Abstract
The visual process in the vertebrate eye requires high amounts of metabolic energy and thus oxygen. Oxygen supply of the avian retina is a challenging task because birds have large eyes, thick retinae, and high metabolic rates but neither deep retinal nor superficial capillaries. Respiratory proteins such as myoglobin may enhance oxygen supply to certain tissues, and thus the mammalian retina harbors high amounts of neuroglobin. Globin E (GbE) was recently identified as an eye-specific globin of chicken (Gallus gallus). Orthologous GbE genes were found in zebra finch and turkey genomes but appear to be absent in non-avian vertebrate classes. Analyses of globin phylogeny and gene synteny showed an ancient origin of GbE but did not help to assign it to any specific globin type. We show that the photoreceptor cells of the chicken retina have a high level of GbE protein, which accumulates to ∼10 μm in the total eye. Quantitative real-time RT-PCR revealed an ∼50,000-fold higher level of GbE mRNA in the eye than in the brain. Spectroscopic analysis and ligand binding kinetics of recombinant chicken GbE reveal a penta-coordinated globin with an oxygen affinity of P50 = 5.8 torrs at 25 °C and 15 torrs at 41 °C. Together these data suggest that GbE helps to sustain oxygen supply to the avian retina.
Keywords: Evolution, Gene Mapping, Hemoglobin Myoglobin, Kinetics, Oxygen Binding, Respiration, Retinal Metabolism, Chicken, Cytoglobin, Neuroglobin
Introduction
Globins constitute a structurally conserved family of heme proteins with the ability to reversibly bind molecular oxygen (O2). A typical globin chain comprises about 140–150 amino acids and includes an iron porphyrin. Until recently, only two types of globins were known in vertebrates (i.e. fishes, amphibians, reptiles, birds, and mammals). Hemoglobin (Hb)3 is certainly the best known globin, which is present in the erythrocytes of the blood and serves for the transport of O2 (1). Hb is a heterotetrameric protein that is composed of two α- and two β-type chains. Myoglobin (Mb) is a monomeric protein located in the myocytes of the heart and the skeletal muscles, which enhances oxygen supply by facilitating diffusion of O2 to the mitochondria or O2 storage (2). Mb may also be instrumental for decomposition of nitric oxide (NO) (2).
Within the past 10 years, additional globin types have been identified in jawed vertebrates (Gnathostomata). Neuroglobin (Ngb) essentially resides in the central and peripheral nervous system (3). Although the true function of Ngb is still a matter of debate (4, 5), there is conclusive evidence that it is associated with the oxidative metabolism (6, 7). High levels of Ngb have been detected in the rodent retina, where it may sustain a high metabolic rate (6, 8). Cytoglobin (Cygb) is expressed in fibroblast-related cell types and distinct neurons (9–11). Cygb may be involved in collagen synthesis or in the function of O2-consuming enzymes (5). Although Hb, Mb, Ngb, and Cygb are widespread among vertebrates (12), other globins appear to be restricted to certain vertebrate taxa. Globin X (GbX) is only present in fishes and amphibians (13, 14). GbX is distantly related to Ngb but is bound to the cell membrane, where it may carry out a protective function.4 Globin Y shows a broad expression pattern in Xenopus tissues (13), but there is no information about its physiological role.
In chicken, an additional globin type was identified. It appears to be preferentially expressed in the eye and therefore has been referred to as eye globin or GbE (15). The presence of a putatively respiratory protein in the chicken eye is of particular interest because the function of the bird eye is constrained by two conflicting demands; on the one hand, the blood vessels should not obstruct the optical path, and on the other hand, a high metabolic rate of the retina requires high O2 supply and thus good vascular perfusion (16). Birds have a high metabolic rate, large eyes, and thick retinae but have an avascular retina without deep retinal and superficial capillaries. Therefore, additional O2 is supplied to the bird retina by the pecten oculi, a unique vascular structure extending into the vitreous chamber (16).
To evaluate the possible role of GbE in O2 supply to the avian retina, we carried out a detailed characterization of chicken GbE, which includes in silico analyses of the gene, molecular, and histological characterization of expression patterns, as well as biochemical analyses of recombinant GbE. Together these data suggest that GbE may actually have an Mb-like role in O2 supply of the bird photoreceptor cells.
EXPERIMENTAL PROCEDURES
Database and Sequence Analyses
The BLAST algorithm (17) was employed to search the databases of genomic DNA sequences available at GenBank (www.ncbi.nlm.nih.gov) and Ensembl. We used Gallus gallus genome Build 2.1 (18), turkey (Meleagris gallopavo) genome UMD 2.0 (19), zebra finch (Taeniopygia guttata) genome Build 1.1 (20), human (Homo sapiens) genome Build 37.2, and zebrafish (Danio rerio) Zv8 to analyze gene synteny. Nucleotide sequences were extracted from the databases and assembled by the aid of Vector NTI 10.3.0 (Invitrogen; Darmstadt, Germany) and GeneDoc 2.6 (21). DNA sequences were translated and analyzed using the programs of the ExPASy Molecular Biology Server. Synonymous and non-synonymous nucleotide substitutions were estimated employing K-estimator 6.1 (22). Interspecific comparison of genes was performed with MultiPipMaker, which computes and visualizes local alignments of two or more sequences based on the BLASTZ algorithm (23). The multiple sequence alignments from PipMaker were visualized as “percent identity plots.”
RNA Extraction
Total RNA was extracted from chicken (G. gallus) brain and eye tissues using the RNeasy mini kit (Qiagen, Hilden, Germany). Tissues were pulverized using a mortar and pestle. 30 mg of tissue samples were supplemented with 600 μl of RLT buffer and further purified using the silica column method according to the manufacturer's instructions (Qiagen). The quality and integrity of RNA were evaluated by reading the absorption ratio at 260 versus 280 nm and by agarose gel electrophoresis.
cDNA Cloning
The full-length chicken Ngb (accession number: NW_060388) and GbE (NM_001008786) coding sequences were amplified from brain and eye with Taq DNA polymerase (Invitrogen) and purified with the QIAquick PCR purification kit (Qiagen). The following oligonucleotide primers were applied, which introduced NdeI and BamHI sites at the 5′ and 3′ ends of the GbE and Ngb coding sequences: GbE, forward, 5′-TACTCACATATGTCTTTCTCTGAAGCGGG-3′, and GbE, reverse, 5′-TACTCTGGATCCTCACCAACCTGCTTCTTTGT-3′; Ngb, forward, 5′-TACTCACATATGGAGAGCGGGATGCTGTCT-3′, and Ngb, reverse: 5′-TACTCTGGATCCTTAGTCCCCCTCCGGGGGAC-3′. After cloning into standard vectors (pGEM® T-Easy vector systems, Promega, Mannheim, Germany; TOPO TA Cloning® kit, Invitrogen), plasmids were sequenced by GENterprise Genomics (Mainz, Germany). The Ngb and GbE cDNA fragments were then cloned into the pET-3a vector, making use of the NdeI and BamHI restriction enzyme sites.
