Abstract
Hepatocellular carcinoma (HCC) is inherently resistant to the majority of clinical anticancer drugs. To obtain drugs that can circumvent or evade such inherent drug resistance of HCC, we investigated the effect of the marinely derived steroid methyl spongoate (MESP) on HCC cells. MESP displayed potent cell killing against a panel of six HCC cell lines, independent of their expression of drug transporters. MESP did not change the function of the drug transporters, and its cell killing was not impaired in multidrug-resistant cancer cells overexpressing the transporters. The cell killing of MESP was irrelevant to estrogen or androgen signaling and was not associated with cell cycle progression, inhibition of microtubules, and topoisomerases. In contrast, MESP potently induced apoptosis via activation of a proapoptotic caspase cascade and relief of the suppression of antiapoptotic signal transducers and activators of transcription 3 (STAT3) signaling. MESP inhibited the phosphorylation of STAT3, a critical survival signaling factor that reduced the expression of the antiapoptotic protein x-linked inhibitor of apoptosis protein but enhanced the expression of the proapoptotic protein Bax, thus promoting caspase-dependent apoptosis. These data reveal that MESP may well serve as an important candidate drug lead for HCC therapy.
Keywords: Apoptosis, Caspase, Drug Resistance, Liver, Tumor, Bax, STAT3, XIAP
Introduction
Hepatocellular carcinoma (HCC)3 is one of the most common and most aggressive cancers worldwide. The majority of patients have a poor prognosis because of their advanced-stage disease, which is inherently resistant to clinical anticancer drugs, largely because HCC cells express drug transporters, including P-glycoprotein (P-gp), multidrug resistance protein (MRP), and/or breast cancer resistance protein (BCRP) (1–3). Therapy with current anticancer drugs could hardly change the natural history of inoperable HCC (over 80% of HCC cases) (2, 4). Discovery of new types of anticancer drugs, especially those rising superior to drug transporters, is primarily urgent to improve the therapy for HCC.
Natural products are extremely important sources of novel anticancer drugs. In fact, over 70% of anticancer drugs have a natural origin (5). The novelty and diversity of chemical structures of natural products give us opportunities to discover potential anticancer candidates with special activities. Several naturally derived drugs in clinical investigation, like salvicine (6–9) and ecteinascidin 743 (10, 11), show prominent actions in disrupting drug-resistant mechanisms by impairing the expression or functions of drug transporters in tumor cells of acquired multidrug resistance (MDR). These drugs set up a paradigm that guides the discovery of anti-HCC drugs from natural products that can evade or circumvent the mechanisms of intrinsic drug resistance, although not particularly against innate MDR as in liver cancer cells.
Steroids are important anticancer drug classes, especially for cancers of sex-related organs such as breast, ovary, and prostate. Those drugs, by mimicking or antagonizing estrogens, prolactin (12), or androgen (13), exert anticancer effect generally in a hormone (receptor)-dependent manner. Such a mode of action limits therapy to the cancers expressing the related hormone receptors or depending on the related hormone signaling pathways. Current steroid anticancer drugs in clinic are mainly synthetic or modified from the related hormones. As anticancer drug candidates, unfortunately, naturally derived steroids, either from land or from the sea, seem to have long been neglected.
MESP (cholest-1-en-3-one-20(R)-oic acid methyl ester, Fig. 1A) is a new, marinely derived steroid from the Sanya soft coral Spongodes sp. (14) and has been synthesized (15, 16). MESP is a unique steroid because of its rare 21-oic acid methyl ester moiety with 20R configuration (16). Such a uniqueness seems to be translated into its special HCC cell killing in a drug transporter-independent manner, as shown in this study. Although the precise structure-activity relationship remains to be clarified further, MESP provides an important drug lead for HCC therapy.
FIGURE 1.
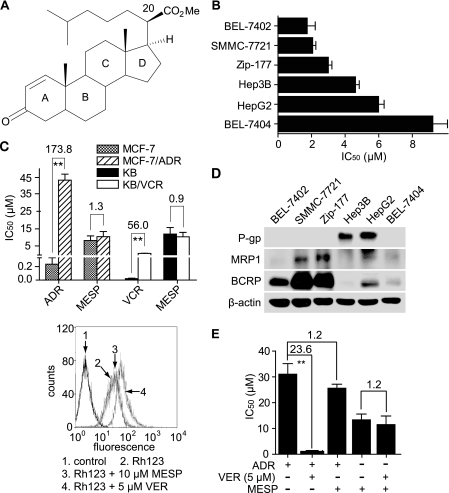
MESP inhibited HCC cell proliferation independent of drug transporters. A, chemical structure of MESP. B, MESP inhibited HCC cell proliferation assessed by SRB assays after 72-h treatments. C, MESP equally inhibited proliferation of MDR cells and their parental cells. IC50 and resistance factor were calculated as described under “Experimental Procedures.” **, p < 0.01. D, the expression of drug transporter proteins in HCC cell lines was determined by Western blotting. E, MESP did not affect the function of drug transporters. Left panel, Rh123 export assays; right panel, VER did not change the cellular sensitivity to MESP. MDR MCF-7/ADR cells were treated with 5 μm VER in combination with ADR or MESP for 72 h. IC50 and resistance factor were calculated as described under “Experimental Procedures.” **, p < 0.01.
EXPERIMENTAL PROCEDURES
Cell Proliferation Assays
The compound MESP was synthesized as described previously (16), with the purity over 99%.
