Abstract
In mammalian cells, multiple cellular processes, including gene silencing, cell growth and differentiation, pluripotency, neoplastic transformation, apoptosis, DNA repair, and maintenance of genomic integrity, converge on the evolutionarily conserved protein KAP1, which is thought to regulate the dynamic organization of chromatin structure via its ability to influence epigenetic patterns and chromatin compaction. In this minireview, we discuss how KAP1 might execute such pleiotropic effects, focusing on genomic targeting mechanisms, protein-protein interactions, specific post-translational modifications of both KAP1 and associated histones, and transcriptome analyses of cells deficient in KAP1.
Keywords: Chromatin Histone Modification, Chromatin Immunoprecipitation (ChIP), Epigenetics, Transcription Factors, Transcriptional Repressor, Zinc Finger, ChIP-seq, KAP1, TIF1B, TRIM28
Introduction
Several independent studies in 1996 identified KAP1 as an interaction partner of members of the family of KRAB (Krüppel-associated box) domain-containing zinc finger transcription factors, variously naming the protein KAP1 (KRAB-associated protein 1), KRIP1 (KRAB-A-interacting protein 1), transcription intermediary factor (TIF)2 1β, or TRIM28 (tripartite motif-containing protein 28) (1–4). KAP1 is a member of a family of ∼60 human TRIM genes (5) and is highly related to three other TRIM proteins, TIF1α, TIF1γ, and TIF1δ (Fig. 1). Although the TIF1 subfamily shares many structural features, there is a high degree of specificity for homo-oligomerization and little functional overlap between TIF1 family members (6). For example, TIF1γ, but not the other related TRIM proteins, is a ligand-dependent co-regulator for nuclear hormone receptors, and TIF1γ, but not other family members, plays a role in signaling by transforming growth factor and in hematopoiesis (7–9). Expression patterns of the TIF1 family members also differ. Studied only in mice to date, TIF1δ is restricted to the testis during the elongating spermatid stage (10), TIF1α is preferentially expressed in the central and peripheral nervous systems early in development (11), and KAP1 is ubiquitously expressed throughout development (12).
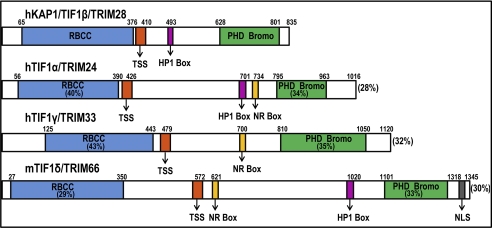
FIGURE 1.
Schematics of the human KAP1 protein (also called TIF1β and TRIM28) and other related proteins, including TIF1α/TRIM24, TIF1γ/TRIM33, and TIF1δ/TRIM66. The overall sequence identity between KAP1 and the other proteins is shown next to the C termini of the other proteins; the percentage sequence identity of the other proteins to the KAP1 protein in the RBCC domain and in the C-terminal PB domain is also shown. The TSS domain, the HP1 box, a domain that has been shown to bind nuclear receptors (NR Box), and a nuclear localization sequence (NLS) domain are also indicated. h, human; m, mouse.
KAP1 is a critical regulator of normal development and differentiation (see TRIM28 in the Transcription Factor Encyclopedia Database); mice deficient in KAP1 die prior to gastrulation (12), whereas mice with KAP1 specifically deleted in the adult forebrain exhibit heightened levels of anxiety and stress-induced alterations in learning and memory (14). KAP1 is also involved in maintaining pluripotency (15), is required for terminal differentiation of mouse embryonic stem cells (16, 17), and has been associated with promoting and inhibiting differentiation of different adult cell types. For example, KAP1 antagonizes erythroid differentiation (18) but promotes differentiation of U937 cells into macrophages (19). Several studies have also implicated KAP1 in tumor development. KAP1 protein levels are increased in liver, gastric, lung, breast, and prostate cancer, and gastric cancer patients with high levels of KAP1 show a significantly lower survival rate (see TRIM28 in The Human Protein Atlas Database) (20, 21, 23). Reduction of KAP1 in gastric cancer cells causes impairment in cell growth with an accumulation of cells in the G1 phase of the cell cycle, and reduction of KAP1 in cells exposed to radiation increases p53 levels, suggesting that KAP1 may promote neoplastic transformation via suppression of apoptosis (24). Such studies have led to the proposal that anti-KAP1 drugs should be developed for anticancer therapy (25). Clearly, KAP1 plays a critical role in proliferation and differentiation of both normal and tumor cells. Presented below is a summary of how studies to date in part support current models of KAP1 function. In addition, we discuss findings that suggest that certain aspects of the current model should be reconsidered.
KAP1 Protein Structure
All TIF1 family members have a similar overall architecture that includes an N-terminal tripartite motif (TRIM), which is a protein-protein and oligomerization interface containing an RBCC (Ring (really interesting new gene) finger, two B-box zinc fingers, and a coiled coil) domain, a central TIF1 signature sequence (TSS) domain consisting of a 25-amino acid tryptophan- and phenylalanine-rich sequence, and a C-terminal combination plant homeodomain (PHD) and bromodomain (1, 8). However, only KAP1, TIF1α, and TIF1δ share a central HP1 (heterochromatin protein 1)-binding domain (Fig. 1). Experiments performed over the last 15 years have shown that KAP1 is highly modular in structure, with separate domains mediating nuclear localization, interaction with transcription factors, oligomerization, and regulation of transcription (26).
The N terminus of KAP1 contains the RBCC domain, a high affinity protein interaction domain stretching from amino acids 20 to 377. The RBCC domain is necessary and sufficient for interaction of KAP1 with the KRAB repression module of KRAB-ZNFs; all three RBCC subdomains contribute to interaction with KRAB modules (6). Although all TIF1 family members contain an RBCC domain, only KAP1 can bind to the KRAB repression modules of KRAB-ZNFs (6). A number of biochemical and biophysical experiments have revealed that the KAP1 RBCC domain binds as a homotrimer to a single KRAB domain. This oligomerization promotes folding of the KRAB domain and encapsulates it in a protease-resistant core (27). The Ring subdomain is a double zinc-binding C3HC4 motif found in >200 proteins that are components of macromolecular complexes with diverse functions in oncogenesis, RNA transport, cell cycle, ubiquitination, and signal transduction (28). The B-box is a cysteine-rich zinc-binding motif of the form CHC3H2 and, together with the coiled-coil domain, provides an extended hydrophobic α-helical region that presents a strong interface for protein-protein interactions (27). Adjacent to the RBCC is the TSS domain; deletion of this domain abrogates transcriptional repression mediated by TIF1γ (8).
