Abstract
Dendritic cells (DCs) and macrophages (MFs) are important multifunctional immune cells. Like other cell types, they express hundreds of different microRNAs (miRNAs) that are recently discovered post-transcriptional regulators of gene expression. Here we present updated miRNA expression profiles of monocytes, DCs and MFs. Compared with monocytes, ∼50 miRNAs were found to be differentially expressed in immature and mature DCs or MFs, with major expression changes occurring during the differentiation. Knockdown of DICER1, a protein needed for miRNA biosynthesis, led to lower DC-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) and enhanced CD14 protein levels, confirming the importance of miRNAs in DC differentiation in general. Inhibition of the two most highly up-regulated miRNAs, miR-511 and miR-99b, also resulted in reduced DC-SIGN level. Prediction of miRNA-511 targets revealed a number of genes with known immune functions, of which TLR4 and CD80 were validated using inhibition of miR-511 in DCs and luciferase assays in HEK293 cells. Interestingly, under the cell cycle arrest conditions, miR-511 seems to function as a positive regulator of TLR4. In conclusion, we have identified miR-511 as a novel potent modulator of human immune response. In addition, our data highlight that miRNA influence on gene expression is dependent on the cellular environment.
Keywords: Dendritic Cells, Gene Expression, Microarray, MicroRNA, Toll-like Receptors (TLR), CD80, Mannose Receptor
Introduction
Dendritic cells (DCs)2 and macrophages (MFs) are important immune cells both capable of pathogen recognition, phagocytosis, antigen presentation, and cytokine production. However, DCs are more specialized for the presentation of antigens, and therefore they have the ability to migrate from tissues to local lymph nodes where they interact with T cells (1, 2). Compared with DCs, MFs act more locally and are considered the main phagocytic cells, removing pathogens and cell debris that is generated either during tissue remodeling associated with apoptosis or during inflammation (1, 3).
In human peripheral blood, DCs represent only ∼0.5% of leukocytes. However, DCs and MFs are relatively easy to differentiate ex vivo from human blood monocytes (MOs), which represent ∼10% of human blood mononuclear cells (4, 5). Depending on locations and progenitors, there exist many different DC and MF populations. Ex vivo, in the presence of granulocyte macrophage colony-stimulating factor (GM-CSF) and IL4 or GM-CSF only, human blood MOs differentiate into the cells, which resemble inflammatory DCs or classically activated MFs and possess MHCII+DC-SIGN(CD209)+CD14− and MHCII−DC-SIGN−CD14+ phenotypes, respectively. After stimulation through pathogen recognition receptors, such as toll-like receptors (TLRs), they achieve activated characteristics and produce higher levels of CD86, CD83, and pro-inflammatory cytokines. Remarkably, the ex vivo differentiated DCs are efficient in stimulation of CD4- and CD8-positive T cells and are also the most common type of DCs used in immunotherapeutic approaches (6, 7).
MicroRNAs (miRNAs) are 21–23-nucleotide-long single-stranded RNAs, which together with partner proteins mainly cause gene silencing by degradation of target mRNAs or inhibition of translation (8–11). However, upon cell cycle arrest, miRNAs can up-regulate the translation as it was first shown for miR-369-3, which functions as positive regulator of the TNF-α protein response to quiescence (12, 13).
Several miRNAs have been shown to be important in differentiation and functioning of immune cells (14, 15). For instance, miR-146 has been confirmed to regulate inflammatory responses in several different cell types (16–18) and is implicated in the multiple cancers and inflammatory diseases (14, 19). The other well studied miRNA in immune cells is miR-155, which is required for normal function of mouse DCs, MFs, and B and T cells (20–22). In DCs and MFs, miR-155 is up-regulated during maturation and influences both the differentiation and immune responses (23–26). More recently, miR-34 and miR-21 were shown to be important for human MO-derived DC differentiation by targeting the mRNAs encoding Jagged1 and WNT1 (27). miR-21 also targets PDCD4 and thereby acts as negative regulator of TLR4 in mouse MFs (28). In addition, several other miRNAs are involved in regulation of components of the TLR signaling system (29).
Based on early releases of miRBase, the expression profiles of miRNAs in human blood MOs, DCs, and MFs have been determined (23, 27, 30). However, the expression and roles of more recently discovered miRNAs in DCs and MFs are not described. The aim of this study was to find novel miRNAs important in MO-derived DC and MF differentiation and functions. Consistent with the previous studies, we found that MOs, DCs, and MFs have distinct miRNA expression profiles, whereas the major changes in miRNA expression occurred during differentiation of MFs and DCs. The most highly up-regulated miRNAs were subjected to further studies, which point to the role of miR-511 in DC differentiation, and importantly, in the regulation of the TLR4 protein level.
EXPERIMENTAL PROCEDURES
Differentiation of DCs and MFs
This study on human blood-derived MOs is approved (Approval 166/T-10) by Ethics Review Committee on Human Research of the University of Tartu. All of the human participants gave written informed consent. Human MOs and MO-derived DCs and MFs were essentially prepared according to Ref. 4 and as described. MOs were purified from freshly collected “buffy coats” obtained from Blood Centre of Tartu University Hospital. PBMCs were prepared by density gradient centrifugation on Ficoll-PaqueTM Plus (GE Healthcare), and MOs were further purified by positive sorting using anti-CD14-conjugated magnetic microbeads using two runs through LS columns (both from Miltenyi Biotech). MOs were differentiated into MFs using 50 ng/ml GM-CSF and into DCs using 50 ng/ml GM-CSF and 25 ng/ml IL4 (both from PeproTech) by growing 6 days at a concentration of 1 million cells/ml in RPMI 1640 supplemented with 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FCS (all from PAA Laboratories). For maturation, 1 μg/ml of LPS (InvivoGen) or 0.1 mg/ml of curdlan (Wake Chemicals) was added to the growing media for 24 h.
