Abstract
Thrombin-catalyzed activation of coagulation factor V (FV) is an essential positive feedback reaction within the blood clotting system. Efficient processing at the N- (Arg709-Ser710) and C-terminal activation cleavage sites (Arg1545-Ser1546) requires initial substrate interactions with 2 clusters of positively charged residues on the proteinase surface, exosites I and II. We addressed the mechanism of activation of human factor V (FV) using peptides that cover the entire acidic regions preceding these cleavage sites, FV (657-709)/ (FVa2) and FV(1481-1545)/(FVa3). FVa2 appears to interact mostly with exosite I, while both exosites are involved in interactions with the C-terminal linker. The 1.7-Å crystal structure of irreversibly inhibited thrombin bound to FVa2 unambiguously reveals docking of FV residues Glu666-Glu672 to exosite I. These findings were confirmed in a second, medium-resolution structure of FVa2 bound to the benzamidine-inhibited proteinase. Our results suggest that the acidic A2-B domain linker is involved in major interactions with thrombin during cofactor activation, with its more N-terminal hirudin-like sequence playing a critical role. Modeling experiments indicate that FVa2, and likely also FVa3, wrap around thrombin in productive thrombin·FV complexes that cover a large surface of the activator to engage the active site.
Introduction
Blood clotting occurs as the result of a complex network of kinetically controlled proteolytic reactions, which includes several positive and negative feedback loops. These loops are essential for the rapid, yet controlled, amplification of the triggering stimulus under physiologic conditions, so that the clot is confined to sites of vessel damage. Thrombin-catalyzed activation of the protein cofactors, factors V and VIII, initiates one of these major positive feedback loops (reviewed by Davie et al1; see Figure 1 for a schematic representation of the FV activation pathway). In the presence of Ca2+ ions and phosphatidylserine-containing membranes, activated factors VIII (FVIIIa) and V (FVa) bind to homologous serine proteinases, FIXa and FXa, respectively, to form critically important macromolecular complexes. Because factor X and prothrombin are their only natural substrates, FVIIIa·FIXa and FVa·FXa complexes are commonly referred to as the Xase and prothrombinase complexes, respectively.
Figure 1.
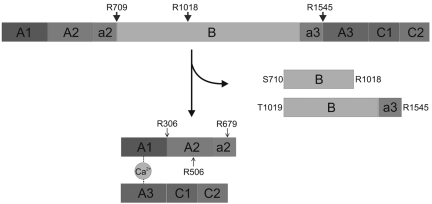
Mechanism of thrombin-catalyzed FV activation. Schematic representation of the human factor V activation pathway. Cleavage of the a2-B domain linker after Arg709 is followed by processing at the Arg1018-Thr1019 site, liberating the so-called fragment E (Ser710-Arg1018). Finally, slower proteolysis of the Arg1545-Ser1546 peptide bond releases fragment C1 (Thr1019-Arg1545), and generates a fully functional cofactor.48,49 Activated protein C (APC) cleavages at Arg306-Asn307, Arg506-Gly507, and Arg679-Lys680 that inactivate FVa are also indicated with arrowheads.
FV and FVIII are large multidomain glycoproteins with a shared A1-A2-B-A3-C1-C2 domain architecture. Relatively long polypeptide stretches rich in acidic residues termed a1, a2, and a3 connect FVIII domains A1 and A2, A2-B, and B-A3, respectively, and the domain organization of the cofactor can thus be described as A1-(a1)-A2-(a2)-B-(a3)-A3-C1-C2. Similarly, the domain structure of FV can be represented as A1-A2-(a2)-B-(a3)-A3-C1-C2. (Alignments of the sequences of FVa2 and FVa3 linkers from various mammalian species are given in supplemental Figure 1A and B, respectively). Physiologic FV/FVIII activation occurs on thrombin-mediated cleavage of 3 scissile peptide bonds, 2 of which, located at the a2-B and a3-A3 junctions, are topologically equivalent in both cofactors. These cleavages liberate the heavily glycosylated B domains and expose binding sites for the cognate proteases, FIXa and FXa, respectively.2–4
Recognition and processing of many physiologically relevant thrombin substrates requires initial interactions with one or both positively charged regions on the proteinase surface, exosites I and II5 (for a review on the general role of exosites in regulating blood coagulation reactions, see Bock et al6). Exosite I, also known as the fibrinogen-recognition exosite,7,8 interacts in addition with natural inhibitors (eg, hirudin)9,10 and cofactors, such as thrombomodulin.11 Exosite II is the major binding site for acidic glycosaminoglycans (eg, heparan sulfate), and for platelet glycoprotein Ib.12,13 Several biochemical and biophysical studies have demonstrated that both exosites are involved in FV/VIII activation.14–18 The electrostatic complementarity between the acidic a1-a3 regions of FV/FVIII and thrombin exosites immediately suggests that direct thrombin-peptide interactions mediate substrate recognition and regulate processing by efficiently presenting Arg-Xxx activation peptide bonds to the thrombin active site. Indeed, recent investigations have implicated a2 peptides in FV19,20 and FVIII activation.21 Similarly, FVa3 is essential for processing at the a3-A3 domain junction of human FV.22
Here we have studied the binding of human FVa2 and FVa3 to immobilized thrombin using surface plasmon resonance (SPR). In addition, we report 2 crystal structures of human α-thrombin bound to the a2 linker of coagulation FV. Against expectations, the structural analysis indicates that the more N-terminal, hirudin-like region Glu666-Glu672 interacts with thrombin exosite I. (Numbers for FV residues refer to the mature human protein). Modeling experiments strongly suggest a binding mode in which the more C-terminal part of the FVa2 bows along the “upper” thrombin surface and runs close to residues that form exosite II before entering the active-site cleft.
Methods
Materials
Human factor V was obtained from Cryopep or Abcam. The B-PER reagent was from Pierce Biotechnology. Nickel-nitrilotriacetic (Ni-NTA) agarose matrix was from QIAGEN GmbH. C4 columns and guard columns for HPLC were from Grace. Centrifugal filter devices were obtained from Millipore. Mini-PROTEAN precast gels were from Bio-Rad, and prestained molecular-weight markers for SDS-PAGE were purchased from Invitrogen. Crystal screen kits were from Hampton Research or Molecular Dimensions. All other chemicals, of the highest purity grade available, were purchased from Sigma-Aldrich or Merck.
Peptides
FV-derived peptides, H-IPDDDEDSYEIFEPPESTVMATR-OH (FV(657-679)), H-RKMHDRLEPEDEESDADYDYQNR-OH (FV(679-701)), H-DYDYQNR-OH (FV(695-701)), and H-DYDYQ-OH (FV(695-699)) were custom synthesized at the Parc Científic de Barcelona. The peptides were made via automated solid-phase methodology using 9-fluorenylmethoxy carbonyl (FMOC) N-protected amino acids, and purified by reverse-phase HPLC (RP-HPLC). Peptide purity (> 95%) was verified by HPLC and mass spectrometry.
Cloning, overexpression, and purification
Human FV regions Pro657-Arg709 and Pro1481-Arg1545 were cloned as fusion proteins with an N-terminal His6-tag followed by a tobacco etch virus (TEV) proteinase cleavage site. Synthetic genes encoding for these fragments were generated via oligonucleotide assembly, digested with restriction enzymes NdeI and BamHI, and ligated into the corresponding sites of the pET3a plasmid (Novagen). Oligonucleotide synthesis, assembly, and cloning into the expression vector were performed at Entelechon.
