Abstract
Non-opsonic phagocytosis is a primordial form of pathogen recognition that is mediated by the direct interaction of phagocytic receptors with microbial surfaces. In the fruit fly Drosophila melanogaster, the EGF-like repeat containing scavenger receptor Eater is expressed by phagocytes and is required to survive infections with Gram-positive and Gram-negative bacteria. However, the mechanisms by which this receptor recognizes different types of bacteria are poorly understood. To address this problem, we generated a soluble, Fc-tagged receptor variant of Eater comprising the N-terminal 199 amino acids including four EGF-like repeats. We first established that Eater-Fc displayed specific binding to broad yet distinct classes of heat- or ethanol-inactivated microbes and behaved similarly to the membrane-bound, full-length Eater receptor. We then used Eater-Fc as a tool to probe Eater binding to the surface of live bacteria. Eater-Fc bound equally well to naive or inactivated Staphylococcus aureus or Enterococcus faecalis, suggesting that in vivo, Eater directly targets live Gram-positive bacteria, enabling their phagocytic clearance and destruction. By contrast, Eater-Fc was unable to interact with live, naive Gram-negative bacteria (Escherichia coli, Serratia marcescens, and Pseudomonas aeruginosa). For these bacteria, Eater-Fc binding required membrane-disrupting treatments. Furthermore, we found that cecropin A, a cationic, membrane-disrupting antimicrobial peptide, could promote Eater-Fc binding to live E. coli, even at sublethal concentrations. These results suggest a previously unrecognized mechanism by which antimicrobial peptides cooperate with phagocytic receptors to extend the range of microbes that can be targeted by a single, germline-encoded receptor.
Keywords: Antimicrobial Peptides, Bacteria, Cell Surface Receptor, Drosophila, Innate Immunity, Pattern Recognition Receptor, Phagocytosis, Class F Scavenger Receptor, Eater, Non-opsonic Phagocytosis
Introduction
Phagocytosis is an evolutionarily ancient mechanism by which cells internalize particles (1, 2). It requires cell surface receptors that bind non-self or altered-self molecules displayed on microbes or dying and aberrant cells (3). Phagocytosis plays a major role in innate immunity as a first line of defense against invasive microbes and by mobilizing and instructing adaptive immunity. Phagocytes must constantly monitor their environment to quickly recognize, ingest, and destroy foreign intruders or altered cells.
To carry out this complex task, the plasma membrane of phagocytes is adorned with a great variety of receptors (3). These recognize their targets either directly or indirectly via opsonins (4) such as antibodies, complement, collectins, or pentraxins. The direct interaction of phagocytes with microbes is carried out by germline-encoded receptors, so-called pattern recognition receptors (5, 6). The term implies that a host organism has only a limited repertoire of receptors for recognizing invariant, conserved molecules (or “patterns”) that are synthesized by many potential pathogens.
The high level of complexity of the phagocyte surface makes it difficult to determine the contributions of individual receptors to host defense (3). Over the past decade, the fruit fly Drosophila melanogaster has emerged as a simpler model system to study phagocytosis in the absence of adaptive immunity (7). Fruit flies have a primitive blood cell system with phagocytic cells that resemble mammalian macrophages (8). These Drosophila phagocytes are long-lived (7, 8) and devoid of neutrophil-like granules (9, 10). They play essential roles in tissue remodeling during development (11) and in immunity during infection (11–14).
Drosophila phagocytes express multiple phagocytic receptors, many of which were identified in recent RNA interference screens in the macrophage-like S2 cells (7). Our laboratory identified Eater, a receptor expressed by S2 cells and primary hemocytes (15). Eater plays a critical, non-redundant role in host survival after bacterial infections (11, 13, 15, 16).2 Flies lacking the eater gene display impaired phagocytosis and increased bacterial loads yet intact immune signaling via the NFκB-like pathways Toll and Imd (15, 16).
Eater belongs to an emerging superfamily of pattern recognition receptors with EGF-like repeats (7, 17–19). Several superfamily members have been linked to phagocytosis of microbes or apoptotic cells, for example Caenorhabditis elegans CED-1, Drosophila Draper, SIMU, Nimrod C1, and the mammalian class F scavenger receptors SCARF1 and MEGF10 (7, 20–25). The Eater ectodomain consists of 32 EGF-like repeats preceded by an N-terminal extension of 40 amino acids that contains a characteristic cysteine-flanked CCXGY motif (15, 19, 26).
The four N-terminal EGF-like repeats of Eater display a higher level of variation in amino acids, repeat length, and N-glycosylation than the remainder of the repeats, which may play a structural role as a stalk (15). We previously showed that this region comprising 199 N-terminal amino acids of Eater is involved in direct recognition of diverse bacteria such as Escherichia coli, Serratia marcescens, and Staphylococcus aureus. Binding could be inhibited by the polyanionic scavenger receptor ligands oxidized and acetylated LDL (15). However, it remains unclear what the natural ligands of Eater are and how Eater can recognize different classes of bacteria in vivo because previous bacterial phagocytosis and binding assays were carried out only with dead bacteria.
In this study, we investigated how Eater recognizes live, intact bacteria. We found that Eater can bind to Gram-positive bacteria irrespective of whether they are dead or alive. By contrast, recognition of live Gram-negative bacteria required surface disruption. Our results indicate that antimicrobial peptides (AMPs)3 may play a role in “preparing” Gram-negative bacteria for phagocytosis in vivo. We propose that this may be a previously unrecognized mechanism by which AMPs cooperate with germline-encoded phagocytic receptors to expand the reach of the latter.
EXPERIMENTAL PROCEDURES
Expression and Purification of Eater-Fc
A fragment encoding the endogenous signal sequence of Eater to the end of the fourth EGF-like repeat (amino acids 1–199) was amplified from plasmid pMT/V5His-Eater-1–199 (15) with High Fidelity PCR Master (Roche Applied Science) using primers 5′-ATAGCTCGGTCCGATGTGGATTTGTAGGATAAC-3′ and 5′-GCTTACCTTCGAAGGGCCCTCTAGA-3′. It was cloned into pCR2.1-TOPO (Invitrogen), excised with RsrII and XhoI, and cloned into the baculovirus expression vector pFASTBACtevFc (27) to generate an in-frame fusion with a tobacco etch virus protease and thrombin cleavage site followed by a C-terminal Fc tag (human IgG1). The correct sequence of the entire insert was confirmed on both strands.
Recombinant bacmids were generated by transformation into E. coli DH10Bac (Invitrogen). For production of secreted Eater-Fc protein, Spodoptera frugiperda 9 (Sf9) cells (Invitrogen) were grown at 27 °C in serum-free HyQ-CCM3 medium (HyClone, Thermo Scientific). High-titer bacmid stock was used to infect 7 liters of Sf9 cells (2 × 106/ml) and incubated at 27 °C for 42 h. Cell culture supernatant was harvested by centrifugation at 5,000 × g for 30 min, filtered (0.22-μm low protein binding filter), and loaded onto a 5-ml HiTrap protein A column on an Äkta FPLC (GE Healthcare). Bound protein was washed with 5 column volumes of 20 mm Hepes, 100 mm NaCl, pH 7.0, eluted using the Gentle Ag/Ab elution buffer at pH 6.6 (Pierce, Thermo Scientific), buffer-exchanged into 20 mm Hepes, 150 mm NaCl, pH 7.0 (Zeba desalt spin column; Pierce, Thermo Scientific), and concentrated to 2 mg/ml with Amicon filter devices (Millipore). To assess purity and size, purified protein was analyzed by Laemmli SDS-PAGE under non-reducing or reducing conditions in the absence or presence of 710 mm β-mercaptoethanol followed by Coomassie Blue staining (GelCode Blue; Pierce, Thermo Scientific). For cleavage of Eater-Fc, the thrombin CleanCleave kit (Sigma) was used according to the manufacturer's instructions.
