Abstract
In newborn mice, PLRP2 is essential for fat digestion. In human infants, the role of PLRP2 in fat digestion is unclear, as it has poor activity against long-chain triglycerides in vitro. Also, many infants carry a genetic polymorphism resulting in a truncated protein, PLRP2 W340X, which may impact function significantly. We re-examined the properties of recombinant human PLRP2 and studied the impact of W340X mutation on its function. In the presence of bile salt micelles and colipase, human PLRP2 hydrolyzed long-chain tri-, di-, and monoglycerides. It hydrolyzed triolein at a level much lower than that of pancreatic triglyceride lipase, but close to that of carboxyl ester lipase, after a long lag phase, which could be eliminated by the addition of oleic acids. Human PLRP2 W340X was poorly secreted and largely retained inside the cell. The retention of the mutant protein triggered endoplasmic reticulum stress and unfolded protein responses. Our results show that earlier studies underestimated human PLRP2 activity against triolein by employing suboptimal assay conditions. In vivo, dietary fat emulsions contain fatty acids as a result of the action of gastric lipase. Consequently, PLRP2 can contribute to fat digestion during early infancy. Furthermore, infants with homozygous W340X alleles will not secrete functional PLRP2 and may have inefficient dietary fat digestion, particularly when breastfeeding is unavailable. Additionally, the aberrant folding of W340X mutant may cause chronic cellular stress and increase susceptibility of pancreatic exocrine cells to other metabolic stressors.
Keywords: Enzyme Mutation, Fatty Acid, Lipase, Lipolysis, Pancreas, Protein Synthesis, Triglyceride
Introduction
Pancreatic lipases are critical for efficient dietary fat digestion. Of the multiple pancreatic lipases, PTL2 is the predominant lipase secreted by the exocrine pancreas. PTL is the archetype of a lipase gene family that includes PTL and PLRP2 (1). Although the genes encoding PTL and PLRP2 reside in the same chromosomal location and likely diverged recently by gene duplication, they differ in their pattern of temporal expression during development in rodents and humans (2–4). In each species, mRNA encoding PLRP2 is expressed before birth and into adulthood, whereas the mRNA encoding PTL is not expressed in the fetal pancreas or at birth. The pattern of temporal expression led to the hypothesis that PLRP2 is critical for efficient dietary fat digestion in the newborn. Subsequently, it was shown that PLRP2-deficient mouse pups have significant fat maldigestion that disappears coincident with the expression of PTL thereby confirming the important role of PLRP2 in fat digestion in the newborn mouse (2). Furthermore, efficient fat digestion was dependent on colipase, indicating that PLRP2 requires colipase in vivo (2, 5).
By analogy, PLRP2 should have an important role in dietary fat digestion in humans, but in vitro studies suggest that human PLRP2 may not function as a triglyceride lipase. Like PTL from all species, human PLRP2 activity against the major dietary long-chain triglycerides is inhibited by micellar bile salts (6). Unlike PTL, the activity of human PLRP2 is not restored by the addition of another pancreatic protein, colipase. The poor activity of human PLRP2 against long-chain triglycerides raises questions about its ability to hydrolyze dietary fats, which are mostly long-chain triglycerides, in the duodenum where bile salt micelles reside. It has been suggested that triglycerides are not a physiological substrate for human PLRP2 and that the major role of human PLRP2 is as a galactolipase (7, 8). If correct, the in vitro findings have important implications for the molecular details of in vivo dietary fat digestion in human newborns.
Additionally, a nonsense polymorphism in the gene encoding human PLRP2 has previously been found in different ethnic populations, with the allele frequencies of 0.55, 0.53, and 0.33 for Africans, Caucasians, and Chinese, respectively (9). The polymorphism results in the premature truncation of human PLRP2, W340X. The predicted protein lacks nearly the entire C-terminal domain of human PLRP2. Our earlier work has shown that the C-terminal domain of human PTL is required for its stability, efficient secretion, and full activity (10). Although human PLRP2 W340X may behave similarly to the truncated human PTL, it is unclear how the truncation impacts the biochemical function of human PLRP2. If human PLRP2 W340X has decreased secretion and activity, the role of PLRP2 in efficient fat digestion in newborns would be limited in subjects with the polymorphism.
To reconcile the apparent contradictions among the rodent data, the human expression data, and the in vitro enzymatic properties of human PLRP2, we reexamined the in vitro properties of human PLRP2 to determine whether it can hydrolyze bile salt emulsions of long-chain triglycerides and if colipase increases activity under appropriate conditions. As a measure of the likelihood that any activity we detected with human PLRP2 was likely to be physiologically important, we compared the activity of human PLRP2 with that of human CEL, another lipase that contributes to fat digestion in the newborn (11–13). We also determined the impact of the W340X mutation on the cell biology and activity of human PLRP2. We expressed the truncated protein in both yeast and cultured mammalian cells and measured the ability of the cells to secrete an active mutant. Because the majority of the mutant protein remained within the cells, we determined whether the UPR was activated in the transfected cells. Our findings provide new insights into the role of PLRP2 in dietary fat digestion in human newborns.
EXPERIMENTAL PROCEDURES
DNA Manipulations
All DNA manipulations were done by standard methods unless stated otherwise. The cDNA encoding for mature human PLRP2 protein was amplified by PCR and subcloned into the yeast protein expression vector pPICZαA, in which the α-factor-mating signal peptide was used. The full-length human PLRP2 cDNA was amplified by PCR and subcloned into the mammalian protein expression vector pcDNA3. The mutation W340X was introduced by QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The full-length human CEL cDNA was excised from pCMV-SPORT6/human CEL (clone ID 5187959, OpenBiosystems, Huntsville, AL) and subcloned into pPICZA. The recombinant human CEL was designed as a secretory protein under its native secretion signal peptide. The sequence of all plasmid DNA constructs was verified by dideoxynucleotide sequencing.
Production and Purification of Recombinant Proteins in Pichia pastoris
Recombinant human PLRP2, PLRP2 W340X, and human CEL were produced in protease A-deficient P. pastoris, SMD1168, following the manufacturer's instructions (Invitrogen). The competent yeast cells were transformed with linearized plasmid DNAs by electroporation, and transformants were screened by immunoblot and/or lipase activity assay as described previously (14, 15).
The large scale production of secreted recombinant proteins in yeast was carried out as described previously (15). The cultures were allowed to grow under methanol induction for ∼16 h for human PLRP2 and its mutant W340X and for ∼72 h for CEL, with daily 1% methanol supplementation. For PLRP2 and its mutant W340X, the clarified media were concentrated to ∼50 ml over a Pellicon XL Biomax 10 membrane (Millipore, Bedford, MA). All protein samples were dialyzed at 4 °C overnight against double distilled H2O containing 2 mm benzamidine.
The dialyzed protein samples for human PLRP2 and its mutant W340X were adjusted to contain 10 mm MES, pH 5.5, and then applied to an equilibrated Mono S FPLC column (GE Healthcare) with 10 mm MES. The bound protein was eluted from the column between 60 and 110 mm NaCl using a linear concentration gradient from 0 to 150 mm NaCl within 60 min. Recombinant human CEL was purified as described previously (16). Briefly, Na3PO4 and EDTA solution was added to the dialyzed sample to obtain 20 and 1 mm final concentrations, respectively, and pH was adjusted to 7.4. The buffered sample was loaded onto an equilibrated heparin-Sepharose column. The bound protein was eluted off from the column at around 300 mm NaCl using a linear salt gradient ranging from 0 to 1 m NaCl within 60 min. Chromatography for all purifications was controlled with an ÄKTAExplorer system (Amersham Biosciences), and the flow rate was set at 1 ml/min. Fractions containing recombinant protein were identified by A280, gel staining, and lipase activity assay and were pooled. The buffer was exchanged to 20 mm Tris-Cl, pH 8.0, and 25 mm Na3PO4, pH 8.0, for human PLRP2 and CEL, respectively. The protein concentration was determined by UV light spectrophotometry at A280 using an extinction coefficient of E1%1 cm = 1.2 and 1.37 for human PLRP2 and CEL, respectively.