Cell proliferation was assessed by sulforhodamine B (SRB) assays (17) for solid tumor cells or by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assays (18) for suspension cells as described previously (6, 8). IC50 was calculated with the logit method. The resistance factor (RF) for each drug was calculated as the ratio of the IC50 value of MDR sublines to that of the parental sublines.
Flow Cytometry
Flow cytometry was used to assess rhodamine 123 (Rh123) export, cell cycle progression, apoptosis (19), and mitochondria membrane potential (MMP) (20). For Rh123 export assays, MCF-7/ADR cells were pretreated with 5 μm verapamil (VER) (Sigma) or 10 μm MESP at 37 °C for 2 h and then treated with Rh123 (Beyotime Institute of Biotechnology, Haimen, China) in the dark for another 15 min. For cell cycle analyses, cells were treated with MESP for 24 h and subsequently harvested, fixed, and stained with 10 μg/ml propidium iodide (PI). For apoptosis assays, cells were incubated with MESP and/or caspase inhibitors, Z-VAD-FMK, AC-LEHD-FMK, and AC-IETD-FMK (Keygen Biotech, Nanjing, China), and then treated with an annexin V/PI apoptosis detection kit (Keygen Biotech). For MMP assays, cells were resuspended in 500 μl of PBS and incubated with 1 μm 5,5′,6,6′-tetrachloro-1,1′,3,3′- tetraethylimidacarbocyanine iodide (Beyotime Institute of Biotechnology) for 20 min at 37 °C. All samples were analyzed using a FACSCalibur cytometer (BD Biosciences). At least 10,000 events were counted for each sample.
Western Blotting
Cells were exposed to MESP for the indicated time. Protein levels were assessed by the standard Western blotting using whole cell lysates except cytochrome C, which was detected using the cytosol fraction isolated with a Pierce mitochondria isolation kit (Thermo Scientific, Rockford, IL).
Antibodies against α-tubulin, cytochrome C, phospho-STAT3 (Tyr-705), STAT3, and Bax were from Cell Signaling Technology (Beverly, MA). Antibodies against MRP1, BCRP, XIAP, death receptor 4 (DR4), and death receptor 5 (DR5) were from Abcam (Cambridge, MA), and antibody against β-actin was from Sigma. All other antibodies came from Santa Cruz Biotechnology (Santa Cruz, CA). Z-DEVD-FMK was from Keygen Biotech.
Immunofluorescence
Cells were exposed to 5 μm MESP, 0.5 μm taxol, or 0.5 μm vincristine (VCR) for 24 h and then collected for standard immunofluorescence assays using a BX51 fluorescence microscope (Olympus, Tokyo, Japan).
Topoisomerase I-mediated Supercoiled pBR322 Relaxation Assays and Topoisomerase II-mediated Kinetoplast DNA (kDNA) Decatenation Assays
TUNEL Assays
TUNEL assays were performed with an in situ cell death detection kit (Roche).
RNA Interference
Cells were transfected using the Lipofectamine RNAiMAX transfection reagent (Invitrogen) for 48 h. Two pairs of stat3 siRNAs (siRNA-1, 5′-GAG UUG AAU UAU CAG CUU ATT-3′; siRNA-2, 5′-GUU UGG AAA UAA UGG UGA ATT-3′) and the negative control were synthesized by GenePharma (Shanghai, China).
Real-time PCR
Total RNA extracted with the TRIzol reagent (Invitrogen) was reverse-transcribed into cDNA with the PrimeScript RT reagent kit (TaKaRa, Dalian, China). cDNA was amplified with the SYBR Premix EX TaqII Kit (TaKaRa, Dalian, China) in a 7500 fast real-time PCR system (Applied Biosystems, Carlsbad, CA). The PCR program was as follows: 95 °C, 30s; 40 cycles (for each cycle 95 °C, 5s; 64 °C, 20s; 72 °C, 15s); 72 °C, 10 min. All primers were synthesized by Sangon (Shanghai, China) as follows: (5′-3′) CCT GAA GCT GAC CCA GGT AGC (forward) and CAC CTT CAC CAT TAT TTC CAA ACT G (reverse) for stat3; TGC TTC AGG GTT TCA TCC AG (forward) and GGC GGC AAT CAT CCT CTG (reverse) for bax; TGG GAC ATG GAT ATA CTC AGT TAA CAA (forward) and GTT AGC CCT CCT CCA CAG TGA A (reverse) for XIAP.
Statistical Analyses
Data were presented as mean ± S.D., and differences were considered significant when the p value was <0.05 as determined by Student's t test. All data were obtained from three independent experiments.
RESULTS
MESP Kills HCC Cells Independently of Drug Transporters
The cytotoxicity of MESP was evaluated by SRB assays against a panel of six HCC cell lines. MESP displayed potent cell killing with an averaged IC50 of 4.4 μm in these cell lines but without apparent selectivity. BEL-7402 cells seemed to be the most sensitive to MESP, with the lowest IC50 of 1.7 μm (Fig. 1B).
