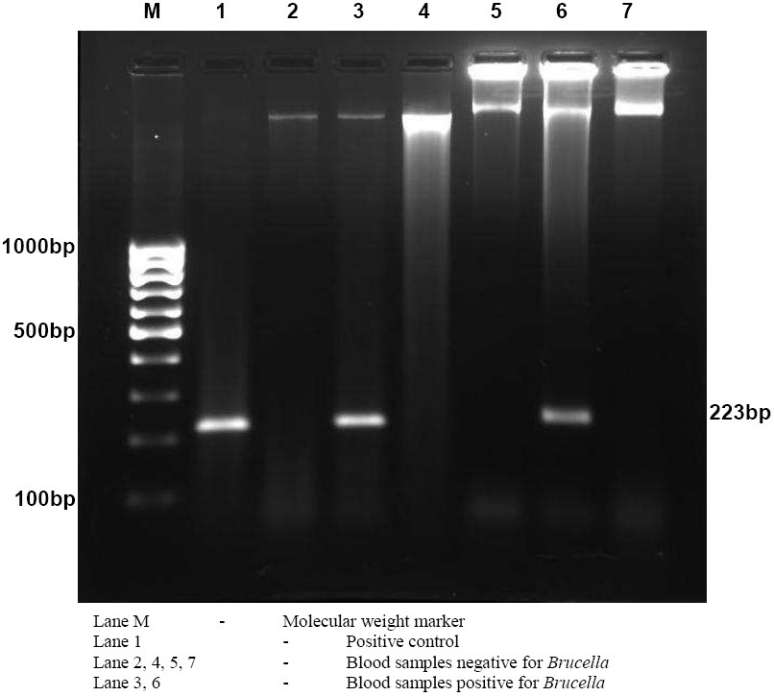
Agarose gel electrophoresis of PCR products obtained by amplification using B4/B5 primer set.
Figure 2.
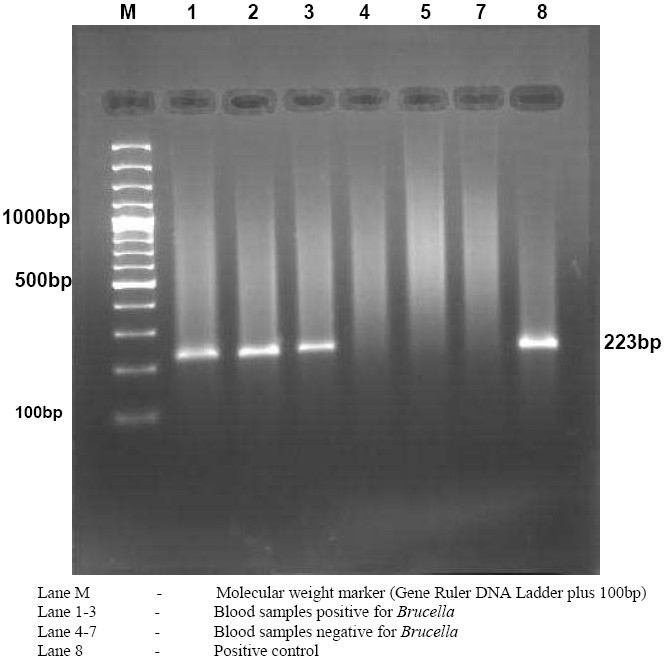
Gel electrophoresis of PCR amplified product showing 223bp band
Table 3.
Comparison of Brucella antibody titre and PCR

The findings on five serologically negative as well as PCR negative blood samples in duplicate for determination of detection limit of PCR assay using primer pair B4/B5 is shown in Table 4. The concentration and purity of extracted DNA from these experimentally inoculated blood samples were found to vary between 10.5-457 ng/μl and 1.72-1.86, respectively. PCR was positive only in spiked blood sample containing 715 cfu/ml (Figure 3).
Table 4.
Results of inoculated blood for detection threshold
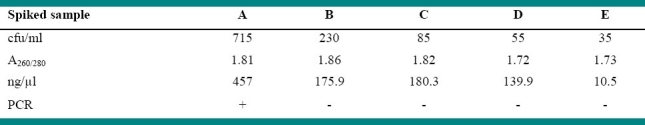
Figure 3.