Determination of Protein Purity and Integrity
The purified proteins were analyzed by gel staining to confirm their homogeneity and integrity. Six μg of purified recombinant proteins human PLRP2 and CEL were resolved by 10 and 7.5% SDS-PAGE, respectively. The gels were stained with GelCode Blue Stain Reagent (Pierce) and scanned by Molecular Imager VersaDoc MP 4000 System (Bio-Rad).
Lipase Activity Assay
Lipase activity of human PLRP2 was determined at 25 °C by using the standard pH-stat method with the emulsion of tributyrin (114 mm), trioctanoin (27 mm), triolein (6.9 mm), diolein (5.1 mm), or monoolein (10.7 mm) in a total volume of 15 ml as described previously (17, 18). Unless otherwise stated, the reaction was carried out over a 5-min period in the presence or absence of a 5-fold molar excess of colipase at the indicated sodium taurodeoxycholate (NaTDC) concentration. To study the effect of free fatty acids on lipase activity, 1 μm oleic acid was added into the emulsion prior to the addition of colipase. Human CEL lipase activity was also assayed by the pH-stat method using 10 μg of purified CEL, with the assay buffer containing 150 mm NaCl, 1 mm Tris-HCl, 2 mm CaCl2, pH 8.0, 3.75% defatted BSA, and varying concentrations of sodium cholate or sodium taurocholate (20). The lipolytic activities are expressed in international lipase units per mg of enzyme. One unit corresponds to 1 μmol of fatty acid released per min.
Colipase Titration
The interaction of human PLRP2 and PTL with colipase was assayed as described previously (19). The assay was performed by measuring lipase activity over a range of colipase concentrations (0–250 nm) with a constant concentration of lipase (3.9 nm) and excess substrate (54 mm trioctanoin) in 4 mm NaTDC assay buffer.
Binding Assays
The interfacial binding of human PLRP2 and PTL was assayed by mixing the enzyme with trioctanoin or triolein emulsion and then measuring the lipase activity remaining in the water phase after the separation of the oil phase. The ability of colipase to anchor lipases to trioctanoin or triolein was measured as described for intralipid (17).
Culture and Transfection of Mammalian Cells
HEK 293T and COS7 cells were purchased from ATCC (Manassas, VA) and maintained in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin and streptomycin at 37 °C in a 5% CO2-humidified incubator. Prior to transfection, the cells were grown to 100% confluency and harvested by trypsinization. Cells were then seeded at ∼50% confluency and allowed to adhere for 16 h, and transfected using FuGENE 6 according to the manufacturer's manual (Roche Diagnostics). A mixture of 5 μl of FuGENE 6 and 1.65 μg of plasmid DNA pcDNA3, pcDNA3/PLRP2, or pcDNA3/PLRP2 W340X in 100 μl of Opti-MEM I reduced serum medium (Invitrogen) per well in 6-well plates was added; for each 10-cm dish used for RNA extraction from HEK 293T cells, a mixture of 25 μl of FuGENE 6 and 10 μg of plasmid DNA pcDNA3, pcDNA3/PLRP2, or pcDNA3/PLRP2 W340X in 500 μl of Opti-MEM I reduced serum medium was added. Forty eight hours after transfection, culture medium and cells were harvested following routine procedures.
MG132 treatment was carried out using HEK 293T cells in 6-well plates. Forty two hours post-transfection, the culture media were aspirated, and the cells were gently washed once with PBS. The cells were incubated with the Opti-MEM I reduced serum medium containing 20 μm MG132 (Sigma) in DMSO or DMSO alone for 6 h prior to harvesting.
Preparation of Cell Lysates
To study the total intracellular PLRP2 and W340X mutant protein levels, harvested cell pellets were resuspended in ice-cold PBS with EDTA-free Complete protease inhibitor mixture (Roche Diagnostics), with 200 μl for HEK 293T and 50 μl for COS7 cells, then mixed with equal volume of 2× Laemmli buffer, followed by 10-min boiling. Otherwise, cells were resuspended in RIPA buffer (150 mm NaCl, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mm Tris, pH 8.0) supplemented with EDTA-free Complete protease inhibitor mixture and Halt phosphatase inhibitor (Thermo Scientific, Rockford, IL), followed by a 30-min incubation on ice. The cell lysates were clarified by centrifugation at 15,000 × g for 15 min at 4 °C. The cell lysate protein concentration was determined using BCA protein assay kit (Thermo Scientific).
Western Blot Analysis
Aliquots of conditioned media (20 μl) or cell lysates (10–50 μg) were electrophoresed on 10% Tris-HCl or 4–15% TGX minigel (Bio-Rad) and transferred onto Immobilon-PVDF membranes (Millipore, Billerica, MA). Membranes were blocked in blocking reagent (LI-COR, Lincoln, NE) for 1 h and incubated with primary antibody overnight at 4 °C and with secondary antibody for 1 h at room temperature. The immunoblots were scanned by ODYSSEY INFRARED IMAGER, and the band density was quantified by Rectangular Method (LI-COR).
Antibodies used in this study were as follows: rabbit polyclonal antibody against human PTL (1:3000) was produced in our laboratory; rabbit anti-BiP (3177; 1:1000), GRP94 (2104; 1:1000), PDI (3501; 1:500), calnexin (2679; 1:2000), and mouse anti-tubulin (3873; 1:3000) were purchased from Cell Signaling; mouse monoclonal antibody against GAPDH (G8795; 1:1000) was purchased from Sigma. IRDye 680 goat anti-rabbit IgG (926-32221; 1:10,000) and IRDye 800 goat anti-mouse IgG (926-32210; 1:5000) were purchased from LI-COR.
RT-PCR Analysis
Total RNA was isolated from HEK 293T cells transfected with given plasmid DNAs using TRIzol (Invitrogen), and 1.5 μg of RNA was then reverse-transcribed with Moloney murine leukemia virus reverse transcription kit from Ambion (Austin, TX) at 44 °C for 1 h in a 20-μl reaction volume. Semiquantitative measurements were performed by PCR of the X-box binding protein-1 (XBP1) cDNA and its spliced form, using the following primers that amplify both forms as follows: XBP1 sense primer, 5′-CCT TGT AGT TGA GAA CCA GG-3′, and XBP1 antisense primer, 5′-GGG CTT GGT ATA TAT GTG G-3′. The XBP1 primers amplify 441- and 415-bp amplicons from the unspliced and spliced cDNAs, respectively. As an endogenous control, a 261-bp fragment of GAPDH was also amplified, using the following primers: GAPDH sense primer, 5′-GTC-CAC TGG CGT CTT CAC CA-3′, and GAPDH antisense primer, 5′-GTG GCA GTG ATG GCA TGG AC-3′. PCR products were run on 2% agarose gels, and the bands were visualized by ethidium bromide staining.