Transfection of Anti-miR Inhibitors and siRNAs
Predesigned anti-miRNA inhibitors, Silencer Select Validated DICER1 siRNAs s23754, s23756, and respective negative controls (Applied Biosystems) were used in Fig. 2. Locked nucleic acid-based miR-511 inhibitor and unlabeled control A (Exiqon) in Fig. 3 and supplemental Fig. S5. All of the transfections were carried out at the concentration of 120 nm siRNA or miRNA inhibitors using 3 μl of siPORT NeoFX Transfection agent for 106 cells/1 ml medium (Applied Biosystems). After the transfection procedure, MOs were differentiated as usual. Transfection efficiency was controlled by fluorescence microscopy of separate transfections by Cy3-labeled negative control miRNA inhibitor or negative control siRNAs and was estimated to be between 90 and 100%.
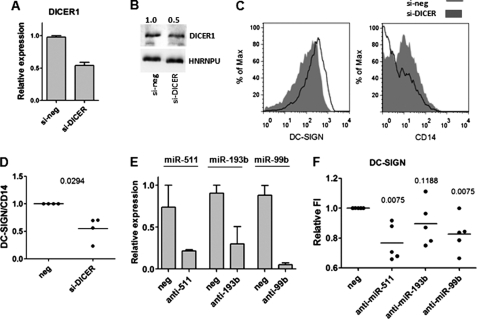
FIGURE 2.
Reduced expression of DICER1 and selected miRNAs results in down-regulation of DC-specific surface marker DC-SIGN. A, inhibition of DICER1 (si-DICER) is shown as an average mRNA expression level with S.E. of three different treatments normalized to average of the negative controls (which equals 1, si-neg). B, Western analysis of differentiating DCs treated with control DICER1 siRNA. The data are normalized to the hnRNPU, the numbers indicate the fold difference compared with the control transfection (which equals 1). C and D, FACS analysis of DCs treated with either si-DICER or control siRNAs. C, histograms represent the geometric mean fluorescent intensities of indicated surface molecules shown as a percentage of maximum in DICER1 siRNA-treated cells (filled gray) relative to control treated cells (black line). D, ratios of DC-SIGN and CD14 mean fluorescence intensity values were calculated and are shown relative to the control transfections (neg, which equal 1), data from four different donors are blotted, and the p value is calculated using Mann-Whitney test. E, inhibition of individual miRNAs is shown as the average miRNA expression level with S.E. of three different treatment normalized to the average levels of control transfections (which equal 1, neg). F, FACS analyses of DCs treated with indicated miRNA inhibitors. Geometric mean fluorescence intensities were normalized to control transfected cells (which equal 1, neg) and are presented as relative fluorescence intensities (FI), data from five different donors are blotted, and the p values are calculated using the Mann-Whitney test.
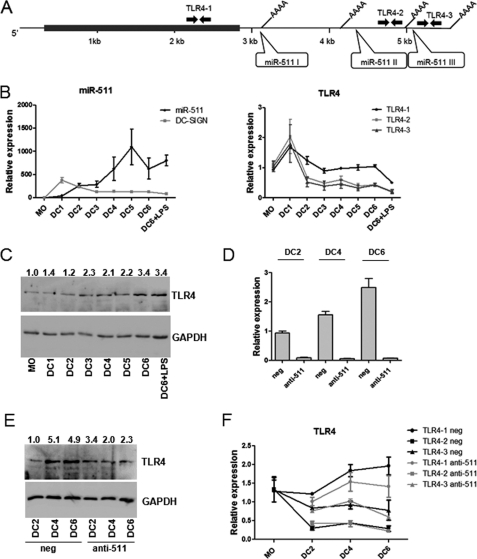
FIGURE 3.
Expression of miR-511 correlates with the enhanced level of the TLR4 protein in differentiating DCs. A, schematic of the TLR4 mRNA transcript containing miR-511 target sites, alternative polyadenylation sites, and positions of RT-PCR primers (designated with arrows). B, the RNA expression levels of miR-511, DC-SIGN, and TLR4 transcripts in differentiating DCs. The data are one representative of three independent experiments. RT-PCR of specific genes are normalized either to HPRT or let-7a and the expression levels in MOs (which equal 1). C and E, Western analysis of TLR4 in differentiating DCs (C) and in the presence of miR-511 inhibitor (anti-511) or the control inhibitor (neg) (E) are normalized to the GAPDH; the numbers indicate the fold difference compared with the MOs (which equals 1, C) or control transfected cells on day 2 (DC2, neg, E). D, inhibition of miR-511 is shown as average miRNA expression level with S.E. of two parallel treatment normalized to the levels of control transfection on day 2 (DC2, neg, which equal 1).
Cloning and Plasmids
For pGL3–3′-UTR reporters, the following 3′-UTR fragments of TLR4 (NM_138554.3): TLR4-I (3096–3736), TLR4-II (4588–5000), TLR4-III (5028–5384), and CD80 (NM_005191.3.) (2184–2525) were PCR-amplified, digested with FseI (New England Biolabs) and XbaI (Fermentas), and inserted downstream from the luciferase (LUC) coding region into the same restriction sites of pGL3-Control (Promega). The amplification primer sequences are available upon request. The cloned plasmids were verified by sequencing. ARE plasmid was a kind gift of S. Vasudevan and is published before (12). The cloning primers: TLR4-I: forward, ATATCTAGAAAAGACAGAGAAAACAGAAAGAGACA, and reverse, ATAGGCCGGCCTTCCTTCCTGCCTCTAGCCC; TLR4-II: forward, ATATCTAGACCCGGAGGCCATTATGCTAT, and reverse, ATAGGCCGGCCCAATTTGATGAGTTTAGACATAGTCAC; TLR4-III: forward, ATATCTAGAATATCAATTATGTCTGAATGAAGCTAT,and reverse, ATAGGCCGGCCAGAGAACTCATCTCAAACAGCC; and CD80: forward, ATATCTAGACCATAGGGCCTCCTTAGATCCC, and reverse, ATAGGCCGGCCGCAAGGTTTGTGAAGCAGCA.
