Abstract
Telomerase is a multisubunit enzyme that maintains genome stability through its role in telomere replication. Although the Est3 protein is long recognized as an essential telomerase component, how it associates with and functions in the telomerase complex has remained enigmatic. Here we provide the first evidence of a direct interaction between Saccharomyces cerevisiae Est3p and the catalytic protein subunit (Est2p) by demonstrating that recombinant Est3p binds the purified telomerase essential N-terminal (TEN) domain of Est2p in vitro. Mutations in a small cluster of amino acids predicted to lie on the surface of Est3p disrupt this interaction with Est2p, reduce assembly of Est3p with telomerase in vivo, and cause telomere shortening and senescence. We also show that recombinant Est3p stimulates telomerase activity above basal levels in vitro in a manner dependent on the Est2p TEN domain interaction. Together, these results define a direct binding interaction between Est3p and Est2p and reconcile the effect of S. cerevisiae Est3p with previous experiments showing that Est3p homologs in related yeast species influence telomerase activity. Additionally, it contributes functional support to the idea that Est3p is structurally related to the mammalian shelterin protein, TPP1, which also influences telomerase activity through interaction with the Est2p homolog, TERT.
Keywords: Chromosomes, DNA Replication, Protein-Protein Interactions, Telomeres, Yeast, Est1, TPP1, Telomerase Activity
Introduction
Telomeres are protein-DNA complexes that protect chromosome termini from nucleolytic digestion and distinguish natural chromosome ends from internal DNA breaks. Although the majority of telomeric DNA is double stranded, the G/T-rich strand forms a protruding 3′-overhang. In the absence of a counteracting mechanism, telomeres shorten during each cell division, ultimately activating cell cycle checkpoints and cellular senescence (1). If these checkpoints are disrupted or bypassed, end-to-end fusions and bridge breakage cycles can ensue. Telomerase, a ribonucleoprotein complex, promotes telomere maintenance and genomic stability by elongating the 3′-overhang by reverse transcription (2).
In budding yeast, telomerase consists minimally of four dedicated subunits: TLC1 RNA, the template RNA (3); Est1p, an accessory protein important for recruiting and activating telomerase at the telomere (4, 5); Est2p, the reverse transcriptase (2); and Est3p, an additional accessory protein necessary for proper activity in vivo (6). Deletion of any of these components eliminates telomerase function in vivo, yielding the Ever Shorter Telomere (EST)4 phenotype (3, 7, 8).
Although these and other telomerase components have been known for well over a decade, few details of their assembly into the ribonucleoprotein complex are understood. Described interactions are largely confined to protein associations with the RNA template. Est1p and Est2p independently bind distinct regions within the central portion of TLC1 RNA (9–11). Sm proteins facilitate RNA stability through interaction with a site near the 3′-end of the RNA (12), and association of the catalytic core of telomerase with the telomere in G1 phase is mediated by interaction of the yKu heterodimer with TLC1 RNA (13). Protein/protein interactions are less well understood. Est1p and Est2p may interact in an RNA-independent manner (5, 14). When Est1p is tethered to an internal chromosomal site via fusion with the Gal4 DNA binding domain, Est2p is recruited to that site in the absence of TLC1 RNA (14). Furthermore, recombinant Est1p and Est2p can bind directly, although TLC1 RNA enhances this interaction (5).
Both the function of Est3p and the mechanism of its assembly into the telomerase complex are unknown. Est1p stimulates the association of Est3p with telomerase in a cell cycle-dependent manner that requires Est2p (15). Very recently, Est1 protein purified from yeast was shown to bind recombinant Est3 protein, yielding strong evidence for a direct interaction between these two proteins (16). Genetic evidence suggests that Est3p interacts with the N-terminal (TEN) domain of Est2p, but no direct protein/protein or protein/RNA interactions have been reported (6, 17, 18). It has been suggested that Est3p binds nucleic acid and possesses helicase activity (19). However, sensitive analysis by NMR spectroscopy of Est3p in the presence and absence of DNA failed to detect an interaction (18).
Although Est3p has been reported to be dispensable for the catalytic activity of Saccharomyces cerevisiae telomerase (20), recent work in the related species Saccharomyces castelli implicates the Est3p homolog in stimulating nucleotide addition processivity (18). In addition, Est3p from Candida albicans is required for robust telomerase activity in vitro with specific primers (21). These data raise the possibility that Est3p affects enzymatic activity in S. cerevisiae but that technical limitations have, to date, precluded the detection of such effects.
Here we show the first biochemical evidence that Est3p binds directly to the TEN domain of Est2p. We also show that recombinant Est3p stimulates telomerase activity during in vitro primer extension and that the interaction of Est3p with the Est2p TEN domain appears required for this function.
EXPERIMENTAL PROCEDURES
Plasmid and Strain Construction
Plasmids for Protein Expression
The plasmid pET Duet EST3 (for expression of His6-EST3) was made by moving EST3 from YCPlac33 EST3 using primers M090 and M091 (supplemental Table S1) into pET Duet-1 (Novagen) using restriction sites BamHI and SalI. EST3 mutants K71A (lysine 71 to alanine), ETN (glutamate 114, threonine 115, and asparagine 117), and DQ (aspartate 166 and glutamine 167) were introduced into pET Duet EST3 by QuikChangeTM (Stratagene) using primer pairs K71A forward and K71A reverse, ETN114AAK forward and ETN114AAK reverse, and DQ forward and DQ reverse, respectively. A vector for expression of MBP-EST2TEN was made by cloning EST2 TEN (residues 1–162) from pKF404 (22) using primer pair LM204.1 forward and LM204.1 reverse into pLM204a (gift of L. Mizoue) using restriction sites EcoRI and PstI. The MBP-EST2TEN fusion gene was then moved into the EcoRV and KpnI sites of pET Duet-1 using primer pair EcoRV MBP forward and KpnI Est2RI reverse to create pET Duet MBP-EST2TEN. MBP alone was cloned into the EcoRV and KpnI restriction sites of pET Duet-1 from pLM204a using primer pair EcoRV MBP forward and KpnI MBP reverse to create pET Duet MBP.
