Abstract
The region in promoters that specifies the transcription machinery is called the core promoter, displaying core promoter elements (CPE) necessary for establishment of a preinitiation complex and the initiation of transcription. A classical CPE is the TATA box. In fission yeast, Schizosaccharomyces pombe, a new CPE, called HomolD box, was discovered. Collectively, 141 ribosomal protein genes encoding the full set of 79 different ribosomal proteins and more than 60 other housekeeping genes display a HomolD box in the core promoter. Here, we show that transcription directed by the HomolD box requires the RNA polymerase II machinery, including the general transcription factors. Most intriguingly, however, we identify, by DNA affinity purification, Rrn7 as the protein binding to the HomolD box. Rrn7 is an evolutionary conserved member of the RNA polymerase I machinery involved in transcription initiation of core ribosomal DNA promoters. ChIP shows that Rrn7 cross-links to a ribosomal protein gene promoter containing the HomolD box but not to a promoter containing a TATA box. Taken together, our results suggest that Rrn7 is an excellent candidate to be involved in the coordination of ribosomal DNA and ribosomal gene transcription during ribosome synthesis and, therefore, offer a new perspective to study conservation and evolvability of regulatory networks in eukaryotes.
Keywords: Promoters, Ribosomal RNA (rRNA), Ribosomes, RNA Polymerase, Transcription, GTF, TATA-less
Introduction
Ribosome biogenesis in eukaryotes is a highly coordinated process involving three different RNA polymerases. RNA polymerase III (RNAPIII)3 synthesizes the small 5 S RNA. RNAPI synthesizes the large rRNA precursor. Transcription at ribosomal RNA core promoters is initiated by the TATA binding protein (TBP) and TBP-associated factors (TAF1s) forming a preinitiation complex (PIC). In human cells, this complex is called SL1 and contains TBP and the TAF1s TAF1110, TAF163, TAFI48, and TAFI41. The TAF1 equivalents in fission and budding yeast are called Rrn6, Rrn7, and Rrn11, respectively, and are found with TBP in a complex called core factor (1, 2). The minor subunits of SL1 have no equivalents in yeast. RNAPII, finally, transcribes the ribosomal protein-encoding genes (3). RNAPII-dependent transcription also requires a PIC. In a TATA box containing core promoters, the first step in the formation of the PIC is the binding of TBP to the TATA box. TBP binds to the TATA box complexed with TFIID (TAF11s) followed by the other GTFs TFIIB, TFIIF, TFIIE, and TFIIH, respectively, to complete PIC formation for the recruitment of pol II (4–6). However, it is also discussed that a preassembled holoenzyme including pol II, TBP, and the GTFs are targeted with the help of the mediator complex onto the DNA in the core promoter region for PIC formation (7, 8).
The 141 ribosomal protein genes of fission yeast encoding the full set of 79 ribosomal proteins are TATA-less promoters. Instead, they all contain the highly conserved sequence CAGTCACA or its inverted form, TGTGACTG, within 100 bp upstream of the ATG start codon. This sequence was termed the HomolD box. It was shown to mediate transcription initiation in vivo, but even more interesting, it also determines the transcriptional start sites, thus functioning as a TATA analog in a core promoter (9–12). On the basis of these experimental findings and followed by computational analyses, it was suggested that Schizosaccharomyces pombe genes involved in ribosome synthesis contain a core promoter in which the HomolD box is the CPE, which in some of these promoters is joined immediately upstream by a highly conserved sequence called the HomolE box (9–12). Indeed, a computational analysis comparing the regulatory modules in ribosomal protein (RP) gene promoters of many yeast species confirms these observations and presents the HomolD and HomolE elements found in S. pombe promoters as a remarkable example of regulatory divergence from the budding yeast Saccharomyces cerevisiae, where the RAP1- and the IFHL sites represent the regulatory modules in RP gene promoters (13).
Interestingly, in the last few years of intensive computational analysis of genomic sequences from different organisms, it turned out that the percentage of classical TATA box-containing core promoters of protein-encoding genes identified in genomes is low. In fact, it has been described that in human and mice promoters, the prevalence of TATA-less promoters is 90% (14–16) Therefore, we searched the Eukaryotic Promoter Database for HomolD box sequences in regions that are defined as core promoters. There are several core promoter regions that contain a HomolD box, including human core promoters, found upstream of several protein-encoding genes such as ATPase and G-protein subunits (unpublished data4). Although the total number of HomolD box-containing core promoters in genomic sequences is small, it appears that promoters with a HomolD box are found throughout the genomes of eukaryotic organisms.
In this report we demonstrate for the first time that HomolD-directed transcription is RNAPII-dependent and requires GTFs from the RNAPII machinery. We identify a protein fraction with binding activity to a HomolD box affinity column. This protein fraction contained Rrn7, which is a transcription initiation factor of the RNAPI machinery operating at core rDNA promoters. We demonstrate that Rrn7 has HomolD box-specific binding activity in vitro and in vivo. These results clearly make Rrn7 an excellent candidate to be the mediator of the cross-talk between rDNA and ribosomal protein gene transcription during ribosome biogenesis.
