Abstract
The sterol-sensing domain (SSD) is a conserved motif in membrane proteins responsible for sterol regulation. Mammalian proteins SREBP cleavage-activating protein (SCAP) and HMG-CoA reductase (HMGR) both possess SSDs required for feedback regulation of sterol-related genes and sterol synthetic rate. Although these two SSD proteins clearly sense sterols, the range of signals detected by this eukaryotic motif is not clear. The yeast HMG-CoA reductase isozyme Hmg2, like its mammalian counterpart, undergoes endoplasmic reticulum (ER)-associated degradation that is subject to feedback control by the sterol pathway. The primary degradation signal for yeast Hmg2 degradation is the 20-carbon isoprene geranylgeranyl pyrophosphate, rather than a sterol. Nevertheless, the Hmg2 protein possesses an SSD, leading us to test its role in feedback control of Hmg2 stability. We mutated highly conserved SSD residues of Hmg2 and evaluated regulated degradation. Our results indicated that the SSD was required for sterol pathway signals to stimulate Hmg2 ER-associated degradation and was employed for detection of both geranylgeranyl pyrophosphate and a secondary oxysterol signal. Our data further indicate that the SSD allows a signal-dependent structural change in Hmg2 that promotes entry into the ER degradation pathway. Thus, the eukaryotic SSD is capable of significant plasticity in signal recognition or response. We propose that the harnessing of cellular quality control pathways to bring about feedback regulation of normal proteins is a unifying theme for the action of all SSDs.
Keywords: Cholesterol Regulation, Endoplasmic Reticulum (ER), Isoprenoid, Protein Degradation, Ubiquitin, ERAD, HMG-CoA Reductase, HRD Pathway, SSD
Introduction
The sterol pathway is the source of diverse and essential lipid molecules in all eukaryotes. These products include the iterated isoprenes produced early in the pathway and sterols, such as cholesterol, made in the later steps (see Fig. 1A). Feedback regulation of the sterol pathway is highly conserved and multifaceted (1–3). The sensing of sterol pathway activity requires detection of lipid intermediates, including extremely insoluble molecules such as cholesterol itself. An understanding of the cellular mechanisms for detecting sterol pathway activity would be of great basic and biomedical utility.
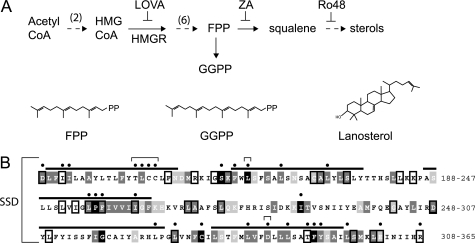
FIGURE 1.
A, abbreviated sterol synthesis pathway. Inhibitors that increase (ZA) or decrease (Lova) the FPP-derived degradation signal for Hmg2 are indicated. Lova inhibits HMGR, and ZA inhibits squalene synthase. Ro48 inhibits oxidosqualene epoxidase, inducing an alternative oxysterol-producing pathway (23) Structures of the 15-carbon FPP and 20-carbon GGPP isoprenes and lanosterol are depicted. B, the Hmg2 SSD conservation and mutants. Membrane spans are over-lined. The extent of conservation for each residue in five different SSDs (alignments shown in supplemental Fig. S1) is indicated by color: black for identical residues, dark gray for conservation, and light gray for semi-conservation. Boxes represent identity in four of five different SSD sequences examined. Alignments and conservation were determined by the T-Coffee multiple sequence alignment program. Hmg2 (YLR450W ORF) protein sequence is from the Saccharomyces Genome Database. Thirty mutants were made in this study. Black dots mark mutated residues; brackets mark residues in Hmg2 that correspond to sterol-insensitive mutants in HMGR and SCAP, e.g., the YIYF to AAAA mutant, Tyr-298, Leu-315, and Asp-428.
In the regulation of mammalian sterol pathway activity, a protein transmembrane region known as the sterol-sensing domain (SSD)3 is required for sterols to alter both the transcription of sterol-relevant genes and the stability of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase (HMGR), one of the rate-determining enzymes of the sterol pathway (6). The SSD is found in a variety of sterol-related proteins (7). Conserved residues are primarily found in the bilayer and juxtamembrane positions dispersed over five transmembrane regions. Although the structure of an SSD has yet to be determined, numerous studies indicate that the SSD mediates responses to specific sterol molecules, thus providing a means of communication between sterols and proteins that control aspects of sterol biology (8–10).
In mammals, the SSD is found in several proteins (7). Two are directly involved in control of the sterol synthesis pathway and have been studied extensively (6). The first is called SCAP (for SREBP cleavage-activating protein). SCAP is a multispanning membrane protein that allows cholesterol regulation of the SREBP transcription factor, which activates the transcription of sterol pathway enzymes and the LDL receptor. When sterols are low, SREBP activity is elevated to increase sterol production and uptake. SREBP activation occurs when the full-length, membrane-bound form undergoes SCAP-dependent vesicular trafficking from the ER to the Golgi where concomitant cleavage and activation occurs. The SCAP-dependent movement of SREBP to the Golgi is regulated by cholesterol. When cellular cholesterol is high, SCAP binds cholesterol, which in turn allows binding to the ER-localized protein INSIG, causing ER retention of SCAP and its associated, unprocessed SREBP (11). Sterol-dependent retention of SCAP prohibits SREBP trafficking and activation so that the SCAP SSD mediates cholesterol-dependent inhibition of SREBP activity (12). Although the SSD has been proposed to mediate sterol recognition, it has recently been shown that a lumenal loop directly preceding the SSD directly binds cholesterol with a nm affinity constant (10, 38). Thus, the role of the SSD in detection of and response to pathway signals is an important, and still open, question.
The second SSD protein directly involved in cholesterol synthesis is HMG-CoA reductase (HMGR), a rate-limiting enzyme of sterol synthesis (see Fig. 1A). HMGR levels are directly regulated by feedback regulation of its stability (1). When sterol synthesis is high, HMGR undergoes rapid ubiquitin-dependent degradation; when sterol synthesis is low, HMGR degradation slows, allowing levels of HMGR to increase. Regulated degradation of mammalian HMGR is mediated by the SSD located in the N-terminal, multispanning membrane domain (12, 13). The SSD allows sterol-dependent binding of INSIG, which promotes proximity of the ER ubiquitin ligase gp78 to HMGR and the ubiquitination of the enzyme (14). Importantly, the response mediated by the HMGR SSD is specific for a derivative of the early pathway intermediate lanosterol, thus ensuring that HMGR degradation is keyed to synthesis of sterols rather than levels of sterols (15, 16). In both cases, the binding of INSIG is a key aspect of SSD function. The work below indicates that the SSD has a direct role in response to sterol pathway signals, independent of INSIG action.
