Abstract
Cell cycle dysregulation is a critical event in virus infection-associated tumorigenesis. Previous studies have suggested that hepatitis C virus NS5B modulates cell cycle progression in addition to participating in RNA synthesis as an RNA-dependent RNA polymerase. However, the molecular mechanisms have thus far remained unclear. In this study, a HepG2 Tet-On NS5B stable cell line was generated to confirm the effect of NS5B on the cell cycle. To better understand the role of NS5B in cell cycle regulation, yeast two-hybrid assays were performed using a human liver cDNA library. The cyclin-dependent kinase 2-interacting protein (CINP) was identified. The interaction between NS5B and CINP was further demonstrated by in vivo and in vitro assays, and their association was found to be indispensable for S phase delay and cell proliferation suppression. Further experiments indicated that NS5B relocalized CINP from the nucleus to the cytoplasm. Directly knocking down CINP by specific siRNA resulted in a significant alteration in the DNA damage response and expression of cell cycle checkpoint proteins, including an increase in p21 and a decrease in phosphorylated Retinoblastoma and Chk1. Similar results were observed in cells expressing NS5B, and the effects were partially reversed upon ectopic overexpression of CINP. These studies suggest that the DNA damage response might be exploited by NS5B to hinder cell cycle progression. Taken together, our data demonstrate that NS5B delays cells in S phase through interaction with CINP and relocalization of the protein from the nucleus to the cytoplasm. Such effects might contribute to hepatitis C virus persistence and pathogenesis.
Keywords: CDK (Cyclin-dependent Kinase), Cell Cycle, DNA Damage, Protein-Protein Interactions, Viral Protein
Introduction
At present, there are at least 170 million people infected with the hepatitis C virus (HCV)4 worldwide (1, 2), and the incidence increases yearly, especially in developing countries. HCV infection can easily develop into a chronic infection that progresses to liver cirrhosis and ultimately hepatocellular carcinoma (3, 4). Until now, there have been no effective treatments to prevent or cure HCV infections. Therefore, there is an urgent need to understand the mechanisms of HCV persistence and pathogenesis. HCV is a single-stranded, positive-sense RNA virus that belongs to the Flaviviridae family. Its 9.6-kb genome can directly function as mRNA, and the mRNA is translated into a large polyprotein precursor. The polyprotein is cleaved by both host and viral proteases to yield structural proteins (core, E1, and E2) and non-structural proteins (P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (5, 6).
As an RNA virus, HCV does not integrate into the host genome. However, there is increasing experimental evidence supporting the possibility that HCV promotes the malignant transformation of hepatocytes through more direct pathways. The interaction of HCV-encoded proteins with host factors results in a series of cellular activities, such as cell cycle progression, apoptosis, transcription, membrane rearrangement, and host immune responses. HCV core protein, NS3, NS4B, and NS5A have all been reported to possess in vitro and in vivo transformation potential (7–11). Cell cycle and cell proliferation controls are the major barriers against malignant transformation. Several HCV-encoded proteins have been shown to modulate normal cell cycle regulation (12–15). NS5B is an RNA-dependent RNA polymerase (16), and because of its essential role in HCV RNA replication, previous studies seeking new HCV therapies have focused on the RNA-dependent RNA polymerase activity of NS5B and attempted to identify inhibitors of this activity (17, 18). Notably, there have also been studies suggesting that NS5B is involved in cell cycle regulation (13, 19, 20), but there is still controversy about this phenomenon and the details of the mechanism.
The DNA damage response is usually utilized by viruses or virus-encoded proteins to modulate the cell cycle to promote their own replication and oncogenic effects. The DNA damage response comprises a network of signal transduction cascades, including the activation of a series of protein kinases that recognize and repair damaged DNA (21, 22). Viruses and viral proteins pose a direct threat to host genomic stability through their ability to induce DNA damage. After DNA damage, three cell cycle checkpoints are activated. 1) The G1/S checkpoint prevents cells from entering S phase. 2) The intra-S phase checkpoint inhibits DNA replication. 3) The G2/M checkpoint prevents damaged DNA from undergoing mitosis (23). The activation of these checkpoints allows time for DNA to repair itself. The major sensors and transducers responsible for the checkpoint analysis are ataxia telangiectasia, mutated (ATM) and ATM- and Rad3-related (ATR). ATM and ATR are conserved protein kinases that belong to the phosphoinositide 3-kinase (PI3K)-related protein kinase family (24). These proteins are activated after DNA damage, leading to the phosphorylation of two important downstream substrates, checkpoint kinase 1 (Chk1) and checkpoint kinase 2 (Chk2) (25–27). Phosphorylated Chk1 and Chk2 induce the proteasome-mediated degradation of Cdc25A/B/C (28, 29) and regulate their corresponding cyclin-CDK complexes during each cell cycle checkpoint.
In this study, we confirmed that NS5B induced cell cycle delay in S phase, and for the first time, we identified a novel human protein, CINP, that interacted with NS5B and was involved in the cell cycle dysfunction caused by NS5B. Mechanistic studies revealed that the relocalization of CINP from the nucleus to the cytoplasm in the presence of NS5B might be responsible for S phase delay by invoking the DNA damage response. These results provide new insight into the mechanisms of HCV persistence and pathogenesis.
EXPERIMENTAL PROCEDURES
Cells and Plasmid Construction
Huh7, Huh7.5, 293T, U-2 OS, and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen). All media contained 2 mm l-glutamine, 100 units/ml penicillin G, and 100 μg/ml streptomycin. All cells were cultured at 37 °C in a humidified atmosphere with 5% CO2.
The fragment encoding full-length NS5B was amplified from the genotype 1b strain (BB7) by polymerase chain reaction (PCR) using the PrimeSTAR enzyme (TaKaRa) and then cloned into the EcoRI and XhoI sites of the pcDNA3.1/myc-His-3×FLAG vector to yield pcDNA3.1/myc-His-3×FLAG NS5B. pTRE2hyg/3×FLAG-NS5B was generated from pcDNA3.1/myc-His-3×FLAG NS5B by PCR and inserted into the BamHI/NotI site of the pTRE2hyg vector. NS5B deletion mutants were made by inserting the appropriate sequence into the EcoRI/NotI site of the pCMV-myc (Invitrogen) vector or the EcoRI/XhoI site of the pcDNA3.1/myc-His-3×FLAG vector.
CDK2, CINP, and its truncated mutants were generated by PCR and introduced into their corresponding vectors using the same method as that for NS5B. The pET28bNS5BΔ21 (HCR6, genotype 1b) was provided by Professor Tetsuya Toyoda (Institut Pasteur of Shanghai) and was used as backbone to generate deletion mutants of NS5B (KOD-Plus Mutagenesis kit, Toyobo). All PCR products were confirmed by DNA sequencing (Invitrogen and Biosune).
Establishment of Stable Cell Line
HepG2 Tet-On advanced cells (Clontech) were cultured in minimum essential medium (Invitrogen) containing 15% FBS (Invitrogen), 100 μg/ml G418, 1× non-essential amino acids, and 2 mm l-glutamine. To generate the NS5B tetracycline-regulated stable cell line, HepG2 Tet-On advanced cells grown in 6-well plates were transfected with 1 μg of pTRE2hyg/3×FLAG-NS5B plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Twenty-four hours later, the cells were digested and plated in 10-cm dishes, and 200 μg/ml hygromycin B (Roche Applied Science) was added. Two weeks later, G418- and hygromycin B-resistant clones were isolated and cultured in medium containing doxycycline-free FBS (Clontech). Successful selection of G418- and hygromycin B-resistant clones were verified by Western blot using an anti-FLAG antibody and an anti-NS5B antibody after doxycycline (Sigma) induction.