EXPERIMENTAL PROCEDURES
Promoter Construction
The rpK5 promoter was cloned by PCR using specific primers (forward, 5′-TGCAAAAGAAGTGTCTGCGTTAAAGCATCA-3′; and reverse 5′-GGGGTTGTTGG AGAATACGGTATTGAGGTTGCA-3′) that contain BamHI and SmaI recognition sites, respectively. Genomic DNA purified from strain 972 h−s of fission yeast S. pombe was used as template. The PCR products generated were digested and ligated in the BamHI/SmaI sites of pUC19. The G-minus cassette was amplified by PCR from a vector called pG5, as described previously (17, 18). Specific primers (forward, 5′-CATACCCTTCCTCCATCTATACC-3′; and reverse, 5′-TGGAATGAGAAATGAGTGTGA-3′) were used and combined with the rpK5 promoter fragment from above. The vector containing the rpK5 promoter G-minus cassette was linearized with EcoRI and used as template for PCR amplification using specific primers. Three promoter constructs were made using primers with the HomolD box wild-type sequence (5′-TTTGGATAGGCGAAAACAGTCACAT TTTACAACAACAATTCAC-3′), with two point mutations in the Homol D box (5′-TTTGGATAGGCGAAAACTGTGACATTTTACAACAACAATTCAC-3′), and with a HomolD box inverted (5′-TTTGGATAGGCGAAAAGTCAGTGTTTTTACAACAACAATTCAC-3). The reverse primer was the same for all three constructs (5′-TAGATTTGGGAAATATAGA AGAAG-3′).
Primer Extension and in Vitro Transcription Assays
For primer extension assays, 3 μg of each construct were mixed with 12 μl of p10X buffer (500 mm HEPES (pH 7.9), 900 mm l-glutamic acid potassium salt, 150 mm magnesium acetate, 50 mm EGTA, 25 mm DTT, 10% glycerol), 7 μl of PEG 20K, 4 μl of S. pombe whole cell extract (WCE). The reaction mix was incubated for 10 min at room temperature, and then 12 μl of E10X mixture (4 mm rATP, 4 mm rCTP, 4 mm rGTP, 4 mm rUTP, 80 mm phosphoenole pyruvic acid), 0.8 μl of RNasin (Promega Corp.), and H-O buffer (20 mm HEPES (pH 7.9), 2 mm EGTA, 5 mm DTT, 0.1 mm PMSF, 10% glycerol) were added to a final volume of 120 ml. The reaction mix was incubated for 45 min at 30 °C, and then stop solution (10 mm EDTA, 200 mm sodium acetate, 0.2% SDS, 1 mg/ml DNA salmon sperm) was added, followed by phenol extraction and ethanol precipitation. The RNA pellet was resuspended in 14 μl of water and annealed with the 32P-labeled oligonucleotide 5′-TAGATTTGGGAAATATAGAAGAAG-3′. 100 units of M-MLV reverse transcriptase (Promega Corp.) and dNTPs (0.5 mm of each) were added and incubated for 30 min at 37 °C. cDNA products were ethanol-precipitated, resuspended in 95% formamide, denatured, and separated on an 6% polyacrylamide (40:1 ratio) denaturing gel. Gels were dried and exposed to film. In vitro transcription was performed as described previously using 0.5 μg of DNA in the assay (17). For immunodepletion, WCE was incubated at room temperature for 30 min with 1 μg of the appropriate antibody. Then 0 or 100 ng of the specific general transcription factor was added to the assay.
EMSA
Each binding mix contained 20 mm HEPES (pH 7.9), 50 mm KCl, 5 mm MgCl2, 0.1 mm EDTA, 5% glycerol, 2% PEG 20K, 2 mm DTT, 0.1 mm PMSF, 50 μg BSA, and 500 ng of poly(dI.dC) for semipurified extracts or 25 ng for affinity fractions or recombinant protein. The proteins were incubated with the binding mix for 5 min at room temperature, and then 20–40 ng end-labeled HomolD box probe from the rpK5 gene or HomolD* probe from the rDNA promoter (approximately 20,000 cpm) was added and incubated for 10 min at 30 °C. In competitive binding assays, a 10-, 100-, and 200-fold excess of unlabeled probe was added. The DNA-protein complexes were analyzed at 4 °C in an 8% acrylamide, 0.2% bisacrylamide (39:1 ratio), 50 mm Tris borate (pH 8.3) gel. The gel was prerun at 4 °C for 1 h at 100 V. The gel was dried and exposed to film for 3 days. For Kd determination, three independent assays were performed with a constant amount of recombinant protein and increasing concentrations of labeled probe containing HomolD or HomolD* from 5 to 2000 nm. PhosphorImager was used for quantification, and the amount of protein-bound and protein-free labeled DNA probe in each assay was determined. Data were fitted, and saturation and Scatchard plots were generated using GraphPad Prism version 4.00 software for Mac OSX (GraphPad Software, San Diego, CA).