Bacterial Strains
Live E. coli DH10B/TOP10 was purchased from Invitrogen. E. coli DH5α GFP, Pseudomonas aeruginosa PA14, and S. aureus ALC1435 GFP were gifts of Fred Ausubel and Candida albicans were from Ian Fraser, all at Massachusetts General Hospital, Boston, MA. S. marcescens Db11-GFP and LPS mutant 20C2 (12), Enterococcus faecalis, and Micrococcus luteus CIPA270 were provided by Dominique Ferrandon, Institut de Biologie Moléculaire et Cellulaire du CNRS, Strasbourg, France. Surface protein A-negative S. aureus Wood 46 (ATCC10832) was from ATCC. Bacteria were grown in LB broth Lennox (United States Biological) or brain heart infusion medium (BD Biosciences) (E. faecalis) and inactivated by heat (60 min at 70 °C or 30 min at 95 °C (PA14)), Carnoy's fixative (75% EtOH, 25% glacial acetic acid for 10 min on ice), or formaldehyde (3% for 20 min at room temperature) or used alive. All bacteria were washed in PBS (10 mm sodium phosphate dibasic, 156 mm sodium chloride, 2 mm potassium phosphate monobasic, pH 7.4) before use.
Eater-Fc Binding to Bacteria
Eater-Fc fusion protein or control human IgG1 or IgG Fc fragment (Athens Research and Technology) was biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (Pierce, Thermo Scientific). For flow cytometry, 2 × 106 bacteria in Robb's Drosophila PBS (28) supplemented with 0.5% BSA and 0.01% sodium azide were incubated with biotinylated proteins for 30 min at room temperature, sedimented at 9,000 × g for 5 min, and washed. For detection of bound biotinylated protein, bacteria were resuspended in the presence of 1 μg/ml streptavidin Alexa Fluor 488 conjugate (Invitrogen) and incubated for 20 min before analysis on a FACSCalibur (BD Biosciences). The bacterial population was gated by forward and side scatter, and 10,000 events were recorded. For assessment of bacterial viability by propidium iodide (PI) exclusion, 50 μg/ml PI was added on ice immediately before analysis. Fluorescence emissions were detected in the FL-1 channel (Alexa Fluor 488 emission: 519 nm) and, where indicated, in the FL-3 channel (PI emission: 620 nm).
Cecropin A Exposure of Bacteria
Chemically synthesized cecropin A from the moth Hyalophora cecropia (KWKLFKKIEKVGQNIRDGIIKAGPAVAVVGQATQIAK) was purchased from Sigma. Cationic control peptide 2K1 with the sequence (GK)6AS(GK)6 (29) was synthesized by standard solid phase peptide synthesis. Both peptides were dissolved in PBS at 100 μm and stored frozen in aliquots at −80 °C. Bacteria were grown to mid log phase in LB broth Lennox at 37 °C, centrifuged (3,500 × g, 4 °C), resuspended in PBS, counted, and adjusted to 108/ml. 50 μl of bacteria was added to 50 μl of PBS containing the indicated concentrations of cecropin A and incubated at 25 °C for the indicated times, placed on ice, and analyzed immediately by flow cytometry in the presence of 50 μg/ml PI. For assessment of Eater-Fc binding to cecropin A-exposed bacteria, subsequent bacterial Eater-Fc binding assays were carried out in PBS at 4 °C, a temperature non-permissive for AMP activity. TFA (used as a counterion to maintain the charge balance in solid phase peptide synthesis) (30) showed no effect on bacterial viability at 9-fold molar excess over peptide (one counterion per positive charge) and up to 10 mm. For control CFU counts, samples were split, 50% was plated on LB Lennox agar, and 50% was processed for flow cytometry. Colonies were counted the next day.
S2 Cell Binding to Bacteria and RNA Interference (RNAi)
Flow cytometry-based bacterial S2 cell binding assays and RNAi by soaking were performed, and data were analyzed and presented as described (15, 31). In short, double-stranded (ds) RNA directed against Eater or pBR322 (control) was synthesized from a PCR product using T7 MegaScript RNA polymerase (Ambion). 5 × 105 S2 cells were incubated first with 7.5 μg of dsRNA for ∼60 h and then with bacteria in Schneider's Drosophila medium (Invitrogen) without serum at 4 °C. To facilitate direct comparison, GFP expressing bacteria were used in all cases; because heating destroyed GFP, heat-inactivated bacteria were labeled with FITC (isomer I; Invitrogen).
Anti-Eater Antibodies, Western Blots, and S2 Cell Surface Staining
A polyclonal antibody against Eater-Fc fusion protein was generated in rabbits by a commercial supplier (Pocono Rabbit Farm and Laboratory Inc.). The protein A-purified IgG fraction was used at 1 μg/ml. For Western blots and cell surface staining, S2 cells were treated with dsRNA as described above, harvested, and washed in PBS. For Western blots, 2.5 × 106 S2 cells were lysed for 15 min at room temperature with CelLytic M (Sigma) supplemented with protease inhibitor mixture (Complete Mini; Roche Applied Science). Lysates were centrifuged (21,000 × g for 15 min at 4 °C), and the supernatants were transferred to ice. An equivalent of 3 × 105 cells was mixed with Laemmli buffer, incubated at 95 °C for 5 min, and separated by SDS-PAGE (6% reducing gel) followed by immunoblot analysis using anti-Eater-Fc antibodies and goat-anti-rabbit IgG conjugated to horseradish peroxidase. For cell surface staining, 2.5 × 105 cells were stained with anti-Eater-Fc in PBS supplemented with 0.5% BSA and 0.01% sodium azide for 20 min on ice followed by 1 μg/ml goat-anti-rabbit-IgG Alexa Fluor 488 conjugate (Invitrogen). For flow cytometry, S2 cells were gated by forward and side scatter, and 5,000 events were recorded. For immunofluorescence microscopy, S2 cells were fixed with formaldehyde (3% for 20 min at room temperature).
Peptidoglycan (PGN) Co-sedimentation
A suspension of polymeric, insoluble PGN from E. coli (InvivoGen), Bacillus subtilis, S. aureus, and M. luteus (all from Sigma) was made in PBS (5 mg/ml), aliquoted, and stored at −20 °C. 60 μg of insoluble PGN was mixed with 1 μg of thrombin-cleaved Eater-Fc in 50 μl of PBS supplemented with protease inhibitor mixture (Complete Mini; Roche Applied Science) and 0.5% BSA. At 0 and 15 min, 10 μl was removed from the mixture. After incubation for 15 min at 4 °C, the remaining 30 μl of mixture was centrifuged at 4 °C at 16,000 × g for 15 min (B. subtilis, S. aureus, M. luteus PGN) or 279,000 × g for 1 h (E. coli PGN). The supernatant was removed, and the pellet was washed two times with 200 μl of supplemented PBS and resuspended in 30 μl. All samples were mixed with Laemmli sample buffer immediately after preparation and incubated at 95 °C for 5 min. Equal amounts of samples (corresponding to 10 μl of starting sample) were analyzed by reducing SDS-PAGE and immunoblotting using anti-Eater-Fc antibodies followed by goat-anti-rabbit IgG conjugated to horseradish peroxidase. As control, 60 μg of insoluble PGN was cleaved with 6 μg (55 units) of mutanolysin from Streptomyces globisporus (Sigma) at 37 °C for 16 h before the addition of 1 μg of thrombin-cleaved Eater-Fc.