Bacterial pellets were extracted with B-PER, the supernatants were warmed to 75°C for 30 minutes, and centrifuged to remove denatured bacterial proteins. The supernatant, enriched in recombinant peptides, was incubated with Ni-NTA agarose matrix for 15 minutes with gentle shaking. After thorough washing of the affinity matrix, bound peptides were eluted with 500mM imidazole. The affinity-purified material was then subjected to reverse-phase chromatography on a C4 column (VYDAC) at a flow rate of 1.0 mL/min. The mobile phases were as follows: phase A, 5% acetonitrile, 0.1% trifluoroacetic acid (TFA) and phase B, 95% acetonitrile, 0.085% TFA. We used an optimized gradient for elution; 5% to 36.5% B over 2 minutes, 36.5% to 62.9% B over 7 minutes, and 62.9% to 95% B over 1 minute. Peptides were lyophilized overnight at −80°C and 0.01 mbar using a CryoDos freeze dryer (Telstar), and stored at −20°C until use. Identity of the highly purified peptides was assessed by mass spectrometric analysis of trypsin digests (supplemental Figure 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
FV activation
Human FV was incubated with α-thrombin (250:1 molar ratio) at 20°C in 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4, 150mM NaCl, 5mM CaCl2, in the presence or absence of 50μM recombinant FVa2 or FVa3. Aliquots of the reaction mixtures were taken at different times, immediately mixed with Laemmli sample buffer, and warmed at 95°C for 15 minutes. Samples were separated on 4%-15% acrylamide gradient gels, which were stained with silver following standard protocols.
Equilibrium binding
The affinity of FV peptides for immobilized thrombin was assessed using a Biacore 3000 system (GE Healthcare). Two different immobilization strategies were used, using CM5 and streptavidin (SA) chips, respectively.
CM5 chips.
Various concentrations of α-thrombin (either free or irreversibly inhibited with d-Phe-l-Pro-l-Arg chloromethyl ketone, FPR-CH2Cl) or its degraded forms, β- and γ-thrombin, were coupled to the chips following activation with an 1:1 mixture of 400mM 1-ethyl-3-(3-dimethylpropyl) carbodiimide (EDC) and 100mM N-hydroxysuccinimide (NHS). Trypsin was coupled to the reference channel at approximately the same density, as judged from the measured response units. To modulate the orientation of thrombin molecules toward the chip surface, we first examined the influence of small molecules that specifically target either exosite I or II, Tyr63-sulfated N-acetyl-hirudin(54-65) (Hir-(54-65)(SO3−)), or low-molecular-weight heparin, respectively. Unexpectedly, we were unable to immobilize α-thrombin in the presence of excess heparin, whereas essentially identical results were obtained in the presence and absence of 10μM Hir-(54-65)(SO3−). In view of the coupling chemistry, these findings strongly suggested that covalent coupling to the CM5 chip surface involved basic exosite II residues, most likely Lys235, Lys236, and/or Lys240 (We use the chymotrypsinogen numbering system for the catalytic domains of serine proteinases, in which residues are numbered according to their topologic equivalence to chymotrypsin[ogen]).
SA chips.
Alternatively, to enhance presentation of both exosites to the analytes, human thrombin was derivatized with biotin-labeled FPR-CH2Cl, and immobilized on SA chips. In this case, trypsin (previously inhibited with biotin-labeled FPR-CH2Cl) and human FXa derivatized with biotin-labeled Glu-Gly-Arg chloromethyl ketone (biotin-EGR-CH2Cl) were used as controls.
All binding experiments were conducted at 25°C in HBS-EP buffer (10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4, 150mM NaCl, 3mM EDTA, 0.005% [vol/vol] surfactant P20). Analytes were injected at different flow rates (from 10 to 60 μL/min), and sensograms were recorded and analyzed using BIAevaluation 4.1 software.
Fluorescence spectroscopy analysis
Fluorescence was measured with an Olis-upgraded SLM 8100 spectrofluorometer (On-Line Instrument Systems). Polyethylene glycol (PEG) 20 000-coated acrylic cuvettes were used to prevent protein absorption. Preparation of florescent probes, methodology, and data analysis were essentially as previously described.23–29 All experiments were conducted at 25°C in 50mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 110mM NaCl, 5mM CaCl2, 1 mg/mL PEG 8000, pH 7.4, in the presence of 1μM FPR-CH2Cl to prevent protein degradation. Briefly, human (pro)thrombin (100nM) was incubated in the presence of 5-fluorescein-labeled Hir-(54-65)(SO3−) ([5F]Hir-(54-65)(SO3−); 20nM) and titrated with the FV peptides. Changes in fluorescence intensity measured with excitation at 491 nm and emission at 520 nm (with band passes of 4 and 8 nm, respectively) as a function of peptide concentration were expressed as (Fobs − Fo)/Fo = ΔF/Fo and fitted by the quadratic binding equation to obtain the maximum fluorescence change ([Fmax − Fo]/Fo = ΔFmax/Fo) and the dissociation constant (KD). Nonlinear least squares fitting was performed with SCIENTIST software (MicroMath), with the stoichiometric factor (n) fixed at 1.
Crystallization
Thrombin solutions in 50mM HEPES, 125mM NaCl, pH 7.4, were incubated with molar excess of the FV peptides, and subjected to crystallization trials. Crystals grew in 100mM MOPS/HEPES-Na, pH 7.5, 12.5% (wt/vol) PEG 1000, 12.5% (wt/vol) PEG 3350, 12.5% (vol/vol) 2-methylpentane-2,4-diol (MPD) containing in addition 30mM Ca2+ and 30mM Mg2+ (FPR-thrombin·FVa2 complex), or 20mM each of D-glucose, d-mannose, d-galactose, l-fucose, d-xylose, and N-acetyl-d-glucosamine (benzamidine·thrombin·FVa2).
Structure solution and model quality
Diffraction data were collected using synchrotron radiation at the European Synchrotron Radiation Facility (ESRF; Grenoble, France), integrated with MOSFLM (http://www.mrc-lmb.cam.ac.uk/harry/mosflm/), and reduced and scaled with CCP4i (http://www.ccp4.ac.uk/ccp4i_main.php). Phases were determined with MolRep using the crystal structure of FPR-inhibited α-thrombin as template (1PPB, Bode et al5). For inspection of electron density maps and fitting into the density we used MIFiT (http://code.google.com/p/mifit/). The quality of the models was assessed with WHATIF (http://swift.cmbi.ru.nl/servers/html/index.html) and MOLProbity (http://molprobity.biochem.duke.edu/). Structure figures were prepared with PyMOL (http://www.pymol.org/).
Results
Fragments of the FVa2 region do not bind with measurable affinity to α-thrombin
Based on previously reported data indicating that FVa2 residues Asp695 to Gln699 were critical for thrombin-catalyzed cofactor activation and prothrombinase activity,19,20,30,31 we conducted crystallization trials of human α-thrombin, both free and irreversibly inhibited with FPR, in the presence of excess peptides FV(695-699) and FV(695-701). Two different crystal forms were obtained, which were measured (resolution: 1.8-2.4 Å), and the structures solved by molecular replacement. However, inspection of the resulting electron density maps did not reveal any residual density that could account for the FV peptides. Therefore, we decided to use SPR technology to reassess the affinity of FVa2 fragments for CM5-immobilized thrombin, both free and FPR-inhibited. Neither these short peptides, nor longer fragments that fully cover each of the 2 hirudin-like sequences within the a2 linker, FV(657-679) or FV(679-701) (supplemental Figure 1A), generated significant resonance signals (not shown).
Two features of our experimental setup could have resulted in less pronounced or null SPR signals: the relatively low molecular masses of the FV peptides (2657 and 2912 Da, respectively; although they are well above the theoretical detection limit of 200 Da), and the fact that thrombin is immobilized in these assays most likely via exosite II residues (see “CM5 chips”). To rule out these possibilities, we also assessed the ability of peptides FV(657-679) and FV(679-701) to compete with fluorescein-labeled hirudin54-65 for binding to exosite I residues. This technique has proven useful to quantitatively study binding of several substrates, inhibitors, and cofactors to thrombin.25,26,29 However, and in agreement with both SPR and x-ray crystallography results, only marginal changes in fluorescence intensity were observed with peptide FV(657-679), yielding a KD of 170 ± 7μM (supplemental Figure 3A). Against expectations, FV(679-701) did not displace [5F]Hir-(54-65) from thrombin exosite I up to the maximum peptide concentrations that could be reached (above 200μM), and jellified at higher concentrations (supplemental Figure 3B).