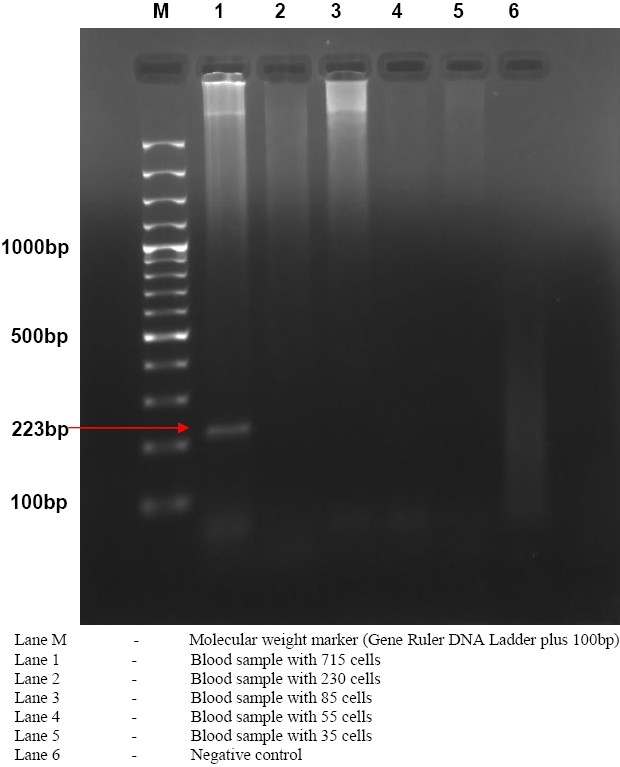
Gel electrophoresis of PCR amplified product of spiked blood samples for PCR limit of detection
The sensitivity and specificity of PCR technique were compared to that of blood culture and STAT titre ≥ 80 IU as gold standard (Table 5). The result revealed 51.3% accuracy for sensitivity of 12.6% and specificity of 100% using STAT as gold standard. Specificity of 88.1% was using blood culture as standard.
Table 5.
Comparative sensitivity, specificity and accuracy of PCR

DISCUSSION
Gender distribution in this study was 97.4% males and 2.6% females. In Egypt Ali et al. studied a sample that males were 72% and reported median age of 32 years (range 13-55) whereas in Spain Queipo-Ortuno et al. reported mean age of 37.9 years (range 14-91).18,19 twenty five percent of the patients had no clinical suspicion of brucellosis but diagnosed as seropositive. Mantur et al. also made diagnosis in 88.7% cases only by routine serology.20
PCR results using B4/B5 primers were positive in 2 out of 14 cases with low positive titre of 80 IU, in 2 out of 26 cases with titre 160 IU and 4 out of the 19 cases with titre 320 IU, while PCR results were negative in 4 and 1 cases with titre 640 and 1280 IU, respectively. This result strongly supports the suggestion by Mantur et al. that SAT titres of < 1: 160 cannot always be disregarded without follow up.20 On the contrary, SAT titres of ≥ 1: 160 do not always signify active infection, especially in Brucella endemic areas. Moreover, Joint FAO/WHO Expert committee on Brucellosis emphasized that in an individual repeatedly exposed to Brucella antigen, such as veterinary surgeons, serological tests are often strongly positive regardless of symptoms.21
In the findings of Ali et al., PCR results using B4/B5 and JPF/JPR primers were reported to be negative in 6 cases with low positive titre of 1: 160, in 7 out of 12 cases with titre 1: 320 and in 2 out of 16 cases with titre 1: 640, while PCR results were positive in 16 cases of titre 1: 1280.18 In Saudi Arabia El-Feki et al. also reported 80% PCR positive in symptomatic cases with a titre of ≥ 1: 80 from blood collected prior to antibiotic treatment.22 In Jordan, Nimri, considered titre of 1: 160 as positive for serodiagnosis and established diagnosis by PCR in 72.7% (120/165) and 12% by blood culture.23 On the contrary, none of these 8 PCR positive samples in our study were culture positive.