The central region of KAP1 includes the HP1-binding domain, which is a hydrophobic PxVxL pentapeptide located between amino acids 486 and 497 (29). The interaction of KAP1 with the chromoshadow domain of HP1 family members is required for repression of reporter genes (30), discussed in greater detail below. Immunofluorescence studies show that the majority of KAP1 has the same staining pattern as HP1γ, which is present throughout the nucleoplasm but excluded from nucleoli. A small percentage of KAP1 is concentrated into dot-like structures that are regions of pericentric heterochromatin, and an even smaller percentage is found in heterochromatic foci and nucleoli in regions co-occupied by HP1β. These studies are consistent with multiple roles for KAP1-HP1 complexes in silencing euchromatic and pericentric heterochromatic regions (26). The remaining central region of KAP1 is least conserved among all of the TIF1 family members and is rich in prolines, glycines, and serines. No well defined structure has been assigned to this domain; rather, the entire central region of KAP1 is in a highly extended and flexible conformation (29). Perhaps this region provides KAP1 with the adaptability needed for interaction with a multitude of protein complexes.
The C-terminal tandem PHD and bromodomain (called the PB domain) of KAP1 lie between amino acids 618 and 835 and function as a highly cooperative unit for transcriptional repression, with both domains being required to obtain maximum levels of repression (31). The PHD finger of KAP1 is a 60-amino acid domain with a C4HC3 arrangement consisting of two zinc atoms cross-braced between antiparallel β-sheets. The bromodomain of KAP1 is a 100-amino acid stretch consisting of four helices bundled in a unique left-turn topology (32). Typically, bromodomains are found in transcriptional activators and are involved in the recognition of acetylated histone tails (33). Similar to other bromodomain-containing proteins, the bromodomain of KAP1 has a conserved hydrophobic core and recognizes the backbone of histone tails. However, unlike the other proteins, KAP1 has lost its ability to contact acetyllysine residues (31). The PB domain of KAP1 can also interact with two chromatin-modifying enzymes: Mi2α, an isoform of the Mi2 protein found in the NuRD (nucleosome remodeling and histone deacetylation) complex, and SETDB1 (SET domain, bifurcated 1), an H3K9me3-specific histone methyltransferase. Because the addition of an inhibitor of histone deacetylases (HDACs) only partially relieves KAP1-mediated repression of reporter genes and because only a small fraction of KAP1 stably associates with Mi2α in vivo, it is thought that Mi2α may play only a minor role in KAP1-mediated repression (31, 34). In contrast, trimethylation of histone H3 at Lys-9 by SETDB1 creates high affinity genomic binding sites for the KAP1-HP1 complex (because of the ability of HP1 to bind to H3K9me3). This, along with the observation that KAP1 and SETDB1 colocalize at thousands of genomic sites in the human genome (35), suggests that SETDB1 may play an important role in KAP1-mediated repression.
The interdependence of the PHD and bromodomain of KAP1 for optimum repression has been recently explained by the elucidation of the individual functions of each subdomain. The bromodomain of KAP1 is the essential interface for mediating interactions with SETDB1, and this interaction occurs in a sumoylation-dependent manner at lysines 554, 575, 676, 750, 779, and 804 of KAP1 (36). Sumoylation of these KAP1 residues also stimulates the histone methyltransferase activity of SETDB1 bound to KAP1 (37). Thus, sumoylated KAP1 is the highly repressive form of KAP1 (37). The PHD of KAP1 contributes by functioning as an intramolecular E3 ligase that sumoylates the adjacent KAP1 bromodomain (37). Thus, the PHD-mediated sumoylation of the KAP1 bromodomain, followed by interaction of SETDB1 with the sumoylated bromodomain, provides a mechanistic basis for the cooperative function of the KAP1 PB domain (36).
Recruitment of KAP1 to the Genome
The biochemical studies described above suggest that KAP1 can coordinate the assembly of a macromolecular complex containing chromatin-remodeling proteins such as Mi2α, SETDB1, and HP1 to create an epigenetically stable and heritable heterochromatic microenvironment (38, 39). However, neither KAP1 nor any of the above-mentioned interaction partners have DNA-binding domains (DBDs). Therefore, other protein partners are required to recruit KAP1 to the genome. KAP1 was originally identified as an interaction partner of two different C2H2 zinc finger proteins, KOX1 and KID-1. C2H2 zinc finger proteins are the largest class of DNA-binding transcription factors encoded in the human genome; about half contain an N-terminal KRAB domain, which interacts with the RBCC domain of KAP1 (40). There are >400 human KRAB-ZNF genes encoding transcripts for 742 different proteins (41). KRAB-ZNFs are postulated to regulate diverse processes such as embryonic development, tissue-specific gene expression, and cancer progression (42). Comparative genomic analyses indicate that the KRAB-ZNF gene family is specific to tetrapod vertebrates, with the repertoire of KRAB-ZNFs differing significantly between species, suggesting that members of this family have evolved to perform species-specific transcriptional regulation; 136 KRAB-ZNFs are primate-specific and may be involved in regulation of the immune and nervous systems (43). The C-terminal regions of KRAB-ZNFs contain tandemly arranged arrays of C2H2 zinc finger modules, comprising from a few to >30 fingers. Individual fingers, each of which can recognize 3 nucleotides of DNA, are separated from each other by a highly conserved linker sequence (44). KRAB-ZNF genes are frequently found in clusters in the human genome, having evolved through duplication and deletion of their zinc finger domains (45). The family of KRAB-ZNF genes has a modest degree of overall coexpression in the human body (46), possibly because the entire family is expressed at low levels in most cells. However, certain family members are highly expressed in several cell types.3