Cell Cycle Analysis
To synchronize cells at the G1/S transition, growing cells were treated with 2 mm thymidine (Sigma) for 16 h, washed with PBS, and placed into fresh medium. Cells were then transfected with the indicated plasmids. Forty-eight hours later, the cells were washed with PBS and digested with 2 mm EDTA. After centrifugation (500 × g, 5 min), the cells were fixed overnight (−20 °C) with 70% precooled ethanol. The fixed cells were washed and then treated with 25 μg/ml propidium iodide (Sigma) and 50 μg/ml RNase A (Sigma) for 30 min (37 °C). DNA content was determined by flow cytometry with a FACSCalibur instrument (BD Biosciences), and data were analyzed with ModFitLT software (BD Biosciences). HepG2 Tet-On advanced NS5B stable cells were treated with and without doxycycline for 48 h. Nocodazole (200 ng/ml; Sigma) was added 7 h before harvesting, and the cells were analyzed by flow cytometry.
Protein Purification, GST Pulldown and Co-immunoprecipitation Assay
NS5BΔ21, CINP, and CDK2 sequences were inserted into a pGEX-4T-1 vector and transformed into Escherichia coli strain BL21/DE3. Proteins were induced by adding 0.4 mm isopropyl β-d-thiogalactopyranoside (Sigma) for 8 h (28 °C). GST pulldown and co-immunoprecipitation were performed as described previously (30). Cells were harvested by centrifugation and then resuspended in buffer (PBS, 1% Triton X-100, and 1 mm dithiothreitol (DTT)) for sonication. After centrifugation of the sonicated lysates, the supernatants were incubated with glutathione-Sepharose 4B beads (GE Healthcare) for purification. Forty-eight hours post-transfection with the indicated plasmids (FuGENE 6, Roche Applied Science), 293T cells were washed three times with ice-cold PBS and suspended in lysis buffer (20 mm Tris-HCl (pH 8.0), 200 mm NaCl, 1 mm EDTA (pH 8.0), 0.5% Nonidet P-40, 1 mm Na3VO4, 25 g/ml phenylmethylsulfonyl fluoride, 1 mm β-glycerophosphate, and 1× protease inhibitor mixture). After shaking for 30 min (4 °C), the cells were centrifuged at 12,000 × g for 10 min (4 °C). The resulting supernatants were divided into three parts. One-tenth of the supernatant was boiled in 40 μl of 2× SDS protein loading buffer and used for input. Equal parts of the remaining supernatant were incubated with GST or GST-fused proteins. After shaking for 2 h (4 °C), the beads were washed with washing buffer (20 mm Tris-HCl (pH 8.0), 200 mm NaCl, 1 mm EDTA (pH 8.0), and 0.5% Nonidet P-40) three times and boiled in 40 μl of loading buffer for SDS-PAGE. For co-immunoprecipitation, the cells were harvested as described above. The supernatant of co-transfected cells was immunoprecipitated with 1 μg of specific antibodies or control IgG (Santa Cruz Biotechnology), shaken for 2 h (4 °C), mixed with 30 μl of protein A/G (Santa Cruz Biotechnology), incubated for another 2 h (4 °C), and washed three times with washing buffer. Proteins bound to the beads were boiled in 40 μl of loading buffer. Purification of His-NS5B and the deletion mutants and the in vitro GST pulldown assay were performed according to previous descriptions (31, 32).
RNA Interference Assay
The siRNA sequences targeting CINP and Chk1 were synthesized by Invitrogen. The effective sequence for CINP siRNA was 5′-UUCUUGCACUGACAGAUAAGACAGG-3′ and for Chk1 siRNA was 5′-CCAGAUGCUCAGAGAUUCUGdTdT-3′. siGFP (Invitrogen) was used as the negative control. When cells reached 30–50% confluence, they were transfected with 60 nm siRNA using Lipofectamine RNAi MAX (Invitrogen) that was diluted in serum-free Opti-MEM (Invitrogen). Five hours later, the medium was replaced with fresh serum-containing medium. Cells were harvested 72 h post-transfection, and CINP was detected using an endogenous anti-CINP antibody (a kind gift from Professor David Cortez, Vanderbilt University).
Cell Proliferation Assay
Cell proliferation was monitored by counting viable cells using Cell Counting Kit-8 (Dojindo). At the time points indicated, 100 μl of the cell suspension (5,000 cells/well) was dispensed into each well of a 96-well plate. After incubation for 24 h (37 °C), 10 μl of Cell Counting Kit-8 solution was added to each well, and the cells were incubated for 1 h (37 °C). Sample absorbance was measured at 450 nm using a microplate reader (Bio-Rad).
RNA Transfection, HCV Infection, and Titration
In vitro synthesis of HCV RNA and electroporation were performed as described previously (33). Briefly, Huh7.5 cells were mixed with in vitro transcribed RNA and electroporated with five pulses lasting 99 μs at 820 V over 1.1 s in an ECM 830 instrument (BTX Genetronics). The culture medium was harvested 13 days post-transfection, and purified viruses were used for infection and titration.
Immunofluorescence Assay
The procedure for immunofluorescence was performed as described previously (34). Briefly, cells were grown on glass coverslips, fixed with 3.5% paraformaldehyde for 15 min, and then stopped by the addition of 30 mm glycine. After washing, the cells were permeabilized with 0.1% Triton X-100 for 15 min and blocked with 3% bovine serum albumin in PBS for at least 1 h (4 °C). Monoclonal anti-myc antibodies were used at a 1:400 dilution, and anti-NS5A 9E10 antibodies (kindly provided by Professor Charles M. Rice, Rockefeller University) were used at a 1:500 dilution. A goat anti-mouse Cy3 secondary antibody (Jackson ImmunoResearch Laboratories) was used at a 1:500 dilution. Nuclei were stained by 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma-Aldrich), and the cells were examined by confocal microscopy (TCS-NT, Leica).
Subcellular Fractionation Analysis
Huh7 cells were grown in 6-cm dishes and transfected with NS5B-, ΔNS5B-, or empty vector-expressing plasmids. Forty-eight hours post-transfection, cytoplasmic and nuclear fractions were obtained using a cytoplasmic and nuclear protein extraction kit (Fermentas) according to the manufacturer's directions. The HCV subgenomic replicon cells and Japanese fulminant hepatitis 1 (JFH-1)-infected Huh7.5 cells were fractionated by a similar process. Extracts were analyzed by Western blot using anti-CINP, anti-FLAG, anti-lamin A/C, and anti-β-tubulin antibodies.
Western Blot Analysis
Cell lysates or beads were prepared in SDS sample buffer (Tris-HCl, SDS, and 20% glycerol). Harvested samples were detected by SDS-PAGE. Proteins from the SDS-PAGE were electrotransferred onto nitrocellulose membrane (Whatman) and probed with the appropriate primary antibody (anti-FLAG (Sigma; 1:5,000), anti-NS5B (Santa Cruz Biotechnology; 1:500), anti-myc (Invitrogen; 1:1,000), the cell cycle regulator antibody kit (Cell Signaling Technology), the DNA damage antibody sampler kit (Cell Signaling Technology), anti-Chk1 (Cell Signaling Technology; 1:1,000), anti-pChk1Ser -317 (Cell Signaling Technology; 1:1,000), anti-His antibody (Cell Signaling Technology; 1:1,000), anti-pRb (Cell Signaling Technology; 1:5,000), anti-pCDK2 (Cell Signaling Technology; 1:500), anti-β-actin (Sigma; 1:5,000), anti-β-tubulin (Sigma; 1:5000), or anti-lamin A/C (Cell Signaling Technology; 1:500)). After incubation with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology; 1:10,000) for 3–4 h, the membranes were washed with PBS with Tween 20. Immunoreactivity was visualized using an ECL system (Western Lightning, PerkinElmer Life Sciences).