Luciferase Assays
8 × 104 HEK293 cells were plated into 24-well plates and transfected after 24 h with 20 ng of Renilla encoding pRL-TK (Promega), 100 ng of pGL3–3′-UTR reporters, and either 50 nm pre-miR-511 precursor or the fluorescein derivative FAM-labeled pre-miR-control (Applied Biosystems) using siPORT NeoFX Transfection Agent (Applied Biosystems). The cells were harvested either after 48 or 96 h in contact inhibition conditions (13) and analyzed using Promega dual luciferase assay. Luminescence was counted with Wallac 1420 (PerkinElmer Life Sciences).
Western Analysis and FACS
For Westerns, rabbit polyclonal anti-human TLR4, (sc-10741; Santa Cruz Biotech) in 2% milk, mouse monoclonal anti-human GAPDH (ab8245; Abcam), mouse monoclonal hnRNPU (ab10297; Abcam), and DICER1 (ab14601; Abcam) in 5% milk were used. Signals were detected with the ECL Advance Western blotting detection kit (GE Healthcare) and captured and quantified by ImageQuantTM-RT ECL image analysis system. For FACS, fluorescence-conjugated antibodies CD83, DC-SIGN, CD86 (BD Biosciences), CD14, and HLA-DR (Miltenyi Biotec) and FACSCalibur (BD Biosciences) were used. The data were analyzed and visualized with FlowJo v. 7.6.
RNA Purification and Quantitative RT-PCR
RNA was purified using TRIzol (Invitrogen) and, when needed, further purified with the RNAeasy mini kit (Qiagen). To maintain small RNA fraction, a 3.5 volume of 100% ethanol was added to the samples before loading on Qiagen mini columns. Alternatively, the miRNAeasy mini kit (Qiagen) was used. The concentration and quality of RNA was assessed with NanoDrop ND-1000 spectrophotometer and Agilent RNA 6000 Nano Kit on Agilent 2100 Bioanalyzer. For mRNA RT-PCR, cDNA from 200 ng of total RNA was synthesized using oligo(dT) and SuperScript III Reverse Transcriptase (Invitrogen). PCR was carried out with Maxima SYBR green/Rox Master Mix (Fermentas). miRNA expression was analyzed using TaqMan MicroRNA assays, TaqMan MicroRNA reverse transcription kit (Applied Biosystems) and 5X HOT FIREPol Probe RT-PCR Mix Plus (Solis Biodyne). All RT-PCRs were carried out on ABI Prism 7900, and the relative gene expression levels were calculated using the comparative Ct (ΔΔCt) method (Applied Biosystems). The RT-PCR primers were the following: DICER: forward, GGCTGTAAAGTACGACTACC, and reverse, GATCTCCTAAGCTCAGAATCC; TLR4–1: forward, ATCCCCTGAGGCATTTAGGC, and reverse, TCAATTGTCTGGATTTCACACCTG; TLR4–2: forward, TCCCTCCCCTGTACCCTTCT, and reverse, AGCATTGCCCAACAGGAAAC; TLR4–3: forward, ATCCCTGGGTGTGTTTCCAT, and Reverse, TGCGGACACACACACTTTCA; and HPRT, forward, GACTTTGCTTTCCTTGGTCAGG, and reverse, AGTCTGGCTTATATCCAACACTTCG.
RNA Profiling and miRNA Target Prediction
miRNA profiling was carried out on Illumina miRNA Universal-16 BeadChips (miRBase version 12.0) and mRNA profiling by Illumina Human-6 v2 BeadChips in Core Facility at the Department of Biotechnology, University of Tartu. The data were analyzed with BeadStudio Gene Expression Module v3.3.7 (Illumina) using Average Normalization for miRNA data and Illumina's custom rank invariant method for mRNA arrays. Genes with differential expression p value <0.05 were considered differentially expressed. Further analyses and visualizations were carried out using Microsoft Excel and Multi Experiment Viewer 4.0. Unsupervised hierarchical clustering was done using Euclidean distance and average linkage analysis. The miRNA and mRNA microarray data are available at ArrayExpress as E-TABM-968 and E-TABM-976, respectively. The compiled miRNA target lists were generated based on Diana microT (v3.0), miRanda (downloaded in September 2008), PicTar (downloaded in August 2009), rna22 (downloaded in August 2009), and Targetscan conserved targets (5.1).
RESULTS
Updated miRNA Profile of Human DCs and MFs
Human blood-derived MOs were differentiated into DCs and MFs as described earlier (4) and stimulated on day 6 with LPS, as TLR4 ligand, or curdlan, as Dectin-1 (CLEC7A) ligand. Flow cytometric analysis showed a well established phenotype of immature DCs (DC-SIGN+CD14−) and MFs (DC-SIGN−CD14+) on day 6 (supplemental Fig. S1A, left panels). Following stimulations, DCs had a significantly higher expression of CD86, HLA-DR (MHCII), and CD83. For activated MFs, enhancement of CD86 and CD83 was detected (supplemental Fig. S1A, histograms). Consistent with the previous studies (4, 16, 18, 19, 23, 27, 31), the expression of miR-155 and miR-146a was strongly induced in response to LPS and curdlan both in DCs and MFs, but the expression of miR-132 was mainly induced during the differentiation phase (supplemental Fig. S1B).