Plasmids for in Vivo Characterization
pKF441 (CEN EST3 URA3) was created by PCR amplification of the EST3 promoter and frameshift-corrected gene from pVL901 (gift of V. Lundblad) using primer pair UEprimer1 and UEprimer4 to create an EcoRI site upstream of the promoter and a KpnI site immediately before the EST3 stop codon. The endogenous EST3 termination sequence was amplified using primer pairs UEprimer5 and UEprimer6 to create a HindIII site downstream of the terminator. UEprimer1 and UEprimer6 were then used to amplify the full-length insert using these two PCR products as templates. The resulting fragment was cloned into YCplac33 (CEN URA3) using EcoRI and HindIII. pKF442 (CEN EST3HA URA3) was created by PCR amplification of the HA3 tag from pVL901 using primer pair KpnI forward and XbaI reverse and ligation into pKF441 using KpnI and XbaI. EST3 and EST3HA were subsequently moved into pRS315 or pRS425 (CEN LEU2 and 2 μm LEU2, respectively) (23, 24) using the PvuII sites of pKF441 or pKF442. The ETN and K71A alleles were subcloned from pET Duet-1 (see above) using restriction sites MscI and XhoI. DQ was created by QuikChangeTM using the same primers as above and subcloned using SpeI and XmaI. All point mutations were verified by sequencing.
Yeast Strains
YKF122 (AVL78 est3::KANR) was created by standard one-step gene replacement. The kanamycin-resistance gene was amplified by PCR from pFA6a-kanMX6 using primers Est3Kan forward and Est3Kan reverse. The PCR product was cloned into the SacI and KpnI sites of pKF441 to create pKF441 est3::KAN. The deletion construct was transformed into AVL78 using a standard lithium acetate protocol. YKF126 (AVL78 est3::KANR EST2-G8-Myc18 (G: glycine)) was created by linearizing pRS304-Est2-G8-Myc18 (gift of V. Zakian) with SwaI and transforming it into YKF122 + pKF441. Strains and plasmids used in this study are shown in supplemental Table S2.
Complementation and Growth Assay
Functional complementation of the est3 mutant alleles was tested in YKF122 (AVL78 est3::KANR) using mutant constructs created in pRS315 (CEN LEU2). YKF122 was complemented with pKF442 (CEN EST3HA URA3); loss of the complementing plasmid was selected on 5-fluoroorotic acid plates, and the wild-type or mutant pRS315 plasmids were subsequently transformed using the standard lithium acetate method. Resulting single colonies were restreaked three times on plates lacking leucine. Cell viability was assessed visually, and telomere length was determined by Southern blotting using XhoI as described previously (17).
Protein Purification
BL21 cells containing pET Duet EST3 were grown in 6 liters of standard Luria broth (LB) with 50 μg/ml ampicillin at 37 °C to an A600 0.3–0.4. After shifting the culture to 16 °C for 1 h, protein expression was induced with 500 μm isopropyl 1-thio-β-d-galactopyranoside overnight with moderate shaking (110 rpm). Cells were harvested by centrifugation at 4 °C and resuspended in 10 ml of TG buffer (+100 mm NaCl) per liter of original culture (TG: 50 mm Tris, pH 7.5, 10% glycerol, and 3 mm β-mercaptoethanol). Cells were lysed using an EmulsiFlex (Avestin) by passing cells three or four times through the machine at 20,000 psi. The resulting extract (60 ml) was incubated with 7.5 ml of Talon® resin (Clontech) for 1 h at 4 °C with gentle agitation and gravity-packed into an empty glass EconColumnTM (Bio-Rad). Resin was washed with 10 column volumes of TG + 300 mm NaCl and 10 mm imidazole. Protein was eluted with 5 column volumes TG + 100 mm NaCl and 100 mm imidazole. The eluate was dialyzed (Spectra/Por® no. 7; 10,000-kDa (Spectrum Laboratories)) at 4 °C overnight into TG buffer, bound to a SourceTM 15Q HR16/10 column (GE Healthcare), and eluted using a linear salt gradient (TG + 0 mm NaCl to TG + 1 m NaCl). The purest fractions were pooled and applied to a SuperdexTM 200 26/60 gel filtration column (GE Healthcare) using TG + 100 mm NaCl buffer. Again, His6-Est3p fractions were pooled and concentrated to 3 ml using a 15-ml, 10-kDa cutoff Amicon® ultraconcentrator (Millipore) and dialyzed into TG + 100 mm NaCl + 50% glycerol. The protein was stored at −20 °C. Mutant proteins were purified in the same manner except that ETN and DQ were purified at pH 8.0. The Est3 protein sequence was verified by mass spectrometry.
Three-liter cultures of BL21 cells containing pET Duet MBP-EST2TEN were grown and harvested as described above. Cells were lysed as above in 10 ml of TEG-200 per liter of original culture (TEG-200: 20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 1 mm EDTA, 10% glycerol). Extract was incubated with 10 ml of amylose resin for 2 h at 4 °C and was packed similarly as above. Resin was washed with 15 column volumes of TEG + 500 mm NaCl. MBP-Est2pTEN was eluted with TEG-200 + 5 mm maltose. Fractions containing MBP-Est2pTEN were further purified over an S200 gel filtration column using TG + 100 mm NaCl buffer. MBP-Est2pTEN was concentrated as described for His6-Est3p and dialyzed into TG + 100 mm NaCl, 50% glycerol. Protein was stored at −20 °C. The Est2TEN protein sequence was verified by mass spectrometry.
All purified proteins were separated by 12% SDS-PAGE to determine purity. All protein preparations used were judged by Bio-SafeTM Coomassie (Bio-Rad) staining to be at least 95% pure. Protein concentration was determined either by spectrophotometry or by comparison of a serial dilution with a known concentration of protein standard. Maltose-binding protein (MBP) was purchased from New England Biolabs and diluted to 20 μm in TG + 100 mm NaCl + 50% glycerol.