Quantitative Real-time Reverse Transcription PCR
Reverse transcription was performed with 300 ng of total RNA, oligo(dT)18 oligonucleotides (10 μm), and 200 units of SuperScriptTM II RNase H− reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Quantitative real-time RT-PCR experiments were carried out on an ABI 7300 real-time PCR system (Applied Biosystems, Darmstadt, Germany) with the ABI Power SYBR Green master mix (Applied Biosystems). Levels of mRNA of Ngb and GbE were evaluated. To avoid amplification of genomic DNA, primer pairs included intron-spanning oligonucleotides: GbE, 5′-GCAGTGCTGGTCAGGATGTT-3′ and 5′-CTCTGACCTGATCCGACTG-3′ (amplicon: 104 bp); Ngb, 5′-AGGAGTGCCTGGCTGCC-3′ and 5′-CACCAACAGCCTGATGCTTC-3′ (amplicon: 144 bp). Oligonucleotides were obtained from Operon Biotechnologies (Cologne, Germany) and tested in advance for amplification efficiency and specificity.
15 ng of cDNA were applied as template in a reaction volume of 25 μl. Primer concentrations were 200 nm for each oligonucleotide. Quantitative real-time PCR was performed with the SYBR Green mix according to the manufacturer's instructions, applying 40 amplification cycles (95 °C/15 s, 60 °C/15 s, 72 °C/30 s). Fluorescence was measured at the end of each amplification cycle. Reactions were run in triplicate or duplicate (negative controls). Specificity of the amplification reaction was analyzed in dissociation curves with a temperature range from 60 to 95 °C. First evaluation of quantitative real-time RT-PCR data was performed with the ABI 7300 Sequence Detection software V.1.3.1 (Applied Biosystems). Efficiency of reaction was measured by the slope of a standard curve, deriving from 10-fold dilutions of plasmids. Expression data were normalized according to the amount of total RNA. Further analyses were carried out employing the Microsoft Office Excel spreadsheet program.
Antibody Preparation
A polyclonal antibody against GbE was produced using a commercial service (Sigma Genosys Ltd.). The antibody was raised in rabbits against synthetic GbE peptides, which cover amino acid positions 42–56 (H2N-SYFTHFKGMDSAEEM-CONH2) and positions 96–109 (H2N-KHATQLKIDPKNFR-CONH2)of chicken GbE, coupled to keyhole limpet hemocyanin. Specific anti-GbE antibodies were affinity-purified using recombinant GbE coupled to a SulfoLink column (Thermo Pierce, Bonn, Germany) according to the instructions of the manufacturer and stored in 50 mm Tris, 100 mm glycine, pH ∼7.4) supplemented with 0.1% NaN3.
SDS-PAGE and Western Blotting
Tissues were homogenized in 10 mm Tris/HCl, pH 7.4, 10 mm NaCl, 5 mm MgCl2, 1 mm DTT, 1 mm Pefabloc SC protease inhibitor (Carl Roth, Karlsruhe, Germany) and CompleteTM protease inhibitor mix (Roche Applied Science). Protein concentrations were determined according to the Bradford method (24).
For SDS-PAGE, protein extracts were heat-denatured in sample buffer (65 mm Tris-HCl, pH 6.8, 1% SDS, 5% β-mercaptoethanol, 10% glycerol) at 95 °C for 5 min and loaded onto a 15% polyacrylamide gel. Semidry electroblotting of proteins from the polyacrylamide gels onto nitrocellulose membranes (A. Hartenstein, Würzburg, Germany) was carried out for 2 h at 0.8 mA/cm2. Nonspecific binding sites were blocked for 1 h with 2% nonfat dry milk in TBS (10 mm Tris/HCl, pH 7.4, 140 mm NaCl). Immunodetection was performed for 2 h at room temperature with affinity-purified polyclonal anti-GbE antibodies diluted 1:100 in 2% milk/TBS. The nitrocellulose filters were washed three times with TBS for 15 min and incubated for 1 h with the goat anti-rabbit antibody coupled with alkaline phosphatase (Dianova, Hamburg, Germany), diluted 1:10,000 in TBS. After the final washing step, detection was carried out with nitro-blue-tetrazolium-chloride and 5-bromo-4-chloro-3-indolyl-phosphate as substrates. Signals were quantified after scanning of the blots with the help of the ImageJ 1.43u program.
Immunohistochemistry
Fresh chicken eyes were fixed in 4% paraformaldehyde in PBS (140 mm NaCl, 2.7 mm KCl, 8.1 mm Na2HPO4, 1.5 mm KH2PO4) for 36 h and stored in PBS at 4 °C until use. The eyes were successively immersed in 10, 20, and 30% sucrose/PBS for ∼24 h for each step. Cryosections of 16-μm thickness were obtained using a Cryostat HM 500 (MICROM International, Walldorf, Germany) and placed on siliconized coverslips (SuperFrost Plus; Thermo Menzel, Heidelberg, Germany). Sections were rehydrated for 10 min with PBS. Proteins were detected either using alkaline phosphatase staining or by immunofluorescence. Purified anti-GbE antibodies were diluted 1:50 in PBS, 0.1% Triton X-100, 1% bovine serum albumin overnight at room temperature. The sections were washed three times for 10 min in PBS. For alkaline phosphatase staining, the sections were incubated for 2 h at room temperature in the dark with goat anti-rabbit antibody coupled with alkaline phosphatase (1:10,000 in PBS; Dianova). Sections were washed three times for 10 min in PBS, and detection was carried out using nitro-blue-tetrazolium and bromochloro-indolyl-phosphate. For immunofluorescence detection, sections were incubated for 2 h with donkey anti-rabbit F(ab′)2 fragment coupled to Cy3 (1:500 in PBS; Dianova) and embedded in 1× PBS/glycerol. The Hoechst dye 33258 (0.3 μg/ml) was added to stain the nuclei. Sections were analyzed using an Olympus BX51 research microscope equipped with a digital camera. Images were combined using Adobe Photoshop CS4 11.0.2.