Purification of Rrn7
Bacterial BL21 (DE3) cells were transformed with pET15b containing the ORF of Rrn7 and plated on Luria-Bertoni-agar containing ampicillin. A single colony was inoculated in 500 ml of Terrific Broth medium supplemented with ampicillin (0.1 mg/ml) and grown at 37 °C to an A600 nm of 0.8. The production of recombinant proteins was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside, and the culture was grown at 37 °C for an additional period of 4 h. The cells were harvested by centrifugation and resuspended in 20 ml of STE buffer (100 mm NaCl, 10 mm Tris-Hcl (pH 8.0), 1 mm EDTA) with lysozyme (0.35 mg/ml) and incubated for 30 min at room temperature. Then, the cells were sonicated, and the recombinant protein was purified as described previously (17).
ChIP
ChIP was performed with slight modifications, as described by Jbel et al. (19). Chromatin was isolated from a wild-type strain (972 h−s), a strain expressing TAP-Rrn7, and a strain expressing both TAP-Rrn7 and HA-Rrn11. Immunoprecipitations were performed using 2 μg of anti-TAP tag (A00683, GenScript) or anti-HA tag (ab9110, Abcam, Inc.). Primer sequences were designed for the rpK5 gene core promoter, including the HomolD box, TSSs, and the first 11 amino acid residues of the protein. Forward, 5′ GGATGACCGAAAACAGTCAC 3′; and reverse, 5′ CCTCCACTCTTTCTTTGTGC 3′). Primer pairs for amplification of the nmt1 gene core promoter: forward, 5′ GGAATCCGATTGTCATTCGG 3′; and reverse, 5′ GGAAAGTGATCTTGTTAGTAG 3′. Primers pairs for amplification of the rDNA core promoter (HomolD*): forward, 5′ GGAGGGATATGGAAGAAAGG 3′; and reverse, 5′ CTCCTTTCAACCACCACTCC 3′. As an internal control, primers were designed for the amplification of parts of the open reading frame of the act1 gene (20).
Immunoprecipitation
Yeast cells were grown to an A600 nm of 0.5 in 25 ml of YPD medium (1% yeast extract, 2% peptone, 2% D-glucose). Cells were harvested and washed with ice-cold IP buffer (50 mm Tris HCl (pH 8.0), 150 mm NaCl) supplemented with 0.1% Nonidet P-40 and 0.1 mm PMSF (wash buffer). Cells were resuspended in 0.5 ml of wash buffer supplemented with 0.1 mm DTT. Glass beads (G8772, Sigma) were added to the cells, and then the mix was placed on vortex for 30 min at 4 °C. The lysates were transferred to a new tube. Glass beads were washed with 0.5 ml of wash buffer, and supernatant was added to the other 0.5 ml of lysate. 1-ml aliquots were centrifuged at top speed at 4 °C, and supernatants were placed in a new tube. 30 μl of protein A-agarose beads were preincubated with 2 μg of anti-TAP tag (A00683, GenScript) or anti-HA tag (ab9110, Abcam) for 1 h at 4 °C and washed three times with IP buffer plus 0.1% Nonidet P-40. These beads were added to the lysates. After 1 h of incubation, beads were spun down and washed three times with ice-cold wash buffer and twice with only IP buffer. Finally, the beads were resuspended with 50 μl of 1× SDS-PAGE sample buffer, and the supernatants were analyzed by Western blot analysis.
Other Experimental Procedures
The DNA affinity column was constructed with CNBr-activated-Sepharose as described by Kadonaga and Tjian (21). Fission yeast strains expressing TAP-Rrn7 and HA-Rrn11 were constructed as described in Newo et al. (22).
RESULTS
The Transcription Directed in Vitro by a HomolD Box Core Promoter Is Dependent on the Pol II System
We and others have developed a highly purified in vitro transcription system for S. pombe RNAPII transcription. Using typical TATA box containing promoters fused to a G-minus cassette, it was shown that the GTFs TFIID (Tbp1 + TAFs), TFIIB, TFIIE, TFIIF, and TFIIH are necessary and sufficient for proper initiation and basal transcription from these TATA box promoter templates (23–25).
Therefore, to investigate whether HomolD box containing promoters are transcribed in vitro by the RNAPII system, we fused the HomolD box-containing promoter of the ribosomal protein gene rpK5 (GeneDB, SPAC1F7.13c) to a G-minus cassette. Three templates were constructed as shown in Fig. 1A. The HomolD box (CAGTCACA, WT) in this promoter has been shown to initiate basal transcription of the lacZ gene as a reporter in vivo and determines two TSSs 49 and 51 nt downstream of the HomolD box, whereas mutations in the HomolD box to CTGTGACA (mut) abolished expression of the reporter gene in vivo (9). The third template contains the sequence TGTGACTG (inv), which is the inverted form of the HomolD box (WT) found in many genes containing this core promoter (12 and introduction). To study whether basal transcription can be initiated from these templates, we used S. pombe WCE as an in vitro system. As shown in Fig. 1B, the WCE is able to support basal transcription from the template containing the HomolD box WT and inv, respectively, whereas the template containing the mutated HomolD box (mut) does not show an in vitro transcript. Addition of α-amanitine, a strong inhibitor of RNAPII, to the reaction abolished transcription from both HomolD box core promoters (WT, inv) indicating that RNAPII is the enzyme that initiates transcription from these promoters (Fig. 1B). Furthermore, primer extension analysis after the reaction shows that basal transcription in vitro is initiated at two TSSs downstream of the HomolD box (WT and inv) comparable with the situation found in vivo (Fig. 1C, 9).