Full-length FVa2 and FVa3 peptides bind to FPR-inhibited, but not to free α-thrombin
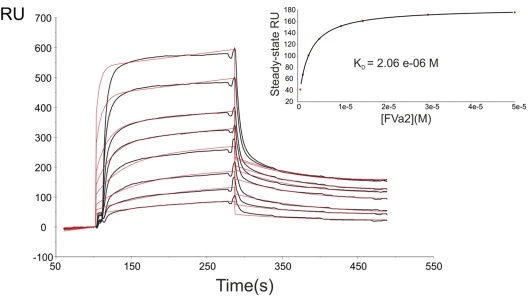
Failure to detect interactions with thrombin using peptides that cover each of the hirudin-like sequences of human FVa2 prompted us to investigate the affinity of the entire acidic linker for the serine proteinase. To roughly estimate the contribution of each thrombin exosite to peptide binding, 2 different coupling strategies were followed to allow interactions of the analytes with either exosite I (CM5 chips) or both exosites (SA chips; see “Equilibrium binding”). Sensorgrams obtained by running various concentrations of FVa2 over the α-thrombin–FPR channel of the CM5 chip (Figure 2) could be straightforwardly fitted by a model of peptide binding to a single site on the immobilized proteinase with a dissociation constant (KD) of 2.06μM (Figure 2 inset and Table 1). Essentially identical results were obtained when the affinity for biotin-labeled, SA-immobilized thrombin was assessed (KD 1.95μM; supplemental Figure 4 and Table 1). On the other hand, no resonance signals were detected when FVa2 samples were passed over CM5 chips coupled with the degraded forms of the proteinase, β-thrombin (Arg75-Tyr76 peptide bond cleaved) or γ-thrombin (Arg75-Tyr76 and Lys149E-Gly150 bonds cleaved).
Figure 2.
The whole A2-B domain linker binds to thrombin with moderate affinity. Representative sensorgrams of the interaction between human FVa2 and immobilized FPR-thrombin are shown (black lines). Recombinant, highly purified FVa2 in HBS-EP buffer (10mM HEPES, pH 7.4, 150mM NaCl, 3mM EDTA, 0.005% [vol/vol] surfactant P20) was flowed over the surface of the FPR–α-thrombin–coated CM5 chip at 30 μL/min. Peptide concentrations from top to bottom were 50, 30, 15, 10, 5, 2.5, 1.2, and 0.6μM. Red lines overlaid with the sensorgrams represent the simultaneous fit by a 1:1 Langmuir binding model using nonlinear least-squares analysis with BIAevaluation 4.1 software. The results of steady-state analysis of the data are given in the inset. Data shown are representative of 4 independent experiments. RU indicates resonance units.
Table 1.
Parameters for FVa2 (FV(657-709)) and FVa3 (FV(1481-1545)) binding to immobilized α-thrombin, measured by SPR
| Analyte | Chip* | Binding model† | ka (M−1s−1) | kd (s−1) | KD, μM‡ |
|---|---|---|---|---|---|
| FV(695-699) | CM5-FPR-T | No binding | — | — | — |
| FV(695-701) | CM5-FPR-T | No binding | — | — | — |
| FV(657-679) | CM5-FPR-T | No binding | — | — | — |
| FV(679-701) | CM5-FPR-T | No binding | — | — | — |
| FV(657-709) | CM5-FPR-T | 1:1 | 3.69 × 103 (± 49.4) | 7.60 × 10−3 (± 1.47 × 10−4) | 2.06 ± 0.05 |
| FV(657-709) | SA-FPR-T | 1:1 | 1.33 × 103 (± 14.4) | 2.38 × 10−3 (± 2.09 × 10−5) | 1.95 ± 0.03 |
| FV(1481-1545) | CM5-FPR-T | 1:1 | 0.999 × 103 (± 8.42) | 1.94 × 10−3 (± 2.98 × 10−5) | 1.96 ± 0.03 |
| FV(1481-1545) | SA-FPR-T | 1:1 | 5.46 × 103 (± 65.3) | 1.60 × 10−3 (± 3.65 × 10−5) | 0.237 ± 0.006 |
— indicates not applicable; FV, factor V; and SPR, surface plasmon resonance.
CM5-FPR-T, FPR-inhibited thrombin immobilized on CM5 chips, most likely via exosite II lysine residues; SA-FPR-T, biotin-FPR–inhibited thrombin immobilized on SA chips; both exosites are presumably accessible.
Binding data was fitted using the BIAevaluation 4.1 software. The binding model chosen represents that with the lowest χ2 value.
Error propagated from experimental errors of ka/kd values.
We next extended our analysis to the COOH-terminal linker, FVa3. Similar observations as with FVa2 were made when using the CM5 chip, and an essentially identical binding constant was determined (KD 1.96μM; supplemental Figure 5A, Table 1). However, analysis of data obtained with the SA chip revealed notably tighter binding to thrombin molecules with an accessible exosite II (KD 0.237μM; supplemental Figure 5B, Table 1), indicating in this case a definite contribution of exosite II residues to complex formation. Also along these lines, competition experiments revealed that FVa3 could still bind to SA chips previously saturated with the A2-B domain linker, while FVa2 was unable to bind to FVa3-saturated surfaces (data not shown).
Unexpectedly, we failed to detect interactions with CM5-immobilized, active-site free α-thrombin using either FVa2 or FVa3 peptides, suggesting that exosite I is not fully developed in free α-thrombin, in line with recent findings (Lechtenberg et al32 and our own unpublished observations).
FVa2 and FVa3 interfere with thrombin-mediated FV activation
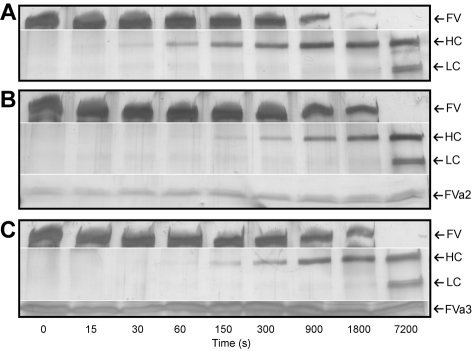
To analyze whether the recombinantly expressed A2-B and B-A3 linkers were able to interfere with thrombin-catalyzed FV activation, we followed the kinetics of FV processing in the presence or absence of FVa2 or FVa3. Indeed, addition of 50μM of either one of the recombinant peptides to the thrombin-FV reaction mixtures delayed disappearance of the band corresponding to the full-length cofactor, and concomitant appearance of the FVa heavy chain (compare Figure 3A-C). These findings indicate that the isolated peptides, albeit unlikely to adopt native-like conformations, were capable of interacting with thrombin in a productive manner, thus interfering with substrate recognition and processing.
Figure 3.
FVa2 and FVa3 interfere with thrombin-catalyzed factor V activation. Human FV was incubated with human α-thrombin at a 250:1 molar ratio, in the presence or absence of the highly purified recombinant fragments of the cofactor (A, thrombin alone; B, thrombin in the presence of 50μM FVa2; C, thrombin in the presence of 50μM FVa3). Aliquots of the reactions were taken at indicated times, separated on 4%-15% SDS-polyacrylamide gels, and silver-stained. The positions of FV, the heavy (HC) and the A3-C1-C2 light-chain doublet (LC) of factor Va are indicated to the right of each panel, along with those of FVa2/FVa3, where appropriate. Notice that FV is processed with slower kinetics in the presence of the recombinant linkers, indicating competition for the activating protease. The figures shown are representative of 4 independent experiments conducted with different preparations of FV and of recombinant FVa2/FVa3.
The crystal structure of the FPR-α-thrombin·FVa2 complex reveals that the more NH2-terminal hirudin-like sequence, Glu666-Glu672, contacts with exosite I
To determine the regions of the cofactor that are involved in important interactions with thrombin, we attempted to crystallize both free and FPR-inhibited α-thrombin in the presence of excess FVa2. Several crystals of free thrombin grown in this manner were measured and processed. However, in line with SPR results, no electron density that could account for the FV peptide was observed in any of these independent crystals (not shown). For this reason, we focused on crystals of inhibited thrombin grown in the presence of excess FVa2.
