Abstract
The inhibitor of apoptosis protein cIAP1 (cellular inhibitor of apoptosis protein-1) is a potent regulator of the tumor necrosis factor (TNF) receptor family and NF-κB signaling pathways in the cytoplasm. However, in some primary cells and tumor cell lines, cIAP1 is expressed in the nucleus, and its nuclear function remains poorly understood. Here, we show that the N-terminal part of cIAP1 directly interacts with the DNA binding domain of the E2F1 transcription factor. cIAP1 dramatically increases the transcriptional activity of E2F1 on synthetic and CCNE promoters. This function is not conserved for cIAP2 and XIAP, which are cytoplasmic proteins. Chromatin immunoprecipitation experiments demonstrate that cIAP1 is recruited on E2F binding sites of the CCNE and CCNA promoters in a cell cycle- and differentiation-dependent manner. cIAP1 silencing inhibits E2F1 DNA binding and E2F1-mediated transcriptional activation of the CCNE gene. In cells that express a nuclear cIAP1 such as HeLa, THP1 cells and primary human mammary epithelial cells, down-regulation of cIAP1 inhibits cyclin E and A expression and cell proliferation. We conclude that one of the functions of cIAP1 when localized in the nucleus is to regulate E2F1 transcriptional activity.
Keywords: Apoptosis, Cell Cycle, Cell Differentiation, Cyclins, E2F Transcription Factor, E3 Ubiquitin Ligase, Transcription Coactivators, IAP (Inhibitor of Apoptosis Proteins), Cell Proliferation
Introduction
Cellular inhibitor of apoptosis protein-1 (cIAP1,3 also named BIRC2, HIAP2) belongs to the IAP family of proteins that all contain at least one copy of the conserved BIR (baculoviral IAP repeat) domain (1, 2). cIAP1 also contains a central CARD (caspase-activating recruitment domain) and a C-terminal RING (really interesting new gene) domain, the latter conferring to the protein an E3 ubiquitin ligase activity. cIAP1 is an important regulator of the signaling pathways activated by the tumor necrosis factor (TNF) receptor superfamily members and modulates nuclear factor-κB (NF-κB) activation (3–6). cIAP1 has the capacity to bind and ubiquitylate several signaling intermediates involved in these pathways, including TRAF2 (TNF receptor-associated factor 2) (1, 5–8), NIK (NF-κB-inducing kinase) (9), ASK1 (apoptosis signal-regulating kinase 1) (10), NEMO (NF-κB essential modulator) (11), and RIP1 (12, 13).
A range of evidence suggests that cIAP1 plays a role in mammalian cancers. cIAP1-encoding Birc2 is a target gene within a chromosome 11q21 amplicon found in cervical, oral, head and neck, lung, esophageal, and hepato-cellular carcinomas (14–18). Independently of the presence of this amplicon, cIAP1 is highly expressed in cancer samples from several origins (18–22). The oncogenic properties of cIAP1 have been demonstrated in p53−/−, c-Myc-expressing mouse hepatocarcinoma cells (18), in p53+/− mouse osteosarcoma (23), and in p53−/− mouse mammary carcinoma (24). We (6, 25, 26) and others (27–29) have shown that cIAP1 was expressed mainly in the nucleus of undifferentiated, proliferating cells and was excluded upon cell differentiation (25) or apoptosis induction (27). cIAP1 is also detected in the nucleus of primary human tumor cells (15, 16, 28, 30). In head and neck squamous cell carcinomas (HNSCCs), the nuclear expression of cIAP1 has been associated with lymph node metastasis and advanced disease stages (16, 30), suggesting that the nuclear function of cIAP1 could account for its oncogenic properties. The nuclear function of cIAP1 remains misunderstood. In the present study, we demonstrate that cIAP1 directly interacts with the transcription factor E2F1 and stimulates its transcriptional activity when recruited on CCNE and CCNA gene promoters. These genes are important regulators of cell cycle progression and cell proliferation (31, 32), promoting the transition from G1 to S phase of the cell cycle (31, 32). E2F1 transcriptional activity is regulated by its dimerization with the co-activator DP1 (Dimerization Partner 1), which favors DNA binding and the recruitment of a number of other proteins (33, 34) that behave as activators or repressors (33–35). One of the functions of cIAP1 in the nucleus appears to be part of the molecular machinery that regulates the transcriptional activity of E2F1 on CCNE and CCNA promoters.
EXPERIMENTAL PROCEDURES
Cell Culture, Chemicals, and Treatments
Human mammary epithelial cells (HMEC) were purchased from Invitrogen (Cergy Pontoise, France) and grown into HUMEC ready medium (Invitrogen). Mouse embryonic fibroblasts (MEF) were provided by J. Silke (Melbourne, Australia). THP1, HT-29, U-2 OS, and CaSki cell lines were grown into RPMI 1640 and MEF and HeLa cells in DMEM medium (Lonza, Verviers, Belgium) supplemented with 10% fetal bovine serum (Lonza). Cells were synchronized using 2 mm thymidine (Sigma-Aldrich) double block. Human CD34+ progenitor cells were prepared from human umbilical cord blood (Etablissement Français du Sang) as previously described (36), cultured over 7 days in StemSpanTM H3000, supplemented with 100 ng/ml rhFlt-3 ligand, 100 ng/ml rhSCF, 20 ng/ml rhIL-3, and 20 ng/ml rhIL-6 (StemCell Technologies, Vancouver, BC, Canada), and then differentiated for 2 weeks into CD14+ monocytic cells by exposure to 25 ng/ml M-CSF (StemCell Technologies) before ChIP experiments. The pan-caspase inhibitor zVAD-fmk was from Sigma-Aldrich.
Transfections, Plasmid Constructs, and siRNA
THP-1 cells were nucleoporated using the AMAXA nucleofector kit V (Amaxa Biosystems, Lonza). Stable THP1 clones expressing cIAP1 antisense were enriched by a 10-day geneticin selection (0.5 mg/ml). Cells were transfected using JET PEI (Polyplus transfection, Ozyme, Saint-Quentin-en-Yvelines, France), Lipofectamine 2000 (Invitrogen) or Interferin (PolyPlus transfection, Ozyme) transfection reagent. DNA constructs used were pcDNA, pcDNA-cIAP1, pcDNA-cIAP1 in antisense orientation (AS), pcDNA-cIAP2, pcDNA-XIAP, pEGFP, pEGFP-cIAP1, pCI, pCI-cIAP1, pCI-cIAP1-H588A, pGL3, pGL-5xE2F-BS, pGL-human CCNA promoter, pGL-human CCNE promoter wt and mutated in E2F binding sites, pCMV-E2F1, pcDNA-E2F2 and E2F-3a. pGEX-based constructs (cIAP1, cIAP1-BIR1–3 (amino acid 1–483), cIAP1-CARD-RING (amino acid 452–618), E2F1, E2F1 amino acid 284–437, E2F1 amino acid 89–191, E2F1 amino acid 41–108, E2F1 amino acid 41–127) were obtained by cloning PCR-generated DNA sequence into pGEX 4T1 (GE Healthcare, Chalfont St. Giles, UK). FLAG-cIAP1 constructs were generated by cloning cIAP1-full length, cIAP1-BIR1–3, cIAP1-CARD-RING in a FLAG-pCR3 vector (Invitrogen). The cIAP1 L47A mutant was generated by site-directed mutagenesis. RNA oligonucleotides used were cIAP1, E2F1, and control siRNA sequence designed and purchased from Qiagen.