Advanced HCC are generally resistant to conventional anticancer drugs because of the expression of drug transporters such as P-gp, MRP, and BCRP (1, 2). To examine whether those proteins affect the cellular sensitivity to MESP, we detected whether MDR tumor cells that overexpress drug transporter proteins were resistant to MESP. For this purpose, we used two MDR tumor cell lines, MCF-7/ADR and KB/VCR. However, both the MDR cells and the parental cells were equally sensitive to MESP (Fig. 1C). In contrast, the MDR cells were highly resistant to the reference anticancer drugs adriamycin (ADR) and VCR that were respectively used to establish the MDR sublines (6). The result indicates that the cellular sensitivity to MESP is not associated with the MDR proteins in those tested cells.
To clarify the relationship between the HCC cell killing of MESP and the expression of drug transporter proteins, we detected the levels of P-gp, MRP1, and BCRP in the tested HCC cells. The data revealed that the expression of those proteins in the six cell lines lacked the relevance to their sensitivity to MESP (Fig. 1, B and D). In particular, the cells (BEL-7404) with the lowest levels of the drug transporter proteins displayed the lowest sensitivity to MESP. Moreover, MESP did not affect the function of drug transporters because it did not change the cellular drug transporter-mediated export of Rh123 (Fig. 1E, left panel), and the classical MDR reversal agent VER did not enhance the cellular sensitivity to MESP (E, right panel) in MCF-7/ADR cells. All the evidence reveals that MESP kills HCC cells regardless of their status of the drug transporters.
Cancer Cell Killing of MESP Is Independent of Hormone Signaling, Cell Cycle, Microtubules, and Topoisomerases
MESP has a steroidal structure (Fig. 1A). As is well known, steroid hormones such as estrogen and androgen play vital roles in the initiation and progression of cancers like breast and prostate cancers. These hormones mediate their actions almost entirely by binding to their respective receptors and then function as transcription factors. Their antagonists, for example, tamoxifen, elicit significant anticancer effects by blocking the binding of hormones to their receptors in those steroid hormone-dependent cancers (23, 24). To clarify whether the anticancer effect of MESP relies on the steroid hormone signaling, we evaluated its proliferation inhibition against a panel of breast cancer cells expressing or not expressing estrogen receptors and prostate cancer cells expressing or not expressing androgen receptors. The data revealed no explicit relevance between the expression of the receptors and the potency of MESP to kill the cancer cells (Fig. 2A).
FIGURE 2.
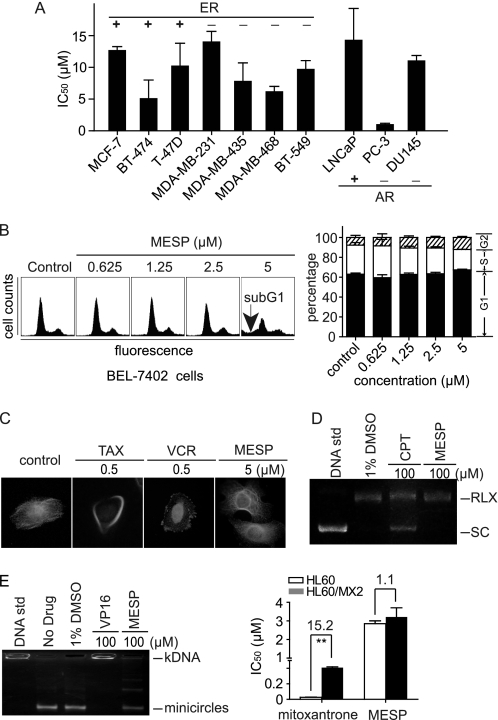
MESP did not affect hormone signaling, cell cycle, microtubules, and topoisomerases. A, MESP inhibited proliferation of breast cancer cells expressing or not expressing estrogen receptor (ER) or prostate cancer cells expressing or not expressing androgen receptor (AR). Cells were treated with MESP for 72 h and then subjected to SRB assays. B, MESP did not affect cell cycle progression. BEL-7402 cells were treated with MESP for 24 h, stained with PI, and analyzed by flow cytometry. Left panel, representative histograms; right panel, mean ± S.D. C, MESP did not interfere with polymerization of cellular microtubules. BEL-7402 cells were treated with different compounds for 24 h. Taxol (TAX) and VCR were used as reference drugs. D, MESP did not inhibit topoisomerase I-mediated supercoiled DNA relaxation. Plasmid pBR322 was incubated with topoisomerase I in the absence or presence of camptothecin (CPT) or MESP at 37 °C for 15 min. DNA samples were separated by electrophoresis on a 1% agarose gel. SC, supercoiled DNA; RLX, relaxed DNA. E, MESP did not inhibit topoisomerase II. Left panel, MESP did not inhibit the topoisomerase II-mediated kDNA decatenation. kDNA was incubated with topoisomerase II in the absence or presence of etoposide (VP16) or MESP at 37 °C for 15 min. DNA samples were separated by electrophoresis on a 1% agarose gel. Right panel, MESP equally inhibited proliferation of topoisomerase II-deficient (HL-60/MX2) and -proficient cells (HL60). Cells were treated with MESP for 72 h and then subjected to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assays. **, p < 0.01.
Cell cycle, microtubules, and topoisomerases are important targets for various conventional anticancer drugs. Flow cytometry showed that MESP could not change the cell cycle progression either in HCC BEL-7402 cells (Fig. 2B) or in SMMC-7721 cells (data not shown). In contrast, MESP at 5 μm induced a typical sub-G1 histogram (Fig. 2B, left panel), indicating the occurrence of apoptosis. Consistent with this result, MESP was shown by immunofluorescence assays not to interfere with the dynamic polymerization of microtubules in BEL-7402 cells (Fig. 2C). MESP did not affect the catalytic DNA cleavage activities of topoisomerases I and II in cell-free systems either (Fig. 2, D and E, left panels). The result was further validated by the equal sensitivity of the topoisomerase II-deficient HL60/MX2 cells and the parental HL60 cells to MESP (Fig. 2E, right panel). The data indicate that those conventional anticancer targets are not targeted by MESP.