Recombinant Expression of Globin E
The coding sequence of GbE was isolated by RT-PCR from total RNA extracted from chicken eye. The GbE cDNA was cloned into the pET3a as vector as described above and expressed in Escherichia coli BL21pLysS host cells. E. coli were grown at 30 °C in L-medium (1% Bacto Tryptone, 0.5% yeast extract, 0.5% NaCl, pH 7.5) containing 10 μg/ml ampicillin, 34 μg/ml chloramphenicol overnight. 1 ml of this culture was applied to 500 ml of L-medium supplemented with 1 mm δ-aminolevulinic acid. The culture was induced at A600 = 0.4–0.8 by the addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 0.4 mm, and expression was continued at 30 °C overnight. Cells were harvested (45 min of centrifugation at 3,500 × g) and resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 1 mm MgCl2, 1 mm dithiothreitol), supplemented with 10 μg/ml DNase, 5 μg/ml RNase, Complete proteinase inhibitor mix (Roche Applied Science), and Pefabloc (Carl Roth). The cells were broken by three freeze-thaw cycles in liquid nitrogen followed by ultrasonication (10 × 30 s). The sample was incubated for 2 h at 37 °C to digest the DNA and RNA. The cell debris was removed by centrifugation for 1 h at 4 °C at 4,000 × g. The supernatant was fractionated by ammonium sulfate precipitation. The reddish 90–100% ammonium sulfate pellet was dissolved in 20 mm Tris-HCl, pH 8.5, and desalted using an Amicon Ultra filter (Millipore, Merck, Darmstadt, Deutschland). Further purification of GbE was achieved by a HiPrepTM 16/10 Q XL ion-exchange column (GE Healthcare, Freiburg, Germany) with a gradient of 0–1 m NaCl in 20 mm Tris-HCl, pH 8.5. Size exclusion chromatography was carried out using a HiLoadTM 26/60 SuperdexTM 75 prep grade (GE Healthcare). The final GbE fractions were analyzed by gel electrophoresis, pooled, concentrated, and stored frozen at −20 °C. Protein concentrations were determined using the Bradford method (24).
Globin E Ligand Binding
Absorption spectra were measured with a Varian Cary 50 or a diode array HP 8453 spectrophotometer. Samples in 50 mm Tris-HCl, 100 mm NaCl, 0.2 mm EDTA buffer at pH 8.0 were reduced with sodium dithionite to obtain the deoxy spectrum. The oxy form was obtained after the addition of 2 mm DTT to ferric GbE under pure O2. Alternatively, the ferric form was reduced with sodium dithionite under pure CO. Excesses of CO were removed with a Sephadex G25 desalting column (GE Healthcare).
Kinetic measurements of ligand binding were made with a flash photolysis system at 25 and 41 °C, as described previously (25, 26). Kinetics at different CO concentrations and under a mixed CO/O2 atmosphere were used to study the competitive ligand binding. Effectively, the CO protects the iron from oxidation, and photodissociation of CO leads to a transient exposure to O2, allowing a determination of both the association and the dissociation rates for O2. 1 torr of O2 partial pressure corresponds to 1.56 × 10−6 mol/liter dissolved O2 at 25 °C, 1.28 × 10−6 mol/liter dissolved O2 at 37 °C, and 1.21 × 10−6 mol/liter dissolved O2 at 41 °C in our 50 mm Tris-HCl, 100 mm NaCl, pH 8.0 buffer. For the experimental conditions of Ngb, the O2 solubility coefficient was 1.82 × 10−6 mol/liter at 25 °C, and for CO, the solubility coefficient was 1.36 × 10−6 mol/liter for the different experimental conditions.
For measuring the CO dissociation rate (koff), GbE was first reduced with a small excess of sodium dithionite under 1 atm CO, which results in the GbE-Fe2+-CO form. CO dissociation rates were measured by displacing CO with an excess of O2. The limiting reaction is the CO dissociation because the O2 binding occurs on the μs timescale. The same rate was obtained by mixing GbE-Fe2+-CO with a deoxygenated buffer containing an excess of potassium ferricyanide. We confirmed that the detection light of the HP 8453 diode array spectrometer did not induce a photodissociation of the heme-CO complex.
For the oxidation kinetics, the sample was first deoxygenated in a sealed optical cuvette under a stream of N2. A slight excess of sodium dithionite was added to reduce the GbE heme moiety. We obtained a stable deoxy form after the addition of 8 μm dithionite to 2 μm GbE. Finally, the cuvette was equilibrated under air within a few seconds to obtain the oxy reduced species and to allow the depletion of the residual unreacted dithionite through its oxidation with the excess of dissolved O2. The full spectra were measured versus time on an HP 8453 diode array spectrometer with the visible tungsten-halogen lamp for each nm. For the slowest oxidation kinetics, the associated absorption changes were measured at several wavelengths every 10 min with the monochromatic light from the xenon flash lamp of the Varian Cary 50 spectrophotometer.
RESULTS
The Chicken and Zebra Finch GbE Gene
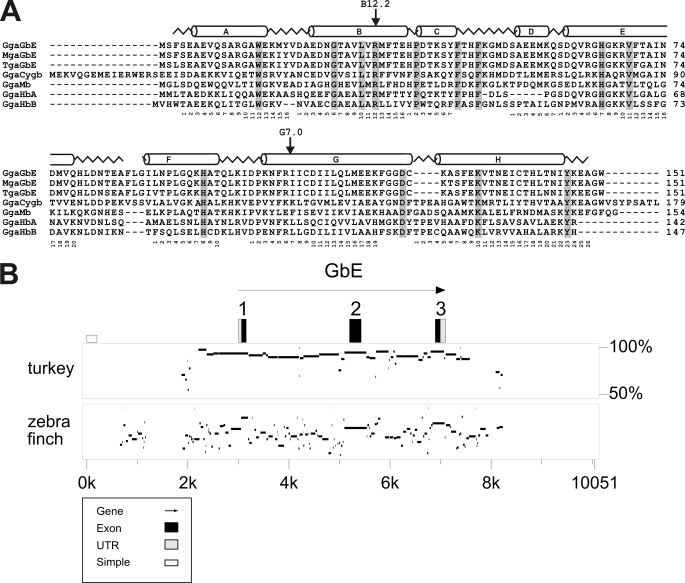
Database analyses showed that the GbE gene resides on the short arm of chicken chromosome 1 at bp 51,039,086–51,043,463 (contig NW_001471513; Build 2.1). We further searched for GbE orthologs in the genomes of other vertebrates. We obtained putative GbE orthologs from the genome assemblies of the turkey M. gallopavo (protein ENSMGAG00000012002) and the zebra finch T. guttata (hypothetical protein LOC100232117, accession number XM_002196350). All three GbE cDNAs cover 453 bp, which translate into proteins of 151 amino acids. On the nucleotide level, chicken GbE (GgaGbE) shares 435 bp (96.0%) with turkey GbE (MgaGbE) and 408 bp (90.1%) with zebra finch GbE (TguGbE). The GgaGbE and MgaGbE proteins are 98.0% identical (99.3% similarity considering isofunctional amino acid replacements), whereas GgaGbE and TguGbE proteins are 92.7% identical and 96.0% similar (Fig. 1A). The turkey GbE gene is on chromosome 1 (UMD 2.0), and the zebra finch GbE resides on chromosome 1A (genome Build 1.1). All three avian GbE genes contain the typical globin introns at positions B12.2 (i.e. between bp 2 and 3 of the 12th codon in helix B) and G7.0.