RESULTS
Production and Purification of Recombinant Proteins in P. pastoris
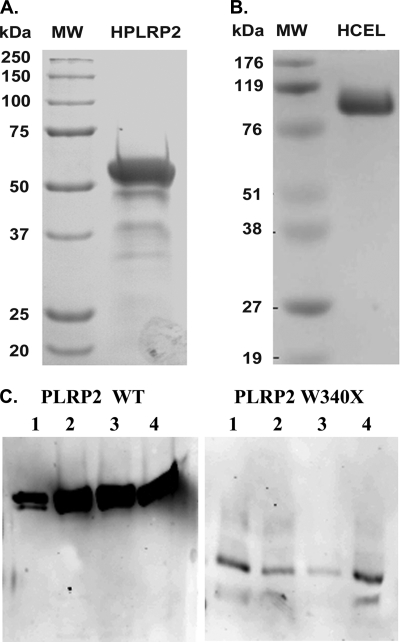
Recombinant proteins were expressed in yeast strain SMD1168 and secreted into the culture medium after methanol induction. The secreted recombinant proteins human PLRP2 and CEL were purified by one-step Mono S and heparin-Sepharose column purification, respectively. The yield was ∼20 mg for human PLRP2 and ∼30 mg for CEL from 1000 ml of yeast culture medium under methanol induction. The purity and integrity of isolated recombinant proteins were assessed by SDS-polyacrylamide gel staining. Purified PLRP2 protein appeared as predominant bands around 53 kDa, accompanied by several minor, faster migrating bands (Fig. 1A). The minor lower bands were recognized with antibody to PTL, indicating that they represent degradation products (data not shown). For human CEL, the predominant band was ∼105 kDa, consistent with post-translational modification by glycosylation (Fig. 1B) (20).
FIGURE 1.
SDS-PAGE and Western blot analysis of the recombinant human lipases produced from Pichia yeast. A and B, 6 μg of purified human recombinant PLRP2 or CEL protein was resolved by 10 and 7.5% SDS-PAGE, respectively. Both gels were stained with Gel-Code blue according to the manufacturer's directions. C, 12 μl of medium aliquots was resolved by 10% SDS-PAGE and transferred onto PVDF membrane, followed by detection with a rabbit anti-human PTL antibody. A, human PLRP2; B, human CEL (HCEL); C, PLRP2 and W340X secreted into the medium by P. pastoris. MW, molecular weight marker.
Western blot analysis revealed that the amount of protein secreted into yeast culture medium was more than 10 times lower for human PLRP2 W340X mutant than for the wild-type (Fig. 1C). This phenomenon remained consistent throughout the screening of more than 120 transformants from three different transformations. We consequently failed to purify the recombinant protein after numerous attempts. Substantial loss of W340X mutant protein consistently occurred during concentration, and the remaining concentrated W340X mutant protein lacked the ability to bind onto the Mono S column under various conditions. Recombinant PLRP2 W340X protein fused with a C-terminal His6 tag also did not bind efficiently to His affinity columns. These findings suggest that in the yeast protein expression system, the secretion of human PLRP2 W340X is markedly reduced, and the secreted mutant protein in the medium is unstable.
Enzymatic Activities of Human PLRP2 and CEL on Various Triglycerides
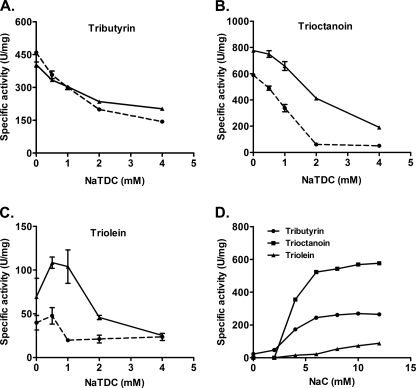
To confirm that the enzymatic properties of our recombinant human PLRP2 were similar to those previously reported by others, we determined its activity against tributyrin, trioctanoin, and triolein using the standard pH-stat assay over 0–4.0 mm NaTDC with and without a 5 m excess of colipase (Fig. 2). Against tributyrin, increasing the bile salt concentration decreased the activity of PLRP2 but did not abolish it. Colipase had little to no effect on activity at any NaTDC concentration (Fig. 2A). Human PLRP2 had just barely detectable activity against trioctanoin in the presence of higher NaTDC concentrations when micelles were present. Colipase had a small stimulatory effect (Fig. 2B). Activity against triolein was similarly low at higher NaTDC concentrations. Colipase had a stimulatory effect at low NaTDC concentrations but had no effect on activity at 4 mm NaTDC (Fig. 2C). Altogether, the enzymatic kinetics of our purified recombinant human PLRP2 were similar to those reported by other groups.
FIGURE 2.
Kinetics of human PLRP2 and CEL against various triglycerides. The effect of bile salts and colipase on human PLRP2 activity against various triglycerides was determined using the standard 5-min pH-stat assay, with 3 μg of PLRP2 at varying concentrations of NaTDC with (solid lines) or without (dotted lines) a 5 m excess of colipase. Human CEL bile salt dependence was determined using 10 μg of CEL on 114 mm tributyrin, 27 mm trioctanoin, or 6.9 mm triolein in the presence of varying concentrations of sodium cholate (NaC). A, PLRP2 activity against 114 mm tributyrin; B, PLRP2 activity against 27 mm trioctanoin; C, PLRP2 activity against 6.9 mm triolein; D, CEL activity against various triglycerides. Each data point is the mean ± S.D. of triplicate experiments.
During early infancy, CEL, another lipase, derived from both mothers' breast milk and the pancreas, is believed to be a major lipase responsible for dietary fat digestion when PTL is absent. To compare the activity of human PLRP2 to human CEL, we measured the lipase activities of human CEL on various triglycerides with several different bile salts. Maximal activity was achieved with 10 mm sodium cholate in the assay, 90 units/mg against triolein, 580 units/mg against trioctanoin, and 260 units/mg against tributyrin (Fig. 2D). Second best activity against triolein was obtained with 12 mm sodium taurocholate, with the activity of 50 units/mg. No activity of human CEL against triolein was detected in sodium deoxycholate or NaTDC (data not shown). Our results for human CEL activity against triolein are similar to those reported in earlier studies by others (20). These initial results suggest that human PLRP2 is less active against medium- and long-chain triglycerides than is CEL.
Lag Time of Human PLRP2 with Triolein
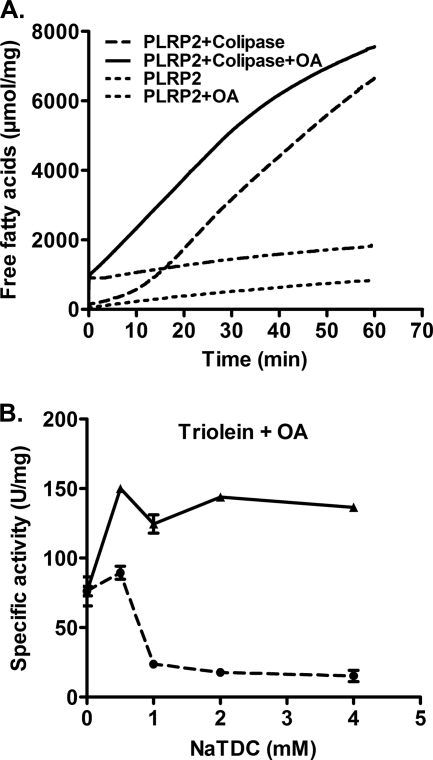
One potential explanation for the decreased activity of human PLRP2 is that it has a lag phase under the assay conditions. Earlier studies demonstrated that PTL has a lag phase under certain conditions, and a burst phase of activity is observed if the assay is allowed to proceed for a longer period (22–26). We sought to determine whether human PLRP2 had similar properties. Consequently, we extended the assays with triolein to 60 min. In the absence of colipase, human PLRP2 displayed very low activity against triolein throughout the 60-min assay (Fig. 3A). In the presence of colipase, a lag phase of ∼17 min was observed with triolein, followed by a burst of activity, with a specific activity of 140 units/mg (Table 1).