RESULTS
Expression and Purification of Eater-Fc
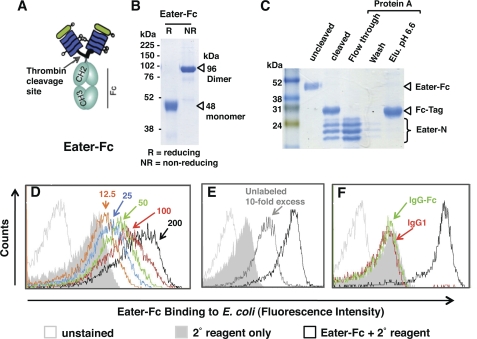
To generate a soluble, secreted Eater receptor variant, we fused the putative N-terminal ligand binding domain (15) to a C-terminal Fc affinity tag using baculovirus expression vector pFASTBACtevFc (27) (Fig. 1A). Eater-Fc could be purified from the supernatants of transfected Sf9 cells with a yield of 2 mg/liter. In agreement with its calculated molecular mass, the purified fusion protein migrated as a single band of about 48 kDa in reducing SDS-PAGE (Fig. 1B). Under non-reducing conditions, it displayed a band size of ∼96 kDa, indicating disulfide bond-mediated dimerization via the C-terminal Fc tag (Fig. 1B). Removal of the Fc tag after thrombin cleavage yielded Eater fragments that migrated in multiple bands corresponding to the calculated molecular mass of 21 kDa and slightly larger (Fig. 1C), consistent with N-glycosylation of the Eater N terminus (15) (supplemental Fig. S1).
FIGURE 1.
Baculovirus-expressed Eater-Fc fusion protein recognizes bacteria. A, schematic depiction of recombinant, secreted Eater-Fc protein. The putative ligand binding domain of Eater (amino acids 1–199 corresponding to two complete tandem EGF-like repeats) was fused to an Fc affinity tag. B and C, Coomassie Blue-stained SDS gels (12%). B, Eater-Fc fusion protein migrates as dimer under non-reducing conditions (NR). Expected molecular mass is indicated by open triangles. R, reducing conditions. C, thrombin-cleaved Eater-Fc and separation of cleavage products by protein A affinity chromatography. Cleaved Eater N-terminal fragment migrated in multiple bands, the lower of which corresponded to the predicted molecular mass (21 kDa). Elu. pH 6.6, elution buffer at pH 6.6. D–F. flow cytometry analysis of direct binding of biotinylated Eater-Fc fusion protein (200 μm; black open curve) to heat-inactivated E. coli. D, concentration-dependent binding (red, 100 μm; green, 50 μm; blue, 25 μm; orange, 12.5 μm). E, inhibition of binding by a 10-fold excess of non-biotinylated Eater-Fc (2 mm; dark gray curve). F, control. No significant binding activity was detected with biotinylated human IgG1 (200 μm; red line) or IgG-Fc (200 μm; green line). All experiments were repeated at least once with similar results.
To assess the biological activity of Eater-Fc, we carried out direct binding assays with heat-inactivated E. coli using flow cytometry (15). Eater-Fc binding was concentration-dependent (Fig. 1D) and could be competed with unlabeled Eater-Fc, precluding that the properties of labeled Eater-Fc had been altered during biotinylation (Fig. 1E). Control IgG1 and IgG-Fc showed no binding to E. coli (Fig. 1F), indicating that the observed binding was not due to the Fc tag. Deglycosylation of Eater-Fc led to a loss of its binding activity (supplemental Fig. S1). Eater-Fc protein was partially refractory to heat denaturation for 5 min at 100 °C, possibly due to a stability-enhancing effect by the Fc tag.
Eater-Fc Binding to Distinct Classes of Non-viable Microbes
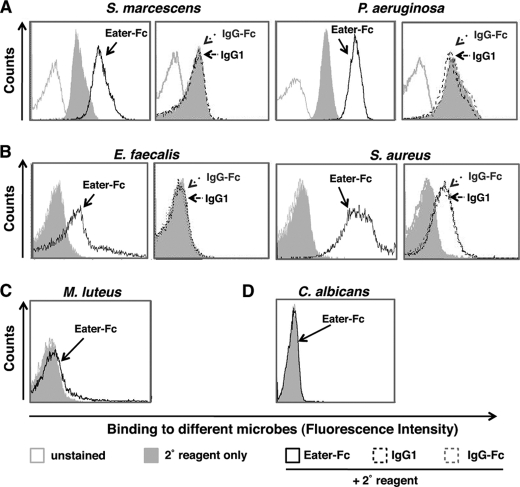
Consistent with the previous characterization of Eater as a scavenger receptor with multiligand specificity (15), Eater-Fc protein displayed broad binding activities toward different classes of heat- or ethanol-inactivated, non-viable bacteria such as S. marcescens and P. aeruginosa (Fig. 2A, supplemental Fig. S2, and data not shown), as well as Gram-positive bacteria of the phylum Firmicutes (E. faecalis, S. aureus) (Fig. 2B). Control IgG1 or IgG-Fc showed no significant binding activity (Fig. 2, A and B). The unrelated Gram-positive bacterium M. luteus (phylum Actinobacteria) and the fungal pathogen C. albicans were not recognized by Eater-Fc (Fig. 2, C and D). This result indicates that microbial binding by Eater-Fc is broad, yet to some extent specific, as would be expected for a pattern recognition receptor (5, 6).
FIGURE 2.
Eater-Fc binds to broad yet distinct classes of heat- or ethanol-inactivated bacteria. Shown is flow cytometry analysis of binding by 200 μm biotinylated Eater-Fc fusion protein (open black curve) or control biotinylated IgG1 and IgG-Fc (broken black or broken gray curves, respectively) when compared with secondary reagent only (gray filled curve) and unstained microbes (open gray curve). A, binding by Eater-Fc to heat-inactivated S. marcescens or ethanol-inactivated P. aeruginosa (Gram-negative Proteobacteria). B, binding by Eater-Fc to heat-inactivated E. faecalis and S. aureus (Gram-positive Firmicutes). C and D, no binding by Eater-Fc to heat-inactivated M. luteus (phylum Actinobacteria) (C) or heat-inactivated C. albicans yeast (D). Experiments were always run in parallel with positive and negative controls and repeated at least once with similar results.
Taken together, our data show that Eater-Fc protein was biologically active and that its binding behavior recapitulated known binding properties of the native Eater receptor (15). It also behaved similarly to a previously described N-terminally truncated ectodomain of Eater expressed in Drosophila S2 cells (Eater1–199His) (15). However, because the latter could be purified only in small amounts (tens of μg), baculovirus-expressed Eater-Fc represents a significant advance and provides a useful tool to study the interaction of Eater with microbes.