In addition, we further used a panel of 15 tyrosine kinases, including EGF receptor, ErbB2/4, kinase domain receptor, fms-like tyrosine kinase, PDGF receptor, insulin-like growth factor 1 receptor, c-Met, and Abelson tyrosine kinase to detect the inhibition of MESP at 10 μm against their enzymatic activity by ELISAs, but no inhibition was found (data not shown).
Taken together, all results indicate that MESP kills cancer cells independent of hormone signaling, cell cycle, microtubules, topoisomerases, and some tyrosine kinase signaling.
MESP Induces Apoptosis
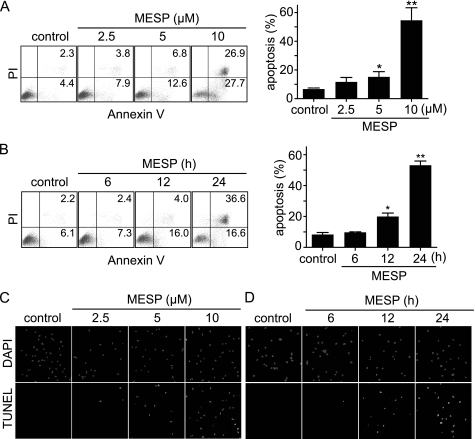
Apoptotic induction is one of the most important ways to kill tumor cells. As shown above, MESP could induce a typical sub-G1 histogram (Fig. 2B), with an apoptotic percentage of 17.1% in BEL-7402 cells at 5 μm, indicating its capability of apoptotic induction. To further characterize this capability, we conducted flow cytometry following annexin V/PI double staining. Fig. 3, A and B, showed that BEL-7402 cells treated with MESP underwent typical apoptosis in a concentration- and time-dependent manner. TUNEL assays also confirmed such a capability of MESP by revealing progressively rising fractions of the positive TUNEL-labeled BEL-7402 cells (Fig. 3, C and D). Consistently, MESP elicited typical apoptotic morphological changes in BEL-7402 cells, such as forming apoptotic bodies (data not shown) that were subsequent to the fragmentation of chromatins as reflected in the TUNEL assays.
FIGURE 3.
MESP induced apoptosis. A and B, MESP increased the percentage of annexin V-positive cells. Cells were treated with gradient concentrations of MESP for 24 h (A) or 10 μm MESP for the indicated time (B). Then the cells were stained with an annexin V/PI apoptosis detection kit for flow cytometry. Left panel, representative histograms; right panel, mean ± S.D. *, p < 0.05; **, p < 0.01. C and D, MESP increased TUNEL-positive cells. BEL-7402 cells were treated with MESP for 24 h (C) or 10 μm MESP for the indicated time (D).
MESP Activates an Apoptotic Caspase Cascade
Caspase-3 is the most important effector caspase in mammalian cells. Once activated via cleavage of procaspase-3 by the initiators caspase-8 and/or caspase-9, it cleaves many proteins, especially DNA repair proteins like poly (ADP-ribose) polymerase, leading to apoptosis (25, 26). Caspase-8, the initiator of the extrinsic apoptosis pathway, is activated by the death receptor signaling, whereas caspase-9, the initiator of the intrinsic apoptosis pathway, is activated via cytochrome C released from mitochondria triggered by irreparable genome damage or cellular stress. Cross-talk between the two pathways could be mediated by the caspase-8-mediated cleavage of Bid (Fig. 4A) (25). Additional cross-talk could also occur downstream of the effector caspase. In particular, active caspase-3 can activate caspase-8, which subsequently cleaves Bid (Fig. 4A) (27, 28). To investigate how MESP induces apoptosis, we detected and found that the treatment with MESP resulted in the cleavage of procaspase-3 and poly (ADP-ribose) polymerase (Fig. 4B).
FIGURE 4.