Next, total RNA from two or three different donors for each condition was analyzed on Illumina miRNA arrays containing probes for 858 human mature miRNAs based on miRBase 12.0 (32) (Fig. 1A). Altogether, 307–380 miRNAs with detection p values < 0.05 were found to be expressed in analyzed cells populations, whereas almost half of the detected miRNAs had relatively high signal intensities (supplemental Table S1). Compared with MOs, 39 miRNAs were up-regulated, and 10 were down-regulated with a differential p value < 0.05 in immature or mature DCs and MFs (Fig. 1A and supplemental Table S2). miR-99b, miR-212, and miR-511 were detected as the three most highly up-regulated miRNAs both in DCs and MFs. Some miRNAs, for instance miR-193b, miR-125b, and miR-99a, were preferentially expressed in DCs, whereas miR-139–5p, miR-1, and miR-218 had higher expression in MFs. Hierarchical clustering showed that according to miRNA expression pattern, MOs are more similar to MFs than DCs (supplemental Fig. S2). Changes in miRNA expression were found to occur mainly during the differentiation. After stimulation, relatively mild increased expression was detected for 16 miRNAs. No miRNAs specific for either LPS or curdlan stimulation were detected (supplemental Table S3). Consistent with quantitative RT-PCR results carried out during the initial cell differentiation verification, miRNAs miR-146a and miR-132 were detected as differentially regulated. However, for miR-155, we could not detect statistically significant expression change on the Illumina arrays (supplemental Fig. S1, A and B).
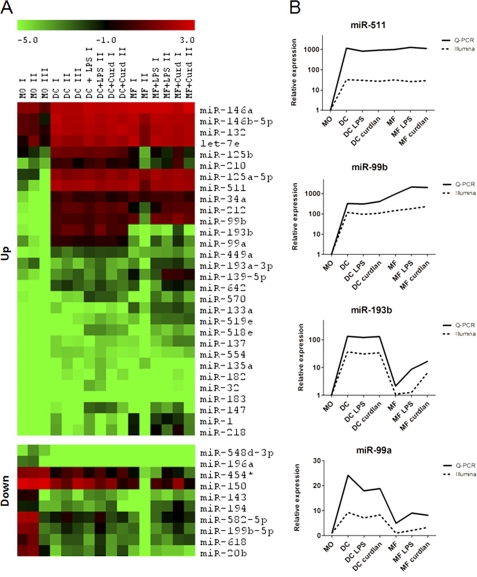
FIGURE 1.
MOs, DCs, and MFs have distinct miRNA expression profiles. A, heat maps of miRNAs that are up- or down-regulated (differential p < 0.05) in DCs and/or MFs to compare with the levels in MOs. The miRNAs, which are shown, have an average expression level of over 100 in at least one condition. Each column represents expression levels of miRNA (shown at right) in each sample (shown above). Log2 expression values for each miRNA are mean-centered across all the analyzed miRNA expression values. The color scale from green (lower) to red (higher) represents deviation from the mean (black). B, verification of Illumina array results with RT-PCR for four example miRNAs. The data from one representative donor are shown and are normalized to the value in MOs (which equals 1).
To further verify Illumina array results, we carried out quantitative RT-PCR for 12 selected miRNAs, which confirmed that miR-193b and -99a were the most highly up-regulated in DCs and miR-511, miR-99b, and miR-212 in both DCs and MFs (Fig. 1B and supplemental Fig. S3 and Table S4). In conclusion, we have described updated miRNA expression profiles of human blood MOs and MO-derived immature and mature DCs and MFs and found several miRNAs, including the most highly up-regulated miR-511 and DC specific miR-193b, not reported in these cells earlier.
siRNA Knockdown of DICER1 and Inhibition of Up-regulated miRNAs Results in Reduced DC-SIGN Expression
To further prove that miRNAs are essential for DC differentiation, we carried out siRNA knockdown of DICER1, a protein that is indispensable for miRNA processing (33). DCs were differentiated in the presence of control siRNAs or two different siRNAs and s23754 s23756 for DICER1. Better knockdown efficiency of DICER1 was achieved with s23754, typically ∼50% on both mRNA and protein level (Fig. 2, A and B), and this inhibition caused lower DC-SIGN and higher CD14 protein levels (Fig. 2C), which is an indication of delayed maturation phenotype in DCs. To further analyze the effect of DICER1 knockdown on DC differentiation, the DC-SIGN/CD14 ratios from four experiments carried out on cells from different donors were calculated and found to be significantly different (Fig. 2D).
Next, we performed ex vivo DC differentiation in the presence of sequence-specific miRNA inhibitors for miR-511 and miR-99b, as the two most highly up-regulated miRNAs in both DCs and MFs, and for DC specific miR-193b (Fig. 1, A and B, and supplemental Table S2). The transfection of miRNA inhibitors resulted in 50–90% reduced expression of specific miRNA if measured by RT-PCR (Fig. 2E), which led to reduced DC-SIGN protein levels, if the inhibitors for miRNA-511 and miR-99b were used (Fig. 2F). The levels of CD14 and HLA-DR surface markers were not influenced by specific miRNA inhibitors (data not shown). In conclusion, the data from DICER1 knockdown and specific miRNA inhibition experiments show that miRNAs in general and miR-511 and miR-99b in particular have impact on DC-SIGN expression and thus presumably also on DC differentiation.
Up-regulated miRNAs Potentially Target Genes with Immune Function
Next, we searched the potential targets for miR-511, miR-99b, and miR-193b. Studies on miR-181 and miR-223 involving different computational and experimental approaches have shown that different programs have overall similar target prediction capability. However, the top third TargetScan predictions have been shown to correlate significantly better with protein down-regulation than predictions of the other programs or the bottom third of TargetScan predictions (34, 35). Thus, we first selected all TargetScan predicted targets with a total context score of less than −0.2, corresponding to the top one-third of the lists. To search for immunologically important targets, we only included 4274 human genes related to immune function according to the Immport Database. As another approach, we compiled lists of potential targets based on five different algorithms: Targetscan 5.1 (36), Miranda (37), DIANA-microT (38), Pictar (39), and rna22 (40). The putative targets were ranked by their position in any of the input lists, and the top 500 genes from this ranking were used in subsequent analysis. Genes with very low expression and average signal intensity <50 based on the Illumina mRNA expression data (E-TABM-976) were excluded. The remaining working lists, containing 135–247 targets per each studied miRNA, were analyzed using the g:GOSt tool at g:Profiler website, which retrieves most significant gene ontology terms and KEGG and REACTOME pathways (41). According to this search, miR-511 showed the greatest potential to target several pathways important for DC and MF differentiation, maturation, and functions (Table 1).