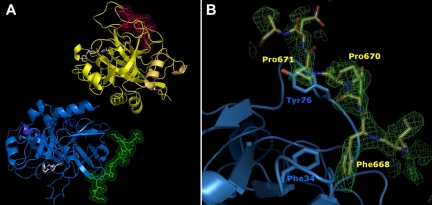
Crystals of the FPR–α-thrombin·FVa2 complex diffract to near-atomic resolution using synchrotron radiation (1.7 Å); these crystals belong to the triclinic space group, and contain 2 complex molecules per asymmetric unit (Figure 4A). No residues with either bad bonds or angles are present in the refined model, and all residues lie within most favored or additionally allowed regions of the Ramachandran plot (Table 2). Major contacts between the 2 complexes are formed by the heavy chains of the 2 thrombin moieties. Noteworthy, dimer formation compromises basic side chains of several exosite II residues from one monomer (eg, Arg93 and Arg233), and, in a less marked manner, from the other monomer (Lys107, Lys236; supplemental Figure 6). This interface also contains the NH2-terminal residues from the light chain of this second thrombin molecule, which are not defined N-terminally of Glu1C.
Figure 4.
High-resolution crystal structure of the thrombin FPR–α-thrombin·FVa2 complex. (A) The 2 thrombin monomers in the asymmetric unit are represented by their secondary structure elements, colored blue and yellow, respectively. Their cognate FV peptides are given with all their nonhydrogen side chain atoms in green and pink, respectively. The FPR moieties covalently bound to thrombin active-site residues His57/Ser195 are depicted color-coded (carbon, gray; nitrogen, blue; oxygen, red). The thrombin molecule to the bottom left is shown in standard orientation, that is, macromolecular substrate residues would run from left to right in productive complexes with the proteinase. (B) Detail of the initial electron density map calculated after rigid-body refinement of the 2 thrombin molecules is shown around exosite I (contoured at 1 σ), with the final FVa2 model superimposed. The side chains of a few critical residues are labeled.
Table 2.
X-ray data collection, refinement statistics, and model analysis for the FPR–α-thrombin·FVa2 complex
| Summary | |
|---|---|
| Data collection | |
| X-ray source | ESRF ID23-1 |
| Wavelength, Å | 0.9725 |
| Space group | P1 |
| Cell constants | a = 52.21 Å, b = 62.26 Å, c = 67.51 Å; α = 99.38°, β = 110.46°, γ = 92.26° |
| Resolution (outer shell, Å) | 62.14-1.69 (1.79-1.69) |
| Total number of observations | 268 275 (38 322) |
| Number of unique reflections | 81 788 (11 372) |
| I/σ(I) | 12.3 (2.4) |
| Completeness (%) | 94.2 (89.6) |
| Redundancy | 3.3 (3.4) |
| Rmerge* (%) | 8.4 (38.6) |
| Refinement and model analysis | |
| Resolution (outer shell, Å) | 31.92-1.696 (1.740-1.696) |
| Rfactor† (%) | 19.96 (36.90) |
| Rfree‡ (%) | 23.88 (41.60) |
| rmsd bonds, Å | 0.023 |
| rmsd angles, ° | 2.032 |
| Ramachandran plot (favored/allowed), % | 97.02/2.98 |
| Clashscore (all atoms)§ | 11.80 (58th percentile)‖ |
| MolProbity score¶ | 2.13 (44th percentile)‖ |
Rmerge = ΣĥΣî|I(hî) − <I(h)>|/ΣĥΣî I(hî), where I(hî) is the intensity of the hth reflection as determined by the ith measurement, and <I(h)> is the average value.
Rfactor = Σ(|Fobs| − |Fcalc|)/Σ|Fobs|, where |Fobs| and |Fcalc| are observed and calculated structure factor amplitudes, respectively.
Rfree: Rfactor calculated for a randomly selected test set comprising 2047 reflections (2.5% of all unique reflections), which were excluded from refinement.
Clashscore is the number of serious steric overlaps (> 0.4 Å) per 1000 atoms.
The 100th percentile is the best among structures of comparable resolution (1.70 ± 0.25 Å); 0th percentile is the worst.
MolProbity score is defined as 0.42574*log(1 + clashscore) + 0.32996*log(1 + max(0,%RotOut-1)) + 0.24979*log(1 + max(0,100-%RamaFavored-2)) + 0.5, where %RotOut is the percentage of bad side-chain rotamers and %RamaFavored is the percentage of residues in favored regions of the Ramachandran plot.
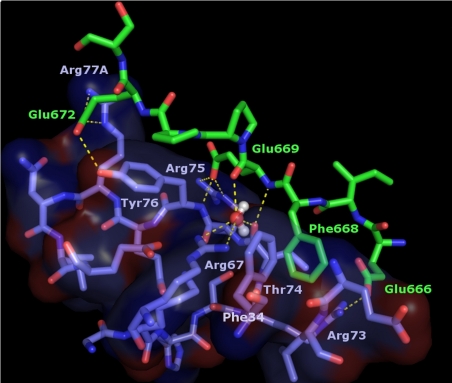
Strong, readily interpretable additional electron density around both exosite I areas was immediately observed in maps calculated after positioning the 2 thrombin molecules (Figure 4B). This extra electron density could be straightforwardly modeled by the Glu666-Glu672(Ser673) peptide from human FV. In particular, the aromatic side chain of Phe668 is fully anchored in thrombin exosite I, occupying a hydrophobic pocket created by both main chain and/or side chain atoms of Phe34, Leu40, Arg67, Arg73, and Thr74 (Figure 5). The phenyl ring of Phe668 engages in strong Van der Waals, edge-to-face interactions with thrombin Phe34; all 6 carbon atoms of this ring are located between 3.7 and 4.1 Å from the Phe34 Cϵ atom (Figure 5).
Figure 5.
The more N-terminal hirudin-like sequence of FVa2, Glu666-Glu672, binds across thrombin exosite I. Close-up around the thrombin exosite I region in the FPR–α-thrombin·FVa2 complex. For simplicity, only residues of thrombin 70-80 loop, a few side chains of the neighboring 37 loop, and a water molecule trapped at the thrombin-FVa2 interface are included in the image. Thrombin residues are color-coded blue-purple (carbon), dark blue (nitrogen) and red (oxygen), and are overlaid with a transparent surface; FV atoms are green (carbon), dark blue (nitrogen) or red (oxygen). Electrostatic interactions are indicated with dashed yellow lines.
The carboxylate group of the following residue, Glu669, accepts a hydrogen bond from the main chain N atom of Tyr76, and forms in addition a strong salt bridge with the guanidinium group of another thrombin-specific basic residue, Arg75. Glu669 is also connected to the proteinase moiety through a water molecule trapped between its oxygen carbonyl atom and that of Thr74; this water is also coordinated by the guanidinium group of Arg67. Further, cofactor residues Pro670 to Ser673 form an arc that covers the side chain of thrombin Tyr76 with Pro670 engaging in important Van der Waals contacts with the phenolic ring. Finally, in one of the complex molecules the carboxylate group of residue Glu672 forms a salt-bridge with the guanidinium of Arg77A, and accepts an H-bond from the hydroxyl of Tyr76. However, electron density for the Glu672/Arg77A side chains is poor, and the orientation of these side chains in the other copy precludes this direct contact. Thus, this electrostatic interaction might seem to play a secondary role in FV recognition.
Unfortunately, the low affinity of peptide regions downstream of Ser673 for cognate thrombin together with packing constraints in these crystals (supplemental Figure 6) preclude definition of most of the path followed by the acidic linker before entering the active-site region of the proteinase.