RESULTS
HCV NS5B Delays Cell Cycle Progression in S Phase
Although still controversial, HCV NS5B has been reported to modify cell cycle progression. Here, we first investigated the effect of NS5B on the cell cycle. U-2 OS cells were transfected with NS5B-expressing plasmids or empty vectors for 48 h, and cell cycle distribution was analyzed by flow cytometry. As shown in Fig. 1A, a higher percentage of cells in S phase (52.13%) was detected in NS5B-expressing cells than in control cells (34.62%). We confirmed NS5B expression in transfected cells by Western blot (Fig. 1A). To verify whether NS5B delayed cell cycle progression in S phase in hepatocytes, we constructed a tetracycline-regulated NS5B stable cell line in HepG2 Tet-On advanced cells. After simultaneously screening with G418 and hygromycin B, we obtained a highly inducible clone with low background. As shown in Fig. 1B, NS5B was detected at low concentrations (0.01 μg/ml) of doxycycline (Dox) 24 h post-induction as assessed using anti-NS5B and anti-FLAG antibodies with peak NS5B expression observed at a concentration of 1 μg/ml Dox. The successful establishment of a stable cell line was further confirmed by the sustained expression of NS5B 2 h after the addition of 1 μg/ml Dox (Fig. 1C). We also compared the NS5B protein level between the HCV subgenomic replicon (a selected genotype 1b cell clone that contains HCV RNA sequences from NS3 to NS5B) and HepG2 Tet-On NS5B cells after Dox induction. The results showed that NS5B expression levels induced by 1 μg/ml Dox in HepG2 Tet-On cells were similar to those in the replicon (Fig. 1D). Next, we used the HepG2 Tet-On NS5B cell line to study the effect of NS5B on the cell cycle. Fig. 1E shows that the presence of NS5B markedly delayed cell cycle progression in S phase. In contrast, most control cells without Dox induction were arrested in G2/M phase by nocodazole, a microtubule inhibitor that synchronizes the cell cycle in G2/M phase. Collectively, these results indicate that NS5B induces S phase delay in U-2 OS cells and HepG2 stable cells.
FIGURE 1.
Effect of NS5B on cell cycle progression. A, cell cycle analysis in U-2 OS cells. U-2 OS cells were grown in 6-well plates and transfected with 1 μg of pcDNA3.1/myc-His-3×FLAG-NS5B or related empty vectors. Forty-eight hours later, cell cycle distribution was monitored by flow cytometry, and protein expression was detected by Western blot using an anti-FLAG antibody. B and C, establishment of a HepG2 Tet-On NS5B stable cell line. The successful construction of a stable cell line was confirmed by Western blot. Cells induced by 0, 0.01, 0.1, 0.2, 0.5, 1, or 2 μg/ml Dox (B) and cells 0, 2, 4, 8, 12, 24, and 48 h after 1 μg/ml Dox induction (C) were examined using an anti-FLAG antibody or anti-NS5B antibody. D, after induction with different concentrations of Dox for 48 h, the expression of NS5B in HepG2 Tet-On NS5B stable cells was compared with that of the replicon cells (R) using an anti-NS5B antibody. E, cell cycle analysis of a HepG2 Tet-On NS5B stable cell line. Cells were induced with and without 1 μg/ml Dox for 48 h and treated with 200 ng/ml nocodazole 7 h before harvesting and cell cycle analysis. Each experiment was repeated three times. Error bars indicate the S.D.
NS5B/CINP Interaction in Vitro and in Vivo
To determine the mechanism of the effect of NS5B on the cell cycle, we assessed NS5B interaction with host proteins using a yeast two-hybrid assay with proteins from a human liver cDNA library. A novel protein, CINP, was identified.
To verify the results of the yeast two-hybrid screening, a GST pulldown assay was performed. GST-fused NS5B that lacked the C-terminal 21 amino acid residues (NS5BΔ21) was expressed in E. coli (BL21/DE3) and purified on glutathione-Sepharose 4B beads. After incubation with myc-CINP-transfected 293T cell lysates, the precipitated proteins were separated by SDS-PAGE and detected by Western blot using an anti-myc antibody. The results showed that CINP bound to GST-NS5BΔ21 but not to GST alone (Fig. 2A). A reciprocal experiment using GST-CINP and FLAG-NS5B gave a similar result (Fig. 2B). Purified GST, GST-CINP, and GST-NS5BΔ21 were detected by Coomassie Brilliant Blue staining (Fig. 2C). To show that NS5B interacted with CINP in cells, 293T cells were ectopically co-transfected with FLAG-NS5B- and myc-CINP-expressing plasmids. The interaction between NS5B and CINP was observed by co-immunoprecipitation with anti-FLAG or anti-myc antibody (Fig. 2, D and E). A similar result in Huh7 cells indicated that this interaction also occurred in the hepatocyte cell line (Fig. 2F). We also observed the interaction between CINP and NS5B of different genotypes (1a and 2a) by GST pulldown assay (supplemental Fig. S1A).
FIGURE 2.
NS5B/CINP interaction. 293T cells were grown in 6-cm dishes and transfected with the pCMV/myc-CINP or pcDNA3.1/myc-His-3×FLAG-NS5B (genotype 1b) plasmids. Forty-eight hours later, the cells were harvested as described under “Experimental Procedures,” and the supernatant was divided into two equal parts and incubated with GST or GST-NS5BΔ21 beads (A) or GST or GST-CINP beads (B) for 2 h. Protein-bound glutathione-Sepharose 4B beads were washed and boiled for 5 min, and CINP and NS5B, respectively, were detected by Western blot using an anti-myc antibody (A) or an anti-FLAG antibody (B). C, Coomassie-stained GST and GST-fused proteins are shown. D, equal amounts of pCMV/myc-CINP and pcDNA3.1/myc-His-3×FLAG-NS5B plasmids were co-transfected into 293T cells grown in 10-cm dishes. Forty-eight hours post-transfection, protein complexes were isolated by anti-FLAG immunopurification, and co-precipitating proteins were detected by Western blot with an anti-myc antibody. E, cell lysates were immunoprecipitated by an anti-myc antibody, and the co-precipitated protein was identified with an anti-FLAG antibody. The same experiment was performed in Huh7 cells, and the results are shown in F. G, pCMV/myc-CINP was co-transfected with GFP- or GFP-NS5B-expressing plasmids into 293T cells. Forty-eight hours post-transfection, cells were fixed for immunofluorescence analysis. The green represents empty GFP vector or NS5B, and CINP was detected using an anti-myc primary antibody and goat anti-mouse Cy3 secondary antibody (red). Representative cells from the same field for each experimental group are shown. H is the quantification of G. A minimum of 200 GFP-positive cells per sample were counted, and the percentage of cells with cytoplasmic CINP in GFP-positive cells was calculated. Results represent the means of data from three independent experiments. Error bars indicate the S.D. IP, immunoprecipitation; IB, immunoblot.
To further demonstrate the interaction between CINP and NS5B, we co-transfected 293T cells with GFP-NS5B and myc-CINP or a GFP vector and myc-CINP and visualized the staining pattern of NS5B and CINP by indirect immunofluorescence using anti-myc antibody. The results showed that CINP was primarily localized to the nucleus in 293T cells. However, it relocalized to the cytoplasmic perinuclear regions and partially colocalized with NS5B in the presence of GFP-NS5B (Fig. 2, G and H). Taken together, these data suggest that NS5B forms a complex with CINP.