The ability of specific KRAB-ZNFs to bind to the RBCC domain of KAP1 has been studied mainly using artificial assays. Using a mammalian two-hybrid system, the RBCC domain of KAP1 was tested for interaction with KRAB domains from 61 different KRAB-ZNFs (48). The majority of these KRAB-ZNFs could bind KAP1 and were dependent on this interaction for their transcription-repressive abilities. One of the tested KRAB-ZNFs was KOX1. The KRAB domain of KOX1, which was initially used to purify KAP1, is often used as a positive control for KAP1 protein interactions and when testing KAP1-mediated repression (1, 2, 35, 49–51). Interestingly, when full-length KOX1 was tested for its ability to bind endogenous KAP1, it showed a much weaker interaction than was previously observed in vitro, suggesting that, although the KRAB domains of many KRAB-ZNFs are capable of binding KAP1 in vitro, this does not necessarily indicate that the KRAB-ZNF is a major KAP1 interaction partner in the cell (48). KAP1 can also interact with KRAB domains that are not associated with zinc finger domains. For example, the KRAB-O (KRAB only) protein serves as a bridge between the DNA-binding protein SRY (sex-determining region Y) and KAP1, recruiting KAP1 to SRY-binding sites (52, 53). Similarly, others have identified a protein called VHLaK (pVHL-associated KRAB-A domain-containing protein), which serves as a bridge between KAP1 and the von Hippel-Lindau tumor suppressor protein (54). Interestingly, both KRAB-O and VHLaK are alternatively spliced versions of KRAB-ZNF genes that produce proteins containing the KRAB domain but lacking the DNA-binding zinc fingers. In certain cell types, KAP1 has also been shown to associate with the transcription factors MM1, E2F1, MDM2, STAT (signal transducers and activators of transcription) family members, HNRNPAB, TEL/ETV6, CCAAT/enhancer-binding protein β, and NGFI, and through these interactions, KAP1 takes part in numerous processes such as intestinal homeostasis, epithelial-mesenchymal transition, and the immediate-early stress response (18, 19, 24, 55–59).
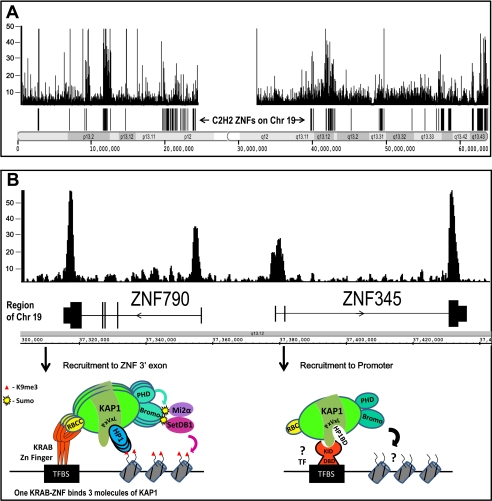
To determine which, if any, of the KRAB-ZNFs and/or other DNA-binding proteins recruit KAP1 to the genome, KAP1-binding sites were identified using ChIP, followed by microarrays (ChIP-chip) or by sequencing (ChIP-seq), which allows the identification of binding sites for a protein to be identified on a genome-wide scale (60–62). More than 7000 binding sites for KAP1 were identified in NTera2 cells by performing whole genome ChIP-chip experiments (63). Subsequently, using a combination of ChIP-chip and ChIP-seq, KAP1 targets were identified in numerous normal and tumor cells. In all cell types, KAP1 displays a unique genomic localization pattern (Fig. 2A). The strongest KAP1-binding sites are the 3′-coding exons of ZNF genes, whereas the other KAP1-binding sites are either near transcription start sites or in intragenic regions. To determine whether KRAB-ZNFs are involved in recruitment of KAP1 to the target sites, ChIP-seq experiments were performed using a series of mutant KAP1 proteins. These studies showed that KAP1 deleted for the RBCC domain was no longer recruited to the 3′-coding exons of ZNF genes, thus providing strong in vivo support for KRAB-ZNF-mediated recruitment of KAP1 (64). Further studies revealed that ZNF274 colocalizes with KAP1 at 3′-coding exons of ZNF genes (35). It has not yet been possible to demonstrate that other KRAB-ZNFs that show positive in vitro interactions with KAP1 bind to KAP1 genomic sites.4 However, the association of KAP1 with specific KRAB-ZNFs may be highly cell type-specific. Interestingly, KAP1 deleted for the RBCC domain can still bind to promoter regions, indicating that KAP1 is recruited to these sites by a novel mechanism independent of a KRAB-ZNF. Thus, there are at least two mechanisms (Fig. 2B) for recruiting KAP1 to the genome, one involving KRAB-ZNFs and one involving other DNA-binding proteins (64). Although the factor that recruits KAP1 to promoters has not yet been identified, mutational analyses suggest that KAP1 may be recruited to promoter targets through protein-protein interactions that occur in the central domain stretching from amino acids 380 to 618 but outside of the HP1 box (64).
FIGURE 2.
Recruitment of KAP1 to the genome. A, shown is the KAP1 ChIP-seq binding pattern and position of the C2H2 ZNF genes for chromosome (Chr) 19 in HEK293 cells. A similar pattern has been observed in numerous cell types. B, shown is KAP1 binding at the 5′- and 3′-ends of two ZNF genes. (The genes are transcribed in the opposite direction, as indicated by the arrowheads.) Under the ZNF790 gene is a model illustrating recruitment of KAP1 and associated proteins to 3′-coding exons of ZNF genes. This recruitment is dependent upon interaction of the RBCC domain of KAP1 with a KRAB-ZNF that is bound to its recognition motif (indicated as TFBS); 3 molecules of KAP1 interact with a KRAB-ZNF. The PHD domain sumoylates the bromodomain, leading to recruitment of SETDB1 and Mi2α and creation of the H3K9me3 mark on nearby nucleosomes. HP1 can bind to KAP1 at the PxVxL motif and also to H3K9me3, stabilizing the bound KAP1-containing complex. Under the ZFN345 gene is a model illustrating recruitment of KAP1 to promoters. This recruitment is dependent upon interaction of KAP1 with a non-KRAB-ZNF DNA-binding protein (indicated by ? TF) that has a KAP1-interacting domain (KID) and a DBD. KAP1 interacts with this non-KRAB-ZNF DNA-binding protein via a region of KAP1 near the HP1-binding domain (HP1BD). KAP1 bound to cellular promoters does not recruit SETDB1 or result in H3K9me3. See text for details.