FIGURE 1.
Conservation of GbE protein and gene. A, comparison of eye globins of chicken (GgaGbE), turkey (MgaGbE) and zebra finch (TguGbE) with chicken Cygb (GgaCygb), myoglobin (GgaMb), hemoglobin α (GgaHbA), and β (GgaHbB). The sperm whale myoglobin structure is superimposed in the upper row, and residues conserved in all these globins are shaded. B, percent identity plot showing the comparison chicken GbE with turkey and zebra finch GbE genes and their ± 3 kb flanking regions. The G. gallus GbE was used as reference; in the upper row, the exons are boxed, with the black boxes representing the coding sequences. The transcriptional orientation is indicated by the arrow. Interspecies sequence identities are shown as horizontal bars on a 50–100% scale.
Comparison of GbE coding sequences allows the evaluation of nucleotide and amino acid evolution. The lineages leading to chicken and turkey diverged between 25 and 50 million years ago (27). The last common ancestor of Galliformes (turkeys and chickens) and zebra finch lived between 66.0 and 86.5 million years ago, near the base of avian radiation (28). This translates into an evolutionary rate of GbE protein of 0.43 ± 0.15 × 10−9 amino acid substitutions per site per year. Mean dS (synonymous substitutions) values were 2.04 ± 0.49 × 10−9, and mean dN (nonsynonymous substitutions) values were 0.23 ± 0.09 × 10−9 replacements per site per year. The degree of sequence conservation ± 3 kb upstream and downstream of the glob1 genes was evaluated with PipMaker, using the chicken GbE as reference (Fig. 1B). Chicken and turkey GbE show strong conservation also in intronic regions and ∼1,000 bp of the upstream region. In the chicken-zebra finch comparison, the putative coding exons are still clearly discernable. Except for scattered sections in the introns, only about 200 bp 5′ of the transcription start point, which may harbor regulatory sequences, are conserved.
Evaluation of chromosomal gene order, which included eight proximal and eight distal genes, showed one-to-one synteny of chicken and zebra finch genes adjacent to GbE (supplemental Table 1). Although the four and six, respectively, immediate neighbors of GbE are also conserved in turkey, there are some rearrangements in this region in the current assembly. GbE appears to be absent in vertebrates other than birds (15). No GbE gene was detected in the preassembly genome of the reptile Anolis carolinensis (WGS contigs). We identified putative orthologs and paralogs of the neighboring genes in the human genome (Build 37.2). Putative human orthologs of five of the proximal genes (POLDIP3, PRP7A, SERHL2, NFAM1, LOC417980) reside on chromosome 22q13, with conserved gene order. Orthologs or paralogs of four distal genes (TTC26, UBN2, LOC770530, LUC7L2) are in synteny on human chromosome 7q34. There is no association of these genes with any globin locus in the human genome.
We further analyzed the positions of paralogous genes relative to globin loci. In chicken, the Mb gene also resides on short arm of chromosome 1, about 3 Mbp proximal of GbE. However, analysis of gene synteny showed no conserved order of the neighboring genes of Mb and GbE. The gene LUC7L2 resides in a head-to-head orientation 4,393 bp distally to GbE. A paralogous LUC7L gene is present on chicken chromosome 14 (positions: 12,896,737–12,919,842), 189,346 bp proximal of the HbA locus. However, LUC7L and HBA are separated by six other genes (Q8UWG9, Q8UWG8, AXN1, Q9PVJ0, ITFG3, ENSGALT00000039666). In the human genome, LUC7L gene is a direct neighbor of the human HbA locus on chromosome 16q13.3 and is ∼6,000 bp adjacent to HBθ1 in a conserved head-to-head orientation. In the zebrafish D. rerio, a LUC7L gene is situated on chromosome 1, 14 kb distal to the Mb gene in a head-to-head orientation, separated by the pane1 gene. Otherwise the zebrafish genome is not informative in terms of the evolution of the GbE locus.
Expression of GbE in Chicken Brain and Eye
Quantitative real-time RT-PCR was carried out for chicken Ngb and GbE, employing total brain and eye tissue (Fig. 2A). Because no standard gene for reliable normalization was available, the mRNA copy numbers were normalized according to the total RNA content. mRNA levels of GbE were higher than those of Ngb in both eye and brain. GbE mRNA was also detected in brain tissue, but GbE mRNA levels were ∼50,000 times higher in the total eye samples.
FIGURE 2.
Expression of GbE protein and RNA. A, expression of GbE and Ngb mRNA in chicken brain and eye, as measured by quantitative real-time RT-PCR. Values are means of three independent replicates (n = 3), and the standard deviations are given. Amounts of globin mRNA are given as copy number per μg of total RNA. Note the logarithmic scale of the y axis. B, Western blot detection of GbE protein in chicken eye. On the left lane, 0.5 μg of purified recombinant GbE was applied; on the right lane, 100 μg of total proteins extracted from eye were loaded. Protein was detected employing a specific anti-GbE antibody. GbE content was estimated by ImageJ to be 1.6 ± 0.1 μg GbE/mg of total eye proteins (n = 3).
We raised a polyclonal antibody against two distinct synthetic peptides of the chicken GbE protein. The antibody was purified from the serum by the aid of the appropriate peptides. The antibody was applied on Western blots, showing a distinct band of about 17 kDa that corresponds to the expected mass of GbE (Fig. 2B). In the recombinant GbE samples, a second band was observed, which has a mass of about 36 kDa. This band may represent a GbE dimer, similar to those found e.g. for Ngb (29). GbE content in total eye proteins was measured by comparison with known amounts of recombinant GbE. We estimated that GbE makes up about roughly 0.16% of total eye proteins, which translates into about 1.6 ± 0.1 μg of GbE/mg of total protein.
Localization of GbE in the Chicken Retina
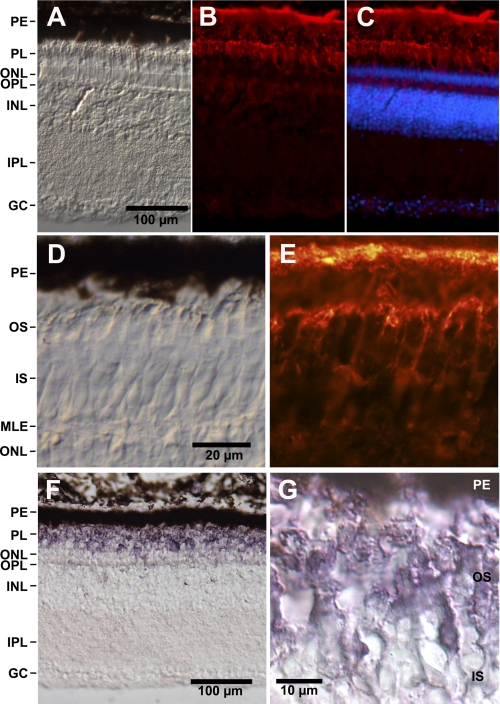
In immunohistochemical studies, the anti-GbE antibody (evaluated in Fig. 2B) showed cytoplasmic staining on cryosections of chicken retina (Fig. 3). Bright anti-GbE immunofluorescence was found in the pigment epithelium and the outer segments of the photoreceptor cells (Fig. 3, B and C). Weak staining was observed in the outer plexiform and the ganglion cell layers. At higher magnification (Fig. 3E), bright anti-GbE immunoreaction was found in the cytoplasm surrounding the stacks of membrane-enclosed disks in the outer segments of the photoreceptor cells. The same distribution of GbE protein was found by employing a secondary antibody coupled with alkaline phosphatase (Fig. 3, F and G).