Primer set B4/B5 used in this study was able to detect 715 cfu/ml. Baddour and Alkhalifa, studied detection limit in three primer pairs and reported primer pair B4/B5, JPF/JPR and F4/R2 was also able to detect 700, 7 × 105 and 7 × 107 cfu/ml, respectively.25 Their study also revealed that B4/B5 primer pair was able to detect the smallest number of bacteria (700 cfu/mL). This finding corroborates well with the fact that some of the PCR false negatives in this study may be because the number of bacteria in blood sample was below 715 cfu/ml.
PCR results were 51.3% accurate for sensitivity of 12.6% and specificity of 100% using STAT as gold standard. Ali et al. reported accuracy of 82% for sensitivity of 79% and specificity of 100% using STA titre > 1: 160 as a standard, whereas 100% sensitivity using blood culture as standard for diagnosis.18 Nimri reported 100% sensitivity and specificity.23 The sensitivity of PCR results in our study was less compared to the findings of other researchers may be because the number of bacteria in peripheral circulation at the time of specimen collection was below the detection limit of primer pairs used in this study. Moreover, the case history of subjects included in this study revealed that some of the serologically positive patients had received treatment either for brucellosis or other non-specific complication with history of PUO. This may be considered as one of the reasons for lower sensitivity of both PCR and blood culture results in this study.
A review of Brucella bacteraemia by Pappas and Papadimitriou indicated that bacteraemia may be transient, initial event in human disease, followed by macrophage invasion, which is the central pathological event.24 Following intracellular replication, bacteraemia may reappear continuously or intermittently. As the disease evolves over time, bacteraemia tends to be absent, as is true for majority of chronic brucellosis cases. Moreover, it has been emphasised that in brucellosis extremely low bacterial load is needed to induce infection. This means that the initial bacteraemic course may run undetected due to the low number of circulating bacteria.
CONCLUSION
In conclusion, early-case reporting is possible by rapid tests like PCR. Thus, PCR is a promising diagnostic tool for routine investigation and surveillance of brucellosis which is the key element for management of prevention and control programmes. Although PCR is going to be the ultimate diagnostic tool for rapid diagnosis of human brucellosis, the results of the present study clearly indicate that patient condition before testing, optimal clinical specimen, sample volume used, simple and efficient DNA extraction protocol that can exclude PCR inhibitors are the points of concern for PCR to be used as a routine test in clinical laboratory practice. To our knowledge, PCR based diagnosis of human brucellosis have not been attempted in India, therefore further studies on these concerns needs to be explored before large scale application of this diagnostic tool for surveillance of the disease.
Acknowledgments
The authors would like to acknowledge Ethiopian Ministry of Education for partially sponsoring the principal investigator, Dr. Moti Yohannes, of this study as part of his Master's research during his stay in India. In addition, we would like to appreciate the technical assistance rendered by GADVASU and IVRI administrative as well as laboratory staffs in the realization of this project.
Footnotes
Conflict of interest statement: All authors declare that they have no conflict of interest.
Source of funding: None.
REFERENCES
- 1.Renukaradhya GJ, Isloor S, Rajasekhar M. Epidemiology, zoonotic aspects, vaccination and control/eradication of brucellosis in India. Vet Micro-biol. 2002;90(1-4):183–95. doi: 10.1016/s0378-1135(02)00253-5. [DOI] [PubMed] [Google Scholar]
- 2.Food and Agriculture Organization of the United Nations, World Organisationfor Animal Health, World Health Organization. Brucellosis in humans and animals. WHO. 2006 [Google Scholar]
- 3.University of Navarra. Pamplona, Spain: Brucellosis 2003 International Research Conference; 2003. Disease Spectrum and Laboratory Diagnosis of Human Brucellosis. [Google Scholar]