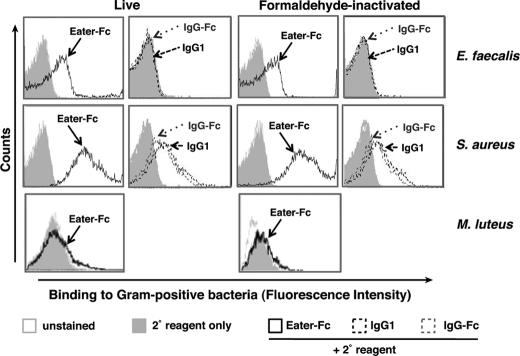
Eater Binding to Live, Naive Gram-positive Firmicutes
To address a longstanding question, namely whether Eater can bind to the surface of live, naive bacteria, we used Eater-Fc and our flow cytometry-based binding assay in conjunction with live bacteria. Heat-, ethanol- or formaldehyde-inactivated bacteria served as controls. Fig. 3 shows that Eater-Fc bound well to live or formaldehyde-inactivated E. faecalis and S. aureus (Fig. 3, first and second rows). Control IgG1 and IgG-Fc did not show significant binding, although elevated background staining was observed with S. aureus. In contrast to S. aureus and E. faecalis, M. luteus was not recognized by Eater-Fc in any condition (Fig. 3, third row).
FIGURE 3.
Eater-Fc binds to live Gram-positive Firmicutes. Shown is flow cytometry analysis of binding by 200 μm biotinylated Eater-Fc fusion protein (open black curve) or control biotinylated IgG1 and IgG-Fc (broken black or broken gray curves, respectively) when compared with secondary reagent only (gray filled curve) or unstained microbes (open gray curve). Upper two rows, Eater-Fc bound to live, as well as to formaldehyde-inactivated, E. faecalis and S. aureus (Phylum Firmicutes). Third row, Eater-Fc did not bind to M. luteus in any condition (Phylum Actinobacteria). These experiments were repeated two times with similar results.
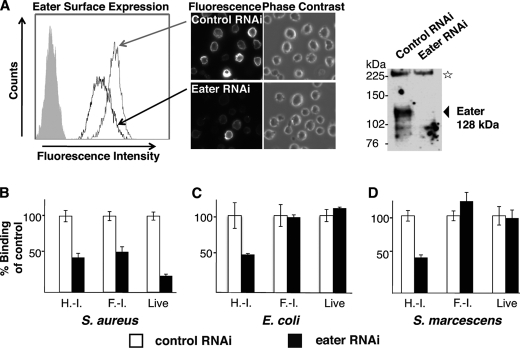
This result was confirmed by using Drosophila S2 cells that express native Eater receptor (15). We first established that native Eater is expressed on the cell surface; S2 cells could be surface-labeled with antibodies directed against the Eater ectodomain. RNAi-mediated knockdown of eater decreased this labeling (Fig. 4A). We then used RNAi-mediated knockdown of eater to address S. aureus binding to S2 cells. eater RNAi lead to a significant decrease of S. aureus binding by S2 cells irrespective of whether the bacteria were heat- or formaldehyde-inactivated or alive (Fig. 4B). The remaining S2 cell binding activity to S. aureus is likely due to a combination of incomplete eater knockdown and binding by other phagocytosis receptors (15). Our results confirmed that Eater is a major phagocytosis receptor for S. aureus on S2 cells (15).
FIGURE 4.
Membrane-bound, native Eater receptor behaves similarly to Eater-Fc. A, specific cell surface staining of Eater on S2 cells. Polyclonal rabbit anti-Eater-Fc antibodies reacted with S2 cells by cell surface staining of live S2 cells (left panel, histogram of flow cytometry analysis; filled gray curve, secondary reagent only) or formaldehyde-fixed (non-permeabilized) S2 cells (central panels, microscopy, 40× magnification). Cell surface staining strongly diminished after Eater RNAi knockdown. In a Western blot of S2 cell lysate, anti-Eater-Fc antibodies recognized a specific band consistent with the predicted molecular mass of Eater (128 kDa; arrow) that disappeared after Eater RNAi knockdown (upper right panel). The star marks a nonspecific band serving as loading control. B–D, binding of bacteria to S2 cells, normalized to dsRNA-treated controls. B, binding of S2 cells to heat- or formaldehyde-inactivated and live S. aureus is partially Eater-dependent because the signal decreased after RNAi knockdown of Eater. C and D, binding of S2 cells to heat-inactivated Proteobacteria (E. coli, S. marcescens) was partially Eater-dependent (signal decrease after Eater-specific RNAi), whereas binding to formaldehyde-inactivated or live Proteobacteria was not Eater-dependent (no change in signal after Eater-specific RNAi). H.-I., heat-inactivated; F.-I., formaldehyde-inactivated. All experiments were repeated at least once with similar results.
The finding that Eater is able to bind to live S. aureus and E. faecalis provides a simple, straightforward explanation for the in vivo protective role of Eater against S. aureus and E. faecalis infections (11, 13, 16). It suggests that Eater may directly target naive Gram-positive Firmicutes in the host, leading to their phagocytic clearance and destruction and to the effective control of bacterial loads.
Absence of Eater Binding to Live, Naive Gram-negative Bacteria
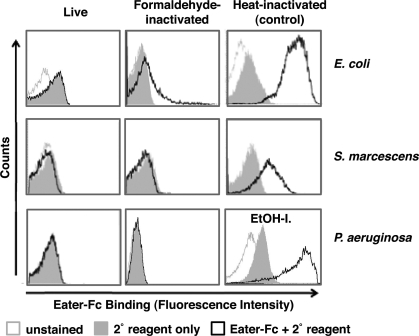
Eater also plays a critical role in the host defense to invasive Gram-negative pathogens such as S. marcescens (15) and P. aeruginosa.2 Surprisingly, we could not detect any Eater-Fc binding to naive or formaldehyde-inactivated E. coli, S. marcescens, or P. aeruginosa (Fig. 5, left and middle panels). By contrast, heat- or ethanol-inactivated Gram-negative bacteria bound well to Eater-Fc (Fig. 5, right panels, supplemental Fig. S2, and data not shown). Rough LPS mutant S. marcescens behaved indistinguishably from wild type (supplemental Fig. S3). Consistent results were obtained for membrane-bound Eater on the surface of S2 cells. As shown previously (15), binding of heat- or ethanol-inactivated Gram-negative bacteria was to a large extent Eater-dependent (Fig. 4, C and D). In contrast to this, binding of live and formaldehyde-inactivated Gram-negative bacteria to S2 cells was not dependent on native Eater (Fig. 4, C and D).
FIGURE 5.
Eater-Fc does not bind to live and formaldehyde-inactivated Gram-negative bacteria. Shown is flow cytometry analysis of binding by 200 μm biotinylated Eater-Fc fusion protein (open black curve) when compared with secondary reagent only (gray filled curve) or unstained microbes (open gray curve). Eater-Fc did not bind to live or formaldehyde-inactivated Proteobacteria (left and middle panels). Right panels, control binding to heat- or ethanol-inactivated Proteobacteria. EtOH-I., ethanol-inactivated. These experiments were repeated two times with similar results.
Taken together, our data suggest that Eater ligands are buried beneath the surface of live and formaldehyde-fixed Gram-negative bacteria or in other ways masked. Membrane-disrupting treatments such as heat or ethanol inactivation lead to unmasking of normally inaccessible ligands. These results raise the intriguing question of how Eater ligands may become accessible in vivo in the host during an infection.