Cell Extracts, Immunoprecipitation, and Western Blot Analysis
Cell lysates and immunoblot analysis were performed as described (6). Nuclear- and cytoplasm-enriched fractions were obtained as described (25).
Primary antibodies used were rabbit anti-human cIAP1 (R&D Systems, Lille, France), GFP (BD Biosciences, Le Pont de Claix, France), TRAF2 (Stressgen), PARP, E2F1, E2F2 and E2F3 (Santa Cruz Biotechnology, Santa Cruz, CA), goat anti-human cIAP1, XIAP (R&D systems), GST (Rockland, PA), mouse anti-human cIAP2 (R&D systems), cyclin A, cyclin E (BD Biosciences), cyclin B, Rb (Cell signaling Technology, Danvers, MA), and HSC70 (Santa Cruz Biotechnology) monoclonal antibodies. Secondary antibodies included goat HRP-conjugated anti-mouse, anti-rabbit, anti-rat, or rabbit anti-goat immunoglobulins (Jackson ImmunoResearch Laboratories, Bar Harbor, ME).
For immunoprecipitation, cells were lysed in a buffer containing 50 mm Tris-HCl, pH 7.4; 150 mm NaCl; 20 mm EDTA; 0.5% Nonidet P-40; 1 mm DTT, and protease inhibitors and incubated overnight at 4 °C under shaking in the presence of anti-E2F1 Ab (C-20, Santa Cruz Biotechnology) or anti-cIAP1 Ab (R&D Systems) coupled to Sepharose G-protein beads (Amersham Biosciences, GE Healthcare) or ANTI-FLAG® M2 Affinity Agarose Gel (Sigma-Aldrich). Beads were washed and resuspended in Laemmli 1× buffer before immunoblot analysis. The co-precipitation experiments were performed in HeLa cells transfected with FLAG constructs and pCMV-E2F1.
Antibody Array
HeLa cells were transfected with pEGFP-NES*-cIAP1 (25). The cell lysate was deposited onto a Cell Cycle antibodyArrayTM (Hypromatrix, Worcester, MA) containing 60 specific antibodies against cell cycle-related proteins following the manufacturer's instructions and immunoblotted with anti-GFP biotin (USBiological, Swampscott, MA) and biotin-HRP (Invitrogen) antibodies.
GST-Pull-down Assay
GST fusion proteins were produced in Escherichia coli, immobilized on glutathione-Sepharose (Amersham Biosciences), and incubated with either HeLa cell lysates or in vitro translated [35S]methionine-labeled cIAP1 protein or recombinant human E2F1 protein (Protein One, Bethesda, MD). The bound proteins to GST-cIAP1 constructs were revealed by immunoblotting. The bound proteins to GST-E2F1 constructs were revealed by immunoblotting or by 10% SDS-PAGE and autoradiography.
Gene Reporter Assay
Cells were transfected with 500 ng of pGL-based constructs, 50 ng of pCMV β-gal reporter vector, and 500 ng of IAP constructs, and/or 100 ng of E2F constructs. Cells were harvested 48 h after and analyzed for luciferase activity using the luciferase assay reagent (Promega, Madison, WI) and a luminometer (Lumat LB9507, Berthold, Thoiry, France). Results were normalized to the β-galactosidase activity using the β-Galactosidase Enzyme Assay System kit (Promega).
RNA Purification, Reverse Transcription, PCR, and Real-time PCR (qPCR)
Total RNA was isolated using the Nucleospin RNA II kit (Macherey-Nagel, Hoerdt, France) or TRIzol Reagent (Invitrogen), reverse transcribed by MMLV reverse transcriptase with oligo(dT) primers (Promega). Specific cDNAs were amplified on an iCYCLER thermocycler (Bio-Rad) or a 7500 FAST thermocycler (Applied Biosystems, Foster City, CA) using the SyBr Green detection protocol. Results were compared with the cyclophilin or HPRT DNA amplification. Primers used for the specific amplification are available upon request. For RT-PCR, the one-step RT-PCR kit (Qiagen) and the iCYCLER thermocycler (Bio-Rad) were used.
Chromatin Immunoprecipitation Assay
Cells were formaldehyde cross-linked, and DNA was isolated and sonicated. Samples were immunoprecipitated using rabbit or goat anti-human cIAP1 (R&D systems), rabbit anti-human E2F1 (C-20, Santa Cruz Biotechnology), anti-AcH3, or DiMeH3K9 pAbs (Upstate, Millipore, Saint-Quentin-en-Yvelines, France), washed, and reverse cross-linked using the chromatin immunoprecipitation kit EZ ChIP from Upstate (Millipore). For the sequential ChIP experiment, samples were first immunoprecipitated with anti-E2F1 pAb. The antibody-bound protein/DNA complexes were eluted using elution buffer (1% SDS, 0.1 m NaHCO3), and a second ChIP was performed using the anti-cIAP1 pAb. PCR and real-time PCR were performed as described above using primers flanking the E2F binding site in CCNE and CCNA promoters.
Cell Cycle Analysis
Cells were incubated for 30 min in the presence of 3 mm BrdU (Sigma Aldrich). Cells were fixed at 4 °C and resuspended in 30 mm HCl and 0.5 mg/ml pepsin for 30 min, then in 2 m HCl over 15 min, stained with primary anti-BrdU Ab, and with secondary anti-mouse Alexa Fluor 488 Ab (Molecular Probes, Invitrogen) and propidium iodide (PI, 10 μg/μl). Cell cycle repartition was assessed by LSRII flow cytometry using FlowJo® Softwares (Tree Star, Inc. Ashland, OR).
Proliferation Analysis
We used CellTraceTM CFSE Cell Proliferation kit (for HeLa Cells) or Click-iTTM EdU Cell Proliferation Assay (for HMEC and MEF) (Molecular Probes, Invitrogen) to measure cell proliferation using a LSRII flow cytometer (BD Biosciences) according to the manufacturer's instructions. The index of proliferation was measured using ModFIT Software (Verity Software House Topsham, ME). THP1 cells were plated at the same density and counted each day.
Statistical Analysis
Student's t test was used for statistical analysis.