Role of KAP1 in Transcriptional Regulation
The genomic recruitment studies of KAP1 have not addressed the functional consequences of KAP1 binding. As described above, KAP1 can interact with HDAC and histone methyltransferase complexes, and it has been suggested that KAP1 regulates transcription via changes in histone modifications at specific target sites. In support of this hypothesis, a recent genome-wide study showed that ZNF 3′-ends that are bound by KAP1 are also bound by SETDB1 and marked by H3K9me3 (35, 64). Also, reporter-promoters bound by inducible KRAB fusion proteins have H3K9me3 (39, 66). Therefore, both when KAP1 is artificially brought to promoters that have been engineered to bind many copies of a KRAB fusion protein and when KAP1 localizes under normal physiological conditions to the 3′-ends of ZNF genes, it recruits SETDB1, which trimethylates histone H3 at Lys-9. It has been proposed that this creates a localized alteration in chromatin structure and/or relocalizes the target regions to domains of heterochromatin (38, 64). Based on these studies, it is reasonable to propose that KAP1 functions as a transcriptional repressor.
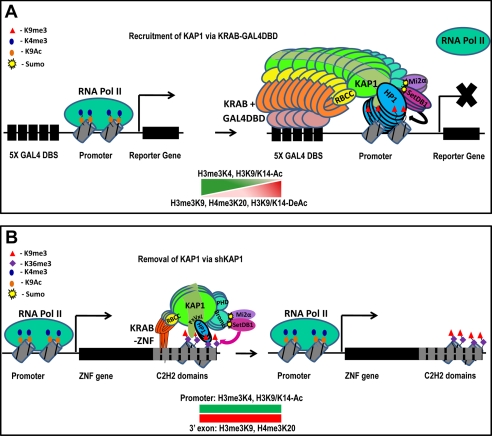
Most of the evidence supporting a role for KAP1 as a transcriptional regulator comes from experiments in which an isolated KRAB domain has been fused to a DBD of another transcription factor. For example, the KOX1 KRAB domain fused to the PAX3 DBD can recruit KAP1 and repress a stably integrated PAX3 site-containing promoter, and a Gal4-KRAB fusion protein can repress the activity of a reporter-promoter with five Gal4 sites (34, 39). Others have used inducible KRAB fusion proteins to demonstrate KAP1-mediated repression (38, 66, 67). Studies of mutant KRAB domains also provide support for a role for KAP1 in repression; substitution mutations in the KRAB domain of various KRAB-ZNFs at two highly conserved residues that are critical for interaction with KAP1 result in diminished repression activity (1, 49, 68, 69). Additionally, KAP1 can repress transcription of reporter genes when directly tethered to DNA as a Gal4-KAP1 fusion protein (39). All of these studies reinforce the idea that KRAB-ZNFs mediate repression in a KAP1-dependent manner, which leads to the creation of a heterochromatic epigenetic profile at the targeted locus (Fig. 3A). KAP1 has also been associated with transcriptional activation; using reporter-promoter assays, KAP1 was shown to function as a coactivator for NGFI-B and CCAAT/enhancer-binding protein β (19, 55, 58). The mechanisms by which KAP1 can activate transcription are not known, but perhaps KAP1-associated HDACs and histone methylases function to modify and release repressor proteins bound to the target promoters.
FIGURE 3.
Transcriptional regulation by KAP1. A, KAP1 can repress transcription when recruited to promoters by a Gal4 DBD-KRAB fusion protein. Before binding of KAP1, the promoter is bound by RNA polymerase II (RNA Pol II) and by active chromatin marks such as H3K4me3 and H3K9Ac. Upon binding of a fusion protein consisting of a Gal4 DBD and a KRAB domain, KAP1 and associated proteins are recruited to the promoter. This recruitment results in the loss of RNA polymerase II, the loss of active chromatin marks, and the creation of the repressive H3K9me3 mark, leading to transcriptional repression. B, reduction of KAP1 has little effect on the expression of endogenous ZNF genes. Under normal conditions, the promoters of ZNF genes are covered by active chromatin marks (H3K9Ac and H3K4me3), and the exons are covered by the transcriptional elongation mark H3K36me3, even though the 3′-coding exons are bound by KAP1, SETDB1, and H3K9me3. Thus, KAP1 target genes are covered by both active and repressed marks, and the genes are transcribed. Both the promoters and 3′-exons of ZNF genes retain their normal epigenetic profile after removal of KAP1 by shRNA. See text for details. DBS, DNA-binding site.
The artificial recruitment experiments clearly demonstrate that KAP1 can influence transcriptional activity in reporter assays. However, whether KAP1 regulates the expression of endogenous cellular genes is less clear. Most cellular promoters bound by KAP1 are not bound by SETDB1 or H3K9me3 (46, 64). However, KAP1 may use alternative methods, perhaps changes in histone acetylation (30), to regulate transcription at cellular promoters. For example, two complexes, one containing KAP1, MM1, and Myc and another containing KAP1 and ZNF160, have been shown to repress transcription from cellular promoters in an HDAC-dependent manner in specific cell types (59, 70). Also, KAP1 can interact with STAT3 (57, 71), and reduction of KAP1 levels results in modest increases in the levels of several STAT3-regulated cellular RNAs. However, the mechanism by which KAP1 mediates these effects is not known. No studies were performed to examine KAP1 occupancy of the regulated promoters; therefore, it is not known if KAP1 directly binds to these promoters. In fact, evidence was presented that KAP1 influences the subnuclear localization of STAT3, suggesting that the effects of KAP1 may have been through protein-protein interactions that occur off the DNA. KAP1 can also bind to the E2F1 protein, and ChIP assays showed KAP1 and E2F1 binding at an E2F target promoter (56). However, it was only in the presence of overexpressed proteins that binding of KAP1 could be detected on the target promoter; further studies in which additional promoters are analyzed are required to address the possibility that E2F1 may recruit KAP1 to the genome. Interestingly, KAP1 seemed to increase the interaction of E2F1 with HDAC1 and to decrease the acetylation on the E2F1 protein, suggesting that KAP1 may affect the regulation of E2F target genes by decreasing the activity of the E2F1 protein.