FIGURE 1.
Requirements of transcription directed by a HomolD-box. A, the HomolD box-containing promoter of the rpK5 gene of S. pombe was fused to a G-minus cassette, and transcription was evaluated by in vitro assays. −94 and −10 indicate the positions upstream of the ATG codon. Three templates were generated: HomolD box (CAGTCACA, WT), mutant (CTCTGACA, mut) and the inverted form (TGTGACTG, inv). B, the in vitro transcription assays were carried out with WCE and the indicated cassettes as template. Transcription products are indicated with an arrow. Addition of α-amanitine to the assays is indicated (+). C, primer extension assays were performed with WCE and a radiolabeled primer for reverse transcription hybridizing to the G-minus cassette. TSSs are indicated with arrows. The sequencing ladder shows the sequence of the inverted template. D, immunodepletion was carried out with WCE using 1 μg of each antibody. Protein-depleted WCE was used in transcription assays as in B. The GTFs are indicated above the lanes. Addition of the appropriate GTF is indicated by + below the lanes. The HomolD box WT cassette was used as a template. Preimmune serum was added to the WCE, indicated by + above the lane. Addition of antibodies to the assays is indicated by + below the lanes.
To determine whether the GTFs of the RNAPII system are involved in the initiation of transcription from the HomolD box template (WT), WCEs were depleted of the GTFs TBP (Tbp1), TAF1, TFIIB, TFIIE, TFIIH, and TFIIF using antibodies against each GTF. Antibodies were made against whole proteins (Tbp1, TAF1, and TFIIB) or specific subunits (TFIIEβ, Tfb1, and TFIIFβ) (supplemental Fig. S2). In the depleted extracts, basal transcription was initiated from the HomolD box template only when the appropriate purified GTF was added (Fig. 1D). These results suggest that HomolD box core promoters in S. pombe are transcribed in vitro by the RNAPII system.
Identification of a Protein Fraction with Binding Activity to the HomolD Box
Our earlier attempts to characterize HomolD core promoters led us to suggest that the HomolD box is most likely the target of a protein with different biochemical features than the TBP in fission yeast called Tbp1 (9, 12). Therefore, we set out to isolate binding activity to HomolD boxes by fractionating WCE as indicated in Fig. 2A. As probes in EMSAs to measure binding activity, we used oligonucleotides containing the HomolD box WT and mut, respectively, of the rpK5 promoter shown in Fig. 1A. Both WCE and the heparin fraction showed specific binding activity. Increasing amounts of unlabeled probe (WT) competed out the shifted complex(es), whereas the unlabeled probe mut does not act as a strong competitor (Fig. 2B and supplemental Fig. S2).
FIGURE 2.
Identification of a protein fraction with specific HomolD box binding activity. A, schematic representation of the purification of the heparin fraction. WCE (400 mg) was loaded onto an S-Sepharose column equilibrated with in S-Q buffer (20 mm HEPES (pH 7.9), 20 mm KCl, 0.1 mm EDTA, 2 mm DTT, 10% glycerol, 0.1 mm PMSF). The proteins that did not interact with the column were recollected and loaded onto a Q-Sepharose column equilibrated in S-Q buffer. The proteins were eluted in a linear gradient from 0.1 to 0.5 m KCl in S-Q buffer, and fractions were analyzed by EMSA. Fractions selected were dialyzed against H buffer (20 mm HEPES (pH 7.9), 10 mm MgCl2, 1 mm EDTA, 50 mm ammonium sulfate, 2 mm DTT, 10% glycerol, 0.1 mm PMSF). These proteins were eluted in a linear gradient from 50 to 700 mm ammonium sulfate in H buffer, and fractions were evaluated by EMSA. This protein fraction was called heparin fraction. B, the heparin fraction was evaluated by EMSA. Aliquots were assayed using a radiolabeled oligonucleotide containing the HomolD box (WT) of the rpK5 gene. For competitive assays, a 10-, 100-, and 200-fold molar excess of unlabeled WT or mut probes were added (Fig. 1A). -, EMSA without proteins added; +, EMSA without unlabeled probes added; f.p., free probe. C, EMSA with purified GTFs. Increasing amounts of recombinant Tbp1 and TFIIB and purified TFIID proteins (containing TAFs and Tbp1) were analyzed by EMSA using the HomolD box WT probe. -, EMSA without proteins added; +, EMSA without unlabeled probes added. D, EMSA with proteins of the heparin fraction eluted from the HomolD affinity column. -, EMSA without protein fractions added. The heparin fraction loaded (at 50 mm KCl) onto the column is indicated as input (IN). Material that did not bind to the matrix is indicated as flowthrough (FT). The column was eluted with 0.1 m KCl and 0.5 m KCl, respectively, as indicated. The arrow indicates shifted bands. E, the proteins eluted from the HomolD box affinity column were separated by SDS-PAGE and stained with Coomassie blue. Wash fractions (50 mm KCl) and fractions eluted at 0.1 mm and 0.5 mm KCl, respectively, are indicated (D). The three major protein bands visualized are marked K1, K2, and K3.