RESULTS
cIAP1 Interacts with E2F1 through Its BIR Domains
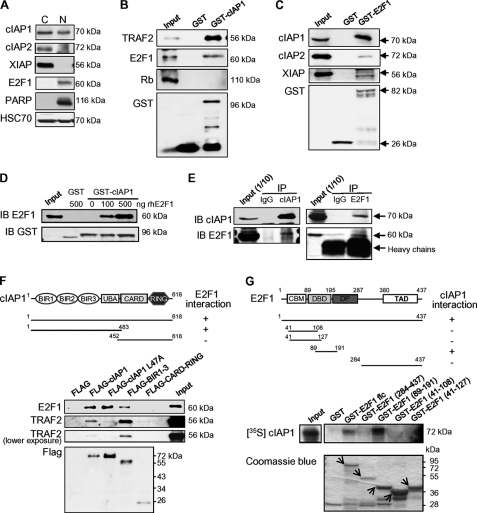
cIAP1 has been identified in the nucleus of human normal cells, e.g. hematopoietic stem cells (25), as well as cancer cells (15, 16, 28, 30). It is also expressed in the nucleus of human colon carcinoma HT-29, human leukemia monocytic THP1 (25, 26), and human epithelial cervix carcinoma HeLa (27) (Fig. 1A) cell lines. To identify nuclear partners of cIAP1, cells were transfected with a GFP-tagged-cIAP1 in which the NES had been mutated to force the nuclear overexpression of the protein (25), and cell lysates were incubated on an antibody array targeting 60 cell cycle-related proteins. Among the 18 detected positive hits, 7 were checked using a higher stringency approach in which GST-cIAP1 is incubated with a cell lysate from untransfected HeLa cells and interactions revealed by immunoblot analysis (Fig. 1B, supplemental Fig. S1A). TRAF2 was used as a positive control (Fig. 1B). Three potential partners were confirmed, namely TTK kinase, Rad52 (supplemental Fig. S1A), and E2F1 transcription factor (Fig. 1B). The GST pull-down experiment also confirmed a negative result of the initial screen, i.e. cIAP1 did not interact with the E2F1 repressor retinoblastoma protein (Rb) (Fig. 1B). The reversed GST-pull-down assay demonstrated a binding of endogenous cIAP1 with GST-E2F1 (Fig. 1C). We also detected a very weak binding of cIAP2 and XIAP on GST-E2F1 (Fig. 1C). GST-cIAP1 can interact with purified human E2F1 protein, indicating a direct interaction (Fig. 1D). cIAP1 was co-expressed with E2F1 in the HeLa cell nucleus-enriched fraction whereas cIAP2 and XIAP were detected in the cytoplasm (Fig. 1A). The in vivo interaction of E2F1 with cIAP1 was confirmed by co-immunoprecipitation (Fig. 1E).
FIGURE 1.
cIAP1 interacts with the transcription factor E2F1. A, immunoblot analysis of cIAP1, cIAP2, XIAP, and E2F1 in cytoplasm (C) and nuclear (N)-enriched fractions. PARP is used to check the nuclear fraction. HSC70: loading control. B–D, GST pull-down analysis of the interaction of GST-cIAP1 (B, D) or GST-E2F1 (C) with indicated proteins from HeLa cell lysate (B, C) or with purified human E2F1 (D). E, endogenous E2F1 (right panel) or cIAP1 (left panel) were immunoprecipitated with anti-E2F1, anti-cIAP1 or irrelevant rabbit Ig (IgG) in HeLa cells before immunoblot analysis of cIAP1 and E2F1. The cIAP1 immunoprecipitation (left) was performed in a nuclear-enriched fraction. F, immunoprecipitation analysis of the interaction of wild type or deletion mutants of cIAP1 with E2F1 and TRAF2. FLAG-conjugated proteins and E2F1 were expressed in HeLa cells and co-immunoprecipitated using anti-FLAG M2-agarose beads, then revealed by immunoblotting using an anti-E2F1, anti-TRAF2, or anti-FLAG specific antibody. A schematic representation of cIAP1 protein structure and deletion constructs used is shown (upper panel). G, GST-pull down analysis of the interaction of in vitro translated [35S]methionine-labeled cIAP1 with indicated GST-E2F1 deletion constructs. The interactions were revealed by autoradiography. E2F1 mutants (arrows) were detected after Coomassie Blue staining of the gel (lower panel). A schematic representation of E2F1 domains and deletion constructs used is shown (upper panel). CBM: cyclin A binding motif; DBD: DNA binding domain; DP: dimerization domain; TAD: C-terminal transactivation domain. Representative experiments are shown.
We then mapped the protein domains required for this interaction by using a co-immunoprecipitation experiment (Fig. 1F) and GST-pull down assay (Fig. 1G and supplemental Fig. S1, B and C). As TRAF2, E2F1 interacted with cIAP1 full-length and with the N-terminal part of the protein that contained BIR 1–3 domains but did not interact with the C-terminal part of cIAP1 that included the CARD and the RING domains (Fig. 1F and supplemental Fig. S1, B and C). Mutation L47A within the BIR1 domain that abolished the cIAP1-TRAF2 interaction (37) did not modify the cIAP1-E2F1 interaction (Fig. 1F). We detected an in vivo interaction of cIAP1 with TRAF2 in the nucleus- and in the cytoplasm-enriched fractions by immunoprecipitation (supplemental Fig. S1D). An in vitro competition experiment indicated that E2F1 and TRAF2 could compete for cIAP1 binding (supplemental Fig. S1E). GST-pull down assay performed by using in vitro translated 35S-labeled full-length cIAP1 and GST-E2F1 or GST-E2F1 deletion constructs demonstrated that cIAP1 interacted with the E2F1 amino acid sequence 89–191 that overlaps its DNA binding domain (DBD) (Fig. 1G).
cIAP1 Stimulates E2F1 Transcriptional Activity
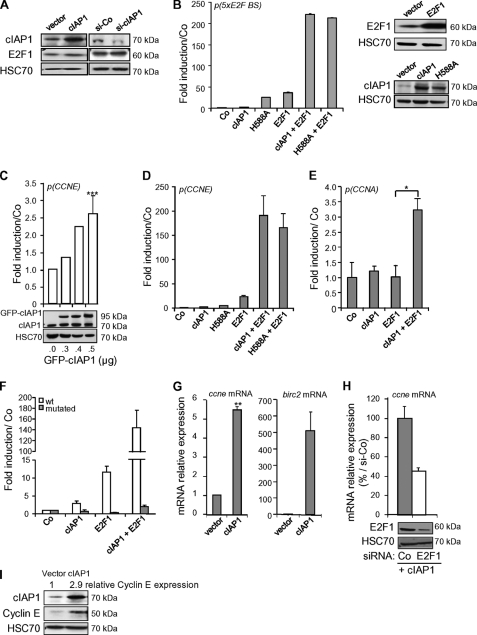
Overexpression or silencing of cIAP1 did not significantly alter the expression level of E2F1 (Fig. 2A). In a luciferase gene reporter assay using a construct containing 5xE2F binding sites upstream of the LUCIFERASE gene, overexpressed cIAP1 dramatically enhanced E2F1 transcriptional activity, which was still observed when a mutation was introduced within the RING domain (H588A) of cIAP1 to abrogate its E3 ubiquitin ligase activity (Fig. 2B). We then analyzed the influence of cIAP1 on CCNE and CCNA promoters, which are two well-identified E2F1 target genes. cIAP1 significantly stimulated CCNE gene expression in a dose-dependent manner on its own (Fig. 2C) and substantially enhanced E2F1 activity on CCNE (Fig. 2D) and to a lower extent CCNA (Fig. 2E) gene promoters. Again, mutation within the RING (H588A) domain or within the BIR1 domain (L47A) that inhibits cIAP1-TRAF2 interaction (Fig. 1F) did not inhibit the capacity of cIAP1 to stimulate E2F1 activity (Fig. 2D and supplemental Fig. S2A). Mutation of E2F binding sites I, II, and III (38) in the CCNE promoter of the reporter construct abolished the transcriptional activity of both cIAP1 and E2F1 and prevented the synergistic effect of cIAP1 and E2F1 (Fig. 2F), suggesting that cIAP1 transcriptional effect depended on intact E2F binding sites. These results were confirmed in HT-29 human colon carcinoma cells that harbor a mutated p53 and in U-2 OS osteosarcoma cells that express wild-type p53 (supplemental Fig. S2, B and C). The capacity of cIAP1 to stimulate cyclin E expression was confirmed by RT-qPCR (Fig. 2G, supplemental Fig. S3) and immunoblotting (Fig. 2I) in HeLa (Fig. 2G) and HT-29 (Fig. 2I, supplemental Fig. S3) and silencing of E2F1 decreased the capacity of cIAP1 to stimulate CCNE mRNA expression (Fig. 2H).