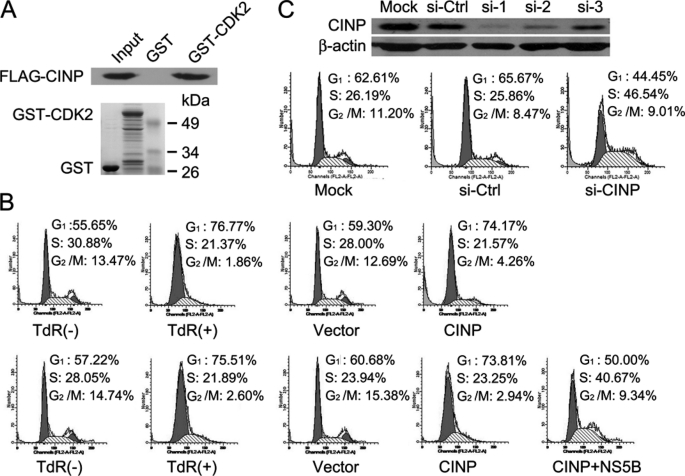
Role of CINP in G1/S Transition
CINP was first identified as a CDK2-interacting protein from a HeLa cDNA expression library. We confirmed this interaction using a GST pulldown assay (Fig. 3A). We then sought to determine whether CINP was also involved in cell cycle regulation. HeLa cells were primarily synchronized in the G1/S transition phase with 2 mm thymidine for 16 h before transfection with CINP or related empty vectors. Following release, cells transfected with empty vectors resumed normal cell cycle progression. However, cells transfected with CINP remained in G1/S phase. This could be reversed by the co-transfection of NS5B with CINP (Fig. 3B). To further evaluate the function of CINP in the cell cycle, CINP expression was knocked down with specific siRNA. Three distinct siRNAs targeting CINP were available, and the knockdown efficiency of each was evaluated with an endogenous anti-CINP antibody 72 h post-transfection (Fig. 3C, upper panel). We chose si-1 for further studies because of its high knockdown efficiency. CINP depletion resulted in an increase in the number of cells in S phase (Fig. 3C, lower panel), which is consistent with the results obtained by overexpression of NS5B. These data indicate the importance of CINP in the regulation of G1/S transition.
FIGURE 3.
Role of CINP in regulation of G1/S transition. A, the CDK2/CINP interaction was confirmed by a GST pulldown assay. B, HeLa cells were synchronized by the addition of 2 mm thymidine (TdR) for 16 h. After release, cells were transfected with CINP or empty vectors or co-transfected with CINP and NS5B. Forty-eight hours later, the cells were harvested and analyzed by flow cytometry. C, HeLa cells were transfected with 60 nm CINP-specific siRNA or control siGFP. Seventy-two hours later, the knockdown efficiency was detected by an anti-CINP antibody (upper panel). HeLa cells were then either mock-transfected or transfected with 60 nm siGFP or siCINP, respectively. Seventy-two hours post-transfection, the cells were analyzed for DNA content by flow cytometry. The results are representative of three independent experiments (lower panel). Ctrl, control.
NS5B and CINP Binding Region Analysis
To better understand the function of the association between NS5B and CINP, it was necessary to map the amino acid sequences required for their binding. To identify the region of CINP that interacted with NS5B, a protein truncation assay was performed. Because CINP has no described domain structure except a predicted coiled coil motif (predicted by SMART software), CINP was truncated at its N-terminal (aa 1–107) and C-terminal (aa 108–212) regions based on a prediction model (Fig. 4A) and fused with GST. The purified proteins were detected by Coomassie Brilliant Blue staining (Fig. 4C). The GST pulldown results showed that only the full-length and C-terminal region, but not the N-terminal region, of CINP bound NS5B (Fig. 4B). Next, we mapped the regions of NS5B responsible for binding CINP. Various NS5B deletion mutants were constructed according to the finger, palm, and thumb domains of NS5B (supplemental Fig. S1B), which were determined by Western blot (supplemental Fig. S1C, left panel). The resulting myc-NS5B deletion mutants were transiently overexpressed in 293T cells. The lysates were incubated with purified GST or GST-CINP for GST pulldown assays, and the bound proteins were immunologically detected by an anti-myc antibody. As shown in supplemental Fig. S1C (right panel), all of the mutants bound GST-CINP. We further truncated NS5B and tested its association with CINP with a yeast two-hybrid assay (data not shown). The results were further confirmed by GST pulldown assay. As shown in Fig. 4D (right panel), mutants NS5B(84–150) and NS5B(378–464) could interact with CINP, whereas NS5B(1–83), NS5B(465–591), and NS5B(96–454) could not, suggesting that the interaction for NS5B on CINP maps to two independent regions: aa 84–95 and aa 455–464. Furthermore, we constructed several NS5B mutants with deletions of aa 84–95 and/or 455–464 in the context of pET28bHCR6NS5BΔ21. The mutations were identified by Western blot with anti-His antibody after induction by isopropyl β-d-thiogalactopyranoside (Fig. 4E), and then in vitro GST pulldown experiments were carried out. The results indicated that deletion of aa 84–95 or aa 455–464 alone did not abolish the binding of NS5B to CINP. Only when both of these sequences were deleted was the binding to CINP abolished (Fig. 4F). In addition, an in vitro de novo transcription experiment indicated that these mutations still possess RNA-dependent RNA polymerase activity, although NS5BΔ455–464 was slightly decreased (supplemental Fig. S2). In conclusion, these results clearly demonstrated that the C-terminal end of CINP and two sites of NS5B (aa 84–95 and 455–464) were required for their binding.
FIGURE 4.
Analysis of NS5B/CINP binding regions. A, schematic diagram of CINP predicted by SMART software. The green bar represents the coiled coil region (amino acids 72–108), and gray bars represent unknown regions. B, 293T cells were grown in 10-cm dishes and transfected with FLAG-NS5B-expressing plasmids. The GST pulldown assay was carried out using purified beads that contained GST, GST-CINP full length, GST-CINP (aa 1–107), and GST-CINP (aa 108–212). Precipitated NS5B was detected by Western blot using an anti-FLAG antibody. C, Coomassie-stained GST and GST-fused proteins are shown. D, deletion mutants of myc-tagged NS5B were transiently transfected into 293T cells and detected by Western blot (left panel). Proteins pulled down by GST-CINP were examined by Western blot with an anti-myc antibody (right panel). E, after induction by isopropyl β-d-thiogalactopyranoside, pET28bHCR6NS5BΔ21 and its deletion mutants (Δ(84–95), Δ(455–464), and Δ(84–95)&(455–464)) were examined by Western blot with anti-His antibody. F, approximately 0.2 μg of His-NS5BΔ21 and mutants were mixed with purified GST- or GST-CINP-containing beads. After washing with PBS with Tween 20, each bound protein was fractionated by SDS-10% PAGE and subjected to Western blot analysis with anti-His monoclonal antibody. IB, immunoblot.
NS5B/CINP Association Is Responsible for S Phase Delay and Suppression of Cell Proliferation
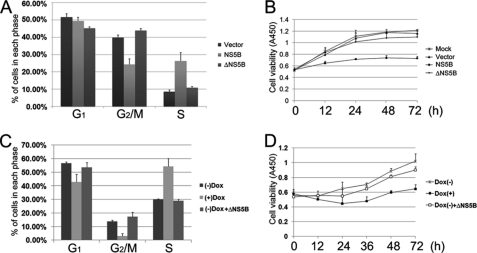
To test whether the NS5B/CINP interaction was involved in cell cycle dysregulation, an NS5B deletion mutant (aa 96–454) that failed to bind CINP (Fig. 4D) was chosen as a control (ΔNS5B). U-2 OS cells were transfected with full-length NS5B, ΔNS5B, or empty vector plasmids, and cell cycle distribution was analyzed by flow cytometry 48 h post-transfection. As shown in Fig. 5A, full-length NS5B induced a higher accumulation of cells in S phase compared with the empty vector and ΔNS5B. We further observed that the growth rate of full-length NS5B-expressing cells was significantly decreased relative to ΔNS5B- or empty vector-transfected cells (Fig. 5B), which is consistent with the results shown in Fig. 1E. Similar results were also observed in HepG2 cells; upon Dox induction, HepG2 Tet-On NS5B stable cells showed a delay in S phase and suppression of proliferation (Fig. 5, C and D). Cells without Dox induction served as a control. We attempted to generate a HepG2 Tet-On ΔNS5B stable cell line but failed to produce a low background, highly inducible clone. Therefore, we transfected constructs expressing ΔNS5B into HepG2 Tet-On NS5B stable cells without Dox induction as a control (Fig. 5, C and D). Taken together, these results indicate that the CINP/NS5B association is required for cell cycle delay and the suppression of cell proliferation.