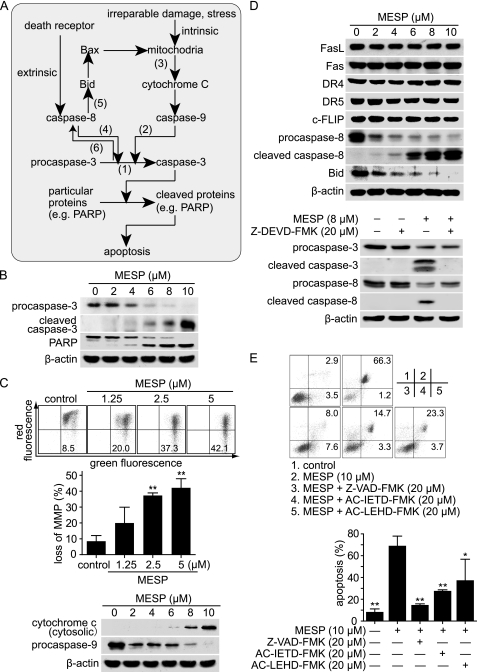
MESP induced apoptosis. A, simplified schematic representation of intrinsic and extrinsic apoptosis pathways and their cross-talk. The numbers (1–6) were labeled for the major stages examined in this section. B, MESP activated caspase-3 and induced the cleavage of poly (ADP-ribose) polymerase by Western blotting. C, MESP triggered mitochondria disruption and caspase-9 activation. Upper and center panels, MESP caused loss of MMP. BEL-7402 cells were treated with MESP for 24 h and then stained with 5,5′,6,6′-tetrachloro-1,1′,3,3′- tetraethylimidacarbocyanine iodide and analyzed by flow cytometry. Upper panel, representative histograms. Center panel, mean ± S.D. **, p < 0.01. Lower panel, MESP promoted the release of cytochrome C from mitochondria and activated caspase-9 analyzed by Western blotting. D, caspase-8 was activated by caspase-3. Upper panel, MESP activated caspase-8, which led to the cleavage of Bid. BEL-7402 cells were treated with MESP for 24 h and then subjected to Western blotting. Lower panel, caspase-3 inhibition abrogated the cleavage of caspase-8 induced by MESP. BEL-7402 cells were pretreated with the caspase-3 inhibitor Z-DEVD-FMK (20 μm) for 2 h followed by the treatment with MESP (8 μm) for an additional 24 h and then subjected to Western blotting. E, caspase inhibitors impaired MESP-induced apoptosis. Cells were pretreated with caspase inhibitors (20 μm) for 2 h followed by the treatment with MESP (10 μm) for an additional 24 h, stained with an annexin V/PI kit, and analyzed by flow cytometry. Upper panel, representative images. Lower panel, mean ± S.D. *, p < 0.05; **, p < 0.01.
To clarify how caspase-3 is activated, we then observed whether MESP affected the major components in apoptotic pathways. MESP was revealed to disrupt the integrity of mitochondria membrane as evidenced by the loss of MMP (20) and the release of cytochrome C into the cytosols, followed by the cleavage of procaspase-9 (Fig. 4C), indicating activation of the intrinsic pathway. MESP also led to activation of caspase-8 and subsequent cleavage of the apoptotic suppressor Bid (Fig. 4D). However, MESP did not throw any apparent impacts on the levels of Fas, FasL, DR4, DR5, and Cellular Fas-associated death domain-like interleukin-1β-converting enzyme inhibitory protein (c-FLIP) (Fig. 4D), all of which are important components in the extrinsic pathway and are located upstream of caspase-8 (29). Therefore, there appears to be a least a possibility of MESP-driven caspase-8 activation via the classical extrinsic apoptosis pathway.
So we asked whether caspase-8 activation driven by MESP was through activated caspase-3, as reported previously in other cases (27, 28). We thus pretreated BEL-7402 cells with the caspase-3 inhibitor Z-DEVD-FMK (20 μm), and this pretreatment resulted in almost complete abrogation of the cleavage of caspase-8 driven by MESP (Fig. 4D), indicating that caspase-3 is required for MESP-mediated caspase-8 activation in this situation.
To confirm the above result, we employed the pan-caspase inhibitor Z-VAD-FMK, the caspase-8 inhibitor AC-IETD-FMK, and the caspase-9 inhibitor AC-LEHD-FMK. The result showed that 20 μm Z-VAD-FMK almost completely abolished, but 20 μm AC-IETD-FMK or 20 μm AC-LEHD-FMK only partly abrogated, the apoptosis induced by 10 μm MESP in BEL-7402 cells (Fig. 4E). The result indicates that MESP induces apoptosis in a caspase-dependent manner in which both caspase-8 and caspase-9 participate.
MESP Inhibits Phosphorylation of STAT3 at Tyrosine 705
The next question is how MESP triggers apoptosis. To answer this question, we have observed but, unfortunately, not found any significant influence of MESP on some important upstream components, including Fas, FasL, DR4, DR5, c-FLIP (Fig. 4D), various kinases (data not shown), microtubules, and topoisomerases (Fig. 2). Those disappointing results made us turn our focus from the conventional proapoptotic factors to newly emerging antiapoptotic proteins.
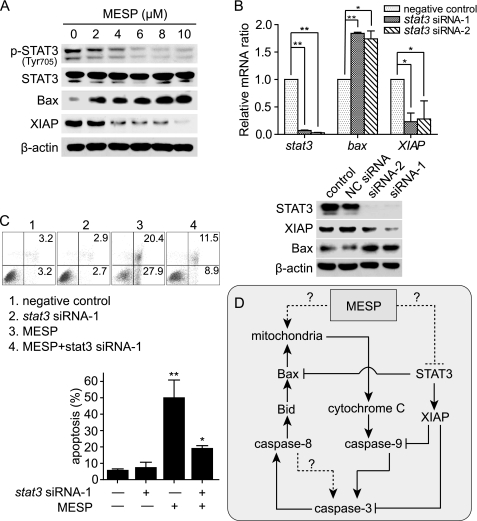
The transcription factor STAT3 has been shown to play important roles in antagonizing apoptosis (30–32). Once phosphorylated at its tyrosine 705, STAT3 is activated and stimulates the expression of antiapoptotic proteins like XIAP (30, 31), and thus STAT3 has been proposed as a promising anticancer target (30). Western blotting revealed that STAT3 was constitutively activated in BEL-7402 cells, and the treatment with MESP inhibited STAT3 phosphorylation at its tyrosine 705 and correspondingly decreased the expression of XIAP protein but did not change the levels of total STAT3 protein in BEL-7402 cells (Fig. 5A). Unexpectedly, however, the levels of Bax protein, an inherent proapoptotic factor, were enhanced (Fig. 5A). To clarify the relationship between the alterations of the levels of both XIAP and Bax proteins and the down-regulation of phospho-STAT3, we used two different pairs of stat3 siRNAs. At the levels of either mRNA or protein, as expected, the stat3 siRNAs resulted in decline of expression of both stat3 and XIAP but in enhancement of expression of bax (Fig. 5B). More importantly, the stat3 siRNA also significantly reduced apoptosis induced by MESP (Fig. 5C). Taken together, the data indicate that MESP induces apoptosis by inhibiting STAT3 and thus strengthening proapoptotic but weakening antiapoptotic factors (e.g. Bax versus XIAP). In contrast, MESP did not affect STAT1 or p53 or their phosphorylated forms (supplemental Fig. S1), suggesting that STAT1 or p53 does not contribute to the apoptotic induction of MESP.