In Vitro Pulldown Assay, Immunoprecipitation, and Western Blot Analysis
MBP or MBP-Est2pTEN (200 pmol) was incubated with 25 μl of amylose resin in buffer I (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 1 mm EDTA, 0.05% Tween 20, and 10% glycerol) for 2 h at 4 °C with gentle agitation. 1 nmol of His6-Est3p or mutant Est3 protein was added and incubated for 3 h at 4 °C. The resin was washed three times for 5 min with 1 ml of buffer I, resuspended in 100 μl of buffer I, loaded into a 700-μl spin cup (Pierce), and washed with 1.5 ml of buffer I. Retained resin was resuspended in 40 μl of buffer I, and 20 μl was separated by 12% SDS-PAGE. For detection of MBP and MBP-Est2pTEN, proteins were blotted onto nitrocellulose (GE Healthcare) and blocked with 5% milk/PBS, pH 7.4, with 0.05% Tween (PBS-T). For detection of His6-Est3, proteins were blotted onto Hybond P membrane (GE Healthcare) and blocked with 5% BSA/PBS-T. HRP-conjugated anti-MBP (New England Biolabs) was used at a dilution of 1:50,000 in 5% milk/PBS-T. Primary antibody for detection of the His6 tag (Santa Cruz Biotechnology) was diluted 1:1,000 in 5% BSA/PBS-T. Secondary antibody was peroxidase-conjugated mouse anti-rabbit (Chemicon) at 1:10,000 in 5% milk/PBS-T. ECL plus Western Blotting Detection system (GE Healthcare) was used for detection.
Yeast protein extract was prepared as described (22). Extracts were normalized to 20 mg/ml and incubated with antibodies as described previously (15, 22). 10 μl of anti-Myc immunoprecipitated material (1/6 of total) or anti-HA immunoprecipitated material was separated by 10–12% step-gradient SDS-PAGE. Proteins were transferred to Hybond P membrane (GE Healthcare). The membrane was blocked with 5% milk/PBS-T followed by incubation with primary antibodies (HA: 1:500 dilution murine monoclonal HA.11 (Covance) or Myc: 1:250 dilution of murine monoclonal Myc Ab-1 (OP10L, EMD Biosciences)) in 5% milk/PBS-T. Secondary antibody was peroxidase-conjugated goat anti-mouse (Chemicon) used at a 1:10,000 dilution in 5% milk/PBS-T. Detection was done as described above.
Telomerase Assay and Northern Blot Analysis
Telomerase was partially purified from wild-type yeast (YPH499) by chromatography over DEAE-Sepharose fast flow resin (GE Healthcare) and Mono Q resin (GE Healthcare) as described previously (5, 25) and used in telomerase DNA extension assays. Briefly, partially purified telomerase extract was incubated with 2 pmol of a 7-base 3′-overhang template (GTGTGTG) immobilized on streptavidin paramagnetic beads (Promega) and extension buffer (50 mm Tris, pH 8.0, 1 mm MgCl2, 1 mm spermidine, 1 mm DTT, 0.5% glycerol, 50 μm dTTP, 10 μCi of [α-32P]dGTP (3,000 Ci mmol−1; Amersham Biosciences)). The 7-base 3′-overhang template was generated by annealing biotin-conjugated Backbone1 primer (supplemental Table S1) to GTG7 base consensus primer. Reactions were incubated for 30 min at 30 °C followed by magnetic collection of DNA-bound beads. The beads were washed twice with 1 × EcoRI buffer (New England Biolabs). Beads were resuspended in 50 μl of 1 × EcoRI buffer, 100 μg/ml BSA, and 10 units of EcoRI and incubated for 1 h at 37 °C. The beads and cleaved DNA fragments were separated magnetically. A PNK (T4 polynucleotide kinase (New England Biolabs)) end-labeled, 27-base oligonucleotide was added, and the DNAs were ethanol-precipitated. DNA was reconstituted in formamide-NaOH loading buffer and run in a 14% acrylamide denaturing gel and subsequently visualized using a PhosphorImager. For the experiments in Fig. 4, BSA or recombinant His6-Est3p was titrated into the extension buffer at varying concentrations before addition of telomerase extract (5).
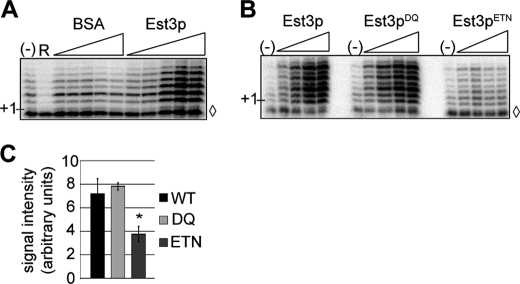
FIGURE 4.
Recombinant Est3p stimulates telomerase activity in vitro in a manner dependent on the Est3p/Est2pTEN interaction. A, partially purified telomerase extracts were prepared from YPH499 cells and incubated with a 7-base 3′-overhang-immobilized DNA primer in the presence of dTTP and [α-32P]dGTP. In lane R, RNaseA was added to telomerase prior to addition of the primer. BSA or recombinant His6-Est3p (0.5, 1, 2.5, 5, or 10 μm) was titrated into DNA extension reactions as indicated. (−) indicates basal telomerase activity. ◊ indicates a 27-mer loading control added prior to DNA precipitation. B, recombinant His6-Est3p (wild type or indicated mutant; 0.25, 0.5, 1, 2.5, and 5 μm) was titrated into DNA extension reactions as described above. (−) indicates basal telomerase activity. ◊ indicates a 27-mer loading control added prior to DNA precipitation. C, recombinant His6-Est3p (wild type or mutant) was added at 2.5 μm to DNA extension reactions. Signal intensity of three replicates was determined using ImageQuant software, and significance was assessed using an analysis of variance (ANOVA) post hoc Dunnett's (control = His6-Est3p). The stimulatory activity of His6-Est3pETN was lower than wild-type His6-Est3p (p = 0.03), whereas His6-Est3pDQ activity was similar to wild type (p = 0.87). Bars represent S.E., and * denotes a p value <0.05.