FIGURE 5.
NS5B/CINP interaction promotes cell cycle dysfunction and suppression of cell proliferation. A, U-2 OS cells were transiently transfected with plasmids expressing NS5B, ΔNS5B, or empty vectors. Forty-eight hours later, the DNA content of the transfected cells was examined by flow cytometry. Error bars represent the S.E. from three independent experiments. B, U-2 OS cells were transfected with the indicated plasmids. Twenty-four hours after transfection, the cells were moved into 96-well plates. At the indicated time points, cells were incubated with 100 μl of DMEM supplemented with 10 μl of Cell Counting Kit-8 solution for 1 h. The 450-nm absorptions were then measured. Each data point represents the mean ± S.D. of three independent measurements. C, HepG2 Tet-On NS5B stable cells with or without 1 μg/ml Dox were analyzed by flow cytometry after 48 h. Cells without Dox induction were transfected with ΔNS5B-expressing plasmids as controls. Error bars represent the S.E. from three independent experiments. D, the cell growth rate was monitored in HepG2 Tet-On NS5B stable cells as described in B.
NS5B Triggers CINP Relocalization from Nucleus to Cytoplasm
To investigate in detail the mechanism(s) responsible for S phase delay upon NS5B/CINP interaction, we first tested whether the CINP protein level was affected by NS5B as Rb has been reported to undergo proteasome-mediated degradation due to its interaction with NS5B (19). Our data showed that NS5B had no significant influence on CINP protein levels (supplemental Fig. S3).
CINP has been found to localize to the nucleus, whereas NS5B primarily localizes to the perinuclear region. Our above data showed a perinuclear colocalization of NS5B with CINP (Fig. 2G), which suggests that NS5B caused the relocalization of CINP from the nucleus to the cytoplasm. To test this hypothesis, GFP-CINP was co-transfected with empty vector, myc-NS5B, or myc-ΔNS5B into Huh7 cells, and cells were analyzed by confocal microscopy after immunological staining with an anti-myc antibody. Consistent with the above results, strong merge signals were observed in the perinuclear regions in GFP-CINP- and myc-NS5B-co-transfected cells. However, most CINP remained localizing to the nucleus in empty vector- and ΔNS5B-transfected cells (Fig. 6A), indicating that NS5B caused CINP to relocalize from the nucleus to the cytoplasm. We then examined the localization of CINP in the HCV subgenomic replicon and JFH-1-infected (JFH-1 is a genotype 2a HCV strain isolate derived from a Japanese individual with fulminant hepatitis) cells. As shown in Fig. 6B, the HCV subgenomic replicon and JFH-1-infected cells also demonstrated a partially cytoplasmic relocalization of CINP.
FIGURE 6.
Immunofluorescence assay to examine CINP relocalization due to NS5B. A, GFP-CINP was co-transfected with equal amount of plasmids expressing myc-NS5B, myc-ΔNS5B, or empty vectors into Huh7 cells. Two days after transfection, the cells were fixed. Immunofluorescence staining was performed using the anti-myc antibody as a primary antibody. The cells were visualized by confocal laser-scanning microscopy. Green fluorescence indicates GFP-CINP, whereas red (Cy3) indicates NS5B or ΔNS5B. Yellow (merged) indicates colocalization. B, Huh7.5 cells, HCV subgenomic replicon cells, and JFH-1-preinfected Huh7.5 cells were transfected with GFP or GFP-CINP. Forty-eight hours later, cells were subjected to immunofluorescence staining as in A with the anti-NS5A antibody used as primary antibody. Green fluorescence indicates GFP or GFP-CINP, and red (Cy3) indicates endogenous NS5A. Yellow (merged) indicates colocalization.
The relocalization of CINP by NS5B was also confirmed by a subcellular fractionation assay. Huh7 cells were transfected with NS5B, ΔNS5B, or empty vector. Forty-eight hours later, cytoplasmic and nuclear fractions were isolated, and CINP distribution was detected by Western blot using an endogenous anti-CINP antibody. The results showed that nuclear CINP was clearly reduced in NS5B-transfected cells with a modest accumulation of CINP in the cytoplasm. However, no alteration was observed in empty vector- or ΔNS5B-transfected cells (Fig. 7A). Furthermore, partially cytoplasmic translocation was also obtained in HCV subgenomic replicons and JFH-1-infected cells (Fig. 7B). In addition, continuously cultured cells after JFH-1 infection were harvested to detect the subcellular distribution of CINP. As shown in Fig. 7C, CINP was primarily localized in the nucleus in uninfected cells; however, cytoplasmic translocation was observed following infection and was maintained to at least passage 4 (Fig. 7C). There was no obvious change in passage 8 cells, which might be due to a decrease in viral titer and protein levels (Fig. 7, C and D). Together, these results clearly indicate that NS5B causes the redistribution of CINP from the nucleus to the cytoplasm.
FIGURE 7.
Subcellular fractionation assay to examine CINP relocalization due to NS5B. A, empty vector-, NS5B-, or ΔNS5B-expressing plasmids were transfected into Huh7 cells. Forty-eight hours later, cytoplasmic and nuclear fractions were isolated and subjected to Western blot with endogenous anti-CINP, anti-lamin A/C (nuclear), anti-β-tubulin (cytoplasmic), and anti-FLAG antibodies. B, the subcellular distribution of CINP in Huh7 cells, HCV subgenomic replicon, or JFH-1-infected cells (multiplicity of infection, 0.1). “Rep” refers to replicon, and “H7” stands for Huh7 cells. C, cells after different passages (P) postinfection (multiplicity of infection, 0.1) were examined by a subcellular fractionation assay as described in A. D, viral titer and protein level were monitored during the experiment in C. Viral titer was represented as 50% tissue culture infective dose (TCID50)/ml, error bars indicate the means ± S.D. of three independent experiments and protein level was determined by Western blot with an anti-NS5A antibody. FL, full length.