FIGURE 3.
Lag phase and effect of oleic acids on PLRP2 activity against triolein. Kinetics of human PLRP2 against 6.9 mm triolein in 4 mm NaTDC was further examined using the prolonged 60-min pH-stat assay with or without 1 μm oleic acids (OA) in the absence or presence of colipase. A, representative continuous tracings of pH-stat assays. The components of the assay are listed in the legend. The statistical analysis of multiple assays is presented in Table 1. B, human PLRP2 activity against 6.9 mm triolein using the standard 5-min pH-stat assay. One μm oleic acids were included in assay buffer. Solid line, 5 m excess of colipase added; dotted line, no colipase. Each point represents the mean ± S.D. of at least three measurements.
TABLE 1.
Lag times and activity of human PLRP2 on triolein in 4 mm NaTDC
All assays were done using 60-min pH-stat method with triolein in 4 mm NaTDC in the presence or absence of 5 m excess of colipase, with or without 1 μm oleic acids (OA). The lag time was determined from the plot of fatty acids released (NaOH consumed) versus time by extrapolating the slope of the burst phase to the time axis and is reported as minutes. Activity (units/mg protein) was determined from the slope of the burst phase and is presented as mean ± S.D. (n = 3).
| Treatment | Lag time | Activity |
|---|---|---|
| min | units/mg | |
| PLRP2 | 0 | 16.2 ± 3.1 |
| PLRP2 + OA | 0 | 15.2 ± 5.6 |
| PLRP2 + colipase | 17.1 ± 0.2 | 141 ± 6.4 |
| PLRP2 + OA + colipase | 0 | 136 ± 2.0 |
To determine whether the lag phase could be eliminated by the addition of digestion products, we repeated the assays with 1 μm oleic acids in the assay reaction mixture. The addition of exogenous oleic acids eliminated the lag phase, and the maximum specific activity was achieved immediately following the addition of human PLRP2 (Fig. 3A). The maximum lipase activity of human PLRP2 on triolein was 135 units/mg, similar to that observed following the lag phase when assayed without the addition of oleic acids (Table 1). Colipase was absolutely required for activity.
Because the standard 5-min pH-stat method underestimated the activity of human PLRP2 on triolein, we next re-examined the effect of bile salts and colipase on human PLRP2 lipase activity against triolein in the presence of oleic acids. The activity of human PLRP2 was inhibited by high bile salt concentrations, with the activity decreasing from ∼150 units/mg at 0.5 mm NaTDC to marginal level at 4 mm NaTDC, and the inclusion of colipase restored the activity to ∼130 units/mg (Fig. 3B). These results indicate human PLRP2 has modest activity against triolein under optimal assay conditions. Although the activity of human PLRP2 against triolein is about 20-fold lower than that of human PTL, under the optimal conditions its activity is comparable with the activity of CEL, suggesting that PLRP2 may play a significant role in dietary fat digestion during early infancy when PTL is absent.
Human PLRP2 Activity Against Diolein and Monoolein
Eydoux et al. (6) previously demonstrated that human PLRP2 exhibited significant lipase activity against monoolein but not triolein and diolein when assayed in 4 mm NaTDC. Because of our results with triolein, we revisited the ability of human PLRP2 to hydrolyze diolein and monoolein. We first measured the activity of human PLRP2 against purified 1,2-diolein and a mixture of 1,3-diolein and 1,2-diolein (85:15%) over 60 min. As shown in Table 2, the lipase activity of human PLRP2 on diolein was quite low when assayed in 4 mm NaTDC without colipase. When colipase was included, human PLRP2 hydrolyzed 1,2-diolein and the diolein mixture at the specific activity of 270 and 220 units/mg, respectively. The addition of oleic acids did not enhance human PLRP2 lipase activity against diolein, and no lag time was observed during the extended 60-min assay (data not shown). Next, we measured the activity of human PLRP2 against monoolein under the similar assay conditions. Human PLRP2 hydrolyzed 1-monoolein with the activity of 150 units/mg, regardless of the presence or absence of colipase (Table 2). Like PTL, human PLRP2 had no detectable activity against 2-monoolein.
TABLE 2.
Activity of human PLRP2 on diolein and monoolein in 4 mm NaTDC
Activities were assayed by 5-min pH-stat method in 4 mm NaTDC in the presence or absence of 5 m excess of colipase. Each result is presented as mean ± S.D. (n = 3).
| Substrate | Activity |
|
|---|---|---|
| −Colipase | +Colipase | |
| units/mg protein | ||
| 85% 1,3-diolein and 15% 1,2-diolein | 18.8 ± 7.4 | 220 ± 7.1 |
| 1,2-Diolein | 13.6 ± 6.6 | 268 ± 9.6 |
| 1-Monoolein | 146 ± 9.5 | 150 ± 3.8 |
| 2-Monoolein | 3.3 ± 3.1 | 4.0 ± 3.6 |
Interaction with Colipase
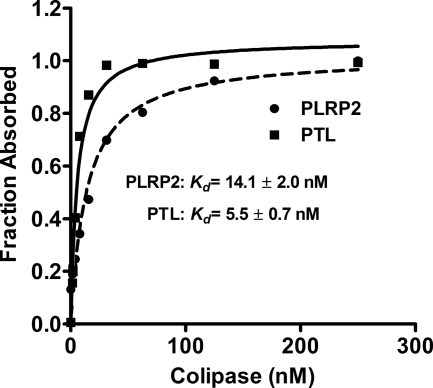
The available data suggest that colipase has a much weaker effect on human PLRP2 than it has on human PTL. To better characterize the apparent differences in colipase interaction between human PTL and PLRP2, we measured the ability of colipase to activate these two lipases over a range of colipase concentrations in the presence of excess trioctanoin and a constant amount of lipase (3.9 nm). From these data, we determined the concentration of colipase that restored half-maximal activity to the lipases (Kd) and the concentration that restored maximal activity (Bmax). The apparent Kd and Bmax values were determined by nonlinear regression with a rectangular hyperbola function. Given the large excess of substrate in the assays, the apparent constants reflect the interaction between colipase and lipase rather than the interaction between colipase and the substrate. The curves for human PTL and PLRP2 are given in Fig. 4. As expected, the apparent Bmax values for human PTL and PLRP2 were similar, and the apparent Kd value was 2.6-fold higher for human PLRP2 than for PTL suggesting PLRP2 has lower affinity for colipase.
FIGURE 4.
Colipase dependence of human PLRP2 and PTL. The assays were done by 5-min pH-stat method. Each assay included 3.9 nm purified recombinant human PLRP2 or PTL, 180 mm trioctanoin, in 4 mm NaTDC with varying concentrations of colipase. The fraction absorbed was calculated from the activity and plotted on the y axis. The symbols are defined on the figure.