Regulated ubiquitination of HMGR was initially discovered in yeast; the Hmg2 isozyme of HMGR undergoes feedback-regulated ubiquitination that in turn leads to proteasomal degradation when sterol pathway activity is high (2, 17, 18). Hmg2 ubiquitination is executed by the Hmg CoA reductase degradation (HRD) pathway, using the ER-localized Hrd1 ubiquitin ligase, of which gp78 is a mammalian homologue (19, 20). Degradation of Hmg2 is keyed to the levels of farnesyl pyrophosphate (FPP), an early 15-carbon isoprene of the sterol pathway (see Fig. 1A); when levels of FPP are high, Hmg2 degradation rate is high, and when FPP levels are low, Hmg2 degradation rate is low (21). Our more recent work has shown that geranylgeranyl pyrophosphate (GGPP), which is structurally analogous to FPP but with 20 carbons, is the likely bona fide signal (see Fig. 1) (22). In addition, we have shown that an oxysterol signal appears to modulate the degradation of Hmg2 as well; increased levels of certain oxysterols enhance the degradation rate controlled by endogenous levels of the FPP-derived signal (23). The overriding theme is that when sterol pathway signals are high, Hmg2 is more prone to enter the HRD degradation pathway and undergo degradation.
The HRD pathway participates in ER-associated degradation (ERAD) and is required for protein quality control, promoting the degradation of a large variety of misfolded or misassembled ER-localized proteins. However, only Hmg2 undergoes sterol pathway-dependent changes in HRD-dependent degradation. We posited that regulation of HRD-dependent degradation was brought about by the FPP-derived signal causing Hmg2 to adopt features of a misfolded quality control substrate, presumably by direct interaction (24). This model is bolstered by in vivo and in vitro limited proteolysis studies, indicating that Hmg2 undergoes the expected structural alterations when exposed to chemical chaperones or isoprenes related to FPP (25). Thus, it appears that the physiological, FPP-derived degradation signal can promote regulated Hmg2 misfolding and can cause regulated destruction by the HRD quality control pathway. We wanted to explore the features of Hmg2 that underlie sensing of sterol pathway molecules required for pathway-regulated degradation.
The SSD has been implicated in the response to sterols in a variety of studies. However, other SSD proteins may be regulated by lipids distinct from canonical sterols (26). Accordingly, we wondered to what extent the highly conserved SSD is involved in regulated degradation of Hmg2 caused by the non-sterol FPP-derived signal. We generated a collection of mutants in which highly conserved residues (usually one in each mutant) were converted to alanine, or in some cases to hydrophobic residues, and evaluated the regulated degradation of each mutant. Because the Hmg2 molecule is “structurally poised” to enter the HRD quality control pathway (24, 25), we paid close attention to the possibility that mutations of the highly conserved SSD residues might simply damage the Hmg2 structure and destabilize it in a manner not directly relevant to regulation. Surprisingly, we found only stabilizing mutations that decreased HRD-dependent Hmg2 degradation. In some cases, single SSD point mutations rendered Hmg2 completely stable. Furthermore, a subset of the SSD mutants specifically altered the response of Hmg2 degradation rate to oxysterols, whereas preserving the response to the main FPP-derived signal. Thus, our data indicate that the SSD allows response to diverse lipid signals, and perhaps, multiple ones in the same protein. Furthermore, the results allow the intriguing idea that the SSD imparts programmed misfolding as an ancient tactic of protein regulation that occurred before the divergence of mammals and yeast. Importantly, the actions of both the FPP-derived signal and the oxysterol that augments Hmg2 response to FPP operate independently of the yeast INSIG proteins. Thus, the studies below indicate a direct role for the SSD in response to sterol pathway signals that control Hmg2 ERAD.
EXPERIMENTAL PROCEDURES
Reagents
Cycloheximide (CHX) stock was stored at 50 mg/ml in DMSO at −20 °C and was used at 50 μg/ml (Sigma-Aldrich). Lovastatin (Lova) stock was stored at 25 mg/ml in methanol ammonia at 4 °C and used at 25 μg/ml. Zaragozic acid (ZA) stock was stored at 10 mg/ml in DMSO at −20 °C and used at 10 μg/ml. Lova and ZA were donated by Merck. Ro48-8071 (Ro48) stock was stored at 40 mg/ml in DMSO and used at 20 μg/ml. Ro48 was a gift from Johannes Aebi at F. Hoffmann-La Roche Ltd.). Mouse monoclonal anti-GFP antibody is from Clontech. Hybridoma supernatant (9E10; obtained from the ATTC) was the source of mouse monoclonal anti-Myc antibody. The monoclonal anti-ubiquitin antibody was a gift from Richard Gardner at the University of Washington.
Yeast Strains and Plasmids
Yeast strains are derived from S288C. All strains (supplemental Table 1) and plasmids (supplemental Table 2) were constructed with standard molecular biology techniques as described previously (27, 28). All SSD mutants were constructed in pRH1581 (TDH3-1mycHMG2-GFP::URA3). Hmg2 SSD mutants were made by the splicing by overlap elongation PCR technique adapted from Horton et al. (29). Primers for mutagenesis are listed in supplemental Table 3. Briefly, Primers A and B and Primers C and D were used in separate PCR reactions to create separate, overlapping fragments containing the mutation. In a second round of PCR, the two fragments were combined with Primers A (oRH2192) and D (oRH2202) to create a single long fragment. This fragment was cut with MscI/SpeI and inserted in pRH1581 cut with the same enzymes. Wild-type and mutant plasmids were cut with StuI to integrate at ura3-52 in RHY519. Strains were grown at 30 °C to mid-log phase (∼0.2 optical density) in synthetic complete medium minus uracil, and then degradation and biochemical assays were performed as described below.