In addition, we examined whether TBP (Tbp1) binds to HomolD box-containing DNA. Because several TAFs of the TFIID complex as well as TFIIB have been shown to bind to DNA and have been described to participate in PIC establishment in TATA-less promoters (26–28), we also checked TFIID and TFIIB for binding activity. As can be seen in Fig. 2C, the HomolD-containing rpK5 promoter does not bind the GTFs TFIIB, TFIID, or TBP alone. Neither of the indicated purified protein fractions led to a shift of the labeled HomolD probe in EMSAs. These results confirm the previous in vivo studies and demonstrate that the HomolD box is not targeted by Tbp1 (9).
To identify the HomolD box binding protein(s), we chose to isolate the protein(s) of the heparin fraction by using an affinity purification step. For this purpose, the HomolD box was coupled to CNBr-activated-Sepharose, and the heparin protein fraction was bound to the HomolD matrix under low-salt conditions (50 mm KCl). Subsequently, the bound material was washed with 0.1 m KCl and then eluted using a buffer with a high salt concentration (0.5 m KCl). All fractions were evaluated for binding activity with EMSA using the WT HomolD box-containing promoter as a probe. The eluate still shows binding activity, as indicated by the shift of the HomolD probe migrating to positions identical with the heparin fraction (I) that was applied to the column (Fig. 2D).
The fractions eluted at 0.1 and 0.5 m KCl, respectively, were separated on SDS-PAGE and stained with Coomassie blue. Three major bands can be visualized in the fractions eluted from the column with 0.5 m KCl (Fig. 2E).
Identification of Rrn7 as the Protein Binding to the HomolD Box
The proteins eluted from the HomolD box affinity column were determined by mass spectrometry. After SDS-PAGE, the three stained bands (Fig. 2E, K1, K2, K3) were individually subjected to in-gel digestion, and the eluted peptides were sequenced and identified by liquid chromatography-coupled tandem mass spectrometry (LC/MS/MS). With this approach, we detected in band K1 with the highest peptide coverage a protein called Sec8 (UniProtKB O74562, located in secretory vessels and involved in cell elongation and septum formation). Band K2 revealed a mannose-1-phosphate guanyltransferase (UniProtKB O74484, Mpg1, involved in cell wall integrity and septum formation). Finally, band K3 contained peptides of a protein called Rrn7 (UniProtKB Q9UST5, RNA polymerase I-specific transcription initiation factor). To the best of our knowledge, there is no report or any other indication that Sec8 or Mpg1 might be DNA binding proteins or involved in transcription. However, Rrn7 has been characterized in S. pombe and has been shown to be the orthologue of human TAF163, which binds in a complex (selectivity factor 1 (SL1) in human and core factor in yeast) with TBP to the rDNA core promoters to mediate transcription initiation by RNAPI. In addition, the core rDNA promoters of S. pombe contain a TATA box element around position −35 upstream of the TSS which, when mutated, impairs but does not prevent transcription initiation of the core rDNA promoter (29, 30).
On the basis of these considerations, we decided to evaluate the heparin fraction for binding affinity to the rDNA core promoter using a 20-bp probe from −46 to −65 nt upstream of the TSS that lacks the TATA element (29). Noteworthy here is that this sequence of 20 bp shows a region of the rDNA core promoter that is broadly similar to the 40-bp HomolD box probe used here, including the HomolD sequence. We call this sequence in the rDNA core promoter HomolD* (Fig. 3A). As can be seen in Fig. 3B, the heparin fraction shows specific binding activity when WT rDNA (HomolD*) is used as a probe. Even at low concentrations, the unlabeled WT rDNA outcompetes the shifted complex easily, whereas the unlabeled probe mut rDNA hardly acts as a competitor.
FIGURE 3.
Rrn7 has HomolD box binding activity. A, 20 bp were used as a WT rDNA probe. When aligned with the 40-bp wt HomolD probe, the best fit shows nine identical nucleotide positions (+), including the HomolD box indicated between asterisks. In mut rDNA and mut HomolD, the mutated nucleotides are shown in gray. In scrambled HomolD, the nucleotides of the HomolD box have been randomized. B, the heparin fraction was evaluated by EMSA to determine rDNA promoter binding activity. Aliquots were assayed using end-labeled double-stranded oligonucleotides containing the region of the rDNA promoter as shown in A. In competitive assays, a 10- and 200-fold molar excess of unlabeled WT rDNA and mut rDNA was added. C, recombinant Rrn7 was analyzed by EMSA to evaluate its binding activity to the rDNA promoter. 100 ng of Rrn7 was added using the same conditions as described in B. D, recombinant Rrn7 was evaluated by EMSA to determine its HomolD box binding activity. A radiolabeled probe containing the WT HomolD box was used. In competitive assays, a 200-fold molar excess of unlabeled probes was added. -, assay without proteins; +, protein and labeled HomolD box probe; WT, cold WT HomolD as competitor added; scrl., scrambled HomolD as competitor added; mut, mut HomolD as competitor added; f.p., free probe. The arrow indicates shifted bands.