FIGURE 5.
MESP inhibited phosphorylation of STAT3, which contributed to the MESP-induced apoptosis. A, MESP inhibited phosphorylation of STAT3 (Tyr-705) in BEL-7402 cells. Cells were treated with MESP for 24 h and then subjected to Western blotting. B, knockdown of stat3 by RNA interference promoted bax but inhibited XIAP expression at both mRNA (top panel) and protein (bottom panel) levels. Cells were transfected with two pairs of siRNAs against stat3 for 48 h. Real-time PCR and Western blotting were done. *, p < 0.05; **, p < 0.01. C, knockdown of stat3 abrogated MESP-induced apoptosis. Cells were transfected with the indicated siRNA for 48 h and then treated with MESP (10 μm) for another 24 h. Annexin V/PI assays were performed to evaluate the apoptosis percentage. Upper panel, representative histograms. Lower panel, mean ± S.D. *, p < 0.05; **, p < 0.01. D, a schematic representation of how MESP induces apoptosis. →, activation; ⊥, inhibition; broken lines, unknown.
DISCUSSION
HCC is one of the most refractory cancers because of its intrinsic resistance to current clinical anticancer drugs. In this study, we present a new prototype compound, MESP, with the capability of killing liver cancer cells regardless of their drug transporter status by apoptotic induction.
As a marinely derived steroid with a unique 21-oic acid methyl ester moiety in 20R configuration (16), in this study, MESP displays the following impressive characteristics: 1) MESP possesses a potent capability of nonselectively killing HCC cells with the IC50s of several μm, an ideal anticancer potency. 2) although of natural origin, MESP equally kills cancer cells expressing drug transporters (P-gp, MRP1 and BCRP) or not, totally different from the majority of naturally derived anticancer drugs in clinical use. 3) distinct from the clinically used anticancer drugs related to steroids, MESP elicits its cancer cell killing independent of the estrogen or androgen signaling. 4) MESP is likely to have its molecular target(s) that may differ from the conventional anticancer targets, including microtubules, topoisomerases, DNA, proteins involved in cell cycle progression, and several common kinases. The antiapoptotic protein STAT3 could be one of its candidate targets. 5) MESP kills HCC cells via apoptosis driven by a mitochondria disruption-triggered caspase cascade, which is potentiated by the relief of inhibition of the antiapoptotic STAT3 signaling. Together with its unique chemical structure, these features apparently allow MESP to be a promising anticancer prototype, especially against HCC.
Overexpression of drug transporters gives rise to tumor resistance to most naturally derived anticancer drugs in clinic. However, in recent years, several marine compounds such as ecteinascidin 743 (10), sipholenol A (33), and bryostatin-1 (34, 35), have been found to disrupt such drug resistance by differential mechanisms and to circumvent MDR. In contrast to those natural compounds, MESP was revealed not to perturb the expression or function of drug transporters, including P-gp, MRP1, and BCRP, but to evade the actions of those transporters. Thus, MESP displays potent activity in killing HCC cells, even expressing different drug transporters (e.g. SMMC-7721, Zip-177, and HepG2). Physiologically, those drug transporters function to detoxify endobiotics and xenobiotics, especially in the detoxification organs like the liver. Consequently, HCC cells generally express various drug transporters that lead to the intrinsic resistance of HCC to the majority of anticancer drugs in clinic. Disrupting drug transporters is likely to impair the physiological functions of the organs expressing them. Therefore, MESP could have an advantage over anticancer agents like ecteinascidin 743, sipholenol A, and bryostatin-1 because it kills HCC cells but might not affect the physiological functions of drug transporters. From this point of view, MESP might have a relatively low toxicity.
It is interesting that MESP has possibly unconventional molecular target(s) with which its interference induces apoptosis rather than cell cycle arrest. This mode of action indicates that MESP is distinct from many clinical anticancer drugs that kill cancer cells following their arresting cell cycle. Such independence of cell cycle progression suggests a possibility that MESP may exert the least impact on those tissues with rapid turnover, such as bone marrow and small intestine epithelium. This further supports its potentially low systematic toxicity.