Flow Cytometry
The GFP fluorescence of cells was measured with a BD Biosciences FACSCalibur flow cytometer as described in Ref. 22. Following treatment with drugs or control treatment, the fluorescence of 10,000 cells in each sample was measured. CHX (50 μg/ml), ZA (10 μg/ml), and Lova (25 μg/ml) were added separately to cultures and incubated at 30 °C for 2 h. Ro48-8071 (20 μg/ml) was incubated at 30 °C for 4 h. Mutants were tested at least three times in each assay.
In Vivo Ubiquitination
Ubiquitination of Hmg2-GFP variants was assayed as in Ref. 22. Log phase cultures were incubated with ZA (10 μg/ml) or untreated for 10 min, cells were lysed, and Hmg2-GFP was immunoprecipitated with rabbit polyclonal anti-GFP antibody. Following SDS-PAGE and transfer to nitrocellulose, immunoblotting was performed with a monoclonal anti-ubiquitin antibody to detect ubiquitin and a monoclonal anti-GFP antibody to detect Hmg2-GFP proteins.
In Vitro Trypsinolysis
Protease protection assays of Hmg2-GFP variants were performed as detailed in Ref. 22 and 25. Microsomes (untreated or treated with 200 mm farnesol) with Hmg2-GFP were incubated with 150 μg/ml trypsin for 0, 2, and 10 min, and then reactions were stopped with 2× urea sample buffer. Following SDS-PAGE, the Myc epitope was detected by immunoblotting with 9E10 monoclonal anti-Myc antibody.
RESULTS
The SSD of Hmg2 has a number of highly conserved residues that appear in many or all SSDs (Figs. 1B and 7, supplemental Fig. S1). To better understand the Hmg2 SSD, we mutated highly conserved residues (usually to alanine) along the span of the domain, producing over 30 mutants in this manner (Figs. 1 and 7, supplemental Fig. S1). We included mutations reported to function in mammalian SSD proteins, including the YIYF to AAAA mutant that rendered mammalian HMGR insensitive to regulated degradation (14) and three mutants, Y298C, L315F, and D428A, that make mammalian SCAP insensitive to sterols (11, 12, 30). Each Hmg2 SSD mutant was then tested for regulated degradation.
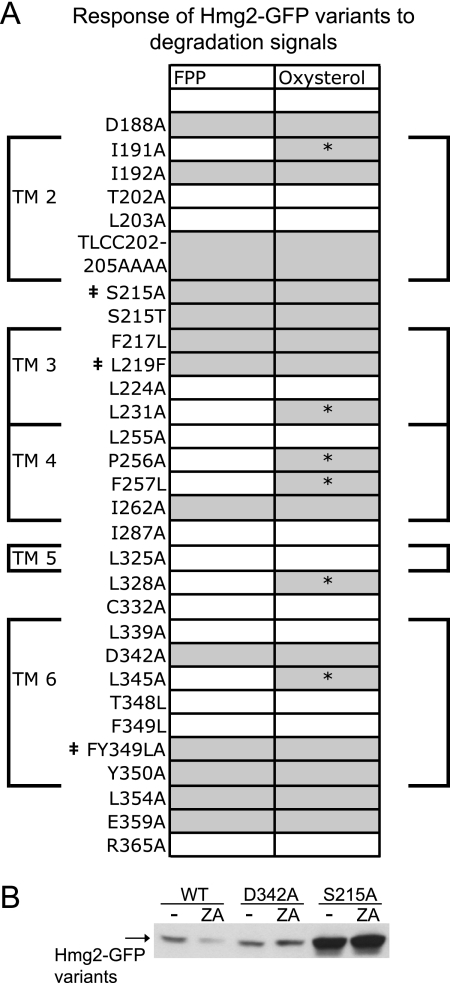
FIGURE 7.
Response of Hmg2 variants to degradation signals. A, results of flow cytometric analyses. Thirty SSD mutants were tested for protein stability and regulated degradation as described in the legend for Fig. 2. Phenotypes are indicated by shading; white signifies wild-type stability and regulated degradation, and gray signifies increased stability and reduced response to degradation signals. A dagger indicates the three stable, non-responsive mutants. Asterisks indicate mutants that show an uncoupled phenotype: normal response to the FPP-derived signal and no response to the oxysterol signal. The membrane spans of the Hmg2 SSD are indicated by large brackets around the table and numbered TM2–TM6. TLCC202–205AAAA, T202A/L203A/C204A/C205A. B, GFP immunoblot following CHX chase assay of WT, partially stable mutant D342A, and stable mutant S215A Hmg2-GFP variants.
In earlier studies of Hmg2 (28), we found that scrambling a five-amino acid region, TFYSA (amino acids 348–352), brought about constitutive degradation that was unresponsive to the FPP-derived signal (25). Several of the residues altered in this NR1 (for non-responsive) mutant are highly conserved between SSDs (Fig. 1, supplemental Fig. S1), suggesting a role for the SSD in regulation. However, the behavior of NR1 brings up an important caveat that informed the studies below. Hmg2 enters the HRD ERAD pathway when FPP-derived signals are elevated, causing structural changes that trigger quality control degradation (2) However, numerous mutants of Hmg2 show constitutive, unregulated entry into the HRD pathway by virtue of their aberrant structure, ranging from subtle to gross alterations (28). NR1 appears to adopt a normal structure by limited proteolysis (25); however, it could also be that NR1 mutant has structural aberrations leading to unregulated degradation. In our analysis of the SSD mutants, this possibility was an important one to consider in understanding the effects of the SSD alterations.
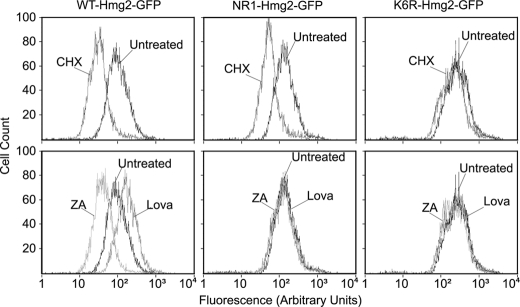
Altering the sequence of Hmg2 can result in three possible extremes of regulatory behavior. A mutant could behave as wild type, responding to changes in FPP levels such that higher FPP levels cause faster degradation. Alternatively, an Hmg2 mutant could undergo constitutive degradation with no response to alterations in FPP, as is the case for NR1. Finally, a mutant Hmg2 could be completely stable and show no response to FPP. Our studies of Hmg2 have provided examples of all three classes of mutants, as shown in Fig. 2.