Eater Binding after Exposure of Gram-negative Bacteria to a Cationic AMP
Efficient phagocytosis, which is critically important in the Drosophila host defense against invasive Gram-negative bacteria (14, 15), happens in the biological context of local and systemic AMP responses (12).2 Because AMPs are well known to destabilize bacterial membranes (32–34), we were interested to determine whether AMPs might be able to unmask Eater ligands.
We first killed E. coli with cecropin A, a prototypic membrane-perturbing cationic peptide conserved from invertebrates to humans (33, 34). Bacterial killing by cecropin A was concentration-dependent and rapid, whereas the cationic control peptide 2K1 (29) had no effect (supplemental Fig. S4). We then used cecropin A-killed E. coli to measure Eater binding by two-color flow cytometry analysis to simultaneously monitor Eater binding and bacterial viability. Fig. 6 shows that Eater-Fc bound well to cecropin A-killed bacteria, whereas control IgG-Fc did not bind. Moreover, the control peptide 2K1 did not increase Eater-Fc binding (Fig. 6). These results suggest that cationic AMPs might play a role in vivo in unmasking Eater ligands on the surface of Gram-negative bacteria. In agreement with this idea, we observed Eater-Fc labeling in EM close to the cell walls of E. coli that had been exposed to cationic AMPs (supplemental Fig. S5).
FIGURE 6.
Eater-Fc binds to E. coli killed by exposure to a cationic AMP. Upper panels, histograms of flow cytometry analysis of binding of 200 μm Eater-Fc (open black curve) or IgG-Fc (green curve) to E. coli when compared with secondary reagent only (gray filled curve) or unstained microbes (open gray curve). Eater-Fc bound to E. coli killed by exposure to 4 μm cecropin A for 10 min at 25 °C but not to E. coli treated with 4 μm control cationic peptide 2K1. Eater-Fc binding to control heat-inactivated E. coli is shown for comparison. Lower panels, dot plots of two-color analysis of the same samples as in the upper row, allowing assessment of Eater binding simultaneously with bacterial viability. The percentage of Eater-Fc binding live or dead bacteria, respectively, is indicated. Experiments were repeated three times with similar results.
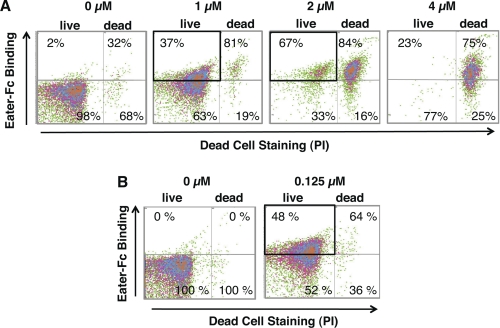
We wondered whether incubating E. coli with cationic AMP under sublethal conditions would be sufficient to promote Eater binding. We used flow cytometry to quantify Eater-Fc binding to live E. coli that had been exposed to increasing amounts of cecropin A. Indeed, Fig. 7A shows that a population of live E. coli became accessible to Eater-Fc after exposure to increasing concentrations of cecropin A (1–4 μm). Because bacterial killing by cecropin A under these conditions was very rapid (less than 10 min), we confirmed this result by performing a prolonged incubation course at a lower concentration of cecropin A (0.125 μm for 2 h; Fig. 7B and supplemental Fig. S4B). CFU counts of the AMP-treated sample were indistinguishable from control and independently confirmed the viability of the bacteria (8 × 104 CFU in 25 μl; one experiment).
FIGURE 7.
Eater-Fc binds to live, AMP-exposed E. coli. A and B, dot plots of two-color flow cytometry analysis of live E. coli exposed at 25 °C to increasing concentrations of cecropin A (0–4 μm) for 10 min (A) or to 0.125 μm cecropin A for 2 h (B). The percentage of Eater-Fc binding of live or dead bacteria, respectively, is indicated. In A and B, bold rectangles highlight Eater-Fc binding to bacteria that were exposed to cecropin A at sublethal concentrations. The boxed bacteria were alive because they excluded PI; this was confirmed by bacterial CFU counts in one repeat experiment. Experiments were repeated twice with similar results.
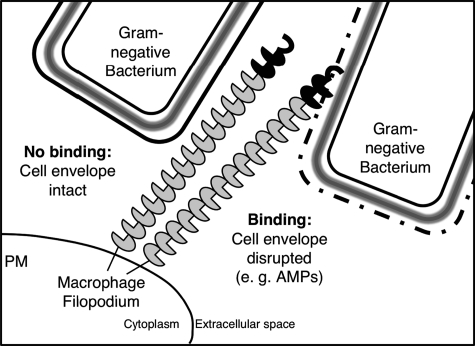
Taken together, our results reveal a novel, previously unrecognized role for AMPs. They indicate that cationic AMPs may be able to alter the surface of Gram-negative bacteria in a way that leads to unmasking of previously inaccessible phagocytic receptor ligands. We propose a model by which AMPs might contribute to the clearance of live Gram-negative bacteria in vivo by perturbing the bacterial surface and making previously inaccessible ligands available for non-opsonic phagocytosis (Fig. 8).
FIGURE 8.
Proposed model for non-opsonic phagocytosis of Gram-negative bacteria by the pattern recognition receptor Eater. Left, Eater ligands on the cell walls of naive (live) Gram-negative bacteria are masked. Right, cationic AMPs destabilize the bacterial outer membrane, disrupt the cell envelope, and lead to exposure of Eater ligands, which renders the bacteria accessible for Eater binding and non-opsonic phagocytosis. Note that bacteria are not drawn to scale here and are larger in reality. PM, plasma membrane.
Differential Binding to PGN by Eater N-terminal Fragment
A nearly ubiquitous cell wall component of bacteria is the murein sacculus, made of PGN, a heteropolymer composed of long glycan chains cross-linked by short peptides (35). It forms a mesh-like exoskeleton outside the plasma membrane of bacteria. On the surface of live Gram-positive bacteria, PGN seems to be at least partially accessible (36). However, in live Gram-negative bacteria, PGN is much less abundant and hidden because it is embedded in the cell envelope under an outer membrane containing LPS (37, 38).
We tested whether polymeric PGN could be an Eater ligand that becomes accessible in E. coli after membrane perturbation. We cleaved Eater-Fc with thrombin to separate the Eater N-terminal fragment from the Fc tag and incubated the mixture with PGN. As shown in Fig. 9, the Eater N-terminal fragment displayed differential binding to different types of polymeric PGN. The peptide stems and cross-linking bridges between the glycan strands are a major source of variation in PGNs (37–39). Bacilli and Gram-negative bacteria synthesize meso-diaminopimelic acid-type PGN (with identical stem peptides and cross-links), whereas S. aureus and M. luteus contain lysine–type PGNs with different peptide bridges (40). Co-sedimentation assays revealed that Eater N-terminal fragment bound to E. coli, B. subtilis, and S. aureus PGN, but not to M. luteus PGN, and was greatly diminished upon cleavage of PGNs by mutanolysin (Fig. 9). The Fc tag displayed partial binding to S. aureus PGN (possibly due to contamination with surface protein A) but no significant binding to the other PGNs. The binding profile of Eater N-terminal fragment toward PGN correlated well with Eater-Fc binding to the corresponding classes of heat- or ethanol-inactivated bacteria (Fig. 2, A–C) and suggested that PGN might be a ligand of Eater.