For Northern blot analysis, RNA was isolated from immunoprecipitation beads and detected by Northern blotting as described previously (15, 22). Whole cell RNA was prepared from 10 ml of mid-log phase cultures (26).
RESULTS
Est2pTEN and Est3p Interact Directly in Vitro
The previously reported allele-specific suppression of temperature-sensitive mutations within the Est2p TEN domain by overproduction of Est3p (17) may reflect a direct interaction between the two proteins. To test this hypothesis, EST3 was fused with an N-terminal His6 tag, and the TEN domain of EST2 (EST2TEN; residues 1–162) was fused to the C terminus of MBP. Tagged proteins were individually expressed in Escherichia coli and purified to >95% apparent homogeneity (Fig. 1A). An 1H-15N heteronuclear single quantum correlation (HSQC) experiment was used to assess His6-Est3p tertiary structure. Unfolding of the protein is expected to cause a collapse of cross-peaks in the region near 8 ppm. Instead, the HSQC of Est3p revealed excellent spectral dispersion with backbone amide peaks that range from ∼6.1 to 9.9 ppm in the proton dimension (supplemental Fig. S1).
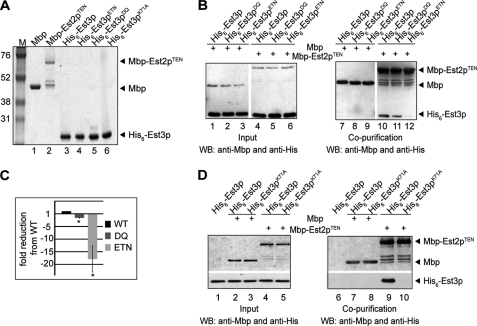
FIGURE 1.
MBP-Est2pTEN interacts directly with His6-Est3p. A, Coomassie-stained gel of recombinant proteins purified from E. coli. Marker sizes (M) are shown in kDa. MBP, MBP-Est2pTEN, and His6-Est3p are 42, 62, and 21 kDa, respectively. The proteins were estimated to be >95% pure. B, 200 pmol of MBP or MBP-Est2pTEN incubated as indicated (+) with 1 nmol of His6-Est3p, His6-Est3pDQ, or His6-Est3pETN and captured on amylose resin. Input (1% of total; left panel) and amylose-bound proteins (right panel) were analyzed by Western blotting (WB) using anti-MBP and anti-His antibodies. Data shown are representative of four independent experiments. C, quantification of data shown in B. -fold reduction in recovery of His6-Est3pDQ and His6-Est3pETN compared with wild type was averaged over four independent experiments. Bars are S.E. Both His6-Est3pDQ and His6-Est3pETN binding are statistically different from His6-Est3p by one-tailed paired t test (p values 0.0002 and 0.0007, respectively) as denoted by *. D, 200 pmol of MBP or MBP-Est2pTEN incubated as indicated (+) with 1 nmol of His6-Est3p or His6-Est3pK71A and captured on amylose resin. Input (1% of total; left panel) and amylose-bound proteins (right panel) were analyzed by Western blotting using anti-MBP and anti-His antibodies. Data shown are representative of three independent experiments.
The individually purified proteins were mixed, and their co-purification was monitored. His6-Est3p did not detectably associate with amylose resin alone (Fig. 1D, lane 6) or when co-incubated with MBP (Fig. 1B, lane 7). In contrast, His6-Est3p robustly co-purified with MBP-Est2pTEN (Fig. 1B, lane 10), indicating that His6-Est3p and MBP-Est2pTEN bind directly in vitro.
To examine the specificity of the interaction between Est3p and Est2pTEN, we created and characterized EST3 mutants known or suspected to influence telomerase assembly and/or function. Based on structural modeling, these mutations are predicted to affect surface-exposed residues and to have minor effects on protein stability (27). Est3pK71A (lysine 71 mutated to alanine) has been shown to disrupt telomere maintenance but retain assembly with telomerase in vivo, as assessed by co-purification of the mutant protein with TLC1 RNA (27). In contrast, individual mutations in residues glutamate 114, threonine 115, or asparagine 117 were shown to shorten telomeres and reduce Est3p association with TLC1 RNA (27). To disrupt this charged region completely, we simultaneously mutated all three residues to create Est3pE114A, T115A, N117K (Est3pETN). Aspartate 166 also contributes to the association of Est3p with TLC1 RNA (27). Because glutamine 167 is conserved in related fungal species, we mutated both residues to create Est3pD166A, Q167A (Est3pDQ).
These mutant proteins were purified as His6 fusion proteins from E. coli and their secondary structure characterized by circular dichroism. Both the wild-type Est3p and each of the altered Est3 proteins displayed strong evidence of secondary structure, suggesting that the amino acid changes did not strongly perturb protein folding (supplemental Fig. S2). Compared with the interaction observed for wild-type His6-Est3p, co-purification of His6-Est3pETN with MBP-Est2pTEN was greatly decreased (Fig. 1, B and C; 17-fold, p = 0.0007). In contrast, the interaction between recombinant MBP-Est2pTEN and His6-Est3pDQ was only slightly decreased compared with wild type (Fig. 1, B and C; 1.5-fold, p = 0.0002). The identification of point mutations that disrupt the Est3p/Est2pTEN interaction in vitro suggests that the association between these two proteins is both direct and specific. Furthermore, residues near Glu114 contribute to the association of Est3p with the TEN domain of Est2p.
Surprisingly, no association between His6-Est3K71A and MBP-Est2pTEN could be detected in the pulldown assay (Fig. 1D, lane 10), even though this mutant can assemble with the telomerase complex in vivo (27) (Fig. 2). This result may suggest that Est3p can assemble with telomerase through additional interactions in vivo. However, we cannot dismiss the possibility that mutation of lysine 71 to alanine disrupts the integrity of the recombinant protein.