There is no indication that the heparin fraction contains, besides Rrn7, other polymerase I transcription factors. Therefore, we used recombinant Rrn7 and determined the binding affinity of the recombinant protein to the HomolD box probe as well as to the rDNA (HomolD*) probe. For this purpose, we performed EMSAs and competition assays. Recombinant Rrn7 binds to the rDNA (HomolD*) probe, causing a mobility shift of the WT rDNA, which cannot be competed with by unlabeled mut rDNA (Fig. 3C). Recombinant Rrn7 also binds to the HomolD box probe, which easily can be competed with by the unlabeled wt HomolD probe. The unlabeled scrambled HomolD probe, in which the nucleotides of the HomolD box have been randomized, does not compete at all for binding. In addition, the unlabeled mut HomolD probe displaying the two point mutations, which abolish in vitro transcription, also does not compete effectively for binding (Fig. 3D). To determine the dissociation constant of recombinant Rrn7, we performed an EMSA with a constant amount of protein while increasing the concentration of the probe. The Kd determined for HomolD was 248.7 ± 35.7 nm and 198.9 ± 44.7 nm for HomolD* (supplemental Fig. S3). These results show that the affinity for both sequences is low but suggest that binding of Rrn7 to the HomolD box as well as to the rDNA (HomolD*) sequence is possible without additional binding partners. We also investigated whether recombinant Rrn7 can direct in vitro transcription from the HomolD box promoter. After depletion of endogenous Rrn7 and adding recombinant Rrn7 to WCE, we recovered in an in vitro transcription assay significant transcription activity (supplemental Fig. S3). This suggests that Rrn7 is acting at HomolD box promoters independently of the other core factor complex members.
Rrn7 Associates with the HomolD Box Core Promoter of the rpK5 Gene in Vivo
In a first attempt to determine whether Rrn7 associates with HomolD box core promoters in vivo, we used the chromatin IP method, in which proteins are cross-linked in vivo to DNA. The presence of individual proteins near specific DNA sequences is monitored by IP of the protein followed by PCR analysis of the coprecipitated DNA region. For this purpose, we fused epitope tags to Rrn7 and replaced the wild-type rrn7+ gene with its TAP-tagged version, now expressing TAP-Rrn7 proteins. We also fused an HA epitope tag to Rrn11 and replaced the wild-type rrn11+gene with the HA-tagged version. Finally, we constructed a strain that coexpresses TAP-Rrn7 and HA-Rrn11. First, we tested whether the fusion protein TAP-Rrn7 still has DNA binding activity. Second, we tested whether TAP-Rrn7 coprecipitates HA-Rrn11. TAP-Rrn7 still binds in vitro to the rDNA core promoter region, and TAP-Rrn7 coprecipitates with HA-Rrn11 and vice versa (supplemental Fig. S4).
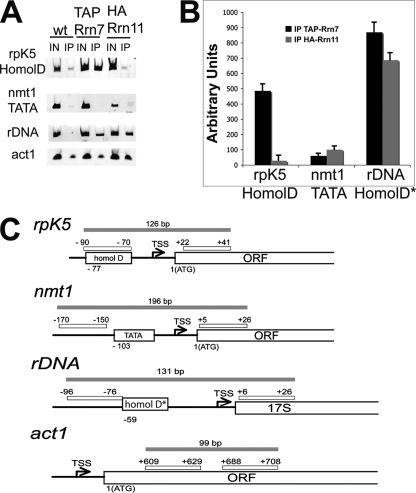
Next, we immunoprecipitated chromatin (ChIP) from cells expressing Rrn7 and Rrn11 (WT) and from cells expressing TAP-Rrn7 and HA-Rrn11 using IgG and anti-HA antibodies, respectively. The immunoprecipitates were tested for the presence of the core HomolD box promoter of the rpK5 gene by PCR using the primer pair as indicated in Fig. 4C. The rpK5 gene is one of three genes encoding ribosomal protein Rpl8, which was used to characterize HomolD promoters in vivo (9). The promoter of the rpK5 gene displays a classical HomolD box promoter. As can be seen in Fig. 4, A and B, IP of TAP-Rrn7 followed by PCR reveals the presence of the rpK5 HomolD promoter, indicating that TAP-Rrn7 is associated with this promoter region. In contrast, IP of HA-Rrn11 followed by PCR clearly shows that Rrn11 is not associated with the rpK5 HomolD promoter region (Fig. 4, A and B). However, when the IPs are followed by PCR using specific primers for the putative HomolD core promoter of an rDNA transcription unit (HomolD* in Fig. 4C), TAP-Rrn7 as well as HA-Rrn11 coprecipitate the core promoter region of the rDNA unit, thus indicating that Rrn7 and Rrn11 bind to the rDNA core promoter. The results presented here clearly demonstrate that TAP-Rrn7, but not HA-Rrn11, associates specifically with the HomolD box promoter of the ribosomal protein gene, whereas no association of Rrn7 can be detected with the classical TATA box core promoter of the nmt1 gene (Fig. 4, A and B).