FIGURE 9.
Eater N terminus (Eater-N) displays differential binding to different types of PGN. A PGN co-sedimentation assay is shown. Thrombin-cleaved Eater-Fc was incubated with different types of insoluble, polymeric PGNs, sedimented by centrifugation, and analyzed by SDS-PAGE followed by immunoblot analysis using anti-Eater-Fc antibodies. T indicates total protein, S indicates supernatant (unbound), and P indicates pelleted (bound) protein fractions. Bands corresponding to intact Eater-Fc or thrombin-cleaved Fc tag and N-terminal fragments are indicated. Upper panel, Eater N terminus bound to E. coli, B. subtilis, and S. aureus PGN, but much less to M. luteus PGN. Results are representative of three independent experiments. Lower panel, control. Eater N terminus could not be detected in the pellet fractions after cleavage of PGNs with mutanolysin, a muramidase that specifically cleaves the glycan backbone of polymeric PGN. DAP-type PGN, diaminopimelic acid-type PGN.
DISCUSSION
We previously proposed that Eater functions as a cell surface-bound pattern recognition receptor in the initial steps of phagocytosis (15). The results obtained in this study support this view. We have shown that Eater is expressed on the surface of phagocytic Drosophila cells and confirmed that it binds directly to dead bacterial particles via its N-terminal domain (40 amino acids followed by four EGF-like repeats). Eater binding covered a broad range of killed bacteria including Gram-negative Proteobacteria as well as Gram-positive Firmicutes but did not extend to the Gram-positive actinobacterium M. luteus and the fungal pathogen C. albicans. These results are also consistent with the recent finding that Eater mediates phagocytosis of E. faecalis and S. aureus, but not M. luteus, by fly hemocytes and S2 cells (16).
The use of live, intact bacteria in this study revealed that recognition of naive bacterial surfaces by Eater was more complex than anticipated on the basis of tests with dead bacterial particles. Live Gram-positive Firmicutes were recognized well by Eater, whereas live Gram-negative Proteobacteria were not, although Eater plays a protective role in both types of infections (11, 13, 15, 16).2 Our results suggest that the outer membrane of Gram-negative bacteria needs to be disrupted for Eater to bind.
The bacterial cell envelope is a highly dynamic organelle that undergoes extensive changes in vivo in response to its host environment (12, 37, 41–44). Therefore, and because of the natural ionic composition of tissue fluids, the exact conditions by which innate immune molecules interact with their targets are hard to reproduce in the laboratory (33, 45). Even so, we were able to demonstrate that pretreatment of live E. coli with the cationic antimicrobial peptide cecropin A was a way to unmask and expose hidden Eater ligands (Figs. 6 and 7). It is unlikely that cecropin A acts as an opsonin that bridges the bacterial surface and Eater because a cationic control peptide that is expected to bind to the bacterial surface via its positive charges (29, 33) did not have any effect (Fig. 6).
Atomic force microscopy has emerged as a powerful tool for direct, non-invasive imaging of the living bacterial surface (46). A recent study measured cationic AMP activity on individual, live, naive E. coli cells (29). Surface corrugation caused by AMP activity correlated with killing kinetics in a two-stage process exhibiting a long lag phase followed by a short “execution” phase. These findings are compatible with a previously unrecognized role for cationic AMPs in non-opsonic phagocytosis: making inaccessible ligands available for phagocytic receptors.
We propose a model (Fig. 8) by which AMP activity under sublethal conditions (conceivably often encountered in vivo, for example in non-inflamed tissues (33, 45)) may promote exposure of previously hidden Eater ligands on the bacterial surface, leading to more efficient clearance and destruction of invasive bacteria. This scenario is supported by an oral-intestinal infection model in which local overexpression of the AMP diptericin in Drosophila midgut epithelium conferred increased protection to invasive S. marcescens (12). One interpretation of these data is that local AMP responses contribute to increased host resistance by preparing bacteria for subsequent Eater-mediated phagocytosis when bacteria manage to cross the gut epithelium.
It remains unclear at present what the mechanistic basis for the opening up of the Gram-negative cell wall by cationic AMPs may be (47, 48). For P. aeruginosa, it was shown that cationic AMPs can displace divalent cations from non-covalent LPS cross-bridges, leading to destabilization and permeabilization of the outer membrane and allowing access of hydrophobic probes or lysozyme (32, 47, 49). This modification of the bacterial surface manifests in membrane blebs observable by electron microscopy (49). The periplasm (the space between the outer and inner membranes of E. coli) is a potentially harmful and highly regulated environment akin to the lysosomes of eukaryotic cells (37, 50). One might speculate that the destructive power of bacterial cell wall remodeling enzymes or lipases could be unleashed upon disruption of outer membrane homeostasis, somehow leading to exposure of normally hidden PGN or PGN-bound molecules (41). It is noteworthy that outer membrane modifications induced by cationic AMP did indeed enhance the non-opsonic phagocytosis of P. aeruginosa by mammalian macrophages (49), as would be predicted by a scenario such as that proposed in the model in Fig. 8.
Our finding that Eater shows binding avidity to polymeric PGNs is consistent with our earlier characterization of Eater as displaying a binding preference for polyanionic ligands (15), reminiscent of scavenger receptors (51) and LPS-binding protein (LBP) (52). It seems that, similar to Eater, several mammalian pattern recognition molecules can bind cell wall components of Gram-negative and Gram-positive bacteria; CD14, Toll-like receptor 2 (TLR2), peptidoglycan recognition proteins (PGRPs), and LPS-binding protein can bind to LPS, lipoteichoic acid, and polymeric PGN, in some cases with overlapping binding sites (39, 52–54). A pattern of multiply iterated anionic charges was suggested to be the common denominator for all these ligands (52).
The much higher avidity of Eater to diaminopimelic acid-type and S. aureus PGN when compared with M. luteus PGN offers a tentative explanation for its inability to bind to the actinobacterium M. luteus and suggests that recognition may be mediated in part by the nature of the peptide stems and cross-links in PGN. However, PGN preparations are often contaminated with other cell wall molecules, some of which are covalently linked to PGN (37, 53, 55). It therefore remains possible that Eater binds to other microbial cell envelope molecules instead of, or in addition to, PGN. The molecular nature of these may be different for different classes of bacteria; moreover, a group of Eater molecules might use a combination of multiple targets. A similar concept has been proposed for the action of cationic AMPs, which have been likened to “dirty drugs” that are able to bind multiple, polyanionic target molecules with moderate affinities (41, 47, 48).
Several ligands have been identified for EGF-like repeat-containing molecules that are related to Eater: LPS for LPS recognition protein (27), lipoteichoic acid for Draper/CED-1 (20), β-glucan for SCARF1/CED-1 (24), and outer membrane protein OmpA for SCARF1 (56). Because Eater-Fc did not bind to naive Gram-negative bacteria (Fig. 5 and supplemental Fig. S3), it seems less likely that Eater recognizes LPS O-antigen or outer membrane proteins such as the mammalian scavenger receptors SR-A and SCARF1 and the phagosomal microbial sensor SLAM (56–60). More likely potential ligands are the strongly negatively charged teichoic acids, which are absent from the cell walls of M. luteus and mycobacterial pathogens (43, 61) but are highly abundant in the cell walls of S. aureus and E. faecalis (37, 43, 62). Recent atomic force microscopy measurements even suggest that teichoic acids may obscure the access to PGN on the surface of naive Gram-positive bacteria (63). We tried to test Eater-Fc binding to LPS and lipoteichoic acids by using flow cytometry-based bacterial binding inhibition assays (with commercially available cell wall components), but ultimately these experiments proved too variable and remained inconclusive.4 Further investigation and different approaches are clearly required to identify biologically relevant Eater ligands.