Colipase Requirement for Lipase Absorption to Emulsion Particles
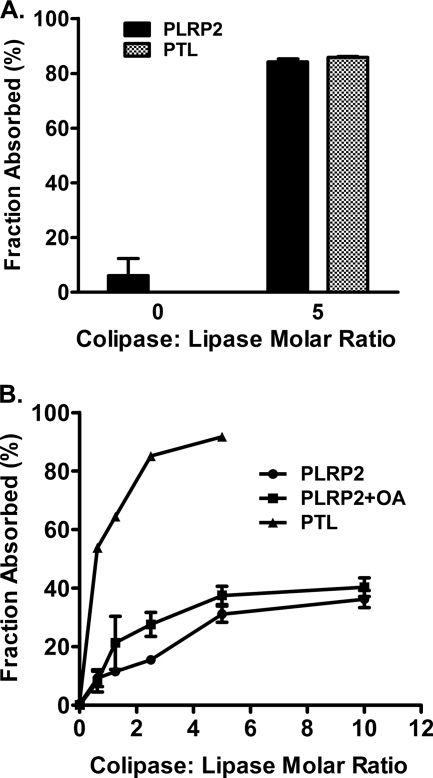
Current models of colipase function hypothesize that colipase restores activity to bile salt-inhibited lipase by anchoring lipase to the substrate. We measured the ability of colipase to aid absorption of human PLRP2 and PTL to trioctanoin and triolein in 4 mm NaTDC. Both human PLRP2 and PTL lacked the ability to absorb onto trioctanoin and triolein without colipase (Fig. 5). The addition of 5 m excess colipase anchored the majority of the lipases absorbed onto trioctanoin (84% for PLRP2 versus 86% for PTL) (Fig. 5A). Absorption of both lipases depended on the colipase in a concentration-dependent fashion. However, the proportion of human PLRP2 absorbed to the emulsion was substantially lower than for PTL (Fig. 5B). Only one-third of human PLRP2 bound to triolein even with 10-fold molar excess of colipase. Furthermore, the addition of oleic acids did not significantly improve the absorption of human PLRP2 to triolein. These results suggest that human PLRP2 requires colipase to absorb to trioctanoin and triolein, although the ability to absorb to triolein emulsions is poor, even in the presence of oleic acids.
FIGURE 5.
Absorption of human PLRP2 and PTL to trioctanoin and triolein. Absorption was determined with 6.0 μg of lipase and expressed as the percentage of the total added lipase activity bound to the lipid phase after centrifugation, as described under “Experimental Procedures.” A, human PLRP2 and PTL absorption to trioctanoin with and without colipase; B, absorption of human PLRP2 and PTL to triolein. OA, 1 μm oleic acids. Each bar represents the mean ± S.D. of at least three measurements.
Secretion of Human PLRP2 W340X in Transfected Mammalian Cells
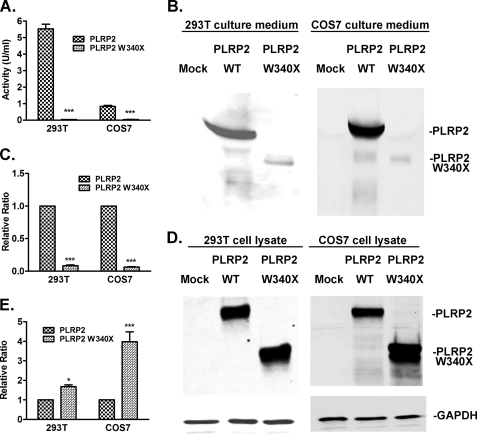
To better define the secretory defect of human PLRP2 W340X observed in the yeast system, we investigated the impact of the naturally occurring W340X truncation on human PLRP2 function in transfected COS7 and HEK 293T cells. As with yeast, the amount of human PLRP2 W340X in the medium of either cell type was minimal by activity measurement or by protein immunoblot, although activity and immunoblot signal were robust for PLRP2 wild-type in both cell lines (Fig. 6, A, B, and D). Conversely, intracellular PLRP2 W340X protein level was >1.6 times that of PLRP2 wild type, particularly in COS7 cells (Fig. 6, C and E).
FIGURE 6.
Cellular secretion and retention of wild-type and W340X human PLRP2 in transiently transfected mammalian cells. Forty eight hours after HEK 293T and COS7 cells were transfected with either pcDNA3 (mock), pcDNA3/PLRP2, or pcDNA3/PLRP2 W340X, conditioned media or the whole cell lysates were collected. The extracellular secretion of wild-type and W340X mutant PLRP2 was measured by lipase activity in the conditioned culture media against tributyrin in 0.5 mm NaTDC (A). The secreted or total intracellular wild-type or W340X mutant protein was measured by protein immunoblot. Protein samples were resolved by 10% SDS-PAGE and transferred onto PVDF membrane and detected with rabbit anti-human PTL polyclonal antibody. GAPDH detected by mouse anti-GAPDH monoclonal antibody was used as a loading control for cell lysate protein samples. A, lipase activity in the media of HEK 293T or COS7 cells; B, protein immunoblot of PLRP2 and W340X in culture media; C, quantitation of relative amounts of PLRP2 and W340X in the culture media; D, protein immunoblot of PLRP2 and W340X in whole cell extracts; E, quantitation of relative amounts of PLRP2 and W340X in the cell lysates. A, C, and E, protein levels of W340X were expressed as the relative ratio to the protein levels of wild-type PLRP2. Each bar represents the mean ± S.D. of at least three measurements. Statistical analysis was performed with paired t test. ***, p < 0.001; *, p < 0.05.
UPR in PLRP2 W340X-transfected Cells
In eukaryotic cells, all newly synthesized secretory proteins must first fold properly in the ER before being delivered to their ultimate destinations. The W340X truncation of PLRP2 led to the elevated intracellular retention of mutant protein, which is likely due to its protein misfolding in the ER. Retention and accumulation of misfolded protein in the ER can result in ER stress, which triggers the UPR. The activation of the UPR ultimately leads to a variety of cellular responses as follows: attenuation of translation to reduce the load of protein synthesis and avoid further accumulation of misfolded proteins in the ER; transcriptional up-regulation of gene-encoding ER chaperones and protein-folding enzymes to increase the folding capacity in the ER; and transcriptional up-regulation of genes encoding components of the ER-associated degradation (ERAD), a process involving the recognition and retrotranslocation of misfolded protein from the ER to the cytoplasm, to be degraded by the proteasome. Within this signal transduction pathway, BiP is the major sensor of ER stress and the master regulator of the UPR.
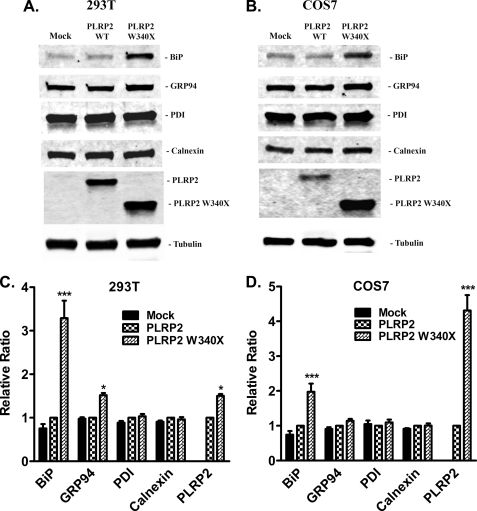
To further study whether PLRP2 W340X mutant causes ER stress and subsequently activates the UPR, we measured the protein levels of BiP, and several other major ER chaperones, such as GRP94, PDI, and calnexin in cell lysates by Western blot (Fig. 7, A and B). The levels of these proteins in cells transfected with wild-type human PLRP2 were similar to the levels detected in mock-transfected cells. Among these ER chaperones, only the level of BiP was significantly increased in both cell lines expressing mutant W340X (Fig. 7, C and D). The level of GRP94 was increased significantly in the HEK 293T cells but only modestly in the COS7 cells transfected with the mutant W340X.
FIGURE 7.