FIGURE 2.
cIAP1 stimulates E2F1 transcriptional activity. A, immunoblot analysis of cIAP1 and E2F1 in HeLa cells transfected with cIAP1 construct or cIAP1 siRNA. HSC70: loading control. B–F, gene luciferase experiments performed in HeLa cells transfected with indicated promoter-luciferase reporter plasmids, along with control (Co) or E2F1-encoding vector and/or 500 ng or indicated amount (C) of empty (Co), cIAP1 or H588A (B, D) encoding constructs. p(5xE2F BS): synthetic promoter containing 5xE2F binding sites (B); p(CCNE) & wt: wt CCNE promoter (B-D, F); p(CCNA): CCNA promoter (E); mutated: E2F binding site-mutated CCNE promoter. Luciferase activity was normalized to β-galactosidase activity and expressed as fold induction of promoter stimulated by empty vector alone. Mean ± S.D. of one representative experiment. Statistical analysis performed using Student's t test. ***: p = 0.0003, n = 10 (C); *: p = 0.013, n = 3 (E). cIAP1 and E2F1 overexpression were checked by immunoblot analysis (B right panels, C lower panel). G and H, quantitative RT-PCR analysis of CCNE or birc2 mRNA in HeLa cells transfected with empty or cIAP1 encoding vectors and/or E2F1-siRNA (si-E2F1). Results are normalized to HPRT mRNA and expressed relative to empty vector (G) or expressed as % of CCNE mRNA induced by cIAP1 in the presence of control siRNA (H). Mean ± S.D. of one representative experiment. Statistically significant differences (**, p = 0.007, n = 3, Student's t test) (G). I, immunoblot analysis of cIAP1 and cyclin E in HT-29 cells transfected with cIAP1 construct. The relative expression of cyclin E in cIAP1-transfected cells compared with empty vector as evaluated after quantification using ImageJ software was shown on the blot. HSC70: loading control.
We then compared the capacity of cIAP1, cIAP2, and XIAP to stimulate E2F1 activity. Overexpression of cIAP1 decreased the expression of cIAP2 and XIAP (Fig. 3A), accordingly to the capacity of cIAP1 to ubiquitinylate and stimulate the degradation of its close relatives (39). cIAP2 and XIAP were much less efficient than cIAP1 for stimulating E2F1 activity on synthetic and CCNE promoters (Fig. 3, A and B). cIAP1 could also stimulate E2F2 and E2F3a, other so-called stimulatory members of the E2F family, although more weakly than E2F1 (Fig. 3C).
FIGURE 3.
Specific activity of cIAP1 on E2F1. Gene luciferase experiments performed in HeLa cells transfected with a synthetic promoter containing 5xE2F binding sites (p(5xE2F BS))(A, C) or CCNE promoter-luciferase reporter plasmid (p(CCNE)) (B) along with E2F1 (A-C), E2F2 or E2F3a constructs (C), and/or 500 ng of empty vector or cIAP1 (A-C), cIAP2- or XIAP (A, B)-encoding constructs. Luciferase activity was normalized to β-galactosidase activity and expressed as fold induction of promoter stimulated by empty vector alone. Mean ± S.D. of one representative experiment is shown. The expression of indicated constructs was checked by immunoblot analysis (lower panels). HSC70: loading control. One representative experiment is shown.
cIAP1 Is Recruited on the CCNE Promoter
To explore whether cIAP1 could bind to the CCNE promoter, we performed chromatin immunoprecipitation (ChIP) using primers that flanked the E2F binding site of the promoter. Primers located inside the CCNE gene sequence are used as a negative control. As expected, E2F1 is recruited on the E2F binding site of the CCNE promoter (Fig. 4, A and B). cIAP1 was detected on the E2F binding site of the CCNE promoter in HeLa, U-2 OS, HT-29, THP1 cell lines and in primary human mammary epithelial cells (HMEC) (Fig. 4, A and C) whereas neither cIAP1 nor E2F1 bound the DNA sequence of the CCNE gene (Fig. 4, B and C). The same approach was used to demonstrate that cIAP1 could also bind the E2F binding site of the CCNA promoter in the cell line and in primary human CD34+ hematopoietic cells (Fig. 4D). ChIP with E2F1 antibody and a Re-ChIP using cIAP1 antibody demonstrated that cIAP1 and E2F1 were recruited at the same promoter region of the CCNE gene (Fig. 4E). These data suggested that cIAP1 was a component of the E2F1 transcriptional complex.
FIGURE 4.
cIAP1 is recruited on E2F binding site of CCN promoters. Chromatin immunoprecipitation experiments performed using an anti-E2F1 (A, B, E), anti-cIAP1 (A, C–E), or an irrelevant antibody (Ig) in HeLa (A–E), U-2 OS, HT-29, THP1, HMEC (C), and CD34+ primary myeloid cells (D). The genomic DNA region encompassing one E2F binding site of the CCNE (p(CCNE) (B, C, E), CCNA (p(CCNA) promoters (D) or a control sequence localized in CCNE gene (B, C left panels) were amplified by PCR (A) or qPCR (B–E). E, ChIP and re-ChIP experiments performed on HeLa cells. The sample was first immunoprecipitated with E2F1 or irrelevant antibody (Ig). The protein-DNA complex was eluted, and a second ChIP was performed using cIAP1 or irrelevant Ab. Results were normalized to input and expressed as relative recruitment compared with irrelevant antibody. Mean ± S.D. of one representative experiment.