The results of this study may have some broader implications. They may point to a general mechanism by which AMPs could cooperate with phagocytic pattern recognition receptors and thereby enlarge the spectrum of microbes that can be recognized by a single germ line-encoded receptor. This may be important in vivo because the efficiency of non-opsonic phagocytosis, especially locally in uninflamed tissues such as lung, is an important determinant for prevention of infection through early clearance of bacteria (2, 64).
AMPs may not be unique in their ability to make previously hidden bacterial ligands accessible or may act synergistically with other defense molecules (33, 47, 65). For an innate immune system, the advantages of extending the microbial ligand repertoire are clear given “the need for thrifty use of a limited set of germ line-encoded receptors” (66). Our findings add a further dimension to this theme: compartmentalization and accessibility of microbial ligands, an emerging topic of increasing importance in the cell biology of innate immune processes in general (67).
Supplementary Material
Acknowledgments
We thank Bol-Luel Lee for the gift of a baculovirus expression plasmid, Janice Lee for synthesizing dsRNAs, Joseph Garlick, Ji-Joon Song, and Robert E. Kingston for help with baculovirus expression, Dominique Ferrandon, Frederick M. Ausubel, and Iain Fraser for microbial strains, David McLaughlin, Lizabeth A. Perkins, and the members of the Ausubel laboratory for support, advice, suggestions, or comments on the manuscript, in particular Brent Cezairliyan, Mark Rosenzweig, and Fred Ausubel. EM was performed by Mary McKee in the Microscopy Core of the Center for Systems Biology/Program in Membrane Biology, which is partially supported by a National Institutes of Health Center for the Study of Inflammatory Bowel Disease Grant DK43351 and a Boston Area Diabetes and Endocrinology Research Center Award DK57521.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 AI44220.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
S. Limmer, S. Haller, J. Lee, R. Feinbaum, C. Kocks, F. M. Ausubel, and D. Ferrandon, manuscript submitted for publication.
Y. S. A. Chung and C. Kocks, unpublished observations.
- AMP
- antimicrobial peptide
- PGN
- peptidoglycan
- PI
- propidium iodide.
REFERENCES
- 1. Metchnikoff I. I. (1908) Ilya Mechnikov-Nobel Lecture. nobelprize.org 28 November 2010. http://nobelprize.org/nobel_prizes/medicine/laureates/1908/mechnikov-lecture.html
- 2. Rabinovitch M. (1995) Trends Cell Biol. 5, 85–87 [DOI] [PubMed] [Google Scholar]
- 3. Stuart L. M., Ezekowitz R. A. (2005) Immunity 22, 539–550 [DOI] [PubMed] [Google Scholar]
- 4. Wright A. E., Douglas S. R., Sanderson J. B. (1989) Rev. Infect. Dis. 11, 827–834 [DOI] [PubMed] [Google Scholar]
- 5. Janeway C. A., Jr. (1989) Cold Spring Harbor Symp. Quant. Biol. 54, 1–13 [DOI] [PubMed] [Google Scholar]
- 6. Janeway C. A., Jr., Medzhitov R. (2002) Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 7. Stuart L. M., Ezekowitz R. A. (2008) Nat. Rev. Immunol. 8, 131–141 [DOI] [PubMed] [Google Scholar]
- 8. Meister M., Lagueux M. (2003) Cell Microbiol. 5, 573–580 [DOI] [PubMed] [Google Scholar]
- 9. Rizki T. M., Rizki R. M. (1984) in Insect Ultrastructure (King R. C., Akai H. eds) pp 579–604, Plenum Press, New York [Google Scholar]
- 10. Lanot R., Zachary D., Holder F., Meister M. (2001) Dev. Biol. 230, 243–257 [DOI] [PubMed] [Google Scholar]
- 11. Defaye A., Evans I., Crozatier M., Wood W., Lemaitre B., Leulier F. (2009) J. Innate Immun. 1, 322–334 [DOI] [PubMed] [Google Scholar]
- 12. Nehme N. T., Liégeois S., Kele B., Giammarinaro P., Pradel E., Hoffmann J. A., Ewbank J. J., Ferrandon D. (2007) PLoS Pathog. 3, e173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charroux B., Royet J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9797–9802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Avet-Rochex A., Perrin J., Bergeret E., Fauvarque M. O. (2007) Genes Cells 12, 1193–1204 [DOI] [PubMed] [Google Scholar]
- 15. Kocks C., Cho J. H., Nehme N., Ulvila J., Pearson A. M., Meister M., Strom C., Conto S. L., Hetru C., Stuart L. M., Stehle T., Hoffmann J. A., Reichhart J. M., Ferrandon D., Rämet M., Ezekowitz R. A. (2005) Cell 123, 335–346 [DOI] [PubMed] [Google Scholar]
- 16. Nehme N. T., Quintin J., Cho J. H., Lee J., Lafarge M. C., Kocks C., Ferrandon D. (2011) PLoS One 6, e14743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sackton T. B., Lazzaro B. P., Schlenke T. A., Evans J. D., Hultmark D., Clark A. G. (2007) Nat. Genet. 39, 1461–1468 [DOI] [PubMed] [Google Scholar]
- 18. Tanaka H., Ishibashi J., Fujita K., Nakajima Y., Sagisaka A., Tomimoto K., Suzuki N., Yoshiyama M., Kaneko Y., Iwasaki T., Sunagawa T., Yamaji K., Asaoka A., Mita K., Yamakawa M. (2008) Insect Biochem. Mol. Biol. 38, 1087–1110 [DOI] [PubMed] [Google Scholar]
- 19. Kurucz E., Márkus R., Zsámboki J., Folkl-Medzihradszky K., Darula Z., Vilmos P., Udvardy A., Krausz I., Lukacsovich T., Gateff E., Zettervall C. J., Hultmark D., Andó I. (2007) Curr. Biol. 17, 649–654 [DOI] [PubMed] [Google Scholar]
- 20. Hashimoto Y., Tabuchi Y., Sakurai K., Kutsuna M., Kurokawa K., Awasaki T., Sekimizu K., Nakanishi Y., Shiratsuchi A. (2009) J. Immunol. 183, 7451–7460 [DOI] [PubMed] [Google Scholar]
- 21. Kurant E., Axelrod S., Leaman D., Gaul U. (2008) Cell 133, 498–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamon Y., Trompier D., Ma Z., Venegas V., Pophillat M., Mignotte V., Zhou Z., Chimini G. (2006) PLoS One 1, e120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plüddemann A., Neyen C., Gordon S. (2007) Methods 43, 207–217 [DOI] [PubMed] [Google Scholar]