NS5B Modifies DNA Damage Response by Relocalizing CINP
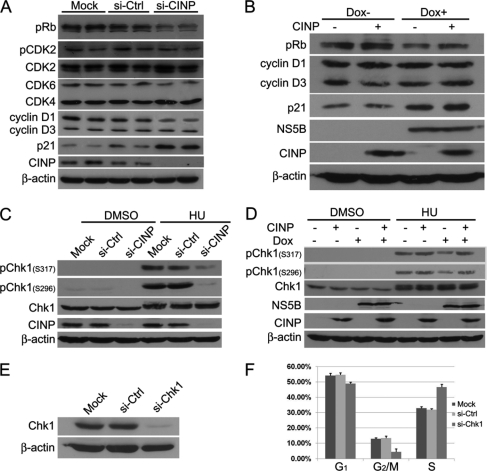
Because CINP translocation by NS5B reduced the amount of CINP available for normal cellular regulation in the nucleus, there might be an alteration of the signaling pathways mediated by CINP. To test this possibility, we used siRNA to specifically inhibit the expression of endogenous CINP in HepG2 Tet-On NS5B stable cells without Dox. First, we examined the expression profile of host proteins, such as cyclin-CDK complexes, pRb, and cyclin-dependent kinase inhibitor molecules, in cells in G1/S transition. Among the molecules investigated, there was a significant decrease in pRb and cyclin D1 and an increase in p21 in CINP-deficient cells compared with controls (Fig. 8A). The down-regulation of pRb by NS5B has been consistently reported by Lemon and co-workers (19, 39, 40), and we confirmed this result in HepG2 Tet-On NS5B stable cells (Fig. 8B). In addition, a slight increase in p21 was also observed in the stable NS5B-expressing cells (Fig. 8B). However, NS5B appeared to have no effect on cyclin D1 or other detected regulators (Fig. 8B and data not shown). As a tumor suppressor, Rb is essential to prevent DNA damage and maintain genomic stability. Studies using Rb-deficient cells have shown the formation of aberrant ploidy and double strand breaks (35, 36). Diminished Rb expression can also lead to inappropriate DNA synthesis following a senescence trigger (37). Of note, CINP has recently been identified as a genome maintenance protein (38), and it is known that the sensing and repairing of DNA damage are involved in cell cycle progression. Therefore, we theorized that the DNA damage response could be exploited by NS5B to delay cells in S phase. Chk1 is a major DNA damage response transducer, and it has been shown to be mainly responsible for the S phase checkpoint. In agreement with previous work (38), we observed a dramatic decrease in phosphorylated Chk1 induced by hydroxyurea in CINP-depleted cells (Fig. 8C). NS5B induction also reduced the phosphorylation level of Chk1 after hydroxyurea treatment although not to the extent seen with CINP silencing (Fig. 8D). Notably, the ectopic overexpression of CINP partially reversed the effect caused by NS5B (Fig. 8, B and D). Then we knocked down Chk1 with specific siRNA (Fig. 8E) and obtained the S phase arrest in Chk1-deficient HepG2 Tet-On cells (Fig. 8F). These results suggest that NS5B hijacks the DNA damage response to induce cell cycle dysfunction, and this might be due to NS5B sequestering cellular CINP and reducing its availability for genomic maintenance.
FIGURE 8.
NS5B modifies DNA damage response. A, HepG2 Tet-On NS5B stable cells were transfected with CINP-specific or control siRNA (si-Ctrl) without Dox addition. Seventy-two hours later, the cells were harvested, and Western blot analysis was performed with the indicated antibodies. siCINP and siGFP transfections were performed in duplicate in each experiment. B, HepG2 Tet-On NS5B stable cells with or without Dox were transfected with or without CINP-expressing plasmids. Forty-eight hours later, the cells were harvested, and Western blot analysis was performed. C, HepG2 Tet-On NS5B stable cells were transfected with CINP-specific or control siRNA without Dox. Forty-eight hours later, the cells were treated with 3 mm hydroxyurea (HU) or an equal volume of DMSO. After incubation for another 24 h, the cells were harvested for Western blot analysis with anti-pChk1 (Ser-296 and Ser-317) and anti-Chk1 antibodies. D, HepG2 Tet-On NS5B stable cells with or without Dox were transfected with or without CINP-expressing plasmids. Twenty-four hours later, the cells were treated with 3 mm hydroxyurea or an equal volume of DMSO. After incubation for another 24 h, the cells were harvested for Western blot with anti-pChk1 (Ser-296 and Ser-317) and anti-Chk1 antibodies. E, HepG2 Tet-On cells were transfected with 60 nm Chk1-specific siRNA or control siGFP. Seventy-two hours later, the knockdown efficiency was detected by an anti-Chk1 antibody. F, after transfection with Chk1-specific siRNA for 72 h, HepG2 Tet-On cells were analyzed for DNA content by flow cytometry. The results are representative of three independent experiments. Error bars indicate S.D.
DISCUSSION
In the present study, we confirmed that NS5B induced S phase delay in U-2 OS cells and HepG2 Tet-On NS5B stable cells. In addition, we showed that NS5B interacted with the human protein CINP and that their association was required for S phase delay. Mechanistic investigations indicated that the cytoplasmic redistribution of CINP by NS5B was the likely cause of cell cycle dysfunction.
Siavoshian et al. (13) first reported the HCV NS5B effect on the cell cycle and cell proliferation. They found that NS5B arrests cells in the G2/M phase in a p53-independent manner, but they did not provide a more detailed mechanism. Recently, other studies on a possible mechanism of cell cycle regulation by NS5B have been reported. Studies performed by Lemon and co-workers (19, 39) demonstrated that HCV NS5B interacts with and down-regulates Rb through a proteasome-dependent pathway. Rb degradation results in the release of E2F and the transcription of a series of genes responsible for DNA replication, promoting entry into S phase and cell division. However, when Rb was knocked down with siRNA, enhanced HCV replication was not observed as expected (40). These results suggest that other mechanisms exist. Another study performed by Naka et al. (20) reported that NS5B delays cell cycle progression by inducing interferon-β. This was based on a previous study indicating that interferon-β up-regulates the nuclear phosphoprotein promyelocytic leukemia (41), leading to S phase delay and the suppression of cell proliferation. In their study, Naka et al. (20) showed that cells expressing NS5B are hypersensitive to DNA-damaging agents, suggesting that the DNA damage response might be involved in the cell cycle delay caused by NS5B.
In this study, an interaction between NS5B and CINP was demonstrated (Figs. 2 and 4). As a newly identified protein, few functions have been assigned to CINP. Its cDNA contains a 636-nucleotide open reading frame that encodes a protein of 212 amino acids. Grishina and Lattes (42) first reported that CINP is phosphorylated by Cdc7 and forms a complex with CDK2-cyclin E and CDK2-cyclin A in the nucleus. The association of CINP with minichromosome maintenance proteins and ORC2 suggests a potential role for CINP in the initiation of DNA replication. In addition, CINP has been identified as a thyroid hormone (triiodothyronine)-responsive gene that might participate in the regulation of cell proliferation (43). Weak CINP sumoylation has been observed by another group through a screening system, suggesting the existence of other modified types of CINP (44). Interestingly, Lovejoy et al. (38) and another group (45) recently identified CINP as a checkpoint protein to maintain genomic stability, thus shedding light on the function of CINP in genomic maintenance and cell cycle regulation.
To maintain genomic integrity and prevent tumorigenesis, recognition and repair of DNA damage are coordinated with the cell cycle in eukaryotic cells. There are numerous examples of viruses exploiting the DNA damage response to dysregulate cell cycle progression, such as Epstein-Barr virus (46, 47), human T-cell lymphotropic virus type 1 (48), Kaposi sarcoma-associated herpesvirus (49), and hepatitis B virus (50). Here, we observed an alteration in DNA damage checkpoint proteins, such as pRb, p21, and pChk1, after CINP knockdown or upon induced expression of NS5B (Fig. 8). Studies have shown that DNA damage responses mediated by Rb inhibit cell cycle progression and cause genomic instability (51, 52). It is noteworthy to mention that DNA damage is always accompanied by p21 expression (53, 54). p21 is a member of the CDK inhibitor family and is activated by another central tumor suppressor, p53. In addition to inhibiting CDKs, p21 also functions as a DNA damage checkpoint protein that regulates cell cycle progression, including intra-S phase regulation (55). Therefore, it is presumed that the loss of Rb coupled with p21 accumulation in the presence of NS5B prevents cell cycle progression from S phase. As viral replication only occurs in proliferating cells, it is not surprising that HCV replication is reduced in Rb-deficient cells (40).