FIGURE 2.
Distinct phenotypes among previously characterized Hmg2 mutants. Strains expressing WT-Hmg2-GFP, NR1-Hmg2-GFP, and K6R-Hmg2-GFP were assayed for protein stability (top row) or regulated degradation (bottom row) by flow cytometry following the appropriate treatments. To assess stability, SC-Ura cultures were grown to log phase and then incubated for 2 h at 30 °C with no drug (Untreated) or 50 μg/ml CHX. To assess regulated degradation (bottom row), similar cultures were incubated with no drug, 25 μg/ml Lova, or 10 μg/ml ZA for 2 h at 30 °C.
To evaluate Hmg2-regulated degradation, we used a flow cytometry method (22, 25) that allows quantitative, reproducible in vivo analyses (31). We analyzed each SSD variant in the context of the GFP fusion Hmg2-GFP, which has the catalytic C terminus replaced with GFP (2). In addition, a Myc tag was included in the first lumenal domain prior to the SSD, in a position that does not affect regulation (25) (see Fig. 9). Each Hmg2-GFP mutant was expressed in strains with identical, high flux through the mevalonate pathway provided by a separately expressed Hmg2 catalytic domain. To examine degradation, we treated a strain with CHX and observed the time-dependent shift to lower cellular fluorescence caused by degradation. To examine regulation, the cells were treated with either HMGR inhibitor Lova to block production of FPP or ZA to cause a buildup of FPP by inhibition of squalene synthase, the enzyme that uses FPP as a substrate (Fig. 1A) (28). Examples of the three extreme regulatory phenotypes (wild type, always degraded, and always stable) described above are demonstrated by flow cytometry in Fig. 2. The effects of CHX on cellular GFP fluorescence are shown in Fig. 2, top row. The effects of lowering FPP with Lova or raising FPP with ZA are shown in the bottom row, in which the steady-state fluorescence of treated and untreated cultures are overlaid. In the wild-type strain, CHX caused the expected time-dependent shift in population fluorescence due to degradation (top, left). Altering the sterol pathway changes cellular fluorescence in the expected ways; lowering FPP with Lova stabilized Hmg2-GFP, increasing its steady state, and increasing FPP with ZA caused increased Hmg2-GFP degradation, lowering steady-state fluorescence (bottom, left) (21, 22). NR1 showed degradation (top, middle) but was unregulated, with no effect of either sterol pathway drug on steady-state levels (bottom, middle). The highly stable K6R point mutant of Hmg2-GFP shows high steady-state fluorescence and no degradation after CHX treatment (top, right). Again, the drugs that alter the sterol pathway did not alter the steady-state level (bottom, right), but in this case, this is because the K6R mutant is always stable at all levels of FPP. We used these assays to evaluate the role of the SSD in Hmg2-GFP regulated degradation.
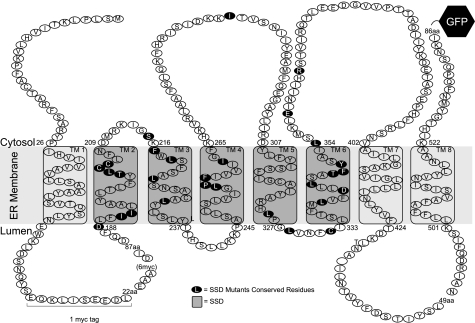
FIGURE 9.
Hmg2 Topology. The SSD is highlighted in dark gray. Residues mutated in this study are highlighted in black. The catalytic domain is substituted with a GFP tag in 1mycHMG2-GFP (pRH1581 and derivatives), and there is a single Myc tag (EQKLISEEDL) between transmembrane domains (TM) 1 and 2 in the first lumenal loop. For past topology studies, we used the unregulated 6myc-Hmg2 variant. In this variant, 16 amino acids (aa) are replaced with six copies of the Myc epitope. Protease protection assays indicated that this portion is in the lumen of the ER.
The SSD mutant phenotypes fell into three classes: wild type, partially stable, and completely stable. The phenotypes of each variant within a group were sufficiently similar that we show data for a few examples in each group and list all in the relevant figure. Members of the first group were comparable to wild type in their degradation rate and response to altering FPP with Lova or ZA (Fig. 3, data not shown).
FIGURE 3.
SSD mutants that behave like wild type. The stability (CHX) and regulated degradation (Lova) of WT and Hmg2-GFP variants were assayed as in Fig. 2. Three examples are shown; all mutants in the class are listed in the table.
The second group showed partial stability indicated by slightly slower degradation with CHX and a blunted response to Lova (Fig. 4). For example, the D342A mutant showed a slightly higher steady-state level in untreated cells and a similar shift caused by CHX (top left panel). This would be consistent with the mutant being less responsive to the ambient levels of FPP in the cells. Lowering the levels of FPP with Lova had little effect, whereas increasing FPP with ZA stimulated degradation that was not as great as that in wild type, also consistent with a blunted response to the FPP-derived degradation signal (top right panel).
FIGURE 4.
SSD mutants that have reduced responses to FPP. A and B, the stability (CHX) (A) and regulated degradation (Lova and ZA) (B) were assayed as in Fig. 2. Two examples are shown; all mutants in the class are listed in the table. TLCC202–205AAAA, T202A/L203A/C204A/C205A.
Members of the third group were extremely stable (Fig. 5), phenocopying the K6R mutant above (Fig. 2). The addition of CHX essentially caused almost no leftward shift. The effects of Lova were either absent due to the protein being completely stable or very small due to slight increases in the already stable construct, such as seen with F349L/Y350A (Fig. 5, bottom right panel). Increasing the degradation signal with ZA did not cause degradation of these versions of Hmg2 (Fig. 8A, data not shown). The affected residues in the FY349LA (F349L/Y350A) mutant are part of the TFYSA cluster in the unregulated, constitutively degraded NR1 mutant (28). In contrast, conversion of Hmg2 to the less extreme F349L/Y350A mutant resulted in high stability and no response to FPP signal, rather than slow degradation with no response to FPP as in the more extreme NR1 mutant.
FIGURE 5.
SSD mutants that are stable and not regulated. The stability (CHX) and regulated degradation (Lova) of WT and Hmg2-GFP variants were assayed as in Fig. 2. The three mutants with this phenotype are shown. FY349LA, F349L/Y350A.