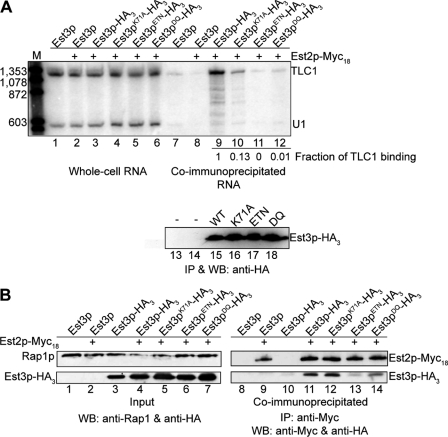
FIGURE 2.
Est3pETN shows reduced association with telomerase in vivo. A, whole cell RNA (lanes 1–6) or anti-HA immunoprecipitations (lanes 7–12) were generated from yeast strains AVL78 (untagged; lanes 1 and 7) or YKF126 (EST2-MYC18 est3::KANR) with pKF441 (CEN EST3 URA3) and pKF448HA (CEN EST3-HA3 LEU2), or pKF449HA (2 μm est3K71A, ETN, or DQ-HA3 LEU2) as indicated. TLC1 and U1 RNA were detected by Northern blotting (upper panel); M is marker in bp. The amount of TLC1 retained in each co-immunoprecipitation (normalized to the wild-type value) is notated under lanes 9–12. This value was determined by first subtracting the amount of TLC1 bound nonspecifically (untagged Est3p; lane 8) from the amount of TLC1 observed in each lane and then dividing by the wild-type value (lane 9). Est3p-HA3 recovery was measured by Western blotting (WB; lower panel; lanes 13–18) in the identical immunoprecipitation. Results are representative of two independent biological replicates (see supplemental Fig. S3). B, protein extracts were isolated from strains AVL78 (lanes 1 and 8), YKF126 containing plasmid pKF441 (lanes 2 and 9), YKF122 (est3::KANR) containing pKF442HA (CEN EST3-HA3 URA3; lanes 3 and 10), or YKF126 containing plasmids pKF441 and pKF448HA or pKF449HA mutants, as indicated. Anti-Myc immunoprecipitations were analyzed by Western blotting with monoclonal anti-Myc and anti-HA antibodies (lanes 8–14). Input (1% of total) was probed with anti-HA and anti-Rap1p antibodies as a loading control (lanes 1–7). Results are representative of two biologically independent experiments.
EST3 Mutant Alleles Alter Telomerase Assembly and Function in Vivo
Because the est3ETN and est3DQ alleles evaluated in this study are different from those investigated previously (27), we characterized their in vivo phenotype. Telomerase assembly was monitored by co-immunoprecipitation of HA-tagged Est3p with TLC1 RNA and with Myc18-tagged Est2p (Fig. 2). The strain background utilized lacked endogenous EST3. EST2 was epitope-tagged at its N terminus by two-step integration of 18 copies of the Myc epitope at the endogenous locus. To avoid complications that might arise from measuring telomerase assembly in senescent strains, the epitope-tagged EST3 variants were expressed from a plasmid in the presence of an untagged CEN EST3-complementing plasmid. Previous work has shown that the untagged Est3 protein does not interfere appreciably with the ability of epitope-tagged Est3p to co-immunoprecipitate with its binding partners (27).
We first verified that TLC1 RNA levels are not affected by the EST3 mutations. As shown in Fig. 2A (lanes 1–6), TLC1 RNA levels detected by Northern blotting from total cellular RNA are equivalent across strains. Est3p was immunoprecipitated from whole cell extract, and an aliquot was analyzed by Western blotting (Fig. 2A, lower panel). Each of the mutant proteins was expressed at a level equivalent to or slightly higher than wild-type Est3p (mutants are expressed from a high copy number 2 μm plasmid). TLC1 RNA was detected by Northern blotting in the identical immunoprecipitates (Fig. 2A, lanes 7–12). After correcting for minor nonspecific recovery of TLC1 RNA from control extract lacking the HA tag on Est3p (Fig. 2A, lane 8), the amount of TLC1 RNA immunoprecipitated with each mutant protein was expressed as a fraction of the association observed for wild-type Est3p. Highly congruent results were obtained in two independent biological replicates (Fig. 2A and supplemental Fig. S3). Est3pETN-HA3 and Est3pDQ-HA3 showed very little if any residual association with TLC1, in agreement with the effects previously observed for the corresponding single amino acid mutations in EST3. Est3pK71A-HA3 consistently retained higher levels of association with TLC1 than the other mutants, but was reduced to ∼10% of the wild-type level (Fig. 2A).
As an alternate measure of telomerase complex assembly, we monitored the co-immunoprecipitation of each Est3p variant with Myc18-Est2p. In agreement with the effects on TLC1 association, Est3pETN-HA3 and Est3pDQ-HA3 were reduced in their ability to co-immunoprecipitate with Myc18-Est2p (Fig. 2B). Interestingly, Est3pK71A-HA3 immunoprecipitated with Myc18-Est2p at a level similar to wild type. The discordance between the ability of Est3pK71A-HA3 to immunoprecipitate with TLC1 and Myc18-Est2p suggests that the interaction between TLC1 RNA and Est3p (likely indirect) is more easily disrupted by the wash conditions than the interaction between Est2p and Est3p. In agreement with previous results (27), we conclude that the residues near glutamate 114 and aspartate 166 are important for Est3p assembly with telomerase, whereas residue lysine 71 plays a less critical role in vivo.