- 24. Means T. K., Mylonakis E., Tampakakis E., Colvin R. A., Seung E., Puckett L., Tai M. F., Stewart C. R., Pukkila-Worley R., Hickman S. E., Moore K. J., Calderwood S. B., Hacohen N., Luster A. D., El Khoury J. (2009) J. Exp. Med. 206, 637–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cuttell L., Vaughan A., Silva E., Escaron C. J., Lavine M., Van Goethem E., Eid J. P., Quirin M., Franc N. C. (2008) Cell 135, 524–534 [DOI] [PubMed] [Google Scholar]
- 26. Somogyi K., Sipos B., Pénzes Z., Kurucz E., Zsámboki J., Hultmark D., Andó I. (2008) Mol. Biol. Evol. 25, 2337–2347 [DOI] [PubMed] [Google Scholar]
- 27. Ju J. S., Cho M. H., Brade L., Kim J. H., Park J. W., Ha N. C., Söderhäll I., Söderhäll K., Brade H., Lee B. L. (2006) J. Immunol. 177, 1838–1845 [DOI] [PubMed] [Google Scholar]
- 28. Robb J. A. (1969) J. Cell Biol. 41, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fantner G. E., Barbero R. J., Gray D. S., Belcher A. M. (2010) Nat. Nanotechnol. 5, 280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roux S., Zékri E., Rousseau B., Paternostre M., Cintrat J. C., Fay N. (2008) J. Pept Sci. 14, 354–359 [DOI] [PubMed] [Google Scholar]
- 31. Rämet M., Pearson A., Manfruelli P., Li X., Koziel H., Göbel V., Chung E., Krieger M., Ezekowitz R. A. (2001) Immunity 15, 1027–1038 [DOI] [PubMed] [Google Scholar]
- 32. Hancock R. E. (1984) Annu. Rev. Microbiol. 38, 237–264 [DOI] [PubMed] [Google Scholar]
- 33. Boman H. G., Hultmark D. (1987) Annu. Rev. Microbiol. 41, 103–126 [DOI] [PubMed] [Google Scholar]
- 34. Bulet P., Stöcklin R., Menin L. (2004) Immunol. Rev. 198, 169–184 [DOI] [PubMed] [Google Scholar]
- 35. Salton R. J. (1984) Molecular Immunology (Atassi M. Z., van Oss C. J., Absolom D. R. eds) pp. 91–116, Marcel Dekker, Inc., New York [Google Scholar]
- 36. Steen A., Buist G., Leenhouts K. J., El Khattabi M., Grijpstra F., Zomer A. L., Venema G., Kuipers O. P., Kok J. (2003) J. Biol. Chem. 278, 23874–23881 [DOI] [PubMed] [Google Scholar]
- 37. Silhavy T. J., Kahne D., Walker S. (2010) Cold Spring Harb. Perspect. Biol. 2, a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mengin-Lecreulx D., Lemaitre B. (2005) J. Endotoxin Res. 11, 105–111 [DOI] [PubMed] [Google Scholar]
- 39. Dziarski R., Gupta D. (2005) J. Endotoxin Res. 11, 304–310 [DOI] [PubMed] [Google Scholar]
- 40. Schleifer K. H., Kandler O. (1972) Bacteriol. Rev. 36, 407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peschel A., Sahl H. G. (2006) Nat. Rev. Microbiol. 4, 529–536 [DOI] [PubMed] [Google Scholar]
- 42. Li M., Lai Y., Villaruz A. E., Cha D. J., Sturdevant D. E., Otto M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9469–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weidenmaier C., Peschel A. (2008) Nat. Rev. Microbiol. 6, 276–287 [DOI] [PubMed] [Google Scholar]
- 44. West N. P., Sansonetti P., Mounier J., Exley R. M., Parsot C., Guadagnini S., Prévost M. C., Prochnicka-Chalufour A., Delepierre M., Tanguy M., Tang C. M. (2005) Science 307, 1313–1317 [DOI] [PubMed] [Google Scholar]
- 45. Finlay B. B., Hancock R. E. (2004) Nat. Rev. Microbiol. 2, 497–504 [DOI] [PubMed] [Google Scholar]
- 46. Dupres V., Alsteens D., Andre G., Dufrêne Y. F. (2010) Trends Microbiol. 18, 397–405 [DOI] [PubMed] [Google Scholar]
- 47. Hale J. D., Hancock R. E. (2007) Expert Rev. Anti. Infect. Ther. 5, 951–959 [DOI] [PubMed] [Google Scholar]
- 48. Hancock R. E., Scott M. G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8856–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sawyer J. G., Martin N. L., Hancock R. E. (1988) Infect. Immun. 56, 693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Duve C., Wattiaux R. (1966) Annu. Rev. Physiol. 28, 435–492 [DOI] [PubMed] [Google Scholar]
- 51. Greaves D. R., Gordon S. (2005) J. Lipid Res. 46, 11–20 [DOI] [PubMed] [Google Scholar]
- 52. Weber J. R., Freyer D., Alexander C., Schröder N. W., Reiss A., Küster C., Pfeil D., Tuomanen E. I., Schumann R. R. (2003) Immunity 19, 269–279 [DOI] [PubMed] [Google Scholar]
- 53. Dziarski R., Gupta D. (2005) Infect. Immun. 73, 5212–5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dziarski R., Tapping R. I., Tobias P. S. (1998) J. Biol. Chem. 273, 8680–8690 [DOI] [PubMed] [Google Scholar]
- 55. Rosenthal R. S., Dziarski R. (1994) Methods Enzymol. 235, 253–285 [DOI] [PubMed] [Google Scholar]
- 56. Jeannin P., Bottazzi B., Sironi M., Doni A., Rusnati M., Presta M., Maina V., Magistrelli G., Haeuw J. F., Hoeffel G., Thieblemont N., Corvaia N., Garlanda C., Delneste Y., Mantovani A. (2005) Immunity 22, 551–560 [DOI] [PubMed] [Google Scholar]
- 57. Areschoug T., Gordon S. (2009) Cell Microbiol. 11, 1160–1169 [DOI] [PubMed] [Google Scholar]
- 58. Berger S. B., Romero X., Ma C., Wang G., Faubion W. A., Liao G., Compeer E., Keszei M., Rameh L., Wang N., Boes M., Regueiro J. R., Reinecker H. C., Terhorst C. (2010) Nat. Immunol. 11, 920–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Peiser L., Makepeace K., Plüddemann A., Savino S., Wright J. C., Pizza M., Rappuoli R., Moxon E. R., Gordon S. (2006) Infect. Immun. 74, 5191–5199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Plüddemann A., Mukhopadhyay S., Sankala M., Savino S., Pizza M., Rappuoli R., Tryggvason K., Gordon S. (2009) J. Innate Immun. 1, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Powell D. A., Duckworth M., Baddiley J. (1975) Biochem. J. 151, 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neuhaus F. C., Baddiley J. (2003) Microbiol. Mol. Biol. Rev. 67, 686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Andre G., Leenhouts K., Hols P., Dufrêne Y. F. (2008) J. Bacteriol. 190, 7079–7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Speert D. P. (1993) Trends Microbiol. 1, 217–221 [DOI] [PubMed] [Google Scholar]
- 65. Ganz T. (2004) J. Leukoc Biol. 75, 34–38 [DOI] [PubMed] [Google Scholar]
- 66. Beutler B. (2003) Immunity 19, 155–156 [DOI] [PubMed] [Google Scholar]
- 67. Barton G. M., Kagan J. C. (2009) Nat. Rev. Immunol. 9, 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.