cIAP1 Recruitment on Cyclin Gene Promoters Is Cell Cycle-regulated
To determine whether cIAP1-E2F1 interaction could be cell cycle-regulated, HeLa cells were synchronized in early S phase by a thymidine double block (supplemental Fig. S4). The G2/M phase reached 8 h after block release was characterized by a decrease in cyclin E and an increase in cyclin B expression (Fig. 5A). The expression of cIAP1 (Fig. 5A), its subcellular localization (not shown), and its interaction with E2F1 (Fig. 5B) did not change significantly along the cell cycle progression. As observed for E2F1 (Fig. 5C, left panel; supplemental Fig. S5A), cIAP1 was mainly recruited on the CCNE promoter in early S phase (Fig. 5D, left panel; supplemental Fig. S5A). Both cIAP1 and E2F1 were also recruited onto the CCNE promoter in late S phase (Fig. 5, C and D, left panels; supplemental Fig. S5A). The recruitment of E2F1 and cIAP1 onto the CCNA promoter was also cell cycle-regulated and occurred later in S phase (Fig. 5, C and D, right panels; supplemental Fig. S5A). cIAP1 and E2F1 were observed to bind the CCNA promoter in primary human CD34+ hematopoietic cells (Fig. 5E). In accordance with the previously described exclusion of cIAP1 from the nucleus in cells undergoing differentiation (25), this recruitment is decreased when CD34+ cells were induced to differentiate into CD14+ monocytes upon M-CSF exposure (Fig. 5E).
FIGURE 5.
The recruitment of cIAP1 on CCN promoters is cell cycle-regulated. A–D, HeLa cells were synchronized into early S phase by a thymidine double block and analyzed 0, 2, 4, 8, and 10 h after block release (see also supplemental Figs. S4 and S5). A, immunoblot analysis of cIAP1, cyclin E and B and E2F1. HSC70: loading control. B, endogenous E2F1 was immunoprecipitated with anti-E2F1 or irrelevant rabbit Ig (IgG) before immunoblot analysis of cIAP1 and E2F1. C and D, ChIP experiments of E2F1 (C) or cIAP1 (D) on CCNE (p(CCNE)) (C and D left panels) or CCNA (p(CCNA)) (C and D, right panels) promoter. E, ChIP of E2F1 and cIAP1 on CCNA promoter performed in undifferentiated (CD34+) and M-CSF-differentiated (CD34−/CD14+) myeloid cells. The genomic DNA region encompassing the E2F-binding site of CCNE or CCNA promoter was amplified by qPCR. Results were normalized to input and expressed as relative recruitment to irrelevant antibody (dotted line). Mean ± S.D. of one representative experiment.
cIAP1 Is Required for E2F1 Binding to the CCNE Promoter
Silencing of cIAP1 in HeLa cells using siRNA (Fig. 6, A–D) inhibited the capacity of E2F1 to stimulate the transcriptional expression of CCNE (Fig. 6A, left panel), and completely abolished the recruitment of cIAP1 and E2F1 on the CCNE promoter (Fig. 6, C and D, left panel). Interestingly, silencing of E2F1 (Fig. 6B) also inhibited the recruitment of cIAP1 (Fig. 6D, right panel). We confirmed these results in the CaSki human epidermoid cervical carcinoma cell line expressing the amplicon 11q21, which contains the birc2 gene (16). As expected, we observed a very high expression of cIAP1 in both nucleus and cytoplasm-enriched fraction compared with HeLa cells (Fig. 6E). cIAP1 was also recruited on the cyclin E promoter, and silencing of cIAP1 prevented the recruitment of E2F1 on the CCNE promoter (Fig. 6F, supplemental Fig. S5B). Moreover, cIAP1 siRNA decreased the acetylation of histone H3 on the CCNE promoter that accompanied the transcriptional activation and increased the dimethylation of histone H3 on lysine 9 (H3K9), which is a feature of transcriptional repression (Fig. 6G) (40).
FIGURE 6.
Contribution of cIAP1 in the transcriptional activity of E2F1. A, quantitative RT-PCR analysis of ccne (right panel), e2f1 (medium panel), and birc2 (left panel) mRNAs in HeLa cells transfected with empty or E2F1-encoding vector and control (Co) or cIAP1-targeted siRNA. Results were normalized to cyclophilin mRNA and expressed relative to empty vector. Mean ± S.D. of one representative experiment. Statistically significant differences (*, p < 0.05, n = 3, Student's t test). B, efficacy of cIAP1 or E2F1-targeted siRNAs was checked by an immunoblot analysis. HSC70: loading control. C, D and F, G, chromatin immunoprecipitation experiments performed using an anti-E2F1 or an anti-cIAP1 (C, D, F), an anti-acetyl histone H3 (Ac H3) or an anti-dimethyl histone H3 on lysine 9 (diMe H3K9) (G) or an irrelevant antibody (Ig) in HeLa (C, D, G) or CaSki (F) cells. The genomic DNA region encompassing the E2F-binding site of the CCNE promoter was amplified by PCR (C) or qPCR (D, F, G). Results are normalized to input and expressed as relative recruitment compared with irrelevant antibody. Mean ± S.D. of one representative experiment (D, F, G) is shown. MW: molecular weight. The efficacy of cIAP1-targeted siRNAs was checked by an immunoblot analysis (F, upper panel). HSC70: loading control. E, immunoblot analysis of cIAP1, XIAP, E2F1, cyclin E, and cyclin A in the cytoplasm (C)- and nucleus (N)-enriched fractions of HeLa and CaSki cells. HSC70: loading control.
cIAP1 Modulates Cyclin Expression and Cell Proliferation
We analyzed the influence of cIAP1 on cyclin expression. siRNA-mediated down-regulation of cIAP1 decreased cyclin E and A mRNA (Fig. 7, A and B) and protein (Fig. 7C) expression in HeLa cells (Fig. 7, A–C) and in primary human mammary epithelial cells (Fig. 7C). A similar effect was observed by down-regulating cIAP1 with an antisense (AS) oligonucleotide construct in THP1 cells (Fig. 7C). Silencing of cIAP1 also decreased the cyclin E transcript in CaSki, B16F10 mouse melanoma and in L929 mouse fibroblast cell lines (Fig. 7D). We did not detect such an effect in murine embryonic fibroblast (MEF) (Fig. 7D), and MEF from deleted mice (MEF cIAP1−/−) did not show a decrease in the cyclin E transcript and protein when compared with wild-type MEF (supplemental Fig. S6, A and B). The cell fractionation experiment revealed that, in contrast to HeLa (Fig. 1A), HMEC (Fig. 7E), CaSki (Fig. 6E), B16F10, and L929 cell lines (Fig. 7E) in which cIAP1 is detected in the nuclear-enriched fraction, the expression of cIAP1 is almost restricted to the cytoplasm compartment in MEF (Fig. 7E), especially in the G0/G1 and S phase of the cell cycle, when E2F1 activity is maximal (supplemental Fig. S6C). Transfection of MEF cIAP1−/− with a cIAP1 construct induced cIAP1 expression in both nuclear and cytoplasm compartments (Fig. 7F) and enhanced E2F1 activity (supplemental Fig. S6D) and cyclin E expression (Fig. 7G). The analysis of cell proliferation and cell cycle repartition showed that down-regulation of cIAP1 (Fig. 7C) slowed down the cell proliferation (Fig. 8, A and B), decreased S phase of cell cycle (Fig. 8C) and increased G0/G1 (Fig. 8, C and D in HeLa (Fig. 7C and Fig. 8, A, C, D) and THP1 (Fig. 7C and Fig. 8, B and D) cells. We did not detect any sign of apoptosis (not shown), and the pan-caspase inhibitor z-VAD-fmk did not affect the capacity of cIAP1-siRNA to decrease HeLa cell growth rate (Fig. 8A). Moreover, the effect of cIAP1-siRNA on cell proliferation could be reverted by co-expression of the cIAP1-encoding vector (Fig. 8A). Silencing of cIAP1 (Fig. 7C) also decreased the proliferation rate as evaluated by 24 h EdU incorporation in primary HMEC (Fig. 8E). Moreover, expression of cIAP1 in the nuclear compartment of MEF cIAP1−/− (Fig. 7F) stimulated cell proliferation (Fig. 8F).