est3ETN Does Not Retain Function in Vivo
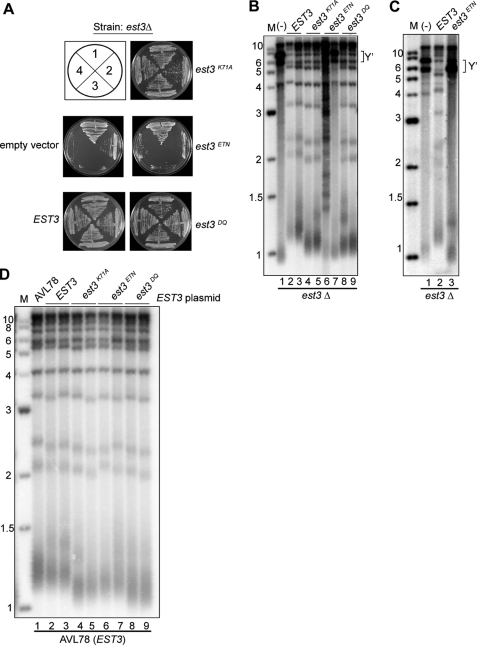
To address the ability of the EST3 variants to support telomere replication, each allele was expressed from a low copy number centromere plasmid in yeast lacking endogenous EST3. The resulting strains were monitored for the ability to support growth over three consecutive restreaks on solid medium. As expected, the yeast expressing wild-type EST3 maintained robust cellular growth, whereas the empty vector control senesced after three restreaks (Fig. 3A). Interestingly, the strain expressing est3ETN resembled the empty vector control for growth (Fig. 3A), whereas est3K71A and est3DQ looked similar to wild type. As a more sensitive measure of telomerase function, the telomere length of cells taken from the final restreak was measured by Southern blotting. In cells with normal telomerase function, XhoI releases a terminal fragment of ∼1.2–1.3 kb from chromosomes possessing one or more subtelomeric Y′ elements (Fig. 3B, lanes 2 and 3). In contrast, cells containing either empty vector or expressing est3ETN displayed hallmarks of telomerase dysfunction (Fig. 3B, lanes 1, 6, and 7). Cells that are unable to utilize telomerase instead use a RAD52-dependent recombination pathway to maintain telomeres, manifested on the Southern blot as stochastic lengthening of the TG-rich telomeric repeats (Fig. 3B, lane 6) or amplification of Y′ elements (brackets, Fig. 3B, lanes 1 and 7) (28). Expression of est3ETN from a high copy number 2-μm plasmid allowed the protein to accumulate at wild-type levels (Fig. 2B, compare lanes 4 and 6) but did not improve complementation for either growth (data not shown) or telomere length (Fig. 3C), indicating that low protein levels alone do not account for the lack of telomere maintenance.
FIGURE 3.
EST3ETN does not complement an est3Δ strain even when overexpressed. A, yeast strain YKF122 complemented with pKF441 was transformed with pRS315 (CEN LEU2 empty vector), pKF448 (CEN EST3 LEU2), or pKF448 mutants (CEN est3K71A, ETN, or DQ LEU2), and loss of the URA3-complementing plasmid was selected on plates containing 5-fluoroorotic acid. Cells were restreaked four times; numbers represent restreaks following loss of the complementing plasmid. B, DNA was extracted from cells grown in liquid culture from the third restreak of strains shown in A. DNA was digested with XhoI, Southern blotted, and probed with a telomeric fragment. (−) indicates the empty vector control. Y′ elements are bracketed. Two independent transformants are shown. M is marker in kb. C, YKF122 (est3::KANR) complemented with pKF441 (CEN EST3 URA3) was transformed with pRS425 alone (2 μm LEU2), lane 1; pKF449 (2 μm EST3 LEU2), lane 2; or pKF449ETN (2 μm EST3ETN LEU2), lane 3. After selection for loss of the complementing plasmid on plates containing 5-fluoroorotic acid, cells were restreaked three times on plates lacking leucine. Telomere blots were performed as described in B. Y′ elements are bracketed. D, AVL78 was transformed with pKF449 (2 μm EST3 LEU2) expressing the indicated EST3 alleles. After three successive restreaks, genomic DNA was digested with XhoI, blotted, and probed with a telomeric fragment. Two biological replicates are shown. The parental wild-type strain (AVL78) is shown for reference.
Although both est3DQ and est3K71A supported normal cellular growth (Fig. 3A), the telomeres in cells expressing these EST3 alleles were maintained at a length shorter than wild type (Fig. 3B), similar to a previous report (27). All of the est3 alleles were dominant negative when overexpressed in the presence of wild-type Est3p, suggesting that they are expressed and retain some function (Fig. 3D). Together, these data reveal that residues Glu114, Thr115, and Asn117 are critically important for telomerase assembly both in vivo and in vitro, whereas Asp166 and Gln167 appear to have a lesser role in vitro. As described previously (27), Lys71 has a less severe effect on telomerase assembly in vivo, although mutation of this residue to alanine disrupts the interaction between Est3p and the TEN domain of Est2p in vitro (see “Discussion”).
Est3p Stimulates Telomerase Activity in Vitro
The function of Est3p in S. cerevisiae has remained elusive since its discovery as a telomerase complex component (6). Although early studies did not detect an obvious loss of catalytic activity upon EST3 deletion (20), more recent studies of S. castelli and C. albicans have demonstrated roles for these Est3p homologs during primer extension in vitro (18, 21). These observations raise the possibility that Est3p from S. cerevisiae may have a similar function but that the particular assay conditions utilized either minimize or mask that effect.
To address this issue, we measured the ability of recombinant Est3p to stimulate the primer extension activity of partially purified yeast telomerase. Biotin-labeled yeast telomeric primers were bound to streptavidin beads and incubated in the presence of radiolabeled nucleotides and wild-type telomerase extract that was partially purified over DEAE and Mono Q resins (5, 29). As previously demonstrated, yeast telomerase adds seven nucleotides (5′-GGTGTGG) to the end of the primer and then terminates elongation. Titration of His6-Est3p into this reaction increased overall telomerase activity in a dose-dependent manner, whereas a BSA control did not (Fig. 4A). Importantly, Est3p does not appear to affect the processivity of telomerase in this assay because neither the maximal length of the extended product nor the relative intensity of each band is altered. We conclude that Est3p (like Est1p) (5) stimulates telomerase activity by increasing the fraction of extended primers.