FIGURE 8.
Analysis of a stable mutant, S215A. A, degradation of Ser-215 variants. B, S215A can enter the HRD pathway when Hrd1 is constitutively overexpressed (OE Hrd1) from the strong TDH3 promoter. EV, empty vector control plasmid. C, S215A cannot be regulated when Hrd1 is overexpressed. The stability (A, CHX) and regulated degradation (C, Lova) of each Hmg2-GFP variant were assayed as in Fig. 2 in strains expressing Hrd1 from the strong TDH3 promoter or in strains carrying an empty vector control plasmid. D, Ser-215 variants show reduced ubiquitination. Log-phase cultures were treated with ZA for 10 min (or untreated). Hmg2 proteins were isolated by anti-GFP immunoprecipitation and separated by SDS-PAGE, and then immunoblotting for ubiquitin and GFP was done. E, S215A structure does not change when degradation signal is present. Dynamics of Hmg2 structure were analyzed by an in vitro trypsinolysis assay (22, 25) whereby microsomes (treated with farnesol (FOH) or untreated) were treated with trypsin, and samples were taken at different times. Proteins were resolved by SDS-PAGE, and then immunoblotting was performed for the lumenal Myc epitope present in the Hmg2-GFP variants.
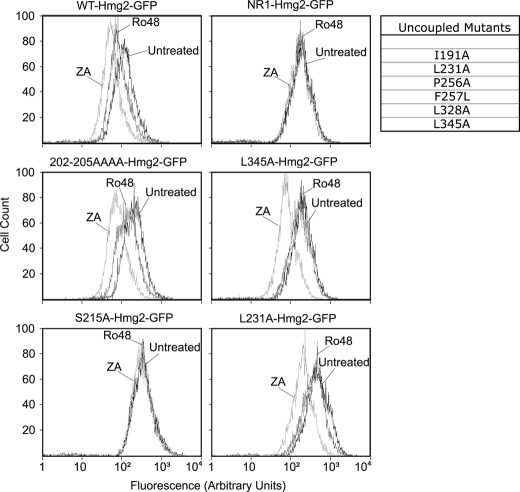
The FPP-derived degradation signal is the principle sterol pathway molecule promoting Hmg2 degradation. However, we have shown that oxysterols can also influence Hmg2 stability. These species are produced in vivo by partial inhibition of the enzyme oxidosqualene epoxidase by selective inhibitors of the enzyme such as Ro48, as described in detail in previous studies (23). Partial inhibition of oxidosqualene epoxidase and the subsequent buildup of oxysterols show reproducible increases in Hmg2 degradation in vivo. This effect is not independent of the FPP signal, leading us to conclude that an oxysterol causes Hmg2 to become more responsive to the FPP-derived signal. Thus, at a given ambient level of FPP, increasing oxysterol levels changes the degradation rate due to this heightened response (23). Oxysterol response can be assayed by treating cells expressing Hmg2-GFP with Ro48 followed by flow cytometry to evaluate lowering of cellular fluorescence (23) (Fig. 6, WT). We wondered whether this oxysterol effect on Hmg2 was also mediated by the SSD. We tested our collection of SSD point mutants with Ro48. Not surprisingly, because the oxysterol effect enhances response to FPP, those mutants that showed altered responses to the FPP signal were similarly affected in their response to Ro48 (Fig. 6, left panels); partially unresponsive mutants were less responsive to increasing oxysterols (e.g. TLCC202–205CCCC(T202A/L203A/C204A/C205A): left, middle), and the fully stable ones were still fully stable (e.g. S215A: left, bottom). Additionally, some of the mutants that showed wild-type responses to FPP alteration were also comparable with wild type in response to oxysterols (Fig. 7 and data not shown). However, a new class of mutants was also detected in these tests. Among the mutants that were comparable with wild-type for FPP responsiveness, there were several that were completely unresponsive to elevating oxysterols. Thus, in these mutants, the FPP response was “uncoupled” from the oxysterol response. Examples of two of these uncoupled mutants are shown in Fig. 6 (right panels, L231A and L345A); there was no shift caused by altering oxysterols with Ro48, whereas the response to ZA was actually accentuated when compared with wild-type. Thus, it would appear that the SSD has information that is specifically required to react to the secondary oxysterol signal, as well as information needed to either respond to or recognize the primary FPP-derived degradation signal.
FIGURE 6.
Response to oxysterol signal. The stability (CHX) and regulated degradation (ZA) were assayed as in Fig. 2. The response to oxysterol signal was measured by flow cytometry following incubation of cultures with no drug (Untreated), 20 μg/ml Ro48, and 10 μg/ml ZA for 4 h at 30 °C. One partially responsive mutant and one non-responsive mutant (S215A) are shown in the left column. Mutants that responded normally to FPP but not to oxysterol signal are termed Uncoupled Mutants and are listed in the table. Data for two examples are shown in the right column.
The results of the above mutant studies (Figs. 3–6) are compiled in Fig. 7 to show the distribution of stabilizing mutants along the linear sequence of the SSD. Each shaded box represents a stabilizing mutant (FPP on left, oxysterol on right), and each shaded box with an asterisk represents one of the mutants that is selectively deficient in oxysterol response. Although it is the case that all mutants with non-wild-type phenotypes are more stable, it is important to note that flow cytometry allowed the resolution of the different degrees of unresponsiveness noted for the various classes. Immunoblotting alone was sometimes not sufficient to resolve these highly reproducible but partial changes in degradation behavior. This is demonstrated in the immunoblots in Fig. 7B showing data for cycloheximide chase assays with wild type, partially stabilized mutant D342A, and fully stable mutant S215A. Although the two mutants are both shown to be more stable than wild type by immunoblotting, the difference in degree of stabilization is only clearly and reliably indicated in the flow cytometric data. As we have shown in our prior work (22, 28), use of flow cytometry adds a level of precise quantitative analysis that is more difficult to attain by biochemical means.