Expression of ER stress markers and protein folding chaperones in mammalian cells transiently transfected with wild-type or W340X mutant human PLRP2. Forty eight hours after HEK 293T and COS7 cells were transfected with either pcDNA3 (mock), pcDNA3/PLRP2, or pcDNA3/PLRP2 W340X, cells were lysed using RIPA buffer, and protein concentrations were then determined by Pierce BCA protein assay kit. Protein samples (30 μg for HEK 293T cell and 15 μg for COS7 cells) were resolved by 4–15% Mini-Protean TGX minigels and transferred onto PVDF membranes, followed by immunoblotting with rabbit anti-BiP, GRP94, protein-disulfide isomerase (PDI), or calnexin antibodies. Tubulin detected by mouse anti-α-tubulin monoclonal antibody was used as loading control, and cellular PLRP2 wild-type or W340X mutant protein levels were used as a reference of transfection. Immunoblots were representative of 10 (HEK 293T cells, A) or 6 (COS7 cells, B) independent experiments. Protein levels were normalized by tubulin and are shown as bar graphs. Each bar represents the mean ± S.D. C, HEK 293T cells (n = 10); D, COS7 cells (n = 6). Statistical analysis was performed with two-way analysis of variance. ***, p < 0.001; *, p < 0.05.
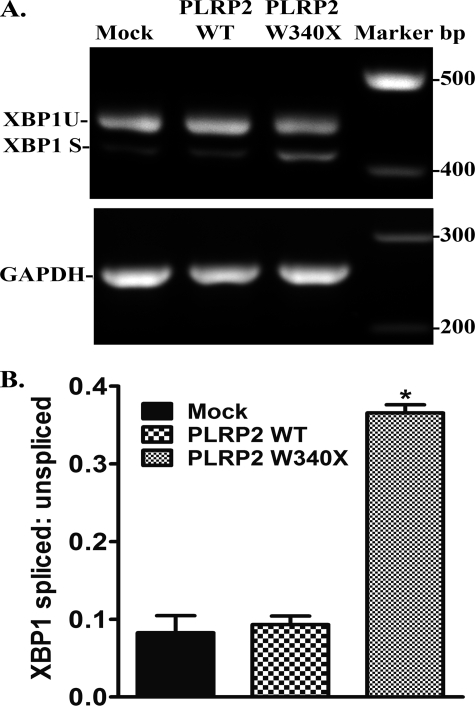
Because BiP protein levels were elevated in cells transfected with W340X mutant PLRP2, suggesting the mutation caused ER stress in the cells, we next determined if there was a downstream effect on XBP1 splicing. In nonstressed cells, BiP binds to the inositol-requiring enzyme 1 (IRE1). During stress, BiP dissociates from IRE1 allowing the activation of IRE1, which in turn catalyzes the cytoplasmic splicing of the XBP1 mRNA to generate a 26-bp shorter mRNA variant. This variant encodes a transcriptional activator for the UPR. We performed RT-PCR analysis of the XBP1 mRNA in mock, human PLRP2 wild-type and W340X mutant-transfected HEK 293T cells (Fig. 8A). The expression of W340X increased the levels of the spliced form of XBP1 mRNA more than 2-fold compared with the levels in mock and human PLRP2-transfected cells (Fig. 8B).
FIGURE 8.
XBP1 mRNA splicing in HEK 293T cells expressing wild-type or W340X mutant human PLRP2. Forty eight hours after transfection, total RNA was isolated from HEK 293T cells transfected with either pcDNA3 (mock), pcDNA3/PLRP2, or pcDNA3/PLRP2 W340X, and RT-PCR was performed to determine the extent of XBP1 mRNA splicing, as a ratio of spliced form to unspliced form. GAPDH was used as a reference control. A, representative ethidium bromide-stained 2% agarose gels; B, quantitation of XBP1 by ImageJ presented as the relative ratio of spliced and unspliced XBP1 mRNA abundance. Each bar represents the mean ± S.D. of at least three measurements. Statistical analysis was performed with two-way analysis of variance. *, p < 0.05.
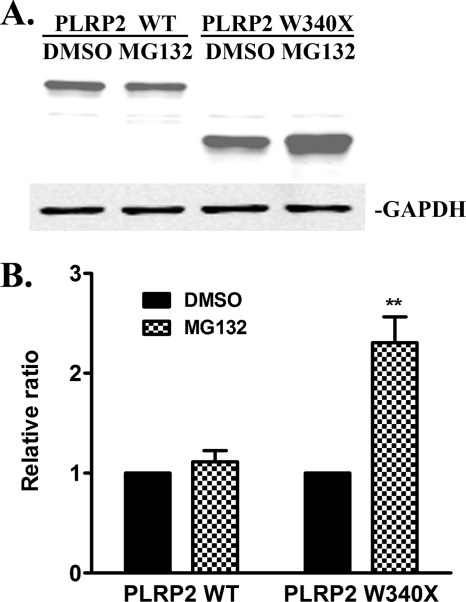
The activation of the UPR may turn on ERAD to clear the misfolded proteins. To investigate whether W340X mutant protein is degraded through ERAD or could be the substrate of ERAD, we next measured the effect of MG132, a proteasome inhibitor, on the intracellular protein levels of human PLRP2 W340X in transfected HEK 293T cells (Fig. 9A). The incubation of cells with 20 μm MG132 for 6 h had little effect on intracellular protein level of wild-type human PLRP2 (Fig. 9B). In contrast, the intracellular level of human PLRP2 W340X was increased more than 2-fold in the presence of MG132, indicating that the proteasomal degradation pathway may be involved in the clearance of PLRP2 W340X mutant protein in the cells. The alternative interpretation that MG132 increases the protein expression of W340X is unlikely because in that case we would expect MG132 to have the same effect on wild-type PLRP2.
FIGURE 9.
Inhibition of intracellular wild-type or W340X mutant PLRP2 protein degradation by MG132 treatment in HEK 293T cells. Forty two hours after HEK 293T cells were transfected with wild-type or W340X mutant human PLRP2, the cells were incubated with DMSO (control) or 20 μm MG132 for 6 h prior to lysis. Proteins were detected in whole cell lysates with rabbit anti human PTL polyclonal antibody using GAPDH as a control. A, protein immunoblot of PLRP2 and W340X. Treatments are above each lane; B, quantitation of total intracellular PLRP2 wild-type or W340X mutant protein level expressed as the relative ratio of the DMSO-treated groups. Each bar represents the mean ± S.D. of three independent measurements. Statistical analysis was performed with paired t test. **, p < 0.01.
DISCUSSION
In this study, we reinvestigated the kinetic properties of human PLRP2 against long-chain substrates. Our results provide a plausible explanation for the differences between the reported in vitro properties of PLRP2 and the in vivo studies that show PLRP2 is essential for dietary fat digestion in newborn mice (5). We found that human PLRP2 can effectively hydrolyze triolein emulsified in bile salt micelles in contrast to previous reports (6, 8). Furthermore, the activity of human PLRP2 was comparable with the activity of CEL, another lipase that contributes to dietary fat digestion in newborns. The apparent discrepancy between our results and prior studies is explained by a long lag time for maximum activity that exceeded the assay time in the previous studies. More importantly, the addition of oleic acids to the assay mixture eliminated the lag phase. In the duodenum, naturally occurring substrate emulsions contain digestion products like oleic acids, which result from digestion by gastric lipase in humans (21). Thus, the partially digested dietary fats are suitable substrates for human PLRP2.
Lag phases have been commonly observed for lipolytic enzymes in the hydrolysis of substrates in emulsions, liposomes, and monolayers (22–26). The increased activity following the lag phase is probably due to increased absorption of PTL and colipase to the substrate interface induced by fatty acids released during the lag phase (22, 27). Although the elimination of the lag phase by fatty acids is well established, the mechanism remains uncertain. Initially, investigators speculated that fatty acids may change the properties of the lipid interface and increase absorption of colipase and PTL to the substrate (22, 27). Subsequently, several lines of evidence suggest that mixed micelles of fatty acid and bile salt can form a high affinity complex with PTL and colipase. This tertiary complex may then absorb efficiently to the lipid interface (26, 28–31).