We also observed a decrease in pChk1 in both CINP-deficient and NS5B-expressing cells. Chk1 is an evolutionarily conserved serine/threonine kinase that is essential for genomic maintenance and serves as a cell cycle checkpoint protein during S and G2/M phases (56–58). Chk1 is phosphorylated by ATR, an S phase-specific sensor/transducer that is activated in response to ultraviolet light, hydroxyurea, and replication stress following genotoxic damage (59). Several previous studies have demonstrated that the down-regulation or inactivation of Chk1 leads to S phase arrest. Jirmanova et al. (60) reported that the inhibition of the ATR/Chk1 pathway by caffeine induces a p38-dependent delay in S phase in mouse embryonic stem cells. A study performed by Shimada et al. (61) showed that Chk1-depleted mouse embryonic fibroblast cells were arrested in S phase, and this was due to transcriptional repression after Chk1 depletion, which led to a loss of histone acetylation. Naruyama et al. (62) also observed S phase arrest after Chk1 depletion that was caused by transcriptional suppression of ribonucleotide reductase 2. The effect of NS5B on pChk1 was not as significant as CINP silencing in this study. There are three possible underlying reasons. First, the specific siRNA dramatically decreases cellular CINP, but NS5B just reduces nuclear CINP to a certain extent. Second, NS5B has been reported in association with ATM and Chk2, key factors of the DNA damage response, that correlated with Chk1 phosphorylation (63). The subsequent effect of their interaction, which might be involved in Chk1 phosphorylation modification, remained unclear up to now. Third, a feedback mechanism to partially restore pChk1 in cells with NS5B present may exist.
The observed influence of NS5B on pRb, p21, and pChk1 in this study indicates that HCV NS5B can modulate the DNA damage response. However, there is no direct evidence that NS5B induces DNA damage and/or impairs DNA repair. To address this question, further studies are required. In fact, there have been many studies focused on the relationship between HCV and the DNA damage response. It is well known that HCV infection induces DNA damage, and studies have revealed that this damage is triggered by the production of inducible nitric-oxide synthase and mitochondrially mediated reactive oxygen species, which are stimulated by HCV core protein and NS3 (64–67). Chromosomal instability induced by the overexpression of NS5A has also been reported by Baek et al. (68), and this instability might be due to cell cycle dysregulation. On the other hand, NS3/4A have been shown to interact with ATM and hinder the efficiency of DNA repair (69). Inhibition of DNA repair by HCV core protein has also been reported: HCV core protein affects ATM activation and inhibits the binding of repair enzymes to damaged DNA through the binding of NBS1 and the inhibition of the formation of the Mre11-NBS1-Rad50 complex (70). Another report has demonstrated that HCV core protein impairs DNA damage repair through the inhibition of DNA glycosylase activity (8). Collectively, these findings suggest that HCV uses a “hit and run” mechanism (70) of modifying cellular genetic materials. Ultimately, this results in chromosomal instability and neoplastic transformation.
Previous studies have indicated that CINP might be a very important host factor because of its association with cyclin-CDK2 complexes, minichromosome maintenance proteins, and ATR-interacting protein in the nucleus. To aid in HCV pathogenesis, the relocalization of CINP by NS5B undoubtedly influences several cellular activities, including DNA replication, cell cycle progression, and genomic maintenance in addition to the effects observed in this study. However, additional data are needed to confirm these results. Nuclear protein relocalization by NS5B has been reported previously: nucleolin has been reported to colocalize with NS5B in perinuclear regions (71), and their interaction is involved in HCV replication (72). NS5B induces the redistribution of the RNA helicase p68 from the nucleus to the cytoplasm where it assists in HCV negative-strand RNA synthesis (34). A striking cytoplasmic relocalization of pRb in JFH-1-infected cells and in cells with NS5B present has also been demonstrated (19, 39). Together with our results, these studies indicate that the NS5B-induced relocalization of nuclear proteins may be a common strategy used by HCV to aid in persistence and pathogenesis.
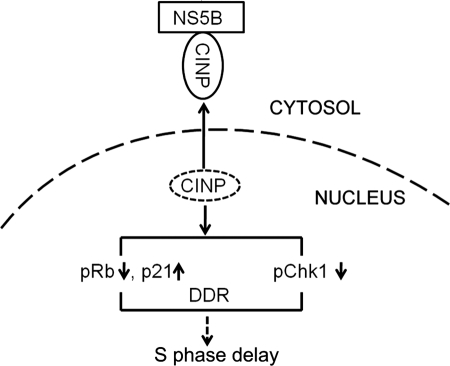
The S phase delay induced by NS5B prevents DNA replication and subsequently inhibits cell proliferation. According to this theory, NS5B plays a negative role in HCV replication. Interestingly, this self-inhibition might be a strategy utilized by HCV to promote low level replication and pathogenesis. Similar phenomena have been reported recently. Theiler murine encephalitis virus RNA-dependent RNA polymerase (3Dpol) has been shown to inhibit not only Theiler murine encephalitis virus itself but also encephalomyocarditis virus and vesicular stomatitis virus infections. The underlying mechanism might be to change some or all host cell types to an antiviral state rather than induce the adaptive immune response (73). HCV NS2 has also been reported to induce endoplasmic reticulum stress and therefore slow down replication by a feedback mechanism (74). We also observed a clear decrease in HCV replication in the subgenomic replicon and JFH-1-infected cells after CINP silencing.5 Based on the results of our study, we propose a mechanistic model in which NS5B is translated in the cytoplasm after HCV infection. The presence of NS5B then traps CINP in the cytoplasm by two possible mechanisms. 1) NS5B interacts with newly synthesized CINP and prevents its nuclear translocation, or 2) CINP is retained by NS5B in the cytoplasm after its nuclear export. Elucidation of the specific mechanism will require additional studies of CINP. CINP relocalization undoubtedly affects the signaling pathway molecules involved in cell cycle regulation as shown by the reduction of pRb and pChk1 and the accumulation of p21. Further studies will be aimed at resolving the detailed cross-talk among pRb, p21, and pChk1 and their alterations. These alterations ultimately resulted in S phase delay via the DNA damage/repair pathway (Fig. 9). Further studies are also needed to determine whether a direct CINP/CDK2 interaction is involved in cell cycle regulation. In conclusion, our results highlight the critical role of CINP in S phase delay induced by NS5B and provide new insights into understanding viral persistence and tumorigenesis following HCV infection.
FIGURE 9.
Postulated mechanism of HCV NS5B-mediated S phase delay through its interaction with CINP. DDR, DNA damage response.
Supplementary Material
Acknowledgments
We thank Professor Charles M. Rice for providing the NS5A antibody and Huh7.5 cells and Professor David Cortez for providing the CINP antibody. We also thank Professor Takaji Wakita for providing the pJFH-1 plasmid and Professor Ralf Bartenschlager for discussions and suggestions.
This work was supported in part by National Megaprojects for Infectious Diseases Grant 2008ZX10203 and the Program for Outstanding Medical Academic Leader of Shanghai.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Y. Wang, Y. Wang, Y. Xu, W. Tong, T. Pan, J. Li, S. Sun, J. Shao, H. Ding, T. Toyoda, and Z. Yuan, unpublished data.
- HCV
- hepatitis C virus
- NS5B
- nonstructural protein 5B of hepatitis C virus
- CINP
- cyclin-dependent kinase 2-interacting protein
- Rb
- Retinoblastoma
- ATM
- ataxia telangiectasia, mutated
- ATR
- ATM- and RAD3-related protein
- Chk
- checkpoint kinase
- JFH-1
- Japanese fulminant hepatitis 1
- aa
- amino acids
- pRb
- phosphorylated Rb
- pChk1
- phosphorylated Chk1
- CDK
- cyclin-dependent kinase
- Dox
- doxycycline.