Having established that recombinant Est3p stimulates primer extension by telomerase, we next determined the effect of each previously characterized mutant Est3 protein in this assay. Est3pETN was significantly reduced in its ability to stimulate telomerase activity compared with wild-type Est3p (Fig. 4, B and C; p = 0.03). Because the association of Est3pETN with the Est2p TEN domain was dramatically reduced in vitro (Fig. 1), this result suggests that the interaction between Est3p and Est2pTEN is required for telomerase stimulation. Consistent with its ability to bind Est2pTEN in vitro, Est3pDQ retained the capacity to stimulate telomerase (Fig. 4, B and C; p = 0.87). Given the reduced assembly and activity of this mutant in vivo, this result suggests that Est3pDQ affects a different, yet uncharacterized function of Est3p. Overall, these data show that S. cerevisiae Est3p can stimulate telomerase activity in a manner dependent on direct interaction with Est2pTEN.
DISCUSSION
Although the major components of yeast telomerase have been known for more than a decade, it has been difficult to determine the details of subunit interactions within the complex. Here we provide the first evidence of a direct Est2p/Est3p interaction in S. cerevisiae. Recombinant Est3p binds the purified TEN domain of Est2p in vitro, and this interaction is largely dependent upon several predicted surface residues of Est3p, including residues glutamate 114, threonine 115, and asparagine 117 (Fig. 1). Additionally, recombinant Est3p stimulates telomerase activity in vitro in a manner dependent upon the Est2p TEN domain interaction (Fig. 4). This result reconciles the effect of S. cerevisiae Est3p with previous experiments showing that Est3p homologs in related yeast species influence the enzymatic activity of telomerase (18, 21).
A Direct Est2p/Est3p Interaction Contributes to the Assembly of Est3p with Telomerase
We showed previously that Est1p stimulates the assembly of Est3p with telomerase (15). However, our failure to detect an Est1p/Est3p interaction by co-immunoprecipitation from cell extract in the absence of Est2p suggested that Est1p might not be the primary binding site for Est3p (15). Additionally, EST1 function is bypassed in a strain expressing a Cdc13-Est2 fusion protein, whereas EST3 remains essential (4). This result implies that Est3p can, in some circumstances, assemble and contribute to telomerase function in the absence of Est1p. Prior to work presented here, genetic results revealed that overexpression of Est3p suppresses mutations located in the Est2p TEN domain, but evidence for a direct interaction between these components was lacking (17).
Here we have combined an in vitro co-purification assay using recombinant proteins with experiments that assess both RNA and protein interactions in the native telomerase complex to gain a clearer understanding of telomerase complex assembly. Using these assays, we demonstrate that the Est2p TEN domain (residues 1–162) interacts directly with Est3p in vitro (Fig. 1). Mutation of three predicted surface-exposed residues (Glu114, Thr115, and Asn117) of Est3p (27) significantly perturbs this interaction in vitro, while also disrupting the assembly of Est3p with telomerase in vivo (Figs. 1 and 2).
Interestingly, the est3DQ mutation decreases the co-immunoprecipitation of Est3p with both TLC1 RNA and Myc18-Est2p in vivo (Fig. 2), but has a fairly minor (< 2-fold) effect on Est2p TEN binding in vitro (Fig. 1, B and C). One interpretation of these data is that the interaction between Est3p and Est2pTEN is insufficient for the assembly of Est3p with the telomerase complex in vivo, consistent with a model in which Est3p makes multiple contacts with telomerase components. This proposal is consistent with the recent report that Est3p interacts directly with Est1p (16).
However, because telomere length is only moderately affected by the DQ mutation, it is also possible that the co-immunoprecipitation experiment overemphasizes the defect of Est3p association in vivo. Indeed, even though the general trends are the same for Est3pETN-HA3 and Est3pDQ-HA3, each mutation has an apparently more severe effect on TLC1 RNA association than on the ability to co-immunoprecipitate with Myc18-Est2p (compare Fig. 2A, lanes 11 and 12, with Fig. 2B, lanes 13 and 14), suggesting that TLC1 RNA might be more sensitive to co-immunoprecipitation conditions than Est2p. This effect is even more pronounced with the Est3pK71A-HA3 mutation. Although the co-immunoprecipitation of TLC1 is reduced to ∼10% of wild-type levels, the association of Est3pK71A-HA3 with Myc18-Est2p is equivalent to that of wild-type Est3p. This difference may arise because Est3p and Est2p interact directly, whereas Est3p and TLC1 do not.
Although Est3pK71A-HA3 and Myc18-Est2p co-immunoprecipitate from cellular extract, the est3K71A mutation abolishes the in vitro interaction between MBP-Est2pTEN and His6-Est3p (Fig. 1D). This result may suggest that a second site of interaction between Est3p and telomerase, such as the interaction with Est1p (16), can mediate Est3p assembly when the TEN domain interaction is compromised. If true, the est3ETN mutation must disrupt both contacts. However, we cannot eliminate the possibility that the Est3K71A protein has defects in vitro (such as a minor disruption in tertiary structure) that are not reflected in vivo.
S. cerevisiae Est3p Contributes to Telomerase Activity in Vitro
Two recent papers have demonstrated that the Est3p homologs in C. albicans and S. castelli influence telomerase activity in vitro (18, 21). We report here that S. cerevisiae Est3p also stimulates basal telomerase activity (Fig. 4). Interestingly, His6-Est3pDQ, the mutant that retains assembly with the Est2p TEN domain, stimulates telomerase activity, whereas two mutants that compromise the Est3p/Est2pTEN interaction (est3ETN and est3K71A) do not (Fig. 4 and data not shown). These results suggest that Est3p requires interaction with the Est2p TEN domain to mediate its stimulatory effect. The requirement for this interaction in the stimulation assay may simply reflect a defect in the recruitment of recombinant Est3p to the telomerase complex in vitro. However, it is also plausible that the stimulatory effect of Est3p requires a specific interaction with the TEN domain that is separable from complex assembly.
Are Est3 and TPP1 Evolutionarily Related?