In the case of human PLRP2, we could not demonstrate increased absorption of human PLRP2 to an emulsion of bile salt and triolein in the presence of oleic acids. The effect of oleic acids on the lag phase likely occurs through another mechanism. One possibility is that oleic acid modifies the physical properties of the interface in a way that facilitates hydrolysis rather than absorption of the colipase-PLRP2 complex. Several models seem plausible. First, the fatty acids may alter the orientation of the colipase-PLRP2 complex on the interface. Second, they may aid diffusion of substrate into the active site. Alternatively, the presence of fatty acids may mediate the transition of human PLRP2 into an active conformation by stabilizing the lid domain in an open position.
A second observation is also consistent with a model of oleic acid activation of human PLRP2 by a mechanism apart from increased absorption of the colipase-PLRP2 complex to substrate. The addition of oleic acids did not increase the effect of colipase on the absorption of human PLRP2 to substrate. In the presence and absence of oleic acids, colipase had a significantly lower apparent affinity for human PLRP2 than for human PTL. Several factors may contribute to the weak human PLRP2 interaction with colipase. Four residues in the human PTL lid domain, Arg-256, Asp-257, Tyr-267, and Lys-268, stabilize the open conformation of the lid domain in human PTL and are essential for a stable lipase-colipase interaction to occur (18, 32). None are conserved in human PLRP2, and substitution mutagenesis of the PTL residues to those of PLRP2 severely impairs the interaction with colipase (8, 18). Also, the open conformation of the lid domain adopted by human PLRP2 likely differs considerably from that of PTL, and it may not be optimally situated to contribute to colipase binding (33). Third, binding of colipase to the C-terminal domain of human PLRP2 may be decreased. Human PTL and PLRP2 share only 55% amino acid identity in the C-terminal domain, and some of these differences may affect colipase absorption to a lipid interface. Although the residues contributing to colipase binding as identified in the crystal structure of the colipase-PTL complex are conserved, other regions may be important (34). For instance, the β5′-loop of human PTL likely influences the interaction of colipase and PTL (28, 35). Six of nine residues differ in this loop between human PTL and PLRP2.
Even though human PLRP2 has decreased affinity for colipase, the presence of colipase is clearly required for activity against trioctanoin and triolein in the presence of 4 mm NaTDC. The effect of colipase on human PLRP2 activity against triolein was most apparent when oleic acids were included in the assay. The colipase dependence correlates with mouse studies showing that colipase is essential for dietary fat digestion in newborn mice and that PLRP2 accounts for the colipase-dependent lipase activity in the newborn mouse pancreas (2, 36).
The human PLRP2 requirement for colipase to effectively hydrolyze trioctanoin and triolein is in distinct contrast to the lack of a colipase requirement for the hydrolysis of tributyrin, phospholipid, galactolipid, and retinyl-palmitate (6, 37, 38). Interestingly, these substrates form mixed micelles with bile salts thereby presenting small aggregates in solution compared with the larger emulsion particles formed by trioctanoin and triolein (6). In the case of retinyl-palmitate, PLRP2 only hydrolyzes this substrate when it is in mixed micelles (38). Clearly, human PLRP2 can interact effectively with substrate presented in a mixed micelle, but like PTL requires colipase to absorb onto substrate emulsions. We provided support for this possibility by directly measuring absorption of human PLRP2 to trioctanoin or triolein emulsions. Little to no human PLRP2 absorbed onto the emulsion unless colipase was present. It has been suggested that the increased mobility of the lid domain in PLRP2 compared with PTL contributes to the increased activity of PLRP2 against substrates that form small aggregates in solution (6, 33). Still, the molecular details of the differences in the interaction of human PLRP2 with mixed micelles and with emulsion particles remain unclear.
We also noted one important difference in the properties of human PLRP2 expressed for this study and that previously reported. In our hands, human PLRP2 hydrolyzed diolein, and the activity depended on colipase in the presence of bile salt micelles. In the previous study, human PLRP2 had poor activity against diglycerides (6). The explanation for the difference is not obvious. The diglycerides were from different sources, but the assay conditions were similar. Regardless, the ability of human PLRP2 to hydrolyze diglycerides in a complex with colipase is consistent with in vivo results of procolipase-deficient mice and PLRP2-deficient mice. In both cases, the feces of the deficient newborns contained abundant amounts of diglycerides (5, 36). Diglycerides were not detectable in the feces of wild-type newborns indicating that both colipase and PLRP2 are required for the in vivo digestion of diglycerides.
Finally, we are the first research group to report the enzymatic properties and cell biology of a commonly occurring mutant of human PLRP2, W340X. Interestingly, in transformed yeast and transfected mammalian cells, the W340X mutant protein was largely retained inside the cells and only a minor fraction appeared in the medium. Although we were unable to purify W340X from the medium of yeast or mammalian cells to accurately characterize its enzymatic kinetics, assays of fresh or concentrated culture media consistently had low to undetectable levels of lipase activity suggesting PLRP2 W340X has little or no activity. The intracellular accumulation of W340X mutant protein further caused ER stress and triggered the UPR activation, as evidenced by elevated BiP protein levels in the cell lysate and XBP1 mRNA splicing. This provides evidence that PLRP2 W340X mutation may contribute to aberrant folding of the protein and may have other cellular effects along with the general loss of lipolytic activity. The ER stress induced by the synthesis of W340X might be partially relieved through degradation by proteosomes, as we showed that MG132 was capable of inhibiting the degradation of the mutant protein W340X but not wild-type human PLRP2.
The W340X polymorphism in human PLRP2 may have important physiological implications. First, the secretory defect of PLRP2 W340X would cause a “loss of function” in dietary fat digestion in humans particularly in the newborn. Compared with other major lipases, PTL and CEL, PLRP2 is suitable to make a substantive contribution to dietary fat digestion during early infancy when PTL is absent and CEL is likely the major lipase. Human PLRP2 may play an even larger role in dietary fat digestion in newborns who consume formula, which does not contain CEL. Formula-fed newborn homozygous carriers of the W340X polymorphism would lack both breast milk CEL and PLRP2 and have less efficient dietary fat digestion. Importantly, the W340X mutation may account for the wide range of fat absorption seen in infants compared with older children and adults (39). Second, the intracellular retention of PLRP2 W340X could place pancreatic acinar cells at greater risk for injury because of prolonged and chronic ER. The cellular consequence of the expression of human PLRP2 W340X needs to be further examined in pancreatic acinar cell lines, which are highly specialized in secretory protein synthesis and folding. Ultimately, for human PLRP2 W340X, both the putative loss of function as a lipase and gain of function as a toxic cellular stressor remain to be verified in vivo.
In conclusion, we have demonstrated that human PLRP2 has sufficient activity against long-chain triglycerides to make a significant contribution to dietary fat digestion in human newborns. The higher activity and larger amounts of PTL secreted from the adult pancreas make it less likely that PLRP2 makes a significant contribution to triglyceride digestion in adults. In fact, PLRP2-deficient adult mice do not have detectable maldigestion of dietary triglycerides (5). More likely, PLRP2 functions to hydrolyze minor dietary fats like galactolipids or vitamin esters in the adult (6, 38). In this regard, a recent report raises a question about the importance of PLRP2 as a galactolipase (40). The investigators found a strong correlation between the gene encoding PLRP2 W340X and populations where cereals are the main dietary component. They speculated that the truncated PLRP2 W340X was more active against plant lipids. Although our data do not address the activity of PLRP2 W340X on plant lipids, the near absence of secreted PLRP2 W340X from transfected cells makes it likely that PLRP2 W340X does not contribute to plant lipid digestion in humans.
Acknowledgment
We thank Rita Miller for thorough proofreading and for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK080820.