Perhaps the most striking aspect of these studies is that every non-wild-type SSD mutant showed a reduced tendency for degradation. None showed unregulated constitutive degradation. This could imply that the SSD either is an integral part of recognition of a degradation signal or is required for entry into the HRD ERAD pathway. To explore some more mechanistic aspects of the role of SSD in Hmg2-regulated degradation, we focused on the highly stable S215A mutant. The serine residue at position 215 in Hmg2 is a highly conserved residue (Fig. 1B, supplemental Fig. S1) Changing Ser to Ala renders Hmg2 extremely stable, essentially phenocopying Hmg2-GFP in a hrd1Δ null strain (Figs. 5 and 8A, and data not shown). The more conservative change of Ser-215 to Thr produces an Hmg2-GFP that is more stable than wild type and less responsive to FPP, as shown in Fig. 4 above and in Fig. 8A. This indicates that the Ser-215 position is very important in regulated degradation because even a conservative alteration has strong effects on the control of Hmg2 stability.
Although we favored a signal response interpretation, it was also possible that the S215A mutation simply rendered Hmg2 unable to enter the HRD pathway under any circumstances. One reason may be that it is simply mistargeted in the cell, away from the HRD pathway; however, S215A (and the other stable mutants) are correctly localized in the ER by fluorescence microscopy (supplemental Fig. S2). The K6R mutant is completely stable and does not undergo degradation even when the Hrd1 E3 ligase, which is rate-limiting for the HRD pathway (20, 32), is overexpressed from the strong TDH3 (GAPDH) promoter (Fig. 8B).4 We have posited that Lys-6 is a primary ubiquitination site (28), which would be consistent with its lack of Hrd1-dependent degradation. We analyzed the behavior of S215A to discern whether it is unable to recognize degradation-enhancing signals or is incapable of entering the HRD pathway.
The S215A point mutant was extremely stable and completely refractory to elevation of FPP by ZA (Fig. 8A). The insensitivity of S215A to elevated signal is as strong as the two “prototype” unregulated mutants: NR1, which shows constitutive degradation unaffected by changes in FPP, and K6R, which is entirely stable. The NR1 mutant still undergoes HRD-dependent degradation but is not affected by alterations in pathway signals. Conversely, the K6R mutant is unresponsive to FPP elevation because it cannot enter the HRD pathway. We tested whether the S215A construct could undergo HRD pathway degradation upon elevating Hrd1 levels in vivo with the strong TDH3 promoter. The K6R mutant remained unresponsive to greatly increased Hrd1 (Fig. 8B, OE Hrd1). Conversely, S215A Hmg2-GFP was degraded upon overexpression of Hrd1, as shown by the leftward shift of the fluorescence histogram, indicating a lowering of steady-state levels (Fig. 8B, OE Hrd1 as compared with EV). Thus, the S215A mutant was capable of undergoing HRD-dependent degradation when Hrd1 was substantially elevated. This degradation was still unresponsive to altering FPP levels with Lova, as shown in the Fig. 8C histograms. Next, we directly tested the ubiquitination status of the S215A mutant in the presence and absence of ZA and compared it with wild-type Hmg2-GFP and the partially stable S215T. As expected, the S215T mutant showed some ubiquitination and a partial response to elevation of FPP, whereas the S215A variant was essentially free of modification in both conditions (Fig. 8D).
Our model was that the S215A mutant was unresponsive to the FPP-derived signal and thus held in a structural state that limited entry into the HRD pathway. We directly tested the S215A mutant for in vitro structural response to farnesol, which we have previously shown to be a reliable indicator of in vivo structural response to the isoprenoid degradation signal (22, 25). Fig. 8E shows the comparison of microsomes expressing wild-type Hmg2-GFP and S215A-Hmg2-GFP, subjected to time-dependent trypsinolysis in the presence and absence of farnesol. Although the wild-type protein showed the expected increased rate of digestion (left) (25), the S215A mutant was totally unresponsive to added farnesol (compare Untreated with FOH, right), consistent with the insensitivity of this point mutant to the in vivo degradation signal.
DISCUSSION
The broadly conserved SSD has been proposed to sense sterol signals (Ref. 7 and references therein and Refs. 12, 14, and 10). However, recent work has also allowed the possibility that the SSD is an effector domain that transduces the sterol signal bound in a different part of the transmembrane domain to engage INSIG proteins (38). Our data indicate that the SSD is directly involved in response to the pathway signals that control Hmg2 stability, including the primary FPP-derived 20-carbon GGPP (21, 22 and the secondary oxysterol signal (23). The studies herein clearly showed that the SSD had a critical and strong role in the response to each of these signals. Because the readout of Hmg2 response to its sterol pathway regulators is the entry into the HRD quality control pathway, care has to be taken in the interpretation of mutant effects on Hmg2-regulated degradation. For example, we know that simply rendering the Hmg2 molecule damaged, either by mutation (28) or through alteration of trans factors such as Spf1 (33), can cause unregulated HRD pathway degradation due to this damage.
Surprisingly, the only phenotypes noted in the tested SSD mutants were varying degrees of increased stabilization. Partially stable mutants were still responsive to the FPP-derived signal, and a few, such as S215A, were completely stable. The highly stable S215A mutant was still able to undergo degradation at elevated levels of Hrd1, meaning that the loss of degradation was due to a lower rate of pathway entry. We believe that the fully stable variants had lost the ability to respond to the FPP-derived signal and thus behaved as though the signal was absent. Our in vitro data indicated that the S215A mutant had lost the ability to undergo the signal-mediated change in structure that renders the protein more susceptible to the HRD pathway. (Fig. 8E) (25). The highly stable F349L/Y350A double mutant was located in the TFYSA sequence altered in NR1. Unlike NR1, the double mutant was highly stable and unresponsive to elevated FPP. Our ability in this case to separate the response to the FPP-derived signal from constitutive degradation in the more subtle mutants lends strong credence to the idea that the Hmg2 SSD is involved in recognition or structural response to degradation signals.
Although our mutagenesis study was not fully saturated, we believe that the uniformity of loss of signaling indicates that this is the primary function of the SSD. Similarly, a recent exhaustive study of the SSD of Schizosaccharomyces pombe SCAP (Scp1) showed a similar loss of sterol sensing without loss of trafficking function (34).
Alteration of the Hmg2 SSD also affected the secondary oxysterol signal produced by the “alternate pathway” with oxidosqualene epoxidase inhibitors (23). Surprisingly, a distinct subset of mutants with normal responses to altered FPP specifically lost the response to oxysterols. Again, the loss of SSD information led to the lessened ability to promote degradation, but in this case, it was the oxysterol-caused enhancement of response to the primary FPP-derived signal.