Most experiments linking KAP1 to regulation of cellular promoters have focused on a small set of genes and address the role of KAP1 in regulating those gene transcripts. Recent ChIP-seq experiments have identified thousands of KAP1-binding sites, and using RNA expression arrays and RNA-seq, it is now possible to address the global effect of KAP1 on transcriptional regulation of its target genes on a genome-wide scale. Because the strongest KAP1-binding sites are the 3′-ends of ZNF genes, one might expect that KAP1 would regulate expression of these genes. Surprisingly, there is no correlation between the level of KAP1 at a ZNF 3′-end and the expression of the ZNF gene; the promoters of the KAP1 ZNF target genes contain the active H3K4me3 and H3K9Ac marks, and the gene bodies are bound by the transcriptional elongation mark of H3K36me3 (Fig. 3B) (72). Also, there is no change in mRNA levels or splicing of KAP1-bound ZNF genes in NTera2 cells upon reduction of KAP1 using shRNAs (64). Thus, the presence of the KAP1-SETDB1-H3K9me3 complex at ZNF 3′-exons does not reduce the levels of transcripts, impede transcriptional elongation, or alter splicing or processing of the mRNAs.5 Although some transcripts show robust changes in expression upon KAP1 knockdown, the majority of the promoters of these genes are not bound by KAP1, suggesting that, in this cell type, the effects of KAP1 on the human transcriptome are mostly indirect (64).
KAP1: Guardian of the Genome
In the KAP1 knockdown experiments described above, only a modest number of genes showed significant changes in RNA levels upon reduction of KAP1. However, we note that KAP1 has been implicated in the repression of endogenous retroviruses (74) and in the regulation of other viruses (75, 76). Interestingly, the loss of KAP1 caused increased expression of endogenous retroviruses in mouse embryonic stem cells but not in mouse embryonic fibroblasts, suggesting that, in certain cell types, KAP1 may not be involved in repressing retroviral transcription or may be functionally redundant with other repressive mechanisms. These findings, coupled with the observations that there are thousands of KAP1-binding sites in the genome but only few cellular genes that respond to loss of KAP1, suggest that a major role of KAP1 may lie outside of transcriptional regulation.
KAP1 has been suggested to regulate apoptosis in a manner independent of its transcriptional activities. KAP1 acts cooperatively with MDM2, a ubiquitin E3 ligase that binds to p53 and marks it for degradation, by recruiting HDAC1 to the MDM2-p53 complex, leading to deacetylation and degradation of p53. Although MDM2 is the major ubiquitin ligase for p53, KAP1 is independently capable of promoting p53 ubiquitination, suggesting that it may encode or recruit a ubiquitin E3 ligase (24). Recently, MAGE proteins, which are highly expressed in various cancers, were shown to be cofactors in KAP1-mediated suppression of p53 activity. MAGE proteins bind to KAP1 and enhance formation of the KAP1-MDM2-p53 complex, leading to suppression of p53-mediated apoptosis and promotion of tumor cell survival (77).
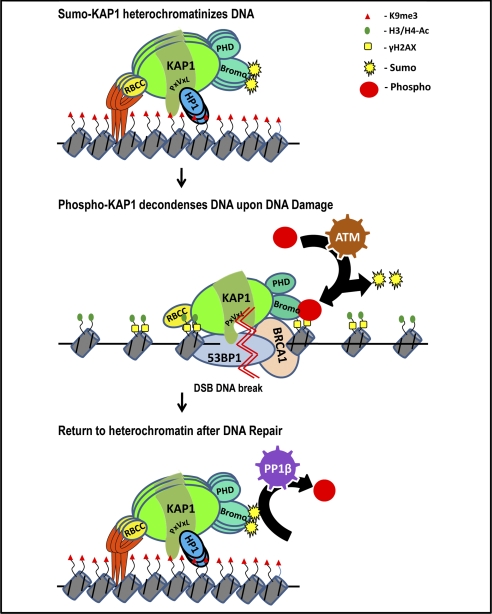
KAP1 has also been implicated in DNA repair. DNA damage such as double-strand break formation induces ATM, which phosphorylates KAP1 (78). It has been proposed that ATM-mediated phosphorylation of KAP1 in response to genotoxic stress results in loss of sumoylated KAP1, leading to derepression of KAP1 target genes involved in promoting cell cycle arrest and apoptosis (79). The KRAB-ZNF ZBRK1 has been shown to repress the GADD45 (growth arrest and DNA damage clone 45) gene in a KAP1-dependent manner (80); perhaps depression of such genes due to a switch from sumoylated to phosphorylated KAP1 is critical for DNA repair. Recent findings implicate protein phosphatase 1β (PP1β) in the recovery of KAP1 repressive function after DNA damage-induced phosphorylation (81). PP1β can interact with the coiled-coil domain of KAP1 and dephosphorylate KAP1, promoting sumoylation of KAP1 and return of its repressive function. Thus, KAP1 exists in a balance between a phosphorylated and a sumoylated state, which influences its repressive abilities (79). Such studies suggest that investigation of the role of KAP1 in regulating the transcriptome should perhaps be repeated under DNA-damaging conditions. However, KAP1 is also thought to have a non-transcriptional role in regulating the DNA damage response (Fig. 4). Upon DNA damage, there is a rapid localization of phosphorylated KAP1 to DNA damage foci, where it colocalizes with numerous DNA damage response proteins (78). Loss of phosphorylated KAP1 renders cells hypersensitive to DNA damage and leads to loss of DNA damage-induced chromatin decondensation, suggesting that KAP1 must play an active role in this process (73, 82). Although phosphorylation of KAP1 is required for the ATM-mediated global chromatin decondensation in response to double-strand breaks (65, 82), the mechanism by which phosphorylated KAP1 mediates this response is still unknown. Perhaps the switch to its phosphorylated form can cause the local chromatin decondensation required for access of DNA repair proteins, and return to its sumoylated form can assist in re-forming condensed chromatin after the DNA is repaired.