A crystal structure of FVa2 bound to benzamidine-inhibited thrombin confirms engagement of exosite I by the NH2-terminal hirudin-like motif
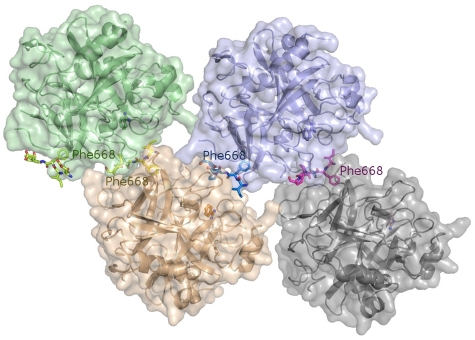
With the aim of confirming these structural results, we performed further crystallization trials using proteinase molecules that are not covalently inhibited. Crystals of benzamidine (Bz)–inhibited thrombin grown in the presence of excess FVa2 contain 4 independent complex molecules in the asymmetric unit (Figure 6), and diffract to 2.55 Å resolution using synchrotron radiation. (Data collection and refinement statistics are given in Table 3).
Figure 6.
The crystal structure of the Bz·α-thrombin·FVa2 complex corroborates FVa2 binding to exosite I residues. The tetramer found in the asymmetric unit is shown, with the 4 independent thrombin molecules represented by their major secondary structure elements colored green, purple, orange, and gray, respectively. The Van der Waals surfaces of the 4 monomers are superimposed in lighter tones of the same colors. The FVa2 peptides are given with all their nonhydrogen atoms, color-coded according to the bound thrombin moiety. The benzamidine molecules that occupy the S1 pockets of thrombin are also shown as color-coded ball-and-stick models. Notice that the phenyl rings of Phe668 dock onto exosite I regions from cognate thrombin molecules.
Table 3.
X-ray data collection, refinement statistics, and model analysis for the Bz α-thrombin·FVa2 complex
| Summary | |
|---|---|
| Data collection | |
| X-ray source | ESRF ID23-2 |
| Wavelength, Å | 0.8726 |
| Space group | P1 |
| Cell constants | a = 62.24 Å, b = 68.23 Å, c = 98.97 Å; α = 72.57°, β = 84.05°, γ = 80.45° |
| Resolution (outer shell, Å) | 64.42-2.55 (2.69-2.55) |
| Total number of observations | 170 648 (24 763) |
| Number of unique reflections | 47 172 (6841) |
| I/σ(I) | 10.8 (2.7) |
| Completeness (%) | 94.6 (94.1) |
| Redundancy | 3.6 (3.6) |
| Rmerge* (%) | 8.0 (42.6) |
| Refinement and model analysis | |
| Resolution (outer shell, Å) | 61.28-2.550 (2.616-2.550) |
| Rfactor† (%) | 20.68 (27.30) |
| Rfree‡ (%) | 27.88 (35.90) |
| rmsd bonds, Å | 0.016 |
| rmsd angles, ° | 1.841 |
| Ramachandran plot (favored/allowed), % | 93.50/5.46 |
| Clashscore (all atoms)§ | 18.74 (78th percentile)‖ |
| MolProbity score¶ | 2.86 (53th percentile)‖ |
Rmerge = ΣĥΣî|I(hî) − <I(h)>|/ΣĥΣîI(hî), where I(hî) is the intensity of the hth reflection as determined by the ith measurement, and <I(h)> is the average value.
Rfactor = Σ(|Fobs| − |Fcalc|)/Σ|Fobs|, where |Fobs| and |Fcalc| are observed and calculated structure factor amplitudes, respectively.
Rfree: Rfactor calculated for a randomly selected test set comprising 5% of all reflections, which were excluded from refinement.
Clashscore is the number of serious steric overlaps (> 0.4 Å) per 1000 atoms.
The 100th percentile is the best among structures of comparable resolution (2.55 ± 0.25 Å); 0th percentile is the worst.
MolProbity score is defined as 0.42574*log(1 + clashscore) + 0.32996*log(1 + max(0,%RotOut-1)) + 0.24979*log(1 + max(0,100-%RamaFavored-2)) + 0.5, where %RotOut is the percentage of bad side-chain rotamers and %RamaFavored is the percentage of residues in favored regions of the Ramachandran plot.
Similar to the FPR-thrombin·FVa2 complex, inspection of the electron density maps revealed that the FV peptide runs across exosite I, with Phe668 and surrounding residues making the most relevant contacts with the proteinase moiety (supplemental Figure 7A). Of note, clear electron density for Glu666 is present in only one of the complex molecules, while Glu672 could not be defined in any of the 4 independent copies (see supplemental Figure 7B for a representation of the final electron density map around exosite I residues). As in the FPR-thrombin·FVa2 complex, extensive thrombin-thrombin contacts preclude definition of the more C-terminal part of the A2-B domain linker. Interestingly, electron density is weak or fragmented for several residues of the 70-80 loop, most notably the apical residues, Arg77A and Asn78, indicating enhanced flexibility.
Discussion
We have addressed the structural basis of thrombin-mediated FV activation using recombinant forms of the acidic linkers that connect domains A2-B and B-A3 in the human cofactor (FVa2 and FVa3, respectively). Using SPR, fluorescence, and crystallographic techniques we could detect, at most, marginally relevant interactions between the proteinase and synthetic peptides that contain each of the hirudin-like motifs within the Ile657-Arg709 stretch of human FV. However, the entire FVa2 region was shown to interact with immobilized thrombin molecules in SPR experiments. A functional and accessible exosite I, and in particular an intact 70-80 loop, was essential for FVa2 binding to thrombin. The practically identical dissociation constants obtained with thrombin-coated CM5 and SA chips (≈ 2μM) suggest that other proteinase regions and in particular exosite II residues contribute little to the recognition of the Arg709-Ser710 activation cleavage site. This dissociation constant is similar to values determined for FV/FVa binding to thrombin based on fluorescence techniques (3.4 and 1.1μM, respectively),16 indicating that the FVa2 linker is responsible for the most relevant thrombin-FV(a) interactions.
By contrast, in the case of FVa3 a markedly lower KD value was obtained with thrombin-coated SA chips, compared with the CM5-immobilized proteinase, which points to a definite contribution of exosite II residues to binding affinity. These findings are in line with a recent study indicating that cleavage of the Arg709-Ser710 peptide bond only depends on exosite I residues, while both exosites are required for processing at the more C-terminal activation cleavage site, Arg1545-Ser1546.18
The more N-terminal hirudin-like sequence of FVa2 mediates thrombin binding
Previous investigations have established an important role for both thrombin exosites in FV activation,14–17 and have in particular shown that exosite I is critical for processing at the more N-terminal site, Arg709-Ser710.18 The current structural work demonstrates that the hirudin-like sequence, Glu666-Glu672, directly interacts with exosite I residues, in particular those that have been identified in a previous mutagenesis study as critical for cofactor activation, Arg73, Arg75, and Tyr76.17 We stress that these contacts were observed in 6 crystallographically independent complex molecules. Our results are difficult to reconcile with reports suggesting an essential role of the Asp695-Gln699 pentapeptide for (pro)thrombin binding, and also cast doubts on its involvement in prothrombinase activity.19,20,31 However, they are fully in line with the fact that truncated FV that lacks a2 residues C-terminal of Thr678 binds normally to agarose-immobilized α-thrombin, while deletion of the whole linker abolishes interactions with thrombin-agarose.33 Perhaps the high negative density of the Asp695-Gln699 peptide, in particular with one or both tyrosine residues sulfated, might have resulted in nonspecific interactions with thrombin exosites. The structural work also explains the observation that an exosite I–directed DNA aptamer, which covers a large area of the “upper” exosite I (Padmanabhan et al34 and Padmanabhan and Tulinsky35; see also supplemental Figure 8A), inhibits cleavage at the Arg709-Ser710 site.18 It is noteworthy that the aptamer does not interact with thrombin residues that build the apolar Phe668-binding pocket, which might explain the relatively high concentrations needed to completely block FV activation.18
FVa2 interacts less extensively with thrombin exosite I than the COOH-terminal hirudin tail
A large number of crystal structures of thrombin bound to hirudin or hirudomimetics (ie, bivalent inhibitors composed of an active site-binding moiety coupled to the C-terminal hirudin tail) have been reported9,36 (recently reviewed in Corral-Rodríguez et al10). Comparison with the current structure of the FPR–α-thrombin·FVa2 complex reveals that 2 important cofactor residues, Phe668 and Glu669, adopt similar conformations as hirudin Phe56H and Glu57H (distances between Cα atoms of topologically equivalent residues below 1.5 Å). Accordingly, these 2 residues engage in similar interactions with exosite I (supplemental Figure 9). Overall, however, the leech-derived inhibitor runs closer to the thrombin surface than FVa2, and the main-chains of the 2 peptides diverge notably COOH-terminal of Pro671/Ile59H. While the FV peptide points away from the proteinase, the last C-terminal hirudin residues form a short α-helix that docks onto the “upper” exosite I region. Side chains of these residues are involved in strong interactions with thrombin, most notably the naturally sulfated Tyr63H, and Leu64H. These additional contacts have no counterpart in the FV linker, explaining why FVa2 possesses a much lower affinity for thrombin than Hir-(54-65)(SO3−) (KD 26-38nM; Bock et al25).