One model for SSD function is that it creates a binding or recognition site for regulatory sterols or other lipids (8–10). In the same vein, we believe the Hmg2 SSD is directly involved in response to the FPP-derived signal. In SCAP and mammalian HMGR, the SSD is required for engagement of INSIG and the subsequent INSIG-mediated regulatory responses. Although the yeast Nsg homologues of INSIG proteins do inhibit the action of the FPP-derived signal, the response of Hmg2 to this signal does not require the INSIGs (35).5 Also, the level of Hmg2-GFP in these studies puts it in excess of the endogenous INSIG proteins (35). Thus, the SSD of Hmg2 is directly mediating the response to two classes of degradation signals, the primary FPP-derived GGPP signal and a secondary oxysterol signal that alters the sensitivity to GGPP. Similarly, S. pombe Scp1 functions in sterol sensing independently of INSIGs (34). It is interesting to note that mammalian HMGR stability appears to be responsive to both a lanosterol-derived sterol and a 20-carbon isoprene derived from geranylgeraniol (14). Although the role of the sterol in mammalian HMGR degradation is well understood, that of the 20-carbon isoprene is not. The requirement of the Hmg2 SSD for recognition of both a sterol and 20-carbon isoprene indicates that this may similarly be the case in mammalian HMGR, but with the constraint that the relative importance of each lipid as a degradation signal is inverted between these two species, with the isoprene signal (GGPP) being the primary determinant in yeast.
The SSD of Hmg2 functions in a manner consistent with our knowledge of other SSD proteins (3, 36). Thus, it is reasonable to imagine that the topology of the SSDs has been preserved in the various proteins that use it in a similar manner. We can use this idea as a previously unrealized constraint on the prediction of Hmg2 topology. Although it is clear that the N terminus is facing the cytosol (28) and that the last hydrophilic loop between transmembrane domains 7 and 8 is also in the lumen, the remainder of the topology is not so clearly defined (although see Ref. 37). If we impose the known topology of the SSD onto our predictions of Hmg2, a modified model emerges (Fig. 9). In this newer SSD-inclusive model, a portion of what was once predicted to be the second cytoplasmic loop is now in the ER lumen. The new model better explains some published observations of the 6myc-Hmg2 variant of Hmg2 (6myc tags replace sequence between two restriction sites (HincII/SpeI)) (19). 6myc-Hmg2 is totally unregulated, showing constitutive HRD-dependent regulation that allowed the first genetic selections for HRD-deficient mutants (19). The older model of 6myc removed a transmembrane domain, whereas the new model places the 6myc tag in a lumenal loop, thus leaving the topology of Hmg2 intact. This new model explains two previously perplexing features of 6myc-Hmg2. First, limited proteolysis of 6myc-Hmg2 in yeast microsomes results in protection of the Myc epitope, consistent with a lumenal location (24). Second, 6myc-Hmg2 is not recognized by the 3A-Hrd1 mutant of the Hrd1 E3 ligase (4). The 3A-Hrd1 mutant is highly selective, only stabilizing Hmg2 (including Hmg2 variants) but not other membrane-bound Hrd1 substrates (4), meaning that 6myc-Hmg2 must have structural features that are very similar to native Hmg2. Taken together, these observations strengthen the model that the Hmg2 molecule has the SSD in the conserved orientation, as would be predicted from its role in signal recognition. Clearly, a structure of the Hmg2 transmembrane domain will be a valuable asset for understanding the regulation of ERAD and the recognition of SSD ligands.
This analysis of the Hmg2 SSD includes both highly conserved residues, such as the broadly conserved serine in S215A, and mutants that were directly discovered to be important in previous studies of sterol-regulating proteins. The YIYF to AAAA mutant was shown to abrogate signal sensing in mammalian HMGR (14), and in our hands, it had a partial insensitivity to the FPP-derived signal (T202A/L203A/C204A/C205A). The Hmg2 L219F mutation is analogous to the L315F mutation in SCAP that renders this protein totally refractory to sensing cholesterol (14). Finally, D342A is analogous to the D428A mutation in SCAP which cannot assume the cholesterol-induced conformational change exhibited by wild-type SCAP (30). These similarities, and the more general observation that some of the most highly conserved residues are responsible for normal Hmg2 regulation, indicate that this domain serves a universal role as a detector or effector of lipid signals.
These studies indicate that the Hmg2 SSD has a broader capacity for mediating response to lipid signals than solely different sterols. GGPP, which we have posited to be the FPP-derived signal, is in the family of isoprenoids. It has been proposed that phosphatidylethanolamine, a phospholipid, may be the lipid signal for regulation of Drosophila SCAP (26). It is possible that the SSD has wide flexibility in binding diverse lipid ligands, as is the case with the G-protein-coupled receptor superfamily (5). Alternatively, it may be that the SSD is a unified effector region that is altered by binding of diverse lipid ligands in distinct parts of each SSD protein. Our model for the action of the Hmg2 SSD is that it permits GGPP-mediated structural changes that render the protein more susceptible to the HRD pathway. We believe that these structural changes amount to partial misfolding from our in vivo and in vitro studies of the effects of test lipids on the structure of the Hmg2 transmembrane domain. Thus, one view of the SSD is that it allows lipid-mediated misfolding that renders the protein more sensitive to the HRD pathway. One readout of this misfolding is the change in trypsin sensitivity caused by farnesol, geranylgeraniol (25), or GGPP in our in vitro limited proteolysis assay (22). Whether the GGPP binds directly to the SSD or to a distinct part of the Hmg2 molecule that allows presentation of SSD information is not yet clear. It may be that the SSDs present in various proteins all act by controlling acquisition of structural traits of QC clients to allow the cell to capitalize on diverse, highly specific QC functions, such as ER retention of SCAP or recruitment of quality control ligases for HMGR or Hmg2. We have proposed that the INSIGs can be viewed as SSD client-specific chaperones, consistent with the idea that the SSD is involved in regulated misfolding. It is most likely the case that both protein QC and the isoprenoid pathway are processes that have evolved very early in life. Perhaps the high specificity of protein QC was at one time the best or even only available tactic for selectively controlling the proteins that produce and transport sterol pathway molecules, and so the paradigm of SSD-caused QC structures, set in the “stone” of the vicissitudes of evolution, remains carved on the edifice of cellular regulation to this day.