TPP1 is a member of the telomere-binding shelterin complex in mammals and influences the processivity of telomerase in vitro (30, 31). Based on prediction algorithms, fungal Est3p has a similar structure to the oligonucleotide/oligosaccharide-binding (OB) fold domain of TPP1, although this observation alone does not imply an evolutionary relationship between the two proteins (27, 32). Intriguingly, deletion of the TPP1 OB fold disrupts telomerase localization to the telomere in human cells (33). Additionally, a sequence-specific interaction between TPP1 and the human TEN domain of TERT is important for telomerase processivity (34). Our observation that the Est2p TEN domain mediates a critical interaction with Est3p that influences both telomerase assembly in vivo and stimulation of telomerase activity in vitro provides an intriguing parallel between the two proteins. The glutamate 114, threonine 115, and asparagine 117 of the Est3p primary structure align near some surface-exposed residues within a small α-helix of the OB fold of TPP1: arginine 159, glutamate 160, aspartate 163, and threonine 164. Determining whether these residues influence telomerase function in higher eukaryotes could give insight into a possible evolutionary relationship between Est3p and TPP1.
Supplementary Material
Acknowledgments
We thank Drs. Victoria Lundblad, Virginia Zakian, and Laura Mizoue for strains and reagents; L. M. and Dr. Markus Voehler for assistance with NMR and CD; L. M. and the Center for Structural Biology at Vanderbilt University for help with protein purification; Dr. David Friedman for mass spectrometry analysis; Margaret Platts for technical assistance; and Jenifer Ferguson and Dr. Christopher Brown for critical reading of the manuscript.
Note Added in Proof
A direct interaction between Est3p and the TEN domain of the Est2p homologs in two Candida species was recently reported by Yan et al. (Yan, W.-F., Chico, L., Lei, M., and Lue, N. F. (2011) Proc. Natl. Acad. Sci. U.S.A. doi: 10.1073/pnas.1017855108).
This work was supported, in whole or in part, by National Institutes of Health Grant GM080393. This work was also supported by Vanderbilt Discovery Grant (to K. L. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
- EST
- ever shorter telomere
- MBP
- maltose-binding protein
- OB
- oligonucleotide/oligosaccharide-binding
- TEN
- telomerase essential N-terminal.
REFERENCES
- 1. Osterhage J. L., Friedman K. L. (2009) J. Biol. Chem. 284, 16061–16065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., Cech T. R. (1997) Science 276, 561–567 [DOI] [PubMed] [Google Scholar]
- 3. Singer M. S., Gottschling D. E. (1994) Science 266, 404–409 [DOI] [PubMed] [Google Scholar]
- 4. Evans S. K., Lundblad V. (1999) Science 286, 117–120 [DOI] [PubMed] [Google Scholar]
- 5. DeZwaan D. C., Freeman B. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hughes T. R., Evans S. K., Weilbaecher R. G., Lundblad V. (2000) Curr. Biol. 10, 809–812 [DOI] [PubMed] [Google Scholar]
- 7. Lendvay T. S., Morris D. K., Sah J., Balasubramanian B., Lundblad V. (1996) Genetics 144, 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lundblad V., Szostak J. W. (1989) Cell 57, 633–643 [DOI] [PubMed] [Google Scholar]
- 9. Livengood A. J., Zaug A. J., Cech T. R. (2002) Mol. Cell. Biol. 22, 2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seto A. G., Livengood A. J., Tzfati Y., Blackburn E. H., Cech T. R. (2002) Genes Dev. 16, 2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chappell A. S., Lundblad V. (2004) Mol. Cell. Biol. 24, 7720–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seto A. G., Zaug A. J., Sobel S. G., Wolin S. L., Cech T. R. (1999) Nature 401, 177–180 [DOI] [PubMed] [Google Scholar]
- 13. Fisher T. S., Taggart A. K., Zakian V. A. (2004) Nat. Struct. Mol. Biol. 11, 1198–1205 [DOI] [PubMed] [Google Scholar]
- 14. Bianchi A., Negrini S., Shore D. (2004) Mol. Cell 16, 139–146 [DOI] [PubMed] [Google Scholar]
- 15. Osterhage J. L., Talley J. M., Friedman K. L. (2006) Nat. Struct. Mol. Biol. 13, 720–728 [DOI] [PubMed] [Google Scholar]
- 16. Tuzon C. T., Wu Y., Chan A., Zakian V. A. (2011) PLoS Genet. 7, e1002060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedman K. L., Heit J. J., Long D. M., Cech T. R. (2003) Mol. Biol. Cell 14, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J., Mandell E. K., Rao T., Wuttke D. S., Lundblad V. (2010) Nucleic Acids Res. 38, 2279–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharanov Y. S., Zvereva M. I., Dontsova O. A. (2006) FEBS Lett. 580, 4683–4690 [DOI] [PubMed] [Google Scholar]
- 20. Lingner J., Cech T. R., Hughes T. R., Lundblad V. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11190–11195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsu M., Yu E. Y., Singh S. M., Lue N. F. (2007) Eukaryot. Cell 6, 1330–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Friedman K. L., Cech T. R. (1999) Genes Dev. 13, 2863–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 25. Toogun O. A., Zeiger W., Freeman B. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5765–5770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chapon C., Cech T. R., Zaug A. J. (1997) RNA 3, 1337–1351 [PMC free article] [PubMed] [Google Scholar]
- 27. Lee J., Mandell E. K., Tucey T. M., Morris D. K., Lundblad V. (2008) Nat. Struct. Mol. Biol. 15, 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chan C. S., Tye B. K. (1983) Cell 33, 563–573 [DOI] [PubMed] [Google Scholar]
- 29. Toogun O. A., Dezwaan D. C., Freeman B. C. (2008) Mol. Cell. Biol. 28, 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Lange T. (2005) Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 31. Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007) Nature 445, 506–510 [DOI] [PubMed] [Google Scholar]
- 32. Yu E. Y., Wang F., Lei M., Lue N. F. (2008) Nat. Struct. Mol. Biol. 15, 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abreu E., Aritonovska E., Reichenbach P., Cristofari G., Culp B., Terns R. M., Lingner J., Terns M. P. (2010) Mol. Cell. Biol. 30, 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zaug A. J., Podell E. R., Nandakumar J., Cech T. R. (2010) Genes Dev. 24, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.