FIGURE 4.
Model for KAP1 involvement in DNA repair. Under normal conditions, sumoylated KAP1 is recruited to the genome via KRAB-ZNFs, resulting in H3K9me3 at nearby nucleosomes. Upon DNA damage (indicated by the double zigzag), there is a switch between the sumoylated and phosphorylated forms of KAP1 (mediated by ATM) and a rapid localization of phosphorylated KAP1 to DNA damage foci, where it may facilitate a local decondensation of chromatin, as indicated by the acetylation of His-3 and His-4 and the presence of H2AX, allowing access of DNA repair proteins such as 53BP1 and BRCA1. A return to the sumoylated form of KAP1 mediated by PP1β may assist in re-forming condensed chromatin after the DNA is repaired. See text for details. DSB, double-strand break.
KAP1 has also been suggested to be involved in suppressing recombination. As noted above, the strongest KAP1 targets are the 3′-coding exons of ZNF genes. ZNF genes are highly homologous, having arisen from genomic duplications (45), and their 3′-coding exons encode tandemly arranged highly repetitive zinc finger domains. Interestingly, binding of KAP1 positively correlates with the number of repeated zinc fingers within the ZNF 3′-exons (72). Based on studies from yeast showing that the Sir2 protein is required to prevent recombination-mediated loss of the ribosomal DNA repeats (47), it has been proposed that heterochromatinization of ZNF 3′-coding exons may prevent recombination-mediated deletion of this large family of highly homologous genes (46, 64, 72). Circumstantial evidence in support of this hypothesis comes from studies showing that the 3′-coding exons of KRAB-ZNF genes are deleted when expression constructs are introduced into cells (13, 22).5 This phenomenon might be due to homologous recombination-mediated deletion of the exogenously introduced 3′-coding exon that has not yet been protected by heterochromatin. If KAP1 can be experimentally linked to suppression of recombination, this would suggest a new function for epigenetic modifications that are currently thought to represent only a repressed transcription state.
Conclusions
KAP1 has been implicated in diverse cellular processes such as development, differentiation, and neoplastic transformation. Although the precise mechanism(s) by which KAP1 influences such processes remains unclear, studies over the past 15 years have revealed several insights into KAP1 function. 1) KAP1 is a scaffold protein that can assemble epigenetic machinery (Fig. 1). Specifically, it interacts with histone methyltransferases and HDACs via a C-terminal PHD and bromodomain. 2) KAP1 binds to thousands of sites in the human genome, including both 3′-coding exons of ZNF genes and promoter regions (Fig. 2). It is recruited to the genome via interaction with KRAB-ZNFs and other transcription factors. 3) KAP1 is a robust transcriptional repressor when artificially recruited in multiple copies to promoters of reporter genes (Fig. 3A) but has very little influence on the transcript levels or epigenetic profiles of its endogenous target genes (Fig. 3B). 4) Post-translational modifications regulate KAP1 function; sumoylated KAP1 is involved in transcriptional repression, whereas phosphorylated KAP1 is involved in DNA repair (Fig. 4).
The modest influence of KAP1 on the human transcriptome and epigenome remains an enigmatic finding, especially considering that mice deficient in KAP1 die prior to gastrulation. Future studies employing specific developmental stages and/or differentiation states may help reveal conditions under which KAP1 plays a key role in transcriptional regulation of cellular genes.
Acknowledgments
We thank Suhas Krishna for help with the figures and Seth Frietze for critical reading of the manuscript.
This work was supported in part by United States Public Health Service Grants CA45240 and HG004558. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
V. X. Jin and P. J. Farnham, unpublished data.
S. Iyengar, S. Frietze, and P. J. Farnham, unpublished data.
S. Iyengar and P. J. Farnham, unpublished data.
- TIF
- transcription intermediary factor
- TSS
- TIF1 signature sequence
- PHD
- plant homeodomain
- HDAC
- histone deacetylase
- DBD
- DNA-binding domain
- PP1β
- protein phosphatase 1β.
REFERENCES
- 1. Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., Rauscher F. J., 3rd (1996) Genes Dev. 10, 2067–2078 [DOI] [PubMed] [Google Scholar]
- 2. Moosmann P., Georgiev O., Le Douarin B., Bourquin J. P., Schaffner W. (1996) Nucleic Acids Res. 24, 4859–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim S. S., Chen Y. M., O'Leary E., Witzgall R., Vidal M., Bonventre J. V. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15299–15304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Le Douarin B., Nielsen A. L., Garnier J. M., Ichinose H., Jeanmougin F., Losson R., Chambon P. (1996) EMBO J. 15, 6701–6715 [PMC free article] [PubMed] [Google Scholar]
- 5. Ozato K., Shin D. M., Chang T. H., Morse H. C., 3rd (2008) Nat. Rev. Immunol. 8, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peng H., Feldman I., Rauscher F. J., 3rd (2002) J. Mol. Biol. 320, 629–644 [DOI] [PubMed] [Google Scholar]
- 7. Le Douarin B., Zechel C., Garnier J. M., Lutz Y., Tora L., Pierrat P., Heery D., Gronemeyer H., Chambon P., Losson R. (1995) EMBO J. 14, 2020–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venturini L., You J., Stadler M., Galien R., Lallemand V., Koken M. H., Mattei M. G., Ganser A., Chambon P., Losson R., de Thé H. (1999) Oncogene 18, 1209–1217 [DOI] [PubMed] [Google Scholar]