Supplementary Material
Acknowledgments
We thank members of the Hampton laboratory for technical support and stimulating discussions about the behavior of the SSD mutants. We thank Michael David (University of California, San Diego) for use of the flow cytometer. C. L. T. thanks the United States Navy Supply Corps for logistics support pertaining to these studies. R. Y. H. acknowledges Robert W. Hampton for fiscal advice and insights about highly significant sequence identity.
This work was supported, in whole or in part, by National Institutes of Health Grants NRSA 5F32GM82195 (to C. L. T.) and 5R01DK051996 to R. Y. H.) through the NIGMS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables 1–3.
R. Garza, unpublished observations.
C. Theesfeld, unpublished observations.
- SSD
- sterol-sensing domain
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated degradation
- HMGR
- HMG-CoA reductase
- SREBP
- sterol regulatory element-binding protein
- SCAP
- SREBP cleavage activating protein
- INSIG
- insulin-induced gene protein
- HRD
- HMG-CoA reductase degradation
- FPP
- farnesyl pyrophosphate
- GGPP
- geranylgeranyl pyrophosphate
- NR1
- non-regulated mutant of Hmg2
- CHX
- cycloheximide
- Lova
- lovastatin
- ZA
- zaragozic acid
- Ro48
- Ro48-8071
- QC
- quality control
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Goldstein J. L., Brown M. S. (1990) Nature 343, 425–430 [DOI] [PubMed] [Google Scholar]
- 2. Hampton R. Y., Garza R. M. (2009) Chem. Rev. 109, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Espenshade P. J., Hughes A. L. (2007) Annu. Rev. Genet. 41, 401–427 [DOI] [PubMed] [Google Scholar]
- 4. Sato B. K., Schulz D., Do P. H., Hampton R. Y. (2009) Mol. Cell 34, 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peng Z. L., Yang J. Y., Chen X. (2010) BMC Bioinformatics 11, 420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 7. Kuwabara P. E., Labouesse M. (2002) Trends Genet. 18, 193–201 [DOI] [PubMed] [Google Scholar]
- 8. Ohgami N., Ko D. C., Thomas M., Scott M. P., Chang C. C., Chang T. Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12473–12478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adams C. M., Reitz J., De Brabander J. K., Feramisco J. D., Li L., Brown M. S., Goldstein J. L. (2004) J. Biol. Chem. 279, 52772–52780 [DOI] [PubMed] [Google Scholar]
- 10. Radhakrishnan A., Sun L. P., Kwon H. J., Brown M. S., Goldstein J. L. (2004) Mol. Cell 15, 259–268 [DOI] [PubMed] [Google Scholar]
- 11. Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. (2002) Cell 110, 489–500 [DOI] [PubMed] [Google Scholar]
- 12. Yabe D., Xia Z. P., Adams C. M., Rawson R. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16672–16677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. (1985) Cell 41, 249–258 [DOI] [PubMed] [Google Scholar]
- 14. Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., DeBose-Boyd R. A. (2003) J. Biol. Chem. 278, 52479–52490 [DOI] [PubMed] [Google Scholar]
- 15. Song B. L., Javitt N. B., DeBose-Boyd R. A. (2005) Cell Metab. 1, 179–189 [DOI] [PubMed] [Google Scholar]
- 16. Nguyen A. D., McDonald J. G., Bruick R. K., DeBose-Boyd R. A. (2007) J. Biol. Chem. 282, 27436–27446 [DOI] [PubMed] [Google Scholar]
- 17. Hampton R. Y., Rine J. (1994) J. Cell Biol. 125, 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hampton R. Y., Bhakta H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12944–12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hampton R. Y., Gardner R. G., Rine J. (1996) Mol. Biol. Cell 7, 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bays N. W., Gardner R. G., Seelig L. P., Joazeiro C. A., Hampton R. Y. (2001) Nat. Cell Biol. 3, 24–29 [DOI] [PubMed] [Google Scholar]
- 21. Gardner R. G., Hampton R. Y. (1999) J. Biol. Chem. 274, 31671–31678 [DOI] [PubMed] [Google Scholar]
- 22. Garza R. M., Tran P. N., Hampton R. Y. (2009) J. Biol. Chem. 284, 35368–35380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gardner R. G., Shan H., Matsuda S. P., Hampton R. Y. (2001) J. Biol. Chem. 276, 8681–8694 [DOI] [PubMed] [Google Scholar]
- 24. Shearer A. G., Hampton R. Y. (2004) J. Biol. Chem. 279, 188–196 [DOI] [PubMed] [Google Scholar]
- 25. Shearer A. G., Hampton R. Y. (2005) EMBO J. 24, 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dobrosotskaya I. Y., Seegmiller A. C., Brown M. S., Goldstein J. L., Rawson R. B. (2002) Science 296, 879–883 [DOI] [PubMed] [Google Scholar]
- 27. Gardner R., Cronin S., Leader B., Rine J., Hampton R. (1998) Mol. Biol. Cell 9, 2611–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gardner R. G., Hampton R. Y. (1999) EMBO J. 18, 5994–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. (1989) Gene 77, 61–68 [DOI] [PubMed] [Google Scholar]
- 30. Feramisco J. D., Radhakrishnan A., Ikeda Y., Reitz J., Brown M. S., Goldstein J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3242–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cronin S. R., Hampton R. Y. (1999) Methods Enzymol. 302, 58–73 [DOI] [PubMed] [Google Scholar]
- 32. Gardner R. G., Swarbrick G. M., Bays N. W., Cronin S. R., Wilhovsky S., Seelig L., Kim C., Hampton R. Y. (2000) J. Cell Biol. 151, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cronin S. R., Khoury A., Ferry D. K., Hampton R. Y. (2000) J. Cell Biol. 148, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hughes A. L., Stewart E. V., Espenshade P. J. (2008) J. Lipid Res. 49, 2001–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Flury I., Garza R., Shearer A., Rosen J., Cronin S., Hampton R. Y. (2005) EMBO J. 24, 3917–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jo Y., Debose-Boyd R. A. (2010) Crit. Rev. Biochem. Mol. Biol. 45, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sengstag C., Stirling C., Schekman R., Rine J. (1990) Mol. Cell. Biol. 10, 672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Motamed M., Zhang Y., Wang M. L., Seemann J., Kwon H. J., Goldstein J. L., Brown M. S. (2011) J. Biol. Chem. 286, 18002–18012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.