FIGURE 4.
Rrn7 associates with the HomolD box core promoter of the rpK5 gene in vivo. A, cross-linked and sonicated extracts from WT-expressing Rrn7 and Rrn11 as well as from cells coexpressing TAP-Rrn7 and HA-Rrn11 were immunoprecipitated using α-TAP and α-HA antibodies, respectively. The coprecipitating DNA was amplified by PCR using gene-specific primer pairs for the HomolD box core promoter of the rpK5 gene, for the TATA-box core promoter of the nmt1 gene, for the putative HomolD box core promoter (called HomolD*) of the rDNA gene, and, as internal control, for the open reading frame of the act1 gene as indicated in C. IN, input WCE; IP, immunoprecipitate. B, the PCR signal of the rpK5 HomolD core promoter DNA fragment, of the nmt1 TATA box fragment and of the rDNA fragment in each immunoprecipitate was normalized to the signal of the act1 PCR product. The signal intensities are expressed in arbitrary units. The mean values of at least three independent experiments were calculated.
DISCUSSION
The data we presented in this report are significant in several respects. First, HomolD boxes found in promoters of the ribosomal protein genes in fission yeast as well as in about 60 other housekeeping gene promoters represent TATA-less promoters. In these promoters, the HomolD box serves to initiate RNAPII-dependent transcription and determines the TSSs. The GTFs of the pol II system, including TBP, are recruited, but neither TFIID nor TBP alone binds directly to the HomolD box promoter sequence (Figs. 1 and 2). Thus, we provide evidence that the HomolD box is a genuine new CPE. In fact, HomolD box promoter elements are found in the genomes of other eukaryotic organisms. Second, we identified Rrn7 with a HomolD DNA affinity purification procedure, and we demonstrated that Rrn7 is the protein that directly binds to the HomolD box promoter in vitro (Fig. 3). Surprisingly, Rrn7 is a protein that has been shown to be involved in transcription initiation of rDNA promoters in fission yeast as well as in other eukaryotes. The orthologues of Rrn7 and Rrn11, human TAF163 and TAF148, respectively, are found complexed in SL1 containing TBP and the TAF1s TAF1110, TAF1 63, TAF148, and TAF141 (2, 30–32). Third, we show that Rrn7 binds directly to rDNA core promoter sequences that show a similarity to HomolD boxes and their sequence context but are not identical with it. We call this region in the rDNA core promoter HomolD*. Noteworthy here is that we show for the first time sequence signatures to which the complex binds and demonstrate by ChIP analysis that, indeed, Rrn7 and Rrn11 are associated in vivo with the core promoter of the rDNA gene, including HomolD* (Figs. 3 and 4). Fourth, in an attempt to demonstrate the involvement of Rrn7 in the transcription of ribosomal protein genes in vivo, we show by ChIP analysis that Rrn7 is associated with the HomolD box core promoter of the ribosomal protein gene rpK5 but is not associated with the TATA box-containing promoter of the nmt1 gene. In addition, Rrn11 is not associated with the HomolD core promoter of the rpK5 gene (Fig. 4).
Taken together, our results presented here and previously support the notion that in fission yeast the regulatory network of ribosome biogenesis is based on promoter modules that clearly differ from those established in S. cerevisiae (13). It has been speculated that the transcriptional system involved in coordinating ribosome synthesis in fission yeast may represent an ancient eukaryotic transcription system (12, 33, 34). Here we provide evidence that in this system the transcription initiation factor Rrn7 is the link between pol I transcription of the rDNA genes and pol II transcription of the ribosomal protein genes. The finding of few but significant numbers of housekeeping genes containing a HomolD box in mammalian genes confirms the notion of the ancient transcription system. Therefore, it will be interesting and worthwhile, besides the further elucidation of the fission yeast system, to investigate the mechanism of transcription of HomolD box promoters in mammalian cells.
Supplementary Material
Acknowledgments
We thank Dr. Lothar Jänsch and his team at the Helmholtz Center for Infection Research, Braunschweig, Germany, for the mass spectroscopy analyses of the heparin fraction. We also thank Dr. Martin Lützelberger for critically reading the manuscript and for valuable suggestions.
This work was supported by Fondo Nacional de Ciencia y Tecnología Grant 1080222.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
J. Contreras-Levicoy, S. Moreira-Ramos, D. A. Rojas, F. Urbina, and E. Maldonado, unpublished data.
- RNAP
- RNA polymerase
- TBP
- TATA binding protein
- TAF
- TATA binding protein-associated factor
- PIC
- preinitiation complex
- Pol II
- polymerase II
- CPE
- core promoter element
- RP
- ribosomal protein
- rDNA
- ribosomal DNA
- WCE
- whole cell extract
- TSS
- transcription start site
- IP
- immunoprecipitation
- inv
- inverted
- mut
- mutated.