FIGURE 7.
Down-regulation of cIAP1 modulates cyclin expression. cIAP1 was down-regulated in HeLa (A–C), HMEC (C) CaSki, B16F10, L929 cells or MEF (D) by using siRNA (A–D) or in THP1 by transfecting an cIAP1 antisense (AS) encoding construct (C). A, RT-PCR analysis of indicated mRNAs. β-2 microglobulin (β-2m) was used as control. B, RT-qPCR analysis of ccne (E) and ccna (A) mRNAs. Results are normalized to cyclophilin mRNA. Statistically significant differences (***, p < 0.005, n = 5, Student's t test). C, immunoblot analysis of indicated proteins in HeLa and HMEC cells transfected with cIAP1 siRNA or in THP1 transfected with an cIAP1 antisense (AS) encoding construct. HSC70: loading control. D, upper panels, RT-qPCR analysis of ccne mRNA in indicated cell lines. Results are normalized to cyclophilin mRNA. Lower panels, immunoblot analysis of cIAP1. HSC70: loading control. E, immunoblot analysis of cIAP1, XIAP, and E2F1 in the cytoplasm (C)- and nucleus (N)-enriched fractions of indicated cell lines. HSC70: loading control. F, immunoblot analysis of cIAP1, XIAP, and E2F1 in the cytoplasm (C)- and nucleus (N)-enriched fractions of MEF cIAP1−/− transfected with cIAP1 construct. HSC70: loading control. G, RT-qPCR analysis of ccne and birc2 mRNA in MEF transfected with the cIAP1 construct. Results are normalized to cyclophilin mRNA.
FIGURE 8.
Down-regulation of cIAP1 modulates cell proliferation and cell cycle repartition. A, flow cytometry analysis of cell proliferation in HeLa cells transfected with control (si-Co) or cIAP1 (si-cIAP1) siRNA and cIAP1-encoding construct in the presence or not of zVAD-fmk 10 μm. Results: mean ± S.D. of at least three independent experiments. Statistically significant differences (**, p < 0.001, n = 5, Student's t test). B, cell proliferation was assessed by cell counting in THP1 clone transfected with empty or cIAP1 antisense (AS)-encoding construct as in Fig. 7C. Results are expressed as mean ± S.D. of at least three independent experiments. C and D, cell cycle analysis in HeLa cell transfected with Co or cIAP1-siRNA. The cell cycle is evaluated in by flow cytometry after BrdU and PI staining of cells. D, percentage of cell in G0/G1 phase of the cell cycle as analyzed by flow cytometry in HeLa cells transfected with control (Co) or cIAP1 siRNA or in THP1 cells transfected with empty vector (V) or an cIAP1 antisense encoding vector (AS). Mean ± S.D. of at least three independent experiments is shown. Statistically significant differences (*, p < 0.05, n = 3, Student's t test). E, flow cytometry analysis of EDu incorporation in HMEC cells transfected with control (si-Co) or cIAP1 (si-cIAP1) siRNA as in Fig. 7C. One representative experiment was shown. F, flow cytometry analysis of EDu incorporation in MEF cIAP1−/− transfected with cIAP1 encoding construct as in Fig. 7F. One representative experiment is shown.
DISCUSSION
In contrast to its closest homologs cIAP2 and XIAP that are localized in the cytoplasm, cIAP1 is expressed in the nucleus of a majority of normal cells until terminal differentiation (25, 27, 29) and in the nucleus of many cancer cells (15, 16, 25–28, 30). The present report identifies a nuclear function for cIAP1. The protein appears to be a co-regulator of E2F1-dependent transcriptional activity.
The E2F family of transcription factors includes 8 members subdivided into subgroups based on structural and functional homologies. These transcription factors are potent cell cycle regulators through their capacity to regulate the expression of genes involved in G1-S phase transition, including CCNE and CCNA genes. E2F proteins promote either activation or repression of gene transcription, depending on the target gene, the pattern of co-regulator partners, and the cellular context (34, 41–43). Molecular partners affect E2F1 transcriptional activity e.g. Rb interaction with E2F1 is associated with transcription inhibition (44, 45). We show that cIAP1 binds a protein sequence of E2F1 (amino-acids 89–191) that overlaps with its DNA binding domain and favors E2F1-mediated transcriptional activation of CCNE and CCNA genes. cIAP1 appears to be important for optimal E2F1 mediated-cyclin E expression. Silencing of cIAP1 inhibits the recruitment of E2F1 on CCNE promoter, suggesting that cIAP1 is required for DNA binding of E2F1. Another molecular partner of E2F whose heterodimerization promotes the transcription factor binding to gene promoters is the co-activator DP1 (32, 46). We did not detect an interaction of cIAP1 with DP1 and DP1 did not affect the capacity of cIAP1 to stimulate E2F1 (not shown). cIAP1 can directly interact with E2F1 in all stage of cell cycle. However, the recruitment of cIAP1 on the cyclin gene promoter is cell cycle-regulated, peaking when the E2F1 activity is maximal. Additional partners or protein modifications may be required for the binding of this heterodimer to DNA at specific phases of the cell cycle.
cIAP1 acts in collaboration with the TRAF2 protein to regulate the TNFR signaling pathway. TRAF2 is also observed to be associated with cIAP1 in the nuclear compartment (supplemental Fig. S1D). However, mutation within the BIR1 domain of cIAP1 that abolishes its binding to TRAF2 does not interfere with the capacity of cIAP1 to interact and stimulate E2F1, suggesting that the transcriptional regulation activity of cIAP1 is independent of TRAF2. Most of the cIAP1 functions identified so far involve its E3 ubiquitin ligase activity (1, 3, 47, 48). The ability of cIAP1 to promote E2F1 transcriptional activity could have been related to the ubiquitination of E2F1 or other E2F1 molecular partners. Actually, we did not detect a ubiquitination of E2F1 by cIAP1 (not shown), and a mutation that suppressed this E3 ligase activity did not abolish the ability of cIAP1 to stimulate the E2F1 transcriptional activity in a luciferase gene reporter assay.