- 9. Doisne J. M., Bartholin L., Yan K. P., Garcia C. N., Duarte N., Le Luduec J. B., Vincent D., Cyprian F., Horvat B., Martel S., Rimokh R., Losson R., Benlagha K., Marie J. C. (2009) J. Exp. Med. 206, 1365–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Khetchoumian K., Teletin M., Mark M., Lerouge T., Cerviño M., Oulad-Abdelghani M., Chambon P., Losson R. (2004) J. Biol. Chem. 279, 48329–48341 [DOI] [PubMed] [Google Scholar]
- 11. Niederreither K., Remboutsika E., Gansmuller A., Losson R., Dollé P. (1999) Mech. Dev. 88, 111–117 [DOI] [PubMed] [Google Scholar]
- 12. Cammas F., Mark M., Dollé P., Dierich A., Chambon P., Losson R. (2000) Development 127, 2955–2963 [DOI] [PubMed] [Google Scholar]
- 13. Wolf D., Goff S. P. (2009) Nature 458, 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jakobsson J., Cordero M. I., Bisaz R., Groner A. C., Busskamp V., Bensadoun J. C., Cammas F., Losson R., Mansuy I. M., Sandi C., Trono D. (2008) Neuron 60, 818–831 [DOI] [PubMed] [Google Scholar]
- 15. Hu G., Kim J., Xu Q., Leng Y., Orkin S. H., Elledge S. J. (2009) Genes Dev. 23, 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cammas F., Oulad-Abdelghani M., Vonesch J. L., Huss-Garcia Y., Chambon P., Losson R. (2002) J. Cell Sci. 115, 3439–3448 [DOI] [PubMed] [Google Scholar]
- 17. Cammas F., Herzog M., Lerouge T., Chambon P., Losson R. (2004) Genes Dev. 18, 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakamura Y., Yamagata T., Maki K., Sasaki K., Kitabayashi I., Mitani K. (2006) Int. J. Hematol. 84, 377–380 [DOI] [PubMed] [Google Scholar]
- 19. Rooney J. W., Calame K. L. (2001) Genes Dev. 15, 3023–3038 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Silva F. P., Hamamoto R., Furukawa Y., Nakamura Y. (2006) Oncogene 25, 5063–5070 [DOI] [PubMed] [Google Scholar]
- 21. Beer D. G., Kardia S. L., Huang C. C., Giordano T. J., Levin A. M., Misek D. E., Lin L., Chen G., Gharib T. G., Thomas D. G., Lizyness M. L., Kuick R., Hayasaka S., Taylor J. M., Iannettoni M. D., Orringer M. B., Hanash S. (2002) Nat. Med. 8, 816–824 [DOI] [PubMed] [Google Scholar]
- 22. Mark C., Looman C., Abrink M., Hellman L. (2001) DNA Cell Biol. 20, 275–286 [DOI] [PubMed] [Google Scholar]
- 23. Yokoe T., Toiyama Y., Okugawa Y., Tanaka K., Ohi M., Inoue Y., Mohri Y., Miki C., Kusunoki M. (2010) Ann. Surg. Oncol. 17, 821–828 [DOI] [PubMed] [Google Scholar]
- 24. Wang C., Ivanov A., Chen L., Fredericks W. J., Seto E., Rauscher F. J., 3rd, Chen J. (2005) EMBO J. 24, 3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okamoto K., Kitabayashi I., Taya Y. (2006) Biochem. Biophys. Res. Commun. 351, 216–222 [DOI] [PubMed] [Google Scholar]
- 26. Ryan R. F., Schultz D. C., Ayyanathan K., Singh P. B., Friedman J. R., Fredericks W. J., Rauscher F. J., 3rd (1999) Mol. Cell. Biol. 19, 4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng H., Begg G. E., Schultz D. C., Friedman J. R., Jensen D. E., Speicher D. W., Rauscher F. J., 3rd (2000) J. Mol. Biol. 295, 1139–1162 [DOI] [PubMed] [Google Scholar]
- 28. Saurin A. J., Borden K. L., Boddy M. N., Freemont P. S. (1996) Trends Biochem. Sci. 21, 208–214 [PubMed] [Google Scholar]
- 29. Lechner M. S., Begg G. E., Speicher D. W., Rauscher F. J., 3rd (2000) Mol. Cell. Biol. 20, 6449–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nielsen A. L., Ortiz J. A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R. (1999) EMBO J. 18, 6385–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schultz D. C., Friedman J. R., Rauscher F. J., 3rd (2001) Genes Dev. 15, 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Capili A. D., Schultz D. C., Rauscher F. J., 3rd, Borden K. L. (2001) EMBO J. 20, 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeng L., Zhou M. M. (2002) FEBS Lett. 513, 124–128 [DOI] [PubMed] [Google Scholar]
- 34. Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J., 3rd (2002) Genes Dev. 16, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frietze S., O'Geen H., Blahnik K. R., Jin V. X., Farnham P. J. (2010) PLoS ONE 5, e15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zeng L., Yap K. L., Ivanov A. V., Wang X., Mujtaba S., Plotnikova O., Rauscher F. J., 3rd, Zhou M. M. (2008) Nat. Struct. Mol. Biol. 15, 626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M. M., Rauscher F. J., 3rd (2007) Mol. Cell 28, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ayyanathan K., Lechner M. S., Bell P., Maul G. G., Schultz D. C., Yamada Y., Tanaka K., Torigoe K., Rauscher F. J., 3rd (2003) Genes Dev. 17, 1855–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sripathy S. P., Stevens J., Schultz D. C. (2006) Mol. Cell. Biol. 26, 8623–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lorenz P., Dietmann S., Wilhelm T., Koczan D., Autran S., Gad S., Wen G., Ding G., Li Y., Rousseau-Merck M. F., Thiesen H. J. (2010) BMC Genomics 11, 206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huntley S., Baggott D. M., Hamilton A. T., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L. (2006) Genome Res. 16, 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Urrutia R. (2003) Genome Biol. 4, 231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nowick K., Hamilton A. T., Zhang H., Stubbs L. (2010) Mol. Biol. Evol. 27, 2606–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Looman C., Abrink M., Mark C., Hellman L. (2002) Mol. Biol. Evol. 19, 2118–2130 [DOI] [PubMed] [Google Scholar]
- 45. Emerson R. O., Thomas J. H. (2009) PLOS Genetics 5, e1000325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vogel M. J., Guelen L., de Wit E., Peric-Hupkes D., Lodén M., Talhout W., Feenstra M., Abbas B., Classen A. K., van Steensel B. (2006) Genome Res. 16, 1493–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gottlieb S., Esposito R. E. (1989) Cell 56, 771–776 [DOI] [PubMed] [Google Scholar]