TABLE 1.
DC- and MF-specific miRNAs potentially target genes from functional pathways or groups important in the differentiation or immune functions
| miRNA | Number of targets | Functional group | Number of genes | ID | Putative targets in functional group | Significancea | Methodb |
|---|---|---|---|---|---|---|---|
| miR-511 | 135 | Toll-like receptor signaling pathway | 107 | KEGG:04620 | TLR4 STAT1 CD80 MAP3K7IP2 CD86 IRAK1 MAP3K7 TIRAP | 2.56E-05 | T |
| Myeloid cell differentiation | 93 | GO:0030099 | BCL6 IRF4 PPARG JAK2 SMAD5 TIRAP | 4.11E-05 | T | ||
| JAK-STAT cascade | 47 | GO:0007259 | NLK STAT1 SOCS2 JAK2 STAT5A SOCS6 STAT4 | 2.84E-08 | T | ||
| Regulation of interleukin-2 production | 28 | GO:0032663 | IRF4 CD80 CD86 STAT5A MAP3K7 | 1.16E-06 | T | ||
| Regulation of transcription from RNA polymerase II promoter | 732 | GO:0006357 | HIPK2 TIAL1 EP300 RYBP BCL6 AHR YY1 HDAC2 IRF4 PPARG STAT5A SRF SMAD5 IRF2 VHL LITAF | 3.73E-05 | T | ||
| 193 | Negative regulation of gene expression | 497 | GO:0010629 | ATBF1 CBX1 DEDD DEDD2 GABPA NAB2 ORC2L POU2F1 RUNX2 RYBP SATB2 SIN3A ZNF281 TARBP2 TGIF1 TNRC6B | 2.29E-05 | C | |
| miR-193b | 234/247 | Chemokine signaling pathway | 189 | KEGG:04062 | CRK CRKL KRAS SOS2 | 1.56E-06 | T |
| Chronic myeloid leukemia | 76 | KEGG:05220 | CRKL CCND1 BCL2L1 RUNX1 CRK KRAS | 5.23E-04 | C | ||
| miR-99b | 225 | β-Catenin binding | 27 | GO:0008013 | NUMB DVL3 SMAD7 C5ORF22 AXIN1 | 8.49E-06 | C |
| Pathways in cancer | 334 | KEGG:05200 | AXIN1 BID CBL CDK6 DVL3 FZD1 HSP90B1 IGF1R IKBKG ITGB1 STAT5B | 7.03E-04 | C |
a The p value form Fisher exact test showing the significance of the overlap between the target list and indicated functional category.
b The target lists used for analysis were generated either based on top third of Targetscan (T) or by compiling five different algorithms, Targetscan 5.1, Miranda, DIANA-microT, Pictar, and rna22 predictions (C).
Expression of miR-511 Accompanies with the Enhanced Level of the TLR4 Protein in Differentiating DCs
Among the Targetscan predictions, TLR4 was found to be one of the best scored (three target sites, context score −0.89) of miR-511 targets expressed in DCs. Thus, we next studied whether TLR4 mRNA and protein levels are influenced by miR-511. We first followed miR-511 and TLR4 expression during the DC differentiation. Because the TLR4 primary transcript has got four different alternative polyadenylation signals, we designed three different primer sets to analyze the levels of alternatively polyadenylated TLR4 mRNAs separately. TLR4–1 recognized all possible TLR4 transcripts; TLR4–2 and TLR4–3 were designed for the transcripts with longer 3′-UTRs containing two or three possible miR-511 target sites, respectively (Fig. 3A and supplemental Fig. S4). As shown on Fig. 3B, the miR-511 level is gradually enhanced during the differentiation from days 1 to 6, whereas the DC-SIGN mRNA level is even higher during the first days if compared with DCs of the later time points (Fig. 3B, left panel). Although the amount of longer TLR4 transcripts seemed to be reduced during the differentiation, the total amount of TLR4 mRNA was relatively constant until DC6 (Fig. 3B, right panel), and the TLR4 protein level was gradually enhanced up to 3.4 times in comparison with MOs (Fig. 3C). We next studied whether reduction of the longer TLR4 transcripts can occur because of the presence of multiple miR-511 sites and whether there is a difference in accumulation of the TLR4 protein when miR-511 is inhibited (Fig. 3D). Interestingly, the TLR4 protein levels were ∼2-fold higher in control-transfected cells in comparison with miR-511 inhibited samples on days 4 and 6 of the differentiation when more miR-511 had accumulated into the cells (Fig. 3, D and E). No significant difference was detected in TLR4 mRNA different transcript levels in miR-511 knockdown samples in comparison with the control transfections (Fig. 3F). Also, the knockdown of miR-511 did not lead to major changes in main DC activation markers CD86 and CD80 and maturation marker CD83 upon LPS stimulation measured by FACS (supplemental Fig. S5A), although reduction of the TLR4 protein was detected in LPS-stimulated DCs when miR-511 was inhibited (supplemental Fig. S5, B–D). These data together show that there is a correlation between high expression of miR-511 and the enhanced level of the TLR4 protein, which suggested that miR-511 is a positive regulator of the TLR4 protein translation in MO-derived DCs.
miR-511 Has Versatile Influence on Its Putative Targets TLR4 and CD80 in Luciferase Assays
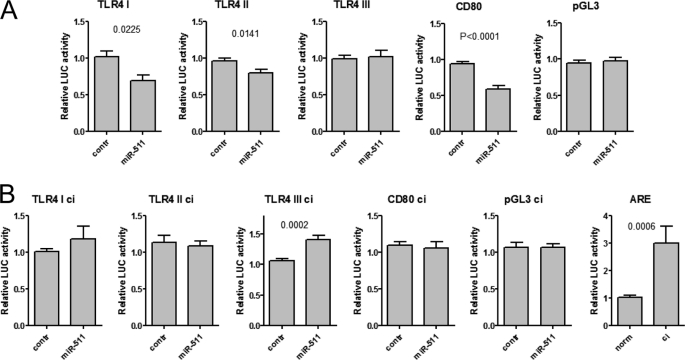
We next studied whether TLR4 is targeted by miR-511 in LUC reporter assays. Because TLR4 contains polyadenylation signals between each potential miR-511 target sequence, three different fragments of TLR4 3′-UTR were inserted downstream the LUC coding region into the pGL3-contr vector. In addition, we cloned the 3′-UTR fragment of CD80 containing two putative miR-511 target sites. Next, the LUC-3′-UTR reporters were transfected either alongside with pre-miR-511 or the control pre-miRNA into the HEK293 cells. It has been shown that in cell cycle-arrested conditions, miRNAs can up-regulate the translation (12, 13). Because human blood MOs do not proliferate during the differentiation and because we observed positive correlation of miR-511 expression and the TLR4 protein level in differentiating DCs, we carried out LUC assays at both 48 and 96 h after the transfection, in normal conditions and in contact inhibited cells where cells are arrested in G0 phase (13). Fig. 4A shows that during normal growth conditions, the expression of the LUC constructs with 3′-UTR fragments containing the first and second predicted TLR4 target sites of miR-511 (TLR4 I and TLR4 II) and 3′-UTR fragment of CD80 containing two miR-511 target sites were down-regulated in the presence of miR-511 when compared with the levels of control transfected cells. No significant influence on the control vector or constructs containing TLR4 III site was detected (Fig. 4A). Interestingly, in contact inhibited cells, in the presence of transfected miR-511, the LUC expression levels for TLR4 I, TLR4 II and CD80 were not higher and for TLR4 III construct, the LUC expression was even significantly enhanced when compared with the control transfected cells. The expression of the ARE plasmid used as a positive control for cell cycle arrest was enhanced in contact inhibited cells when compared with the control plasmid (Fig. 4B). These data together indicate that miR-511, depending on the target mRNA 3′-UTR and cellular environment, can either up- or down-regulate the target gene expression.
FIGURE 4.
The influence of miR-511 to expression of its proposed targets in luciferase reporter assays. A and B, the LUC activity was measured either 48 (A, normal conditions) or 96 (B, contact inhibited conditions, ci) hours after the transfection of the LUC reporters and pre-miR-511 or the control pre-miRNA. The data are means with S.E. of at least eight different transfections. All of the data are normalized to Renilla and the LUC values of control transfections (which equal 1). LUC activity measurements of the ARE reporter used as positive control were done 48 h (norm, which equal 1) and 96 h (ci) after the transfections and are normalized to Renilla and the levels of pGL3-control. Three different target sequences of TLR4 are cloned in separate LUC constructs and are designated as TLR4 I, II, and III. The p values are calculated using the Mann-Whitney test.
DISCUSSION
Ex vivo differentiated human blood MO-derived DCs and MFs are excellent experimental models to study the differentiation and functions of MO-DC cell lineage. In addition, MO-derived DCs are the most common type of DCs used in immunotherapeutic approaches (6, 7). In this study, we first describe updated miRNA expression profiles of human blood MO-derived immature and mature DCs and MFs. Consistent with previous studies (23, 27, 31, 42, 43), we found that these cell types have distinct miRNA expression profiles and detected several novel strongly up-regulated miRNAs, including miR-511. For the activation and maturation of MFs and DCs, we used LPS and curdlan as ligands for TLR4 and Dectin-1, respectively, both leading to activation through the NF-κB pathway (44–46). Concordantly, very similar miRNA expression changes were seen for both inductions, whereas we could not find any novel highly expressed and substantially up-regulated endotoxin-dependent miRNA. We also could not detect miR-155 among LPS stimulated genes from Illumina array, whereas its up-regulation was obvious by RT-PCR. It is possible that up-regulation of miR-155 after LPS stimulation occurs on RNA processing level, because the RT-PCR with TaqMan probes detects only mature miRNAs, whereas the Illumina arrays might also recognize miRNA precursors.
Further, knockdown of DICER1 with siRNAs led to lower expression of DC-SIGN and a higher level of monocyte marker CD14, confirming that miRNAs in general are needed for proper differentiation of DCs. Similarly, specific inhibition of miR-511 and miR-99b resulted in a reduction of DC-SIGN, indicating that these two miRNAs influence DC differentiation. Previously, Hashimi et al. (27) have shown that inhibition of the number of miRNAs, including miR-99b, influences MO-derived DC differentiation. However, based on data from three donors, they found that only inhibition of miR-34a had a statistically significant impact. Similarly to our study, the extent of influence of specific miRNA inhibitors was variable in cells from different donors, which indicates that differences exist in the complicated miRNA expression network in each individual.
The regulation by miRNAs mainly occurs via pairing of miRNA 6–8-nucleotide seed sequences to the target mRNA (35). Therefore, prediction of true miRNA targets by computational approaches has been a challenge. According to the two approaches used in this study, we found many genes from ontology groups and pathways with immune functions to be overrepresented among miRNA-511 potential targets. Interestingly, miR-511-1 and miR-511-2 genes themselves are located within the fifth intron of mannose receptor gene MRC1 and its very recent human specific copy MRC1L, respectively. Therefore, expression of miR-511 is most probably regulated coordinately with MRC1 genes, which are only expressed in MFs, some DCs and subsets of vascular and lymphatic endothelial cells (47). In this study, we examined the possible impact of miR-511 to its proposed targets TLR4 and CD80. Interestingly, we found that miR-511 knockdown results in enhanced TLR4 protein levels in differentiating DCs. However, we could not detect a consistent influence of miR-511 knockdown to DC activation markers CD86 and CD80 as well as to maturation marker CD83 (supplemental Fig. S5). It is possible that in other cell types or in certain disease conditions, miRNA-511 impact is more obvious.
It has been shown that in the cell cycle arrest at G0 phase, certain miRNAs can enhance the target protein levels (12, 13). The alternative possibility would be that miR-511 competes with any more strong degradation factor, as has been recently shown for miR-466, which replaces TTP protein and therefore stabilizes IL10 mRNA (48). In some cases, the mechanism of uncommon expression enhancement effect is not clear as it is for miR-155, which has stabilizing influence on the TNF-α mRNA in alcohol-treated MFs (49). Still, the influence of miR-511 on TLR4 protein expression seems to be direct because in HEK293 cells either reduced LUC levels in normal conditions for TLR I and II or enhanced LUC level for TLR III in contact inhibited cells were determined in the presence of miR-511. In addition, although human MOs can be differentiated in tissue culture conditions, they do not proliferate, and thus, they are cell cycle-arrested. Even though it is generally accepted that many partially and fully differentiated immune cells are quiescent or arrested in G0/resting phase, only a limited number of studies about their entering and leaving from this state are available. Previously has been demonstrated that differentiation of functional DCs from human blood-derived MOs depends on expression of p21 encoded by CDKN1A gene, which is considered a hallmark of the G1 phase (50). According to the mRNA array analysis we performed, p21 is also up-regulated during the DC differentiation. The p27 encoding gene CDKN1B, a hallmark of G0, however, is slightly down-regulated on day 6 of DC differentiation in comparison with MOs. After the stimulation with LPS, it is up-regulated again (E-TABM-976). The similar expression pattern of p27 is confirmed in protein level by a very recent study, which also suggests that miR-221 has an influence on the p27 protein level in differentiating DCs (51). Our data, showing that miR-511 inhibition results in differentiating DCs and the LUC reporter assays in proliferating and contact inhibited HEK293 cells, indicate that regulation of TLR4 by miR-511 can also depend on the cell cycle. Further studies are needed to better understand the influence of miR-511 and miRNAs in general in distinct cellular conditions.
In conclusion, we have determined updated miRNA expression profiles for human blood MOs, DCs and MFs and have found miR-511 as highly expressed MF- and DC-specific miRNAs with strong immune regulatory potential. In addition, our data highlight that the influence of miRNAs can be modulated by cellular environment.
Supplementary Material
Acknowledgments
We thank Tõnis Org (University of Tartu) for help in array analysis, Tarmo Annilo (University of Tartu) and Liam O'Mahony (SIAF) for critical reading of the manuscript, and Shobha Vasudevan (Harvard Medical School) for fruitful discussions.
This work was supported by Tartu University Baseline Funding Grant 08904, Estonian Science Foundation Grants 8350 and 8358, the European Regional Development Fund and Archimedes Foundation, the Estonian Ministry of Education and Research Targeted Funding Grant SF0180021s07, and Tiger University Program of the Estonian Information Technology Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S5.
- DC
- dendritic cell
- MF
- macrophage
- MO
- monocyte
- DC-SIGN
- dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- GM-CSF
- granulocyte macrophage colony-stimulating factor
- LUC
- luciferase
- TLR
- toll-like receptor
- miRNA
- microRNA.
REFERENCES
- 1. Geissmann F., Manz M. G., Jung S., Sieweke M. H., Merad M., Ley K. (2010) Science 327, 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shortman K., Naik S. H. (2007) Nat. Rev. Immunol. 7, 19–30 [DOI] [PubMed] [Google Scholar]
- 3. Mosser D. M., Edwards J. P. (2008) Nat. Rev. Immunol. 8, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sallusto F., Lanzavecchia A. (1994) J. Exp. Med. 179, 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Auffray C., Sieweke M. H., Geissmann F. (2009) Annu. Rev. Immunol. 27, 669–692 [DOI] [PubMed] [Google Scholar]
- 6. Tyagi R. K., Mangal S., Garg N., Sharma P. K. (2009) Expert Rev. Anticancer Ther. 9, 97–114 [DOI] [PubMed] [Google Scholar]
- 7. Melief C. J. (2008) Immunity 29, 372–383 [DOI] [PubMed] [Google Scholar]
- 8. Amaral P. P., Dinger M. E., Mercer T. R., Mattick J. S. (2008) Science 319, 1787–1789 [DOI] [PubMed] [Google Scholar]
- 9. Hobert O. (2008) Science 319, 1785–1786 [DOI] [PubMed] [Google Scholar]
- 10. Makeyev E. V., Maniatis T. (2008) Science 319, 1789–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu J. (2008) Curr Opin Cell Biol. 20, 214–221 [DOI] [PubMed] [Google Scholar]
- 12. Vasudevan S., Tong Y., Steitz J. A. (2007) Science 318, 1931–1934 [DOI] [PubMed] [Google Scholar]
- 13. Vasudevan S., Tong Y., Steitz J. A. (2008) Cell Cycle 7, 1545–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Connell R. M., Rao D. S., Chaudhuri A. A., Baltimore D. (2010) Nat. Rev. Immunol. 10, 111–122 [DOI] [PubMed] [Google Scholar]
- 15. Lodish H. F., Zhou B., Liu G., Chen C. Z. (2008) Nat. Rev. Immunol. 8, 120–130 [DOI] [PubMed] [Google Scholar]
- 16. Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Benedetto A., Agnihothri R., McGirt L. Y., Bankova L. G., Beck L. A. (2009) J. Invest. Dermatol. 129, 14–30 [DOI] [PubMed] [Google Scholar]
- 18. Jurkin J., Schichl Y. M., Koeffel R., Bauer T., Richter S., Konradi S., Gesslbauer B., Strobl H. (2010) J. Immunol. 184, 4955–4965 [DOI] [PubMed] [Google Scholar]
- 19. Williams A. E., Perry M. M., Moschos S. A., Larner-Svensson H. M., Lindsay M. A. (2008) Biochem. Soc. Trans. 36, 1211–1215 [DOI] [PubMed] [Google Scholar]
- 20. Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A. (2007) Science 316, 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Connell M. A., Keegan L. P. (2006) Nat. Struct. Mol. Biol. 13, 3–4 [DOI] [PubMed] [Google Scholar]
- 22. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ceppi M., Pereira P. M., Dunand-Sauthier I., Barras E., Reith W., Santos M. A., Pierre P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2735–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martinez-Nunez R. T., Louafi F., Friedmann P. S., Sanchez-Elsner T. (2009) J. Biol. Chem. 284, 16334–16342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu L. F., Thai T. H., Calado D. P., Chaudhry A., Kubo M., Tanaka K., Loeb G. B., Lee H., Yoshimura A., Rajewsky K., Rudensky A. Y. (2009) Immunity 30, 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Connell R. M., Chaudhuri A. A., Rao D. S., Baltimore D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7113–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hashimi S. T., Fulcher J. A., Chang M. H., Gov L., Wang S., Lee B. (2009) Blood 114, 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sheedy F. J., Palsson-McDermott E., Hennessy E. J., Martin C., O'Leary J. J., Ruan Q., Johnson D. S., Chen Y., O'Neill L. A. (2010) Nat. Immunol. 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 29. O'Neill L. A., Sheedy F. J., McCoy C. E. (2011) Nat. Rev. Immunol. 11, 163–175 [DOI] [PubMed] [Google Scholar]
- 30. Fulci V., Chiaretti S., Goldoni M., Azzalin G., Carucci N., Tavolaro S., Castellano L., Magrelli A., Citarella F., Messina M., Maggio R., Peragine N., Santangelo S., Mauro F. R., Landgraf P., Tuschl T., Weir D. B., Chien M., Russo J. J., Ju J., Sheridan R., Sander C., Zavolan M., Guarini A., Foà R., Macino G. (2007) Blood 109, 4944–4951 [DOI] [PubMed] [Google Scholar]
- 31. Lehtonen A., Ahlfors H., Veckman V., Miettinen M., Lahesmaa R., Julkunen I. (2007) J. Leukocyte Biol. 82, 710–720 [DOI] [PubMed] [Google Scholar]
- 32. Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J. (2008) Nucleic Acids Res. 36, D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perron M. P., Provost P. (2009) Methods Mol. Biol. 487, 369–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008) Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 36. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Genome Res 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) PLoS Biol. 2, e363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maragkakis M., Reczko M., Simossis V. A., Alexiou P., Papadopoulos G. L., Dalamagas T., Giannopoulos G., Goumas G., Koukis E., Kourtis K., Vergoulis T., Koziris N., Sellis T., Tsanakas P., Hatzigeorgiou A. G. (2009) Nucleic Acids Res. 37, W273–W276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 40. Miranda K. C., Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., Lim B., Rigoutsos I. (2006) Cell 126, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 41. Reimand J., Kull M., Peterson H., Hansen J., Vilo J. (2007) Nucleic Acids Res. 35, W193–W200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A. O., Landthaler M., Lin C., Socci N. D., Hermida L., Fulci V., Chiaretti S., Foà R., Schliwka J., Fuchs U., Novosel A., Müller R. U., Schermer B., Bissels U., Inman J., Phan Q., Chien M., Weir D. B., Choksi R., De Vita G., Frezzetti D., Trompeter H. I., Hornung V., Teng G., Hartmann G., Palkovits M., Di Lauro R., Wernet P., Macino G., Rogler C. E., Nagle J. W., Ju J., Papavasiliou F. N., Benzing T., Lichter P., Tam W., Brownstein M. J., Bosio A., Borkhardt A., Russo J. J., Sander C., Zavolan M., Tuschl T. (2007) Cell 129, 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin P., Han T. H., Ren J., Saunders S., Wang E., Marincola F. M., Stroncek D. F. (2010) J. Transl. Med. 8, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trinchieri G., Sher A. (2007) Nat. Rev. Immunol. 7, 179–190 [DOI] [PubMed] [Google Scholar]
- 45. Jin M. S., Lee J. O. (2008) Immunity 29, 182–191 [DOI] [PubMed] [Google Scholar]
- 46. Palma A. S., Feizi T., Zhang Y., Stoll M. S., Lawson A. M., Díaz-Rodríguez E., Campanero-Rhodes M. A., Costa J., Gordon S., Brown G. D., Chai W. (2006) J. Biol. Chem. 281, 5771–5779 [DOI] [PubMed] [Google Scholar]
- 47. Kerrigan A. M., Brown G. D. (2009) Immunobiology 214, 562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma F., Liu X., Li D., Wang P., Li N., Lu L., Cao X. (2010) J. Immunol. 184, 6053–6059 [DOI] [PubMed] [Google Scholar]
- 49. Bala S., Marcos M., Kodys K., Csak T., Catalano D., Mandrekar P., Szabo G. (2011) J. Biol. Chem. 286, 1436–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kramer J. L., Baltathakis I., Alcantara O. S., Boldt D. H. (2002) Br. J. Haematol. 117, 727–734 [DOI] [PubMed] [Google Scholar]
- 51. Lu C., Huang X., Zhang X., Roensch K., Cao Q., Nakayama K. I., Blazar B. R., Zeng Y., Zhou X. (2011) Blood 117, 4293–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.