MEFs from animals in which the studied gene has been deleted is a useful tool to check the function of a studied protein. Unfortunately, these cells could not be used to explore the ability of cIAP1 to promote E2F1 transcriptional activation as cIAP1 is localized in the cytoplasm of these differentiated cells (supplemental Fig. S5), in accord with our previous observation that cIAP1 migrates from the nucleus to the cytoplasm in cells undergoing terminal differentiation (25, 26). We show that nuclear cIAP1 is recruited on the CCNA gene promoter in undifferentiated hematopoietic stem cells and cannot be detected on this promoter in monocytes obtained by M-CSF-induced differentiation of these cells. Redistribution of cIAP1 from the nucleus to the cytoplasm could favor the decrease in cell proliferation and cell cycle exit that characterizes terminal cell differentiation. Additional studies will indicate whether this redistribution of cIAP1 could also favor the repression, inhibition, or degradation of E2F1 that is required for normal occurrence of differentiation in several cellular models (44, 49, 50).
The influence of cIAP1 on tumor development has been well demonstrated in several mouse carcinoma models (18, 23, 24). Down-regulation of cIAP1 decreases tumor cell growth in vivo (18, 23, 24) and decreases the proliferation in human breast cancer cell line MCF-7 (51) and mouse primary carcinoma cells (24). Accordingly, we also observe a decrease in cell proliferation after cIAP1 down-regulation, which is accompanied by a decrease in cyclin E and A expression. Interestingly, the murine hepatocellular carcinoma harboring 9qA1 amplicon, which contains cIAP1-, cIAP2-, and yap1-encoding genes were observed to overexpress cyclin E (18). Moreover, a recent report showed that the 9A1 amplicon could be substituted by an inactivation of the E2F-repressor Rb in p53−/− mouse mammary carcinogenesis (24). The ability of cIAP1 to promote E2F1-mediated transcription activity of CCN genes, whose overexpression was associated with poor prognosis in several tumor types (52), could account for the oncogenic properties of the protein.
Supplementary Material
Acknowledgments
We thank Dr. D. Cress, Dr. B. Henglein, Dr. K. Katula, Dr. G. Leone, Dr. J. Lees, Dr. P. Meier, Dr. R. Pestell, Dr. J. Silke, Dr. R. Weinberg, and Dr. K.M. Yao for kindly providing plasmids and the cell line, Dr. N. Droin for efficient help in real-time PCR analysis, Shweta Tyagi for advice with the ChIP experiment, and Lydie Desoche for technical assistance. We are grateful for the use of the cytometry platform (IFR100, Dijon, France). We are grateful to Pascal Meier for critical reading of the manuscript.
This work was supported by grants from the Ligue Nationale Contre le Cancer (équipe labellisée, ES) and the Comité de Côte d'Or, Nièvre, Saône et Loire and Yonne of the Ligue contre le Cancer (to L. D.), the Association pour la Recherche sur le Cancer (ARC, to L. D.), the association “Cent pour Sang la Vie”, the Association Nationale de la Recherche and the National Institute of Cancer, and fellowships from the “Ministère de l'Enseignement Supérieur et de la Recherche” of France (to J. C., A. D., J. B., A. M., B. L. and S. P.) and ARC (to J. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- cIAP1
- cellular inhibitor of apoptosis protein-1
- CARD
- caspase-activating recruitment domain
- MEF
- mouse embryonic fibroblasts
- RING
- really interesting new gene
- BIR
- baculoviral IAP repeat
- UBA
- ubiquitin-associated domain
- DP1
- dimerization partner 1
- HMEC
- human mammary epithelial cells
- Rb
- retinoblastoma protein.
REFERENCES
- 1. Dubrez-Daloz L., Dupoux A., Cartier J. (2008) Cell Cycle 7, 1036–1046 [DOI] [PubMed] [Google Scholar]
- 2. Gyrd-Hansen M., Meier P. (2010) Nat. Rev. Cancer 10, 561–574 [DOI] [PubMed] [Google Scholar]
- 3. Varfolomeev E., Vucic D. (2008) Cell Cycle 7, 1511–1521 [DOI] [PubMed] [Google Scholar]
- 4. Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]
- 5. Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J. (2007) Cell 131, 682–693 [DOI] [PubMed] [Google Scholar]
- 6. Dupoux A., Cartier J., Cathelin S., Filomenko R., Solary E., Dubrez-Daloz L. (2009) Blood 113, 175–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li X., Yang Y., Ashwell J. D. (2002) Nature 416, 345–347 [DOI] [PubMed] [Google Scholar]
- 8. Vince J. E., Chau D., Callus B., Wong W. W., Hawkins C. J., Schneider P., McKinlay M., Benetatos C. A., Condon S. M., Chunduru S. K., Yeoh G., Brink R., Vaux D. L., Silke J. (2008) J. Cell Biol. 182, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zarnegar B. J., Wang Y., Mahoney D. J., Dempsey P. W., Cheung H. H., He J., Shiba T., Yang X., Yeh W. C., Mak T. W., Korneluk R. G., Cheng G. (2008) Nat. Immunol. 9, 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao Y., Conze D. B., Hanover J. A., Ashwell J. D. (2007) J. Biol. Chem. 282, 7777–7782 [DOI] [PubMed] [Google Scholar]
- 11. Tang E. D., Wang C. Y., Xiong Y., Guan K. L. (2003) J. Biol. Chem. 278, 37297–37305 [DOI] [PubMed] [Google Scholar]
- 12. Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. (2008) Mol. Cell 30, 689–700 [DOI] [PubMed] [Google Scholar]
- 13. Park S. M., Yoon J. B., Lee T. H. (2004) FEBS Lett. 566, 151–156 [DOI] [PubMed] [Google Scholar]
- 14. Snijders A. M., Schmidt B. L., Fridlyand J., Dekker N., Pinkel D., Jordan R. C., Albertson D. G. (2005) Oncogene 24, 4232–4242 [DOI] [PubMed] [Google Scholar]
- 15. Imoto I., Yang Z. Q., Pimkhaokham A., Tsuda H., Shimada Y., Imamura M., Ohki M., Inazawa J. (2001) Cancer Res. 61, 6629–6634 [PubMed] [Google Scholar]