- 4.Mitka S, Anetakis C, Souliou E, Diza E, Kansouzidou A. Evaluation of different PCR assays for early detection of acute and relapsing brucellosis in humans in comparison with conventional methods. J Clin Microbiol. 2007;45(4):1211–8. doi: 10.1128/JCM.00010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bricker BJ. PCR as a diagnostic tool for brucellosis. Vet Microbiol. 2002;90(1-4):435–46. doi: 10.1016/s0378-1135(02)00228-6. [DOI] [PubMed] [Google Scholar]
- 6.Fekete A, Bantle JA, Halling SM, Sanborn MR. Preliminary development of a diagnostic test for Brucella using polymerase chain reaction. J Appl Bacteriol. 1990;69(2):216–27. doi: 10.1111/j.1365-2672.1990.tb01512.x. [DOI] [PubMed] [Google Scholar]
- 7.Navarro E, Casao MA, Solera J. Diagnosis of human brucellosis using PCR. Expert Rev Mol Diagn. 2004;4(1):115–23. doi: 10.1586/14737159.4.1.115. [DOI] [PubMed] [Google Scholar]
- 8.Thakur SD, Thapliyal DC. Seroprevalence of brucellosis in man. J Commun Dis. 2002;34(2):106–9. [PubMed] [Google Scholar]
- 9.Shringi BN, Sharma S, Sharma KN. Comparative study of conventional serological test for the diagnosis of brucellosis. Indian journal of animal sciences. 2002;72(7):553–4. [Google Scholar]
- 10.Mudaliar S, Bhore A, Pandit D. Detection of antibodies to Brucella abortus in animal handlers. Indian J Med Sci. 2003;57(5):181–6. [PubMed] [Google Scholar]
- 11.Mrunalini N, Reddy MS, Ramasastry P, Rao MR. Seroepidemiology of human brucellosis in Andhra Pradesh. Indian Veterinary Journal. 2004;81(7):744–7. [Google Scholar]
- 12.Mutnal MB, Purwar S, Metgud SC, Nagmoti MB, Patil CS. PCR confirmation of cutaneous manifestation due to Brucella melitensis. J Med Microbiol. 2007;56(Pt 2):283–5. doi: 10.1099/jmm.0.46927-0. [DOI] [PubMed] [Google Scholar]
- 13.Alton GG, Jones LM, Pietz DE. Laboratory techniques in brucellosis. 2nd ed. Switzerland: World Health Organisation, Geneva; 1975. [PubMed] [Google Scholar]
- 14.Quinn PJ, Carter ME, Markey BK, Carter GR. Clinical Veterinary Microbiology. Sydney, Toronto: Mosby; 1993. [Google Scholar]
- 15.OIE. Manual of standards for diagnostic tests and vaccines. Office International des. 2004 doi: 10.1016/s0065-3519(99)80052-4. [DOI] [PubMed] [Google Scholar]
- 16.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J. 3rd ed. Vol. 3. New York: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 18.El Kholy AA, Gomaa HE, El Anany MG, Abd El RE. Diagnosis of human brucellosis in Egypt by PCR. Journal of Infection in Developing Countries. 2007;1(2):177–81. [Google Scholar]
- 19.Queipo-Ortuno MI, Morata P, Ocon P, Manchado P, Colmenero JD. Rapid diagnosis of human brucellosis by peripheral-blood PCR assay. J Clin Microbiol. 1997;35(11):2927–30. doi: 10.1128/jcm.35.11.2927-2930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mantur BG, Biradar MS, Bidri RC, Mulimani MS, Veerappa, Kariholu P, et al. Protean clinical manifestations and diagnostic challenges of human brucellosis in adults: 16 years’ experience in an endemic area. J Med Microbiol. 2006;55(Pt 7):897–903. doi: 10.1099/jmm.0.46097-0. [DOI] [PubMed] [Google Scholar]
- 21.Joint FAO/WHO Expert Committee on Brucellosis: Sixth report. 6th ed. USA: WHO Publications Center; 1986. Technical report series / World Health Organization. [distributor] [Google Scholar]
- 22.Elfaki MG, Al Hokail AA, Nakeeb SM, Al Rabiah FA. Evaluation of culture, tube agglutination, and PCR methods for the diagnosis of brucellosis in humans. Med Sci Monit. 2005;11(11):MT69–74. [PubMed] [Google Scholar]
- 23.Nimri LF. Diagnosis of recent and relapsed cases of human brucellosis by PCR assay. BMC Infect Dis. 2003;3:5. doi: 10.1186/1471-2334-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pappas G, Papadimitriou P. Challenges in Brucella bacteraemia. Int J Antimicrob Agents. 2007;30(Suppl 1):S29–31. doi: 10.1016/j.ijantimicag.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Baddour MM, Alkhalifa DH. Evaluation of three polymerase chain reaction techniques for detection of Brucella DNA in peripheral human blood. Can J Microbiol. 2008;54(5):352–7. doi: 10.1139/w08-017. [DOI] [PubMed] [Google Scholar]