FIGURE 3.
Localization of GbE protein and mRNA in the chicken retina. A–C, longitudinal cryosections of the chicken retina. PE, pigment epithelium; PL, layer of outer and inner segments of photoreceptor cells; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GC, layer of ganglion cells. A, bright field microscopy image. Scale bar, 100 μm. B, indirect anti-GbE immunofluorescence. Bright immunofluorescence is visible in the pigment epithelium and the outer segments of photoreceptors; weak staining is visible in the outer plexiform layer and the ganglion cells. C, merged figure showing staining of the nuclei with Hoechst dye 33258. D and E, higher magnification of the photosensitive outer segments (OS) shows anti-GbE immunofluorescence of the cytoplasm surrounding the stacks of membrane-enclosed disks (E). D, bright field image. Scale bar, 20 μm. IS, inner segments; MLE, membrana limitans externa. F and G, immunohistological staining employing a secondary antibody coupled with alkaline phosphatase shows strong staining of the outer segments.
Kinetics of Ligand Binding of the Chicken GbE
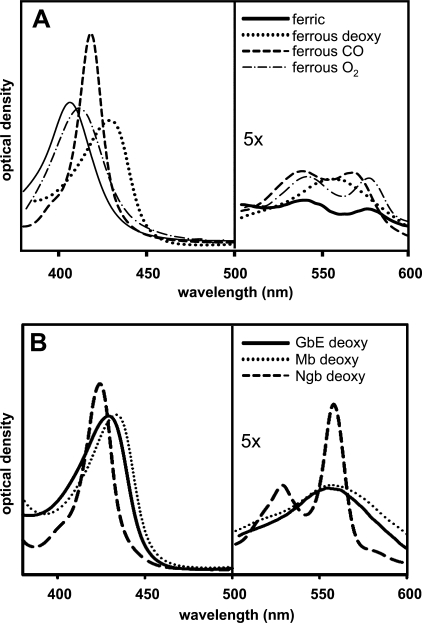
Chicken GbE was recombinantly expressed in E. coli BL21(DE3)pLysS employing the pET system. Purification from the E. coli supernatant was carried out by a three-step protocol, which includes ammonium sulfate precipitation followed by ion-exchange and size exclusion chromatography. The recombinant GbE elutes as a monomer. The absorbance spectrum of oxygenated GbE displays a Soret band at 411 nm, an α-band at 541 nm, and a β-band at 577 nm (Fig. 4A). After reduction with sodium dithionite under nitrogen, the ferrous deoxy form was obtained, and a large amplitude of the Soret band (429 nm) and a maximum in the visible region (556 nm) were observed. The ferric (Fe3+) form has maxima at 406, 540, and 576 nm. The absorption spectrum of deoxy-GbE resembles that of Mb but is clearly distinct from that of Ngb, indicating a penta-coordinated binding scheme (Fig. 4B).
FIGURE 4.
Absorbance spectra of GbE. A, spectral forms of ferric (solid line), ferrous deoxygenated (dotted line), ferrous CO-form (dashed line), and ferrous oxygenated (dot-dash line) of recombinant GbE were read from 380 to 600 nm. The protein concentration was ∼0.15 mg/ml. B, comparison of spectra of deoxygenated GbE (solid line), Mb (dotted line), and Ngb (dashed line).
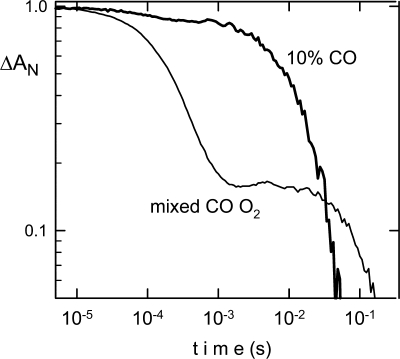
The ligand kinetics of recombinantly expressed chicken GbE after CO photodissociation showed a slight heterogeneity of the binding curve (Fig. 5). We determined a kon(CO) rate of 7 × 105 m−1 s−1 at 25 °C and 10.5 m−1 s−1 at 41 °C (Table 1). The koff(CO) rate was 0.18 s−1 at 25 °C. Application of a mixed CO/O2 atmosphere allows a determination of the on and off rates for O2 (kon = 9 × 106 m−1 s−1, koff = 85 s−1 at 25 °C and kon = 18 × 106 m−1 s−1, koff = 340 s−1 at 41 °C). The half-saturation pressure for O2 binding of chicken GbE was P50 = 5.8 torrs at 25 °C and 15 torrs at 41 °C. The kon(O2) rate is thus ∼10 times faster than kon(CO), and the koff(O2) is ∼500 times faster than koff(CO). This results in a CO affinity about 35 times higher than O2 (CO/O2 partition coefficient for GbE of M = 35). Values of M for a globin are typically in the range 10–100. Note that the addition of DTT did not modify the O2 and CO binding properties.
FIGURE 5.
Flash photolysis kinetics of GbE. After photodissociation of CO, the CO recombination showed a slight heterogeneity (bold line) in the recombination kinetics. The use of a mixed CO/O2 atmosphere (thin line) allows a determination of the oxygen on and off rates. GbE has the characteristic of classical globins, where CO binds more tightly, but O2 binds more rapidly. Photolysis of CO produces the unliganded (penta-coordinated) transient; the rapid phase is essentially oxygen binding, and the slow phase is the replacement of oxygen by CO. Data are presented on a log-log scale as the normalized change in absorption versus time, for curves measured at 25 °C, pH 8.0.
TABLE 1.