- 48. Itokawa Y., Yanagawa T., Yamakawa H., Watanabe N., Koga H., Nagase T. (2009) Biochem. Biophys. Res. Commun. 388, 689–694 [DOI] [PubMed] [Google Scholar]
- 49. Peng H., Gibson L. C., Capili A. D., Borden K. L., Osborne M. J., Harper S. L., Speicher D. W., Zhao K., Marmorstein R., Rock T. A., Rauscher F. J., 3rd (2007) J. Mol. Biol. 370, 269–289 [DOI] [PubMed] [Google Scholar]
- 50. Zhao X. H., Zhu X. D., Liu J., Rao X. J., Huang P. T. (2003) Sheng Wu Gong Cheng Xue Bao 19, 608–612 [PubMed] [Google Scholar]
- 51. Fertey J., Hurst J., Straub E., Schenker A., Iftner T., Stubenrauch F. (2011) J. Virol. 85, 2918–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peng H., Ivanov A. V., Oh H. J., Lau Y. F., Rauscher F. J., 3rd (2009) J. Biol. Chem. 284, 35670–35680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jin V. X., O'Geen H., Iyengar S., Green R., Farnham P. J. (2007) Genome Res. 17, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li Z., Wang D., Na X., Schoen S. R., Messing E. M., Wu G. (2003) EMBO J. 22, 1857–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chang C. J., Chen Y. L., Lee S. C. (1998) Mol. Cell. Biol. 18, 5880–5887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang C., Rauscher F. J., 3rd, Cress W. D., Chen J. (2007) J. Biol. Chem. 282, 29902–29909 [DOI] [PubMed] [Google Scholar]
- 57. Tsuruma R., Ohbayashi N., Kamitani S., Ikeda O., Sato N., Muromoto R., Sekine Y., Oritani K., Matsuda T. (2008) Oncogene 27, 3054–3059 [DOI] [PubMed] [Google Scholar]
- 58. Rambaud J., Desroches J., Balsalobre A., Drouin J. (2009) J. Biol. Chem. 284, 14147–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Satou A., Taira T., Iguchi-Ariga S. M., Ariga H. (2001) J. Biol. Chem. 276, 46562–46567 [DOI] [PubMed] [Google Scholar]
- 60. Farnham P. J. (2009) Nat. Rev. Genet. 10, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schones D. E., Zhao K. (2008) Nat. Rev. Genet. 9, 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gilchrist D. A., Fargo D. C., Adelman K. (2009) Methods 48, 398–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. O'Geen H., Squazzo S. L., Iyengar S., Blahnik K., Rinn J. L., Chang H. Y., Green R., Farnham P. J. (2007) PLoS Genet. 3, e89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Iyengar S., Ivanov A. V., Jin V. X., Rauscher F. J., 3rd, Farnham P. J. (2011) Mol. Cell. Biol. 31, 1833–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Goodarzi A. A., Noon A. T., Jeggo P. A. (2009) Biochem. Soc. Trans. 37, 569–576 [DOI] [PubMed] [Google Scholar]
- 66. Groner A. C., Meylan S., Ciuffi A., Zangger N., Ambrosini G., Dénervaud N., Bucher P., Trono D. (2010) PLoS Genet. 6, e1000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Barde I., Laurenti E., Verp S., Groner A. C., Towne C., Padrun V., Aebischer P., Trumpp A., Trono D. (2009) J. Virol. 83, 5574–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Margolin J. F., Friedman J. R., Meyer W. K., Vissing H., Thiesen H. J., Rauscher F. J., 3rd (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4509–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Witzgall R., O'Leary E., Leaf A., Onaldi D., Bonventre J. V. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4514–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Takahashi K., Sugi Y., Hosono A., Kaminogawa S. (2009) J. Immunol. 183, 6522–6529 [DOI] [PubMed] [Google Scholar]
- 71. Kamitani S., Ohbayashi N., Ikeda O., Togi S., Muromoto R., Sekine Y., Ohta K., Ishiyama H., Matsuda T. (2008) Biochem. Biophys. Res. Commun. 370, 366–370 [DOI] [PubMed] [Google Scholar]
- 72. Blahnik K. R., Dou L., Echipare L., Iyengar S., O'Geen H., Sanchez E., Zhao Y., Marra M. A., Hirst M., Costello J. F., Korf I., Farnham P. J. (2011) PLoS ONE 6, e17121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Goodarzi A. A., Noon A. T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P. A. (2008) Mol. Cell 31, 167–177 [DOI] [PubMed] [Google Scholar]
- 74. Rowe H. M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P. V., Layard-Liesching H., Verp S., Marquis J., Spitz F., Constam D. B., Trono D. (2010) Nature 463, 237–240 [DOI] [PubMed] [Google Scholar]
- 75. Wolf D., Cammas F., Losson R., Goff S. P. (2008) J. Virol. 82, 4675–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chang P. C., Fitzgerald L. D., Van Geelen A., Izumiya Y., Ellison T. J., Wang D. H., Ann D. K., Luciw P. A., Kung H. J. (2009) Cancer Res. 69, 5681–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang B., O'Herrin S. M., Wu J., Reagan-Shaw S., Ma Y., Bhat K. M., Gravekamp C., Setaluri V., Peters N., Hoffmann F. M., Peng H., Ivanov A. V., Simpson A. J., Longley B. J. (2007) Cancer Res. 67, 9954–9962 [DOI] [PubMed] [Google Scholar]
- 78. White D. E., Negorev D., Peng H., Ivanov A. V., Maul G. G., Rauscher F. J., 3rd (2006) Cancer Res. 66, 11594–11599 [DOI] [PubMed] [Google Scholar]
- 79. Li X., Lee Y. K., Jeng J. C., Yen Y., Schultz D. C., Shih H. M., Ann D. K. (2007) J. Biol. Chem. 282, 36177–36189 [DOI] [PubMed] [Google Scholar]
- 80. Zheng L., Pan H., Li S., Flesken-Nikitin A., Chen P. L., Boyer T. G., Lee W. H. (2000) Mol. Cell 6, 757–768 [DOI] [PubMed] [Google Scholar]
- 81. Li X., Lin H. H., Chen H., Xu X., Shih H. M., Ann D. K. (2010) Sci. Signal. 3, ra32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D. C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. (2006) Nat. Cell Biol. 8, 870–876 [DOI] [PubMed] [Google Scholar]