- 16. Imoto I., Tsuda H., Hirasawa A., Miura M., Sakamoto M., Hirohashi S., Inazawa J. (2002) Cancer Res. 62, 4860–4866 [PubMed] [Google Scholar]
- 17. Dai Z., Zhu W. G., Morrison C. D., Brena R. M., Smiraglia D. J., Raval A., Wu Y. Z., Rush L. J., Ross P., Molina J. R., Otterson G. A., Plass C. (2003) Hum. Mol. Genet. 12, 791–801 [DOI] [PubMed] [Google Scholar]
- 18. Zender L., Spector M. S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S. T., Luk J. M., Wigler M., Hannon G. J., Mu D., Lucito R., Powers S., Lowe S. W. (2006) Cell 125, 1253–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gordon G. J., Mani M., Mukhopadhyay L., Dong L., Yeap B. Y., Sugarbaker D. J., Bueno R. (2007) J. Pathol. 211, 439–446 [DOI] [PubMed] [Google Scholar]
- 20. Tamm I., Kornblau S. M., Segall H., Krajewski S., Welsh K., Kitada S., Scudiero D. A., Tudor G., Qui Y. H., Monks A., Andreeff M., Reed J. C. (2000) Clin. Cancer Res. 6, 1796–1803 [PubMed] [Google Scholar]
- 21. Kempkensteffen C., Hinz S., Christoph F., Köllermann J., Krause H., Schrader M., Schostak M., Miller K., Weikert S. (2007) Int. J. Cancer 120, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 22. Krajewska M., Krajewski S., Banares S., Huang X., Turner B., Bubendorf L., Kallioniemi O. P., Shabaik A., Vitiello A., Peehl D., Gao G. J., Reed J. C. (2003) Clin. Cancer Res. 9, 4914–4925 [PubMed] [Google Scholar]
- 23. Ma O., Cai W. W., Zender L., Dayaram T., Shen J., Herron A. J., Lowe S. W., Man T. K., Lau C. C., Donehower L. A. (2009) Cancer Res. 69, 2559–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng L., Zhou Z., Flesken-Nikitin A., Toshkov I. A., Wang W., Camps J., Ried T., Nikitin A. Y. (2010) Oncogene 38, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Plenchette S., Cathelin S., Rébé C., Launay S., Ladoire S., Sordet O., Ponnelle T., Debili N., Phan T. H., Padua R. A., Dubrez-Daloz L., Solary E. (2004) Blood 104, 2035–2043 [DOI] [PubMed] [Google Scholar]
- 26. Didelot C., Lanneau D., Brunet M., Bouchot A., Cartier J., Jacquel A., Ducoroy P., Cathelin S., Decologne N., Chiosis G., Dubrez-Daloz L., Solary E., Garrido C. (2008) Cell Death Differ. 15, 859–866 [DOI] [PubMed] [Google Scholar]
- 27. Samuel T., Okada K., Hyer M., Welsh K., Zapata J. M., Reed J. C. (2005) Cancer Res. 65, 210–218 [PubMed] [Google Scholar]
- 28. Ponnelle T., Chapusot C., Martin L., Bonithon-Kopp C., Bouvier A. M., Plenchette S., Rageot D., Faivre J., Solary E., Piard F. (2003) Pathol. Res. Pract. 199, 723–731 [DOI] [PubMed] [Google Scholar]
- 29. Vischioni B., van der Valk P., Span S. W., Kruyt F. A., Rodriguez J. A., Giaccone G. (2006) Hum. Pathol. 37, 78–86 [DOI] [PubMed] [Google Scholar]
- 30. Tanimoto T., Tsuda H., Imazeki N., Ohno Y., Imoto I., Inazawa J., Matsubara O. (2005) Cancer Lett. 224, 141–151 [DOI] [PubMed] [Google Scholar]
- 31. DeGregori J., Johnson D. G. (2006) Curr. Mol. Med. 6, 739–748 [DOI] [PubMed] [Google Scholar]
- 32. Dimova D. K., Dyson N. J. (2005) Oncogene 24, 2810–2826 [DOI] [PubMed] [Google Scholar]
- 33. Blais A., Dynlacht B. D. (2007) Curr. Opin. Cell Biol. 19, 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tyagi S., Chabes A. L., Wysocka J., Herr W. (2007) Mol. Cell 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 35. Sun A., Bagella L., Tutton S., Romano G., Giordano A. (2007) J. Cell Biochem. 102, 1400–1404 [DOI] [PubMed] [Google Scholar]
- 36. Paggetti J., Largeot A., Aucagne R., Jacquel A., Lagrange B., Yang X. J., Solary E., Bastie J. N., Delva L. (2010) Oncogene 29, 5019–5031 [DOI] [PubMed] [Google Scholar]
- 37. Mace P. D., Smits C., Vaux D. L., Silke J., Day C. L. (2010) J. Mol. Biol. 400, 8–15 [DOI] [PubMed] [Google Scholar]
- 38. Geng Y., Eaton E. N., Picón M., Roberts J. M., Lundberg A. S., Gifford A., Sardet C., Weinberg R. A. (1996) Oncogene 12, 1173–1180 [PubMed] [Google Scholar]
- 39. Conze D. B., Albert L., Ferrick D. A., Goeddel D. V., Yeh W. C., Mak T., Ashwell J. D. (2005) Mol. Cell Biol. 25, 3348–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 41. Wang C., Chen L., Hou X., Li Z., Kabra N., Ma Y., Nemoto S., Finkel T., Gu W., Cress W. D., Chen J. (2006) Nat. Cell Biol. 8, 1025–1031 [DOI] [PubMed] [Google Scholar]
- 42. Ianari A., Gallo R., Palma M., Alesse E., Gulino A. (2004) J. Biol. Chem. 279, 30830–30835 [DOI] [PubMed] [Google Scholar]
- 43. Hallstrom T. C., Nevins J. R. (2006) Genes Dev. 20, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chong J. L., Wenzel P. L., Sáenz-Robles M. T., Nair V., Ferrey A., Hagan J. P., Gomez Y. M., Sharma N., Chen H. Z., Ouseph M., Wang S. H., Trikha P., Culp B., Mezache L., Winton D. J., Sansom O. J., Chen D., Bremner R., Cantalupo P. G., Robinson M. L., Pipas J. M., Leone G. (2009) Nature 462, 930–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sahin F., Sladek T. L. (2010) Int. J. Biol. Sci. 6, 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Polager S., Ginsberg D. (2008) Trends Cell Biol. 18, 528–535 [DOI] [PubMed] [Google Scholar]
- 47. LaCasse E. C., Mahoney D. J., Cheung H. H., Plenchette S., Baird S., Korneluk R. G. (2008) Oncogene 27, 6252–6275 [DOI] [PubMed] [Google Scholar]
- 48. Srinivasula S. M., Ashwell J. D. (2008) Mol. Cell 30, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gibbs J. D., Liebermann D. A., Hoffman B. (2008) Oncogene 27, 98–106 [DOI] [PubMed] [Google Scholar]
- 50. Ivanova I. A., Nakrieko K. A., Dagnino L. (2009) Oncogene 28, 52–62 [DOI] [PubMed] [Google Scholar]
- 51. Xu L., Zhu J., Hu X., Zhu H., Kim H. T., LaBaer J., Goldberg A., Yuan J. (2007) Mol. Cell 28, 914–922 [DOI] [PubMed] [Google Scholar]
- 52. Sutherland R. L., Musgrove E. A. (2004) J. Mammary Gland Biol. Neoplasia 9, 95–104 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.