- PTL
- pancreatic triglyceride lipase
- CEL
- carboxyl ester lipase
- ERAD
- ER-associated degradation
- NaTDC
- sodium taurodeoxycholate
- UPR
- unfolded protein response
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Cygler M., Schrag J. D., Sussman J. L., Harel M., Silman I., Gentry M. K., Doctor B. P. (1993) Protein Sci. 2, 366–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. D'Agostino D., Lowe M. E. (2004) J. Nutr. 134, 132–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Payne R. M., Sims H. F., Jennens M. L., Lowe M. E. (1994) Am. J. Physiol. 266, G914–G921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang Y., Sanchez D., Figarella C., Lowe M. E. (2000) Pediatr. Res. 47, 184–188 [DOI] [PubMed] [Google Scholar]
- 5. Lowe M. E., Kaplan M. H., Jackson-Grusby L., D'Agostino D., Grusby M. J. (1998) J. Biol. Chem. 273, 31215–31221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eydoux C., De Caro J., Ferrato F., Boullanger P., Lafont D., Laugier R., Carrière F., De Caro A. (2007) J. Lipid Res. 48, 1539–1549 [DOI] [PubMed] [Google Scholar]
- 7. Amara S., Lafont D., Fiorentino B., Boullanger P., Carrière F., De Caro A. (2009) Biochim. Biophys. Acta 1791, 983–990 [DOI] [PubMed] [Google Scholar]
- 8. Sias B., Ferrato F., Grandval P., Lafont D., Boullanger P., De Caro A., Leboeuf B., Verger R., Carrière F. (2004) Biochemistry 43, 10138–10148 [DOI] [PubMed] [Google Scholar]
- 9. Cao H., Hegele R. A. (2003) J. Hum. Genet. 48, 443–446 [DOI] [PubMed] [Google Scholar]
- 10. Jennens M. L., Lowe M. E. (1995) J. Lipid Res. 36, 1029–1036 [PubMed] [Google Scholar]
- 11. Andersson Y., Sävman K., Bläckberg L., Hernell O. (2007) Acta Paediatr. 96, 1445–1449 [DOI] [PubMed] [Google Scholar]
- 12. Fredrikzon B., Hernell O., Bläckberg L., Olivecrona T. (1978) Pediatr. Res. 12, 1048–1052 [DOI] [PubMed] [Google Scholar]
- 13. Wang C. S., Martindale M. E., King M. M., Tang J. (1989) Am. J. Clin. Nutr. 49, 457–463 [DOI] [PubMed] [Google Scholar]
- 14. Lowe M. E. (1997) Methods Enzymol. 284, 285–297 [DOI] [PubMed] [Google Scholar]
- 15. Yang Y., Lowe M. E. (1998) Protein Expr. Purif. 13, 36–40 [DOI] [PubMed] [Google Scholar]
- 16. Sahasrabudhe A. V., Solapure S. M., Khurana R., Suryanarayan V., Ravishankar S., deSousa S. M., Das G. (1998) Protein Expr. Purif. 14, 425–433 [DOI] [PubMed] [Google Scholar]
- 17. Cordle R. A., Lowe M. E. (1998) J. Lipid Res. 39, 1759–1767 [PubMed] [Google Scholar]
- 18. Yang Y., Lowe M. E. (2000) J. Lipid Res. 41, 48–57 [PubMed] [Google Scholar]
- 19. Crandall W. V., Lowe M. E. (2001) J. Biol. Chem. 276, 12505–12512 [DOI] [PubMed] [Google Scholar]
- 20. Hansson L., Bläckberg L., Edlund M., Lundberg L., Strömqvist M., Hernell O. (1993) J. Biol. Chem. 268, 26692–26698 [PubMed] [Google Scholar]
- 21. Carriere F., Barrowman J. A., Verger R., Laugier R. (1993) Gastroenterology 105, 876–888 [DOI] [PubMed] [Google Scholar]
- 22. Borgström B. (1980) Gastroenterology 78, 954–962 [PubMed] [Google Scholar]
- 23. Verger R. (1980) Methods Enzymol. 64, 340–392 [DOI] [PubMed] [Google Scholar]
- 24. Verger R. (1984) in Lipases (Borgstrom B., Brockman H. L. eds) 1st Ed., pp. 84–150, Elsevier Science Publishers B.V., Amsterdam [Google Scholar]
- 25. Brockman H. L., Law J. H., Kézdy F. J. (1973) J. Biol. Chem. 248, 4965–4970 [PubMed] [Google Scholar]
- 26. Larsson A., Erlanson-Albertsson C. (1986) Biochim. Biophys. Acta 876, 543–550 [DOI] [PubMed] [Google Scholar]
- 27. Wieloch T., Borgström B., Piéroni G., Pattus F., Verger R. (1982) J. Biol. Chem. 257, 11523–11528 [PubMed] [Google Scholar]
- 28. Freie A. B., Ferrato F., Carrière F., Lowe M. E. (2006) J. Biol. Chem. 281, 7793–7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patton J. S., Donnér J., Borgström B. (1978) Biochim. Biophys. Acta 529, 67–78 [DOI] [PubMed] [Google Scholar]
- 30. Hermoso J., Pignol D., Penel S., Roth M., Chapus C., Fontecilla-Camps J. C. (1997) EMBO J. 16, 5531–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pignol D., Ayvazian L., Kerfelec B., Timmins P., Crenon I., Hermoso J., Fontecilla-Camps J. C., Chapus C. (2000) J. Biol. Chem. 275, 4220–4224 [DOI] [PubMed] [Google Scholar]
- 32. van Tilbeurgh H., Egloff M. P., Martinez C., Rugani N., Verger R., Cambillau C. (1993) Nature 362, 814–820 [DOI] [PubMed] [Google Scholar]
- 33. Eydoux C., Spinelli S., Davis T. L., Walker J. R., Seitova A., Dhe-Paganon S., De Caro A., Cambillau C., Carrière F. (2008) Biochemistry 47, 9553–9564 [DOI] [PubMed] [Google Scholar]
- 34. van Tilbeurgh H., Bezzine S., Cambillau C., Verger R., Carrière F. (1999) Biochim. Biophys. Acta 1441, 173–184 [DOI] [PubMed] [Google Scholar]
- 35. Chahinian H., Bezzine S., Ferrato F., Ivanova M. G., Perez B., Lowe M. E., Carrière F. (2002) Biochemistry 41, 13725–13735 [DOI] [PubMed] [Google Scholar]
- 36. D'Agostino D., Cordle R. A., Kullman J., Erlanson-Albertsson C., Muglia L. J., Lowe M. E. (2002) J. Biol. Chem. 277, 7170–7177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andersson L., Carriére F., Lowe M. E., Nilsson A., Verger R. (1996) Biochim. Biophys. Acta 1302, 236–240 [DOI] [PubMed] [Google Scholar]
- 38. Reboul E., Berton A., Moussa M., Kreuzer C., Crenon I., Borel P. (2006) Biochim. Biophys. Acta 1761, 4–10 [DOI] [PubMed] [Google Scholar]
- 39. Fomon S. J., Ziegler E. E., Thomas L. N., Jensen R. L., Filer L. J., Jr. (1970) Am. J. Clin. Nutr. 23, 1299–1313 [DOI] [PubMed] [Google Scholar]
- 40. Hancock A. M., Witonsky D. B., Ehler E., Alkorta-Aranburu G., Beall C., Gebremedhin A., Sukernik R., Utermann G., Pritchard J., Coop G., Di Rienzo A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, Suppl. 2, 8924–8930 [DOI] [PMC free article] [PubMed] [Google Scholar]