Consistent with its rare chemical structure, MESP induces apoptosis in an apparently unique way. On one hand, MESP disrupted MMP, which led to the release of cytochrome C into cytosols and subsequently activated caspase-9. The active caspase-9 thus activated the effector caspase-3, resulting in a typical apoptotic phenotype. Moreover, the active caspase-3 further activated caspase-8, which cleaved the antiapoptotic protein Bid and thus abrogated its inhibition on the proapoptotic factor Bax. Consequently, the release of cytochrome C and thereby the activation of caspase-9 were potentiated. In this process, feedback activation of a caspase cascade is clearly manifested, which is initiated by MESP-driven cytochrome C release-mediated caspase-9 activation, followed by caspase-3 and then caspase-8 cleavage, and strengthened by the cross-talk between caspase-8 and caspase-9 via Bid and Bax (Fig. 5D). On the other hand, MESP also relieves the apoptotic suppression of STAT3 signaling by reducing its constitutive activation (phosphorylation at Tyr-705) and thus abrogating its inhibition on Bax, caspase-3, and caspase-9 (Fig. 5D), which in turn promotes the activity of the abovementioned caspase cascade. All those characteristics collectively reveal the uniqueness of the MESP-driven caspase cascade because it is apparently different from the caspase cascade via the classical intrinsic and extrinsic apoptosis pathways, although they share caspases (Figs. 4A and 5D).
Our results further indicate that MESP could regulate the balance between the antiapoptotic and the proapoptotic strength in cancer cells, relieving the former (e.g. XIAP) but enhancing the latter (e.g. Bax). In particular, we reveal in this study for the first time that bax is likely to be a target gene of the transcription factor STAT3. STAT3 appears to negatively modulate the expression of the bax gene because reduction of the STAT3 activity, either with MESP or with specific stat3 siRNAs, leads to the increased expression of bax at both protein and mRNA levels. This finding shows that STAT3 promotes cell survival not only via positively regulating antiapoptotic genes but also via negatively regulating proapoptotic genes, giving new insights into the functions of STAT3 and the strategy of targeting STAT3 for cancer therapy. Considering the elevated activity of STAT3 in a wide variety of human tumors and tumor cell lines (36), in particular, HCC (37–39), the ability of MESP to inhibit STAT3 signaling may allow itself to be outstanding for HCC therapy. Therefore, MESP could be a new potent apoptosis modulator characteristic of activating a unique caspase cascade, partly via alleviating the apoptotic suppression of STAT3.
Unfortunately, MESP did not show obvious in vivo anticancer activity in human HCC xenografts in nude mice (data not shown), possibly because of its unsatisfactory pharmacokinetic property. Nevertheless, together with its potent in vitro HCC cell killing independent of drug transporters and distinctive apoptotic induction, the unique chemical structure of MESP provides a promising anticancer drug lead for further modification and optimization from which a new type of drugs for HCC therapy may well arise.
Supplementary Material
This study was supported by National Natural Science Foundation of China Grants 81025020, 81021062, 21072204 and 30730108); National Basic Research Program of China Grant 2010CB934000; Major Program of Knowledge Innovation of the Chinese Academy of Sciences Grant KSCX1-YW-22; Science and Technology Commission of Shanghai Municipality Grant 09540704100; and Chinese Academy of Sciences, Key Project Grant SIMM0907KF-09.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Methods.
- HCC
- hepatocellular carcinoma
- MESP
- methyl spongoate
- P-gp
- P-glycoprotein
- MRP
- multidrug resistance protein
- BCRP
- breast cancer resistance protein
- MDR
- multidrug resistance
- SRB
- sulforhodamine B
- Rh123
- rhodamine 123
- MMP
- mitochondria membrane potential
- VER
- verapamil
- PI
- propidium iodide
- XIAP
- x-linked inhibitor of apoptosis protein
- DR
- death receptor
- VCR
- vincristine
- ADR
- adriamycin
- STAT3
- signal transducers and activators of transcription 3.
REFERENCES
- 1. Papatheodoridis G. V., Lampertico P., Manolakopoulos S., Lok A. (2010) J. Hepatol. 53, 348–356 [DOI] [PubMed] [Google Scholar]
- 2. Thomas M. (2009) J. Gastroenterol. 44, 136–141 [DOI] [PubMed] [Google Scholar]
- 3. Severi T., van Malenstein H., Verslype C., van Pelt J. F. (2010) Acta Pharmacol. Sin. 31, 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee T. K., Castilho A., Ma S., Ng I. O. (2009) Liver Int. 29, 955–965 [DOI] [PubMed] [Google Scholar]