Ligand binding and autoxidation for GbE, Mb, and Ngb
Experimental conditions for GbE and Mb: 50 mm Tris-HCl, 100 mm NaCl, 5 mm DTT, pH 8.0. Human Ngb experimental conditions: 100 mm potassium phosphate, 2 mm DTT, pH 7.0 (25). The autoxidation rate of Ngb is taken from Dewilde et al. (29), and the Mb data are from Sono et al. (45). M is the partition coefficient.
| Protein |
|||||
|---|---|---|---|---|---|
| GbE (Chicken) | GbE (Chicken) | Mb (Horse) | Mb (Horse) | Ngb (Human) | |
| Temperature (°C) | 25 | 41 | 25 | 37 | 25 |
| konCO (μm−1s−1) | 0.7 | 1.05 | 0.65 | 0.95 | 40 |
| koffCO (s−1) | 0.18 | 0.023 | 0.014 | ||
| K CO (nm) | 260 | 45 | 0.35 | ||
| konO2 (μm−1s−1) | 9 | 18 | 18 | 31 | 170 |
| koffO2 (s−1) | 85 | 340 | 27 | 90 | 0.7 |
| K O2 (μm) | 9.2 | 18 | 1.6 | 2.8 | 0.004 |
| M (K O2/K CO) | 35 | 29 | 12 | ||
| konHis (s−1) | 1800 | ||||
| koffHis (s−1) | 0.6 | ||||
| 1/K His | 3000 | ||||
| P50O2 (torr) | 5.8 | 15 | 1 | 2.2 | 6.8 |
| Autoxidation (h−1) | 0.1 | 1 | 0.05 | 5 | |
We noted significant photo-induced oxidation of GbE to the ferric Fe3+ form that is unable to bind O2. Exposure to UV light at 280 nm provokes oxidation within a few minutes (25 °C). Therefore, the exposure to UV was minimized in our measurements. Under these conditions, GbE displays autoxidation (t½ = ∼10 h at 25 °C and ∼1 h at 41 °C). These rates are slower than that of Ngb, which oxidizes in less than 15 min at >25 °C (human Ngb t½ = 12 min at 25 °C, pH 8.0; mouse Ngb: t½ = ∼5 min; at 37 °C, pH 7.0) (29), and within the range of other penta-coordinate globins such as Mb, taking into account the lower O2 affinity for GbE because there is an inverse relation between oxygen affinity and autoxidation rate (30).
DISCUSSION
GbE is unique among vertebrates because it is specifically expressed in the eye (Fig. 2). The essentially eye-specific expression of GbE was further confirmed by five expressed sequence tags (DR425234, DR427888, DR430010, DR430941, CD527046) that derive from an eye-specific cDNA library of 15-day chicken embryos. A single additional GbE expressed sequence tag derives from a brain library (CN224774). An eye-specific globin was identified in the nematode Mermis nigrescens, which exhibits a shadowing function for phototaxis (31). The anatomy and function of the avian eye render such function as highly unlikely for GbE, which may rather function as a typical Mb in O2 supply.
Emergence and Evolution of Globin E
The evolution of a protein may provide clues to its function. GbE appears to be restricted to birds because no ortholog could be identified in the sequences from any other vertebrate class. Surprisingly, no GbE was detected in the genome of Anolis. However, currently we cannot decide whether this failure is due to the actual absence of GbE in reptiles or the fragmentary nature of the available genomic sequences from A. carolinensis. Therefore, GbE may have evolved in the avian, sauropsid, or an earlier lineage. Phylogenetic analyses tentatively suggest an association of GbE with either the Cygb (15) or the Mb (32) clades, but the support for either grouping was poor. A relationship of GbE and Cygb is tentatively supported by a unique insertion of three amino acids (FLG) at the EF corner (Fig. 1).
The position in the genome and gene synteny may provide additional information about globin evolution (33, 34). In this context, LUC2L is the only informative gene (13, 35). Paralogous LUC2L genes are located in head-to-head orientation adjacent to avian GbE, avian and human HbA, and fish Mb. Although this syntenic position does not support any particular relationship of GbE, it suggests an early origin of this gene. In any case, GbE emerged from the Mb/Cygb globin lineage before the radiation of Gnathostomata, at least ∼420 million years ago (28). Thus GbE must have been independently lost in fishes, amphibians, and mammals. This may be due to the emergence of vascularized retinae.
The evolutionary rate of GbE protein (0.43 ± 0.15 × 10−9 amino acid substitutions per site per year) is somewhat faster than the rates of Ngb and Cygb (∼0.3 × 10−9) but slower than those of Hb and Mb (∼1 × 10−9) (12, 34), suggesting a rather conserved gene. Levels of selective constraint may also be measured by dS/dN ratios, which are highly variable among vertebrate globins (34). Strong selective pressure on a coding region favors synonymous substitutions (dS) over non-synonymous substitutions (dN). For GbE, we calculated a ds/dn ratio of 7.41–14.03, which was slightly higher than that of mammalian Mb (7.29) and indicates purifying selection (34). Otherwise, only mammalian Ngb and Cygb have dS/dN ratios greater than 10. Thus GbE appears to be a conserved gene, suggesting an important function.
A Respiratory Function of Globin E
The recently identified “novel” vertebrate globins Ngb, Cygb, and GbX share the common characteristics of being hexa-coordinated in the deoxygenated form, which is characterized by two peaks in the visible spectrum around 530 and 560 nm (Fig. 4B) (25, 26, 29). By contrast, the absorption spectrum and ligand binding kinetics show that GbE is a typical penta-coordinated globin, similar to Mb and Hb (Fig. 4B and Table 1). Although the function of vertebrate hexa-coordinated globins is still poorly understood, there is no doubt that Hb and Mb function in O2 delivery. Thus, by analogy, an Mb-like role may be inferred for GbE. The measured O2 affinity of chicken GbE (P50 = 5.8 torrs) is lower than that of chicken Mb (P50 = 3.6 torrs) (36) but consistent with a function of GbE in O2 supply. The autoxidation rate is consistent with a respiratory function of GbE, too.
The Possible Role of Globin E in the Avian Retina
To support its visual function, the vertebrate retina consumes large amounts of O2. An adequate cellular oxygen environment is crucial for the appropriate function of retinal cells. The vascular retina of most mammals has a dual blood supply in which the outer retina is nourished by choroidal blood vessels that lie immediately behind the pigment epithelium. The inner retina is supported by the deep capillary network located in the outer plexiform layer and the superficial capillaries adjacent to the ganglion cell layer (37, 38). However, birds possess an avascular retina, in which the deep retinal and superficial capillaries are essentially absent. O2 is delivered by the choroidal vascular bed and the pecten, a vascular structure extending from the retina into the posterior chamber (16, 39).
No GbE was detected in the pecten structure of the chicken eye (not shown). By contrast, a high concentration of GbE was found in the neuronal retina, which accumulates to 1.6 μg/mg of total eye protein (about 10 μm GbE). Given the fact that most of the protein resides in the vitreous chamber, local GbE concentration within the retina is certainly much higher and well within the range of a typical Mb (40, 41).