While electrostatically-driven interactions are likely to orient FVa2 toward exosite I, the strongest contacts with thrombin residues are made by aromatic/aliphatic side chains, most notably Phe668. A similar preeminence of nonpolar interactions has long been recognized in thrombin-hirudin complex formation. Despite the large differences between the A2-B domain linker and hirudin/hirudomimetics (supplemental Figure 9), by far the largest contributions to inhibitor binding are provided by aromatic/aliphatic side chains.10 Most notably, replacement of hirudin Phe56H by residues with branched side chains (Ile, Val, or Thr) results in a 30-, 60-, or 115-fold increase in the inhibition constant, respectively.37 Similarly, the Phe56H→Gly analog of a potent hirudomimetic, P552, possesses a 473-fold higher KI value than the unmodified inhibitor.38 A similar role to Phe668 is played by residue Phe55 in PAR1.39
The whole A2-B domain linker wraps around thrombin in a productive thrombin·FV complex
Despite repeated crystallization trials, so far, we have not been able to obtain a crystal form in which the whole FVa2 sequence is defined by electron density. However, the binding mode of residues that immediately precede and follow the activation cleavage sites of other thrombin substrates has been revealed in previous structural investigations. Crystal structures of thrombin bound to fibrinopeptide A residues Asp7-Arg16 or to FXIII residues Thr28-Arg37, corresponding to nonprime sites P10 to P1, have been reported.40–42 Additional interactions of the prime residues P1′ to P3′ were revealed in the structure of bovine thrombin complexed with an uncleavable analog of the longer peptide, Asp7-Arg19.43
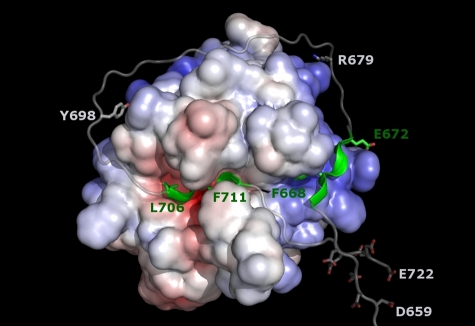
More appropriate for modeling the FVa2 sequence around the Arg709-Ser710 site in a productive complex with thrombin is the recently solved crystal structure of the inactive Ser195Ala thrombin mutant bound to residues Ala33-Glu57 of human PAR1.39 We first modeled the Leu706 (P4)–Phe711 (P2′) stretch of FV essentially following the equivalent PAR1 sequence, and then extended the polypeptide chains without other constraint than observation of standard peptide geometries. A hypothetical, energy-minimized model of human α-thrombin bound to FV residues Ile657-Glu722 is presented in Figure 7.
Figure 7.
Hypothetical model of the productive thrombin·FVa2 complex. The model was generated based on the current structure of FPR–α-thrombin·FVa2 and that of S195A thrombin bound to a fragment from the PAR1 ectodomain that includes the activation cleavage site (PDB code 3LU9).39 Factor V residues Leu706 (P4) to Phe711 (P2′) were modeled according to the conformation of the equivalent PAR1 sequence, and the 2 polypeptide fragments were then extended following standard peptide geometries. The well-defined active-site– and exosite I–binding regions are highlighted in green. Notice that the relatively large distance between these 2 peptides (∼ 40 Å between Cα atoms of residues Glu672 and Leu706) necessarily implies that the intervening sequence has to adopt a rather extended conformation, running close to the thrombin surface. The spatial proximity of clusters of acidic residues, Asp659-Asp663 and Glu719-Glu722, in the Michaelis complex is also noteworthy. The side chain of the arginine residue targeted by APC (Arg679) is also shown, as well as Tyr698, which could contact basic residues at the edge of exosite II, especially if sulfated.
In addition to its excellent stereochemistry, 2 observations underscore the plausibility of this model. First, FVa2 approaches the triplet of arginines at positions 93, 97, and 101 that are the only exosite II residues reported to be involved in FV activation.14,17 Interestingly, a sulfated Tyr698 residue could engage in electrostatic interactions with any of these basic residues, perhaps explaining some reports on the role of the C-terminal FVa2 region in (pro)thrombin binding,19,44 and the relevance of FV sulfation for efficient thrombin-catalyzed activation.45,46 Second, the model offers an explanation for the finding that an exosite II–directed DNA aptamer fails to inhibit cleavage at the a2-B domain border.18 Although the structure of this aptamer has not been reported, an RNA aptamer that also binds exosite II has been structurally characterized in complex with thrombin.47 Comparison with the current thrombin·FVa2 model reveals that simultaneous binding to the proteinase is possible without steric clashes (supplemental Figure 8B), which might also apply to the DNA aptamer.
Two major conclusions can be derived from this modeling exercise, which are independent of the specific details of thrombin-FVa2 interactions. First, the relatively long distance between residues Glu672, the position of which is derived from the current experimental work, and Leu706 (position P4; predicted with high confidence from other thrombin·substrate complexes) necessarily implies that the intervening sequence has to run close to the thrombin surface, and is likely to engage in several, albeit weaker contacts with the proteinase. This would explain the negligible affinity of FVa2 fragments for thrombin. Second, in the Michaelis complex, the N-terminal peptide of the B domain would run close to the FVa2 exosite I–binding region. As a consequence, clusters of negatively charged residues located between the globular A2 domain and the exosite I–binding motif (Asp659-Asp663) and downstream of the cleavage site (Glu719-Glu722) would be arranged in close vicinity (Figure 7). Repulsion between these 2 acidic clusters might contribute to local unfolding of the a2-B region to facilitate thrombin binding and/or promote rapid substrate release after proteolysis of the Arg709-Ser710 site.
Mechanism of human FV activation
The current results suggest an explanation for the well-characterized, kinetically preferred pathway of human FV activation18,48,49 that allows explaining processivity. Recognition of the Arg709-Ser710 site is primarily facilitated by thrombin interactions with the Glu666-Glu672 stretch (Figure 5, supplemental Figure 10A). Once anchored to exosite I, the more C-terminal part of the acidic linker wraps around the proteinase, aided by transient interactions with residues on the border of exosite II. In this manner, the Arg709-Ser710 peptide bond is presented to the catalytic machinery in an optimal conformation for cleavage (Figure 7, supplemental Figure 10B). After processing, electrostatic repulsion of Asp659-Asp663 and Glu719-Glu722 stretches forces the more C-terminal substrate residues to leave the active-site cleft. Conceivably, thrombin remains bound to the cleaved FV molecule via exosite I, and could gain access to the now fully exposed Arg1018-Thr1019 peptide bond, basically driven by interactions with the immediate vicinity of the scissile peptide bond (supplemental Figure 10C). In this regard, it is noteworthy that the residues at positions P2 (Pro1017) and P4 (Leu1015) correspond to those of an ideal thrombin substrate.