REFERENCES
- 1. Kiyosawa K., Sodeyama T., Tanaka E., Gibo Y., Yoshizawa K., Nakano Y., Furuta S., Akahane Y., Nishioka K., Purcell R. H., Alter H. J. (1990) Hepatology 12, 671–675 [DOI] [PubMed] [Google Scholar]
- 2. Hoofnagle J. H. (2002) Hepatology 36, S21–S29 [DOI] [PubMed] [Google Scholar]
- 3. Bowen D. G., Walker C. M. (2005) J. Hepatol. 42, 408–417 [DOI] [PubMed] [Google Scholar]
- 4. Chisari F. V. (2005) Nature 436, 930–932 [DOI] [PubMed] [Google Scholar]
- 5. Bartenschlager R., Lohmann V. (2000) J. Gen. Virol. 81, 1631–1648 [DOI] [PubMed] [Google Scholar]
- 6. Penin F., Dubuisson J., Rey F. A., Moradpour D., Pawlotsky J. M. (2004) Hepatology 39, 5–19 [DOI] [PubMed] [Google Scholar]
- 7. Moriya K., Fujie H., Shintani Y., Yotsuyanagi H., Tsutsumi T., Ishibashi K., Matsuura Y., Kimura S., Miyamura T., Koike K. (1998) Nat. Med. 4, 1065–1067 [DOI] [PubMed] [Google Scholar]
- 8. Machida K., Tsukamoto H., Liu J. C., Han Y. P., Govindarajan S., Lai M. M., Akira S., Ou J. H. (2010) Hepatology 52, 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sakamuro D., Furukawa T., Takegami T. (1995) J. Virol. 69, 3893–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Einav S., Sklan E. H., Moon H. M., Gehrig E., Liu P., Hao Y., Lowe A. W., Glenn J. S. (2008) Hepatology 47, 827–835 [DOI] [PubMed] [Google Scholar]
- 11. Giménez-Barcons M., Wang C., Chen M., Sánchez-Tapias J. M., Sáiz J. C., Gale M., Jr. (2005) J. Interferon Cytokine Res. 25, 152–164 [DOI] [PubMed] [Google Scholar]
- 12. Yang X. J., Liu J., Ye L., Liao Q. J., Wu J. G., Gao J. R., She Y. L., Wu Z. H., Ye L. B. (2006) Virus Res. 121, 134–143 [DOI] [PubMed] [Google Scholar]
- 13. Siavoshian S., Abraham J. D., Kieny M. P., Schuster C. (2004) Arch. Virol. 149, 323–336 [DOI] [PubMed] [Google Scholar]
- 14. Ohkawa K., Ishida H., Nakanishi F., Hosui A., Ueda K., Takehara T., Hori M., Hayashi N. (2004) J. Biol. Chem. 279, 11719–11726 [DOI] [PubMed] [Google Scholar]
- 15. Arima N., Kao C. Y., Licht T., Padmanabhan R., Sasaguri Y., Padmanabhan R. (2001) J. Biol. Chem. 276, 12675–12684 [DOI] [PubMed] [Google Scholar]
- 16. Lohmann V., Körner F., Herian U., Bartenschlager R. (1997) J. Virol. 71, 8416–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deore R. R., Chern J. W. (2010) Curr. Med. Chem. 17, 3806–3826 [DOI] [PubMed] [Google Scholar]
- 18. Vermehren J., Sarrazin C. (2011) Clin. Microbiol. Infect. 17, 122–134 [DOI] [PubMed] [Google Scholar]
- 19. Munakata T., Nakamura M., Liang Y., Li K., Lemon S. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18159–18164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naka K., Dansako H., Kobayashi N., Ikeda M., Kato N. (2006) Virology 346, 348–362 [DOI] [PubMed] [Google Scholar]
- 21. Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 22. Jackson S. P., Bartek J. (2009) Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weitzman M. D., Lilley C. E., Chaurushiya M. S. (2010) Annu. Rev. Microbiol. 64, 61–81 [DOI] [PubMed] [Google Scholar]
- 24. Shiloh Y. (2003) Nat. Rev. Cancer 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 25. Guo Z., Kumagai A., Wang S. X., Dunphy W. G. (2000) Genes Dev. 14, 2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lopez-Girona A., Tanaka K., Chen X. B., Baber B. A., McGowan C. H., Russell P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11289–11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Melchionna R., Chen X. B., Blasina A., McGowan C. H. (2000) Nat. Cell Biol. 2, 762–765 [DOI] [PubMed] [Google Scholar]
- 28. Aressy B., Ducommun B. (2008) Anticancer Agents Med. Chem. 8, 818–824 [DOI] [PubMed] [Google Scholar]
- 29. Boutros R., Dozier C., Ducommun B. (2006) Curr. Opin. Cell Biol. 18, 185–191 [DOI] [PubMed] [Google Scholar]
- 30. Peng L., Liang D., Tong W., Li J., Yuan Z. (2010) J. Biol. Chem. 285, 20870–20881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weng L., Du J., Zhou J., Ding J., Wakita T., Kohara M., Toyoda T. (2009) Arch. Virol. 154, 765–773 [DOI] [PubMed] [Google Scholar]
- 32. Shirota Y., Luo H., Qin W., Kaneko S., Yamashita T., Kobayashi K., Murakami S. (2002) J. Biol. Chem. 277, 11149–11155 [DOI] [PubMed] [Google Scholar]
- 33. Lindenbach B. D., Evans M. J., Syder A. J., Wölk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., Rice C. M. (2005) Science 309, 623–626 [DOI] [PubMed] [Google Scholar]
- 34. Goh P. Y., Tan Y. J., Lim S. P., Tan Y. H., Lim S. G., Fuller-Pace F., Hong W. (2004) J. Virol. 78, 5288–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Harn T., Foijer F., van Vugt M., Banerjee R., Yang F., Oostra A., Joenje H., te Riele H. (2010) Genes Dev. 24, 1377–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Srinivasan S. V., Mayhew C. N., Schwemberger S., Zagorski W., Knudsen E. S. (2007) J. Biol. Chem. 282, 23867–23877 [DOI] [PubMed] [Google Scholar]
- 37. Chicas A., Wang X., Zhang C., McCurrach M., Zhao Z., Mert O., Dickins R. A., Narita M., Zhang M., Lowe S. W. (2010) Cancer Cell 17, 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lovejoy C. A., Xu X., Bansbach C. E., Glick G. G., Zhao R., Ye F., Sirbu B. M., Titus L. C., Shyr Y., Cortez D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19304–19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Munakata T., Liang Y., Kim S., McGivern D. R., Huibregtse J., Nomoto A., Lemon S. M. (2007) PLoS Pathog. 3, 1335–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McGivern D. R., Villanueva R. A., Chinnaswamy S., Kao C. C., Lemon S. M. (2009) J. Virol. 83, 7422–7433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vannucchi S., Percario Z. A., Chiantore M. V., Matarrese P., Chelbi-Alix M. K., Fagioli M., Pelicci P. G., Malorni W., Fiorucci G., Romeo G., Affabris E. (2000) Oncogene 19, 5041–5053 [DOI] [PubMed] [Google Scholar]
- 42. Grishina I., Lattes B. (2005) Cell Cycle 4, 1120–1126 [PubMed] [Google Scholar]
- 43. Miller L. D., Park K. S., Guo Q. M., Alkharouf N. W., Malek R. L., Lee N. H., Liu E. T., Cheng S. Y. (2001) Mol. Cell. Biol. 21, 6626–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jakobs A., Himstedt F., Funk M., Korn B., Gaestel M., Niedenthal R. (2007) Nucleic Acids Res. 35, e109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paulsen R. D., Soni D. V., Wollman R., Hahn A. T., Yee M. C., Guan A., Hesley J. A., Miller S. C., Cromwell E. F., Solow-Cordero D. E., Meyer T., Cimprich K. A. (2009) Mol. Cell 35, 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gruhne B., Sompallae R., Masucci M. G. (2009) Oncogene 28, 3997–4008 [DOI] [PubMed] [Google Scholar]
- 47. Kudoh A., Fujita M., Zhang L., Shirata N., Daikoku T., Sugaya Y., Isomura H., Nishiyama Y., Tsurumi T. (2005) J. Biol. Chem. 280, 8156–8163 [DOI] [PubMed] [Google Scholar]
- 48. Grossman W. J., Kimata J. T., Wong F. H., Zutter M., Ley T. J., Ratner L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1057–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Radkov S. A., Kellam P., Boshoff C. (2000) Nat. Med. 6, 1121–1127 [DOI] [PubMed] [Google Scholar]