The high concentration of Ngb in the rodent retina led to the suggestion that this globin increases the availability of O2 for the respiratory chain (6, 8). In mouse, rat, and Guinea pig, Ngb is associated with the mitochondria and thus cellular respiration. Total concentration of Ngb in the mouse retina was estimated to be ∼100–200 μm. Because no antibody is available that specifically recognizes chicken Ngb, we could not determine Ngb protein concentration in chicken. However, in the chicken eye, Ngb mRNA levels are about 100-fold lower than that of GbE (Fig. 2A), suggesting that Ngb does not play a major role in O2 supply in this tissue.
GbE is mainly localized in the photoreceptor cells, which are in charge for visual perception. High ATP amounts are consumed by the photoreceptor cells for the maintenance of the dark current, for the fast turnover of photosensitive membranes, or for the phototransduction process itself (42). On the other hand, vascularization of the avian retina is poor. It is therefore tempting to assume that GbE enhances O2 supply to the photoreceptor cells, thereby supporting the visual process in the bird eye. However, we cannot formally exclude other functions of GbE, such as protection from reactive oxygen species or a redox reaction, which may be induced by light in the outer segments of the photoreceptor cells (43, 44).
Supplementary Material
This work was supported by the Deutsche Forschungsgemeinschaft (Grants Bu956/11 and Ha2103/3) and INSERM.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
M. Blank, J. Wollberg, F. Gerlach, K. Reimann, A. Roesner, T. Hankeln, A. Fago, R. E. Weber, and T. Burmester, manuscript submitted.
- Hb
- hemoglobin
- Mb
- myoglobin
- Ngb
- neuroglobin
- Cygb
- cytoglobin
- GbE
- eye-specific globin
- GbX
- globin X
- dN
- non-synonymous substitution
- dS
- synonymous substitution.
REFERENCES
- 1. Dickerson A. E., Greis I. (1983) Hemoglobin: Structure, Function, Evolution, and Pathology, Benjamin-Cummings Publishing Company, Inc., Menlo Park, CA [Google Scholar]
- 2. Wittenberg J. B., Wittenberg B. A. (2003) J. Exp. Biol. 206, 2011–2020 [DOI] [PubMed] [Google Scholar]
- 3. Burmester T., Weich B., Reinhardt S., Hankeln T. (2000) Nature 407, 520–523 [DOI] [PubMed] [Google Scholar]
- 4. Burmester T., Hankeln T. (2009) J. Exp. Biol. 212, 1423–1428 [DOI] [PubMed] [Google Scholar]
- 5. Hankeln T., Ebner B., Fuchs C., Gerlach F., Haberkamp M., Laufs T. L., Roesner A., Schmidt M., Weich B., Wystub S., Saaler-Reinhardt S., Reuss S., Bolognesi M., De Sanctis D., Marden M. C., Kiger L., Moens L., Dewilde S., Nevo E., Avivi A., Weber R. E., Fago A., Burmester T. (2005) J. Inorg. Biochem. 99, 110–119 [DOI] [PubMed] [Google Scholar]
- 6. Bentmann A., Schmidt M., Reuss S., Wolfrum U., Hankeln T., Burmester T. (2005) J. Biol. Chem. 280, 20660–20665 [DOI] [PubMed] [Google Scholar]
- 7. Mitz S. A., Reuss S., Folkow L. P., Blix A. S., Ramirez J. M., Hankeln T., Burmester T. (2009) Neuroscience 163, 552–560 [DOI] [PubMed] [Google Scholar]
- 8. Schmidt M., Giessl A., Laufs T., Hankeln T., Wolfrum U., Burmester T. (2003) J. Biol. Chem. 278, 1932–1935 [DOI] [PubMed] [Google Scholar]
- 9. Burmester T., Ebner B., Weich B., Hankeln T. (2002) Mol. Biol. Evol. 19, 416–421 [DOI] [PubMed] [Google Scholar]
- 10. Nakatani K., Okuyama H., Shimahara Y., Saeki S., Kim D. H., Nakajima Y., Seki S., Kawada N., Yoshizato K. (2004) Lab. Invest. 84, 91–101 [DOI] [PubMed] [Google Scholar]
- 11. Schmidt M., Gerlach F., Avivi A., Laufs T., Wystub S., Simpson J. C., Nevo E., Saaler-Reinhardt S., Reuss S., Hankeln T., Burmester T. (2004) J. Biol. Chem. 279, 8063–8069 [DOI] [PubMed] [Google Scholar]
- 12. Burmester T., Haberkamp M., Mitz S., Roesner A., Schmidt M., Ebner B., Gerlach F., Fuchs C., Hankeln T. (2004) IUBMB Life 56, 703–707 [DOI] [PubMed] [Google Scholar]
- 13. Fuchs C., Burmester T., Hankeln T. (2006) Cytogenet. Genome Res. 112, 296–306 [DOI] [PubMed] [Google Scholar]
- 14. Roesner A., Fuchs C., Hankeln T., Burmester T. (2005) Mol. Biol. Evol. 22, 12–20 [DOI] [PubMed] [Google Scholar]
- 15. Kugelstadt D., Haberkamp M., Hankeln T., Burmester T. (2004) Biochem. Biophys. Res. Commun. 325, 719–725 [DOI] [PubMed] [Google Scholar]
- 16. Pettigrew J. D., Wallman J., Wildsoet C. F. (1990) Nature 343, 362–363 [DOI] [PubMed] [Google Scholar]
- 17. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 18. Hillier L. W., Miller W., Birney E., Warren W., Hardison R. C., Ponting C. P., Bork P., Burt D. W., Groenen M. A. M., Delany M. E., Dodgson J. B., Chinwalla A. T., Cliften P. F., Clifton S. W., Delehaunty K. D., Fronick C., Fulton R. S., Graves T. A., Kremitzki C., Layman D., Magrini V., McPherson J. D., Miner T. L., Minx P., Nash W. E., Nhan M. N., Nelson J. O., Oddy L. G., Pohl C. S., Randall-Maher J., Smith S. M., Wallis J. W., Yang S.-P., Romanov M. N., Rondelli C. M., Paton B., Smith J., Morrice D., Daniels L., Tempest H. G., Robertson L., Masabanda J. S., Griffin D. K., Vignal A., Fillon V., Jacobbson L., Kerje S., Andersson L., Crooijmans R. P. M., Aerts J., et al. (2004) Nature 432, 695–716 15592404 [Google Scholar]