Cleavage at this second site releases the Ser710-Arg1018 region (fragment E) and facilitates processing of the C-terminal peptide bond, Arg1545-Ser1546 (Thorelli et al50), probably by increasing its accessibility (supplemental Figure 10D). This final step requires thrombin transfer to the FVa3 region, explaining its slower kinetics,48 perhaps because the multiple 9-residue repeats N-terminal of this site adopt a definite, as yet unknown tertiary structure. Presumably, the more C-terminal hirudin-like sequence of FVa3 binds first to exosite II (supplemental Figure 10E), followed by displacement of FVa2 from exosite I and simultaneous docking of the Arg1545-Ser1546 site into the thrombin active-site cleft. Finally, cleavage of this peptide liberates fragment C1 (Thr1019-Arg1545) and generates the fully functional cofactor (supplemental Figure 10F).
In summary, we have unambiguously established that the more N-terminal, hirudin-like sequence of FVa2 interacts with thrombin exosite I. We are tempted to speculate that this binding mode is shared by FVa3 and FVIIIa1-a3, although lack of significant sequence similarity with FVa2 impedes a straightforward assignment of residues that could occupy positions equivalent to those of, for example, Phe668. Furthermore, and considering the ability of Hir-(54-65) to interfere with FVa-mediated activation of prothrombin,28 it is conceivable that the Glu666-Glu672 region of the activated cofactor interacts with pro- and/or meizothrombin essentially as revealed in complexes with α-thrombin.
Accession numbers
Coordinates have been deposited in the Protein Data Bank with accession numbers 3P6Z and 3P70.
Supplementary Material
Acknowledgments
We thank the ESRF staff and in particular Dr A. Popov for help with data collection.
M.Á.C.-R. was supported by FPI grant BES2005-6841 from Spanish Ministry of Science and Innovation, and E.H.-C. by a grant from Spanish Ministry of Foreign Affairs and Cooperation (MAEC-AECID). The financial support from Spanish Ministry of Science and Innovation (SAF2004-00 543, SAF2007-64 140 and SAF2010-15 668) and of Fundació La Marató de TV3 (P.F.-P.), and from BIO2009-08 983 (R.G.-G.), is gratefully acknowledged. This work was supported in part by National Institutes of Health grant RO1 HL038779 from the National Heart, Lung, and Blood Institute (P.E.B.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.Á.C.-R. and E.H.-C. performed experiments and analyzed data; P.E.B. and R.G.-G. designed experiments and analyzed data; and P.F.-P. designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pablo Fuentes-Prior, PhD, Institute for Biomedical Research, Hospital de la Santa Creu i Sant Pau, Sant Antoni Maria Claret 167, 08025 Barcelona, Spain; e-mail: pfuentes@santpau.cat.
References
- 1.Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991;30(43):10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 2.Steen M, Villoutreix BO, Norstrom EA, Yamazaki T, Dahlbäck B. Defining the factor Xa-binding site on factor Va by site-directed glycosylation. J Biol Chem. 2002;277(51):50022–50029. doi: 10.1074/jbc.M205609200. [DOI] [PubMed] [Google Scholar]
- 3.Toso R, Camire RM. Removal of B-domain sequences from factor V rather than specific proteolysis underlies the mechanism by which cofactor function is realized. J Biol Chem. 2004;279(20):21643–21650. doi: 10.1074/jbc.M402107200. [DOI] [PubMed] [Google Scholar]
- 4.Lenting PJ, van de Loo JW, Donath MJ, van Mourik JA, Mertens K. The sequence Glu1811-Lys1818 of human blood coagulation factor VIII comprises a binding site for activated factor IX. J Biol Chem. 1996;271(4):1935–1940. doi: 10.1074/jbc.271.4.1935. [DOI] [PubMed] [Google Scholar]
- 5.Bode W, Mayr I, Baumann U, Huber R, Stone SR, Hofsteenge J. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J. 1989;8(11):3467–3475. doi: 10.1002/j.1460-2075.1989.tb08511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bock PE, Panizzi P, Verhamme IMA. Exosites in the substrate specificity of blood coagulation reactions. J Thromb Haemost. 2007;5(s 1):81–94. doi: 10.1111/j.1538-7836.2007.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsiang M, Jain AK, Dunn KE, Rojas ME, Leung LL, Gibbs CS. Functional mapping of the surface residues of human thrombin. J Biol Chem. 1995;270(28):16854–16863. doi: 10.1074/jbc.270.28.16854. [DOI] [PubMed] [Google Scholar]
- 8.Pechik I, Madrazo J, Mosesson MW, Hernandez I, Gilliland GL, Medved L. Crystal structure of the complex between thrombin and the central “E” region of fibrin. Proc Natl Acad Sci U S A. 2004;101(9):2718–2723. doi: 10.1073/pnas.0303440101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rydel TJ, Ravichandran KG, Tulinsky A, et al. The structure of a complex of recombinant hirudin and human alpha-thrombin. Science. 1990;249(4966):277–280. doi: 10.1126/science.2374926. [DOI] [PubMed] [Google Scholar]
- 10.Corral-Rodríguez MÁ, Macedo-Ribeiro S, Barbosa Pereira PJ, Fuentes-Prior P. Leech-derived thrombin inhibitors: from structures to mechanisms to clinical applications. J Med Chem. 2010;53(10):3847–3861. doi: 10.1021/jm901743x. [DOI] [PubMed] [Google Scholar]
- 11.Fuentes-Prior P, Iwanaga Y, Huber R, et al. Structural basis for the anticoagulant activity of the thrombin-thrombomodulin complex. Nature. 2000;404(6777):518–525. doi: 10.1038/35006683. [DOI] [PubMed] [Google Scholar]
- 12.Dumas JJ, Kumar R, Seehra J, Somers WS, Mosyak L. Crystal structure of the GpIbalpha-thrombin complex essential for platelet aggregation. Science. 2003;301(5630):222–226. doi: 10.1126/science.1083917. [DOI] [PubMed] [Google Scholar]
- 13.Celikel R, McClintock RA, Roberts JR, et al. Modulation of alpha-thrombin function by distinct interactions with platelet glycoprotein Ibalpha. Science. 2003;301(5630):218–221. doi: 10.1126/science.1084183. [DOI] [PubMed] [Google Scholar]
- 14.Esmon CT, Lollar P. Involvement of thrombin anion-binding exosites 1 and 2 in the activation of factor V and factor VIII. J Biol Chem. 1996;271(23):13882–13887. doi: 10.1074/jbc.271.23.13882. [DOI] [PubMed] [Google Scholar]
- 15.Dharmawardana KR, Bock PE. Demonstration of exosite I-dependent interactions of thrombin with human factor V and factor Va involving the factor Va heavy chain: analysis by affinity chromatography employing a novel method for active-site-selective immobilization of serine proteinases. Biochemistry. 1998;37(38):13143–13152. doi: 10.1021/bi9812165. [DOI] [PubMed] [Google Scholar]
- 16.Dharmawardana KR, Olson ST, Bock PE. Role of regulatory exosite I in binding of thrombin to human factor V, factor Va, factor Va subunits, and activation fragments. J Biol Chem. 1999;274(26):18635–18643. doi: 10.1074/jbc.274.26.18635. [DOI] [PubMed] [Google Scholar]
- 17.Myles T, Yun TH, Hall SW, Leung LLK. An extensive interaction interface between thrombin and factor V is required for factor V activation. J Biol Chem. 2001;276(27):25143–25149. doi: 10.1074/jbc.M011324200. [DOI] [PubMed] [Google Scholar]
- 18.Segers K, Dahlbäk B, Bock PE, Tans G, Rosing J, Nicolaes GAF. The role of thrombin exosites I and II in the activation of human coagulation factor V. J Biol Chem. 2007;282(47):33915–33924. doi: 10.1074/jbc.M701123200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck DO, Bukys MA, Singh LS, Szabo KA, Kalafatis M. The contribution of amino acid region Asp695-Tyr698 of factor V to procofactor activation and factor Va function. J Biol Chem. 2004;279(4):3084–3095. doi: 10.1074/jbc.M306850200. [DOI] [PubMed] [Google Scholar]