- 5. Newman D. J., Cragg G. M. (2007) J. Nat. Prod. 70, 461–477 [DOI] [PubMed] [Google Scholar]
- 6. Miao Z. H., Tang T., Zhang Y. X., Zhang J. S., Ding J. (2003) Int. J. Cancer 106, 108–115 [DOI] [PubMed] [Google Scholar]
- 7. Miao Z. H., Ding J. (2003) Cancer Res. 63, 4527–4532 [PubMed] [Google Scholar]
- 8. Miao Z. H., Tong L. J., Zhang J. S., Han J. X., Ding J. (2004) Int. J. Cancer 110, 627–632 [DOI] [PubMed] [Google Scholar]
- 9. Cai Y., Lu J., Miao Z., Lin L., Ding J. (2007) Cancer Biol. Ther. 6, 1794–1799 [DOI] [PubMed] [Google Scholar]
- 10. Barthomeuf C., Bourguet-Kondracki M. L., Kornprobst J. M. (2008) Anticancer Agents Med. Chem. 8, 886–903 [DOI] [PubMed] [Google Scholar]
- 11. Kanzaki A., Takebayashi Y., Ren X. Q., Miyashita H., Mori S., Akiyama S., Pommier Y. (2002) Mol. Cancer Ther. 1, 1327–1334 [PubMed] [Google Scholar]
- 12. LaPensee E. W., Ben-Jonathan N. (2010) Endocr. Relat. Cancer 17, R91–107 [DOI] [PubMed] [Google Scholar]
- 13. Isaacs J. T. (2010) Expert Opin. Investig. Drugs 19, 1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan X. H., Lin L. P., Ding J., Guo Y. W. (2007) Bioorg Med. Chem. Lett. 17, 2661–2663 [DOI] [PubMed] [Google Scholar]
- 15. Jiang C. S., Huang C. G., Feng B., Li J., Gong J. X., Kurtán T., Guo Y. W. (2010) Steroids 75, 1153–1163 [DOI] [PubMed] [Google Scholar]
- 16. Gong J. X., M. Z., Yao L. G., Ding J., Kurtan T., Guo Y. W. (2010) Synlett, 480–482 [Google Scholar]
- 17. Skehan P., Storeng R., Scudiero D., Monks A., McMahon J., Vistica D., Warren J. T., Bokesch H., Kenney S., Boyd M. R. (1990) J. Natl. Cancer Inst. 82, 1107–1112 [DOI] [PubMed] [Google Scholar]
- 18. Mosmann T. (1983) J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 19. Yin J., Wan Y. J., Li S. Y., Du M. J., Zhang C. Z., Zhou X. L., Cao Y. J. (2011) Acta Pharmacol. Sin. 32, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salvioli S., Ardizzoni A., Franceschi C., Cossarizza A. (1997) FEBS Lett. 411, 77–82 [DOI] [PubMed] [Google Scholar]
- 21. Meng L. H., Zhang J. S., Ding J. (2001) Biochem. Pharmacol. 62, 733–741 [DOI] [PubMed] [Google Scholar]
- 22. Tanabe K., Ikegami Y., Ishida R., Andoh T. (1991) Cancer Res. 51, 4903–4908 [PubMed] [Google Scholar]
- 23. Msaouel P., Diamanti E., Tzanela M., Koutsilieris M. (2007) Expert Opin. Emerg. Drugs 12, 285–299 [DOI] [PubMed] [Google Scholar]
- 24. Peng J., Sengupta S., Jordan V. C. (2009) Anticancer Agents Med. Chem. 9, 481–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grutter M. G. (2000) Curr. Opin. Struct. Biol. 10, 649–655 [DOI] [PubMed] [Google Scholar]
- 26. Timmer J. C., Zhu W., Pop C., Regan T., Snipas S. J., Eroshkin A. M., Riedl S. J., Salvesen G. S. (2009) Nat. Struct. Mol. Biol. 16, 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Slee E. A., Harte M. T., Kluck R. M., Wolf B. B., Casiano C. A., Newmeyer D. D., Wang H. G., Reed J. C., Nicholson D. W., Alnemri E. S., Green D. R., Martin S. J. (1999) J. Cell Biol. 144, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang D., Lahti J. M., Kidd V. J. (2000) J. Biol. Chem. 275, 9303–9307 [DOI] [PubMed] [Google Scholar]
- 29. Giménez-Bonafé P., Tortosa A., Pérez-Tomás R. (2009) Curr. Cancer Drug. Targets 9, 320–340 [DOI] [PubMed] [Google Scholar]
- 30. Al Zaid Siddiquee K., Turkson J. (2008) Cell Res. 18, 254–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fletcher S., Drewry J. A., Shahani V. M., Page B. D., Gunning P. T. (2009) Biochem. Cell Biol. 87, 825–833 [DOI] [PubMed] [Google Scholar]
- 32. Masuda M., Wakasaki T., Suzui M., Toh S., Joe A. K., Weinstein I. B. (2010) Curr. Cancer Drug Targets 10, 117–126 [DOI] [PubMed] [Google Scholar]
- 33. Abraham I., Jain S., Wu C. P., Khanfar M. A., Kuang Y., Dai C. L., Shi Z., Chen X., Fu L., Ambudkar S. V., El Sayed K., Chen Z. S. (2010) Biochem. Pharmacol. 80, 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He X., Fang L., Wang J., Yi Y., Zhang S., Xie X. (2008) Cancer Res. 68, 8678–8686 [DOI] [PubMed] [Google Scholar]
- 35. Shukla S., Ohnuma S., Ambudkar S. V. (2011) Current Drug Targets 12, 621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buettner R., Mora L. B., Jove R. (2002) Clinical Cancer Research 8, 945–954 [PubMed] [Google Scholar]
- 37. Kusaba M., Nakao K., Goto T., Nishimura D., Kawashimo H., Shibata H., Motoyoshi Y., Taura N., Ichikawa T., Hamasaki K., Eguchi K. (2007) J. Hepatol. 47, 546–555 [DOI] [PubMed] [Google Scholar]
- 38. Rajendran P., Ong T. H., Chen L., Li F., Shanmugam M. K., Vali S., Abbasi T., Kapoor S., Sharma A., Kumar A. P., Hui K. M., Sethi G. (2011) Clin. Cancer Res. 17, 1425–1439 [DOI] [PubMed] [Google Scholar]
- 39. Liu Y., Liu A., Xu Z., Yu W., Wang H., Li C., Lin J. (2011) Apoptosis 16, 502–510 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.