REFERENCES
- 1. Laferté A., Favry E., Sentenac A., Riva M., Carles C., Chédin S. (2006) Genes Dev. 20, 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siddiqi I., Keener J., Vu L., Nomura M. (2001) Mol. Cell. Biol. 21, 2292–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rudra D., Warner J. R. (2004) Genes Dev. 18, 2431–2436 [DOI] [PubMed] [Google Scholar]
- 4. Orphanides G., Reinberg D. (2002) Cell 108, 439–451 [DOI] [PubMed] [Google Scholar]
- 5. Thomas M. C., Chiang C. M. (2006) Crit. Rev. Biochem. Mol. Biol 41, 105–178 [DOI] [PubMed] [Google Scholar]
- 6. Sikorski T. W., Buratowski S. (2009) Curr. Opin. Cell Biol. 21, 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hahn S. (2004) Nat. Struct. Mol. Biol. 11, 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esnault C., Ghavi-Helm Y., Brun S., Soutourina J., Van Berkum N., Boschiero C., Holstege F., Werner M. (2008) Mol. Cell 31, 337–346 [DOI] [PubMed] [Google Scholar]
- 9. Witt I., Straub N., Käufer N. F., Gross T. (1993) EMBO J. 12, 1201–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Witt I., Kwart M., Gross T., Käufer N. F. (1995) Nucleic Acids Res. 23, 4296–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gross T., Käufer N. F. (1998) Nucleic Acids Res. 26, 3319–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Witt I., Käufer N. F. (2004) in: The Molecular Biology of Schizosaccharomyces pombe (Egel R. ed), pp. 343–351, Springer, Berlin [Google Scholar]
- 13. Tanay A., Regev A., Shamir R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7203–7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C. A., Taylor M. S., Engström P. G., Frith M. C., Forrest A. R., Alkema W. B., Tan S. L., Plessy C., Kodzius R., Ravasi T., Kasukawa T., Fukuda S., Kanamori-Katayama M., Kitazume Y., Kawaji H., Kai C., Nakamura M., Konno H., Nakano K., Mottagui-Tabar S., Arner P., Chesi A., Gustincich S., Persichetti F., Suzuki H., Grimmond S. M., Wells C. A., Orlando V., Wahlestedt C., Liu E. T., Harbers M., Kawai J., Bajic V. B., Hume D. A., Hayashizaki Y. (2006) Nat. Genet. 38, 626–635 [DOI] [PubMed] [Google Scholar]
- 15. Kim TH., Barrera L. O., Zheng M., Qu C., Singer M. A., Richmond T. A., Wu Y., Green R. D., Ren B. (2005) Nature 436, 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cooper S. J., Trinklein N. D., Anton E. D., Nguyen L., Myers R. M. (2006) Genome Res 16, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tamayo E., Bernal G., Teno U., Maldonado E. (2004) Eur. J. Biochem. 271, 2561–2572 [DOI] [PubMed] [Google Scholar]
- 18. Sawadogo M., Roeder R. G. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4394–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jbel M., Mercier A., Pelletier B., Beaudoin J., Labbé S. (2009) Eukaryot. Cell 8, 649–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du L. L., Nakamura T. M., Russell P. (2006) Genes Dev. 20, 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kadonaga J. T., Tjian R. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 5889–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newo A. N., Lützelberger M., Bottner C. A., Wehland J., Wissing J., Jänsch L., Käufer N. F. (2007) Nucleic Acids Res. 35, 1391–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flores O., Maldonado E., Reinberg D. (1989) J. Biol. Chem. 264, 8913–8921 [PubMed] [Google Scholar]
- 24. Maldonado E., Ha I., Cortes P., Weis L., Reinberg D. (1990) Mol. Cell. Biol. 10, 6335–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Contreras-Levicoy J., Urbina F., Maldonado E. (2008) FEBS J. 275, 2873–2883 [DOI] [PubMed] [Google Scholar]
- 26. Verrijzer C. P., Chen J. L., Yokomori K., Tjian R. (1995) Cell 81, 1115–1125 [DOI] [PubMed] [Google Scholar]
- 27. Burke T. W., Kadonaga J. T. (1997) Genes Dev. 11, 3020–3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee D. H., Gershenzon N., Gupta M., Ioshikhes I. P., Reinberg D., Lewis B. A. (2005) Mol. Cell. Biol. 25, 9674–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boukhgalter B., Liu M., Guo A., Tripp M., Tran K., Huynh C., Pape L. (2002) Gene. 291, 187–201 [DOI] [PubMed] [Google Scholar]
- 30. Russell J., Zomerdijk J. C. (2005) Trends Biochem. Sci. 30, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zomerdijk J. C., Beckmann H., Comai L., Tjian R. (1994) Science 266, 2015–2018 [DOI] [PubMed] [Google Scholar]
- 32. Comai L., Zomerdijk J. C., Beckmann H., Zhou S., Admon A., Tjian R. (1994) Science 266, 1966–1972 [DOI] [PubMed] [Google Scholar]
- 33. Hamada M., Huang Y., Lowe T. M., Maraia R. J. (2001) Mol. Cell. Biol. 21, 6870–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nabavi S., Nazar R. N. (2008) Curr. Genet. 54, 175–184 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.