- 20.Bukys MA, Orban T, Kim PY, Beck DO, Nesheim ME, Kalafatis M. The structural integrity of anion binding exosite I of thrombin is required and sufficient for timely cleavage and activation of factor V and factor VIII. J Biol Chem. 2006;281(27):18569–18580. doi: 10.1074/jbc.M600752200. [DOI] [PubMed] [Google Scholar]
- 21.Newell JL, Fay PJ. Acidic residues C-terminal to the A2 domain facilitate thrombin-catalyzed activation of factor VIII. Biochemistry. 2008;47(33):8786–8795. doi: 10.1021/bi8007824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marquette KA, Pittman DD, Kaufman RJ. The factor V B-domain provides two functions to facilitate thrombin cleavage and release of the light chain. Blood. 1995;86(8):3026–3034. [PubMed] [Google Scholar]
- 23.Bock PE. Active-site-selective labeling of blood coagulation proteinases with fluorescence probes by the use of thioester peptide chloromethyl ketones. I. Specificity of thrombin labeling. J Biol Chem. 1992;267(21):14963–14973. [PubMed] [Google Scholar]
- 24.Bock PE. Active-site-selective labeling of blood coagulation proteinases with fluorescence probes by the use of thioester peptide chloromethyl ketones. II. Properties of thrombin derivatives as reporters of prothrombin fragment 2 binding and specificity of the labeling approach for other proteinases. J Biol Chem. 1992;267(21):14974–14981. [PubMed] [Google Scholar]
- 25.Bock PE, Olson ST, Bjork I. Inactivation of thrombin by antithrombin is accompanied by inactivation of regulatory exosite I. J Biol Chem. 1997;272(32):19837–19845. doi: 10.1074/jbc.272.32.19837. [DOI] [PubMed] [Google Scholar]
- 26.Friedrich R, Panizzi P, Fuentes-Prior P, et al. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425(6957):535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 27.Anderson PJ, Nesset A, Dharmawardana KR, Bock PE. Characterization of proexosite I on prothrombin. J Biol Chem. 2000;275(22):16428–16434. doi: 10.1074/jbc.M001254200. [DOI] [PubMed] [Google Scholar]
- 28.Anderson PJ, Nesset A, Dharmawardana KR, Bock PE. Role of proexosite I in factor Va-dependent substrate interactions of prothrombin activation. J Biol Chem. 2000;275(22):16435–16442. doi: 10.1074/jbc.M001255200. [DOI] [PubMed] [Google Scholar]
- 29.Anderson PJ, Bock PE. Role of prothrombin fragment 1 in the pathway of regulatory exosite I formation during conversion of human prothrombin to thrombin. J Biol Chem. 2003;278(45):44489–44495. doi: 10.1074/jbc.M306916200. [DOI] [PubMed] [Google Scholar]
- 30.Hirbawi J, Bukys MA, Barhoover MA, Erdogan E, Kalafatis M. Role of the acidic hirudin-like COOH-terminal amino acid region of factor Va heavy chain in the enhanced function of prothrombinase. Biochemistry. 2008;47(30):7963–7974. doi: 10.1021/bi800593k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bukys MA, Kim PY, Nesheim ME, Kalafatis M. A control switch for prothrombinase: characterization of a hirudin-like pentapeptide from the COOH terminus of factor Va heavy chain that regulates the rate and pathway for prothrombin activation. J Biol Chem. 2006;281(51):39194–39204. doi: 10.1074/jbc.M604482200. [DOI] [PubMed] [Google Scholar]
- 32.Lechtenberg BC, Johnson DJD, Freund SMV, Huntington JA. NMR resonance assignments of thrombin reveal the conformational and dynamic effects of ligation. Proc Natl Acad Sci U S A. 2010;107(32):14087–14092. doi: 10.1073/pnas.1005255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toso R, Camire RM. Role of hirudin-like factor Va heavy chain sequences in prothrombinase function. J Biol Chem. 2006;281(13):8773–8779. doi: 10.1074/jbc.M511419200. [DOI] [PubMed] [Google Scholar]
- 34.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J Biol Chem. 1993;268(24):17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 35.Padmanabhan K, Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr D Biol Crystallogr. 1996;52(2):272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- 36.Liu CC, Brustad E, Liu W, Schultz PG. Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin. J Am Chem Soc. 2007;129(35):10648–10649. doi: 10.1021/ja0735002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Betz A, Hofsteenge J, Stone SR. Role of interactions involving C-terminal nonpolar residues of hirudin in the formation of the thrombin-hirudin complex. Biochemistry. 1991;30(41):9848–9853. doi: 10.1021/bi00105a006. [DOI] [PubMed] [Google Scholar]
- 38.Cheng Y, Slon-Usakiewicz JJ, Wang J, Purisima EO, Konishi Y. Nonpolar interactions of thrombin and its inhibitors at the fibrinogen recognition exosite: thermodynamic analysis. Biochemistry. 1996;35(40):13021–13029. doi: 10.1021/bi961039c. [DOI] [PubMed] [Google Scholar]
- 39.Gandhi PS, Chen Z, Di Cera E. Crystal structure of thrombin bound to the uncleaved extracellular fragment of PAR1. J Biol Chem. 2010;285(20):15393–15398. doi: 10.1074/jbc.M110.115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishnan R, Sadler JE, Tulinsky A. Structure of the Ser195Ala mutant of human α-thrombin complexed with fibrinopeptide A(7-16): evidence for residual catalytic activity. Acta Crystallogr D Biol Crystallogr. 2000;56(4):406–410. doi: 10.1107/s0907444900001487. [DOI] [PubMed] [Google Scholar]
- 41.Stubbs MT, Oschkinat H, Mayr I, et al. The interaction of thrombin with fibrinogen. A structural basis for its specificity. Eur J Biochem. 1992;206(1):187–195. doi: 10.1111/j.1432-1033.1992.tb16916.x. [DOI] [PubMed] [Google Scholar]
- 42.Sadasivan C, Yee VC. Interaction of the factor XIII activation peptide with alpha-thrombin: crystal structure of its enzyme-substrate analog complex. J Biol Chem. 2000;275(47):36942–36948. doi: 10.1074/jbc.M006076200. [DOI] [PubMed] [Google Scholar]
- 43.Martin PD, Malkowski MG, DiMaio J, Konishi Y, Ni F, Edwards BF. Bovine thrombin complexed with an uncleavable analog of residues 7-19 of fibrinogen Aalpha: geometry of the catalytic triad and interactions of the P1′, P2′, and P3′ substrate residues. Biochemistry. 1996;35(40):13030–13039. doi: 10.1021/bi960656y. [DOI] [PubMed] [Google Scholar]
- 44.Kalafatis M, Beck DO, Mann KG. Structural requirements for expression of factor Va activity. J Biol Chem. 2003;278(35):33550–33561. doi: 10.1074/jbc.M303153200. [DOI] [PubMed] [Google Scholar]
- 45.Hortin GL. Sulfation of tyrosine residues in coagulation factor V. Blood. 1990;76(5):946–952. [PubMed] [Google Scholar]
- 46.Pittman DD, Tomkinson KN, Michnick D, Selighsohn U, Kaufman RJ. Posttranslational sulfation of factor V is required for efficient thrombin cleavage and activation and for full procoagulant activity. Biochemistry. 1994;33(22):6952–6959. doi: 10.1021/bi00188a026. [DOI] [PubMed] [Google Scholar]
- 47.Long SB, Long MB, White RR, Sullenger BA. Crystal structure of an RNA aptamer bound to thrombin. RNA. 2008;14(12):2504–2512. doi: 10.1261/rna.1239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki K, Dahlbäck B, Stenflo J. Thrombin-catalyzed activation of human coagulation factor V. J Biol Chem. 1982;257(11):6556–6564. [PubMed] [Google Scholar]
- 49.Keller FG, Ortel TL, Quinn-Allen MA, Kane WH. Thrombin-catalyzed activation of recombinant human factor V. Biochemistry. 1995;34(12):4118–4124. doi: 10.1021/bi00012a030. [DOI] [PubMed] [Google Scholar]
- 50.Thorelli E, Kaufman RJ, Dahlbäck B. Cleavage requirements for activation of factor V by factor Xa. Eur J Biochem. 1997;247(1):12–20. doi: 10.1111/j.1432-1033.1997.00012.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.