- 19. Dalloul R. A., Long J. A., Zimin A. V., Aslam L., Beal K., Blomberg Le Ann, Bouffard P., Burt D. W., Crasta O., Crooijmans R. P., Cooper K., Coulombe R. A., De S., Delany M. E., Dodgson J. B., Dong J. J., Evans C., Frederickson K. M., Flicek P., Florea L., Folkerts O., Groenen M. A., Harkins T. T., Herrero J., Hoffmann S., Megens H. J., Jiang A., de Jong P., Kaiser P., Kim H., Kim K. W., Kim S., Langenberger D., Lee M. K., Lee T., Mane S., Marcais G., Marz M., McElroy A. P., Modise T., Nefedov M., Notredame C., Paton I. R., Payne W. S., Pertea G., Prickett D., Puiu D., Qioa D., Raineri E., Ruffier M., Salzberg S. L., Schatz M. C., Scheuring C., Schmidt C. J., Schroeder S., Searle S. M., Smith E. J., Smith J., Sonstegard T. S., Stadler P. F., Tafer H., Tu Z. J., Van Tassell C. P., Vilella A. J., Williams K. P., Yorke J. A., Zhang L., Zhang H. B., Zhang X., Zhang Y., Reed K. M. (2010) PLoS Biol. 8, e1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Warren W. C., Clayton D. F., Ellegren H., Arnold A. P., Hillier L. W., Künstner A., Searle S., White S., Vilella A. J., Fairley S., Heger A., Kong L., Ponting C. P., Jarvis E. D., Mello C. V., Minx P., Lovell P., Velho T. A., Ferris M., Balakrishnan C. N., Sinha S., Blatti C., London S. E., Li Y., Lin Y. C., George J., Sweedler J., Southey B., Gunaratne P., Watson M., Nam K., Backström N., Smeds L., Nabholz B., Itoh Y., Whitney O., Pfenning A. R., Howard J., Völker M., Skinner B. M., Griffin D. K., Ye L., McLaren W. M., Flicek P., Quesada V., Velasco G., Lopez-Otin C., Puente X. S., Olender T., Lancet D., Smit A. F., Hubley R., Konkel M. K., Walker J. A., Batzer M. A., Gu W., Pollock D. D., Chen L., Cheng Z., Eichler E. E., Stapley J., Slate J., Ekblom R., Birkhead T., Burke T., Burt D., Scharff C., Adam I., Richard H., Sultan M., Soldatov A., Lehrach H., Edwards S. V., Yang S. P., Li X., Graves T., Fulton L., Nelson J., Chinwalla A., Hou S., Mardis E. R., Wilson R. K. (2010) Nature 464, 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicholas K. B., Nicholas H. B., Jr., Deerfield D. W., 2nd (1997) EMBNEW NEWS 4, 14 [Google Scholar]
- 22. Comeron J. M. (1999) Bioinformatics 15, 763–764 [DOI] [PubMed] [Google Scholar]
- 23. Schwartz S., Zhang Z., Frazer K. A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W. (2000) Genome Res. 10, 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 25. Uzan J., Dewilde S., Burmester T., Hankeln T., Moens L., Hamdane D., Marden M. C., Kiger L. (2004) Biophys. J. 87, 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hamdane D., Kiger L., Dewilde S., Green B. N., Pesce A., Uzan J., Burmester T., Hankeln T., Bolognesi M., Moens L., Marden M. C. (2003) J. Biol. Chem. 278, 51713–51721 [DOI] [PubMed] [Google Scholar]
- 27. Griffin D. K., Robertson L. B., Tempest H. G., Vignal A., Fillon V., Crooijmans R. P., Groenen M. A., Deryusheva S., Gaginskaya E., Carré W., Waddington D., Talbot R., Völker M., Masabanda J. S., Burt D. W. (2008) BMC Genomics 9, 168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Benton M. J., Donoghue P. C. (2007) Mol. Biol. Evol. 24, 26–53 [DOI] [PubMed] [Google Scholar]
- 29. Dewilde S., Kiger L., Burmester T., Hankeln T., Baudin-Creuza V., Aerts T., Marden M. C., Caubergs R., Moens L. (2001) J. Biol. Chem. 276, 38949–38955 [DOI] [PubMed] [Google Scholar]
- 30. Brantley R. E., Jr., Smerdon S. J., Wilkinson A. J., Singleton E. W., Olson J. S. (1993) J. Biol. Chem. 268, 6995–7010 [PubMed] [Google Scholar]
- 31. Burr A. H., Hunt P., Wagar D. R., Dewilde S., Blaxter M. L., Vanfleteren J. R., Moens L. (2000) J. Biol. Chem. 275, 4810–4815 [DOI] [PubMed] [Google Scholar]
- 32. Hoffmann F. G., Opazo J. C., Storz J. F. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14274–14279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gillemans N., McMorrow T., Tewari R., Wai A. W., Burgtorf C., Drabek D., Ventress N., Langeveld A., Higgs D., Tan-Un K., Grosveld F., Philipsen S. (2003) Blood 101, 2842–2849 [DOI] [PubMed] [Google Scholar]
- 34. Wystub S., Ebner B., Fuchs C., Weich B., Burmester T., Hankeln T. (2004) Cytogenet. Genome Res. 105, 65–78 [DOI] [PubMed] [Google Scholar]
- 35. Patel V. S., Cooper S. J., Deakin J. E., Fulton B., Graves T., Warren W. C., Wilson R. K., Graves J. A. (2008) BMC Biol. 6, 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Enoki Y., Ohga Y., Kawase M., Nakatani A. (1984) Biochim. Biophys. Acta 789, 334–341 [DOI] [PubMed] [Google Scholar]
- 37. Osborne N. N., Casson R. J., Wood J. P., Chidlow G., Graham M., Melena J. (2004) Prog. Retin. Eye Res. 23, 91–147 [DOI] [PubMed] [Google Scholar]
- 38. Yu D. Y., Cringle S. J. (2001) Prog. Retin. Eye Res. 20, 175–208 [DOI] [PubMed] [Google Scholar]
- 39. Vasilescu J., Guo X., Kast J. (2004) Proteomics 4, 3845–3854 [DOI] [PubMed] [Google Scholar]
- 40. Wittenberg J. B. (1992) Adv. Comp. Environ. Physiol. 13, 60–85 [Google Scholar]
- 41. Wittenberg J. B., Wittenberg B. A. (1990) Annu. Rev. Biophys. Biophys. Chem. 19, 217–241 [DOI] [PubMed] [Google Scholar]
- 42. Pugh E. N., Jr., Lamb T. (2000) in Molecular Mechanisms of Visual Transduction (Stavenga D. G., DeGrip W. J., Pugh E. N., Jr. eds.) pp. 183–255, Elsevier Science Publishers B.V., Amsterdam [Google Scholar]
- 43. Organisciak D. T., Wang H. M., Xie A., Reeves D. S., Donoso L. A. (1989) Prog. Clin. Biol. Res. 314, 493–512 [PubMed] [Google Scholar]
- 44. Penn J. S., Naash M. I., Anderson R. E. (1987) Exp. Eye Res. 44, 779–788 [DOI] [PubMed] [Google Scholar]
- 45. Sono M., Smith P. D., McGray J. A., Asakura T. (1976) J. Biol. Chem. 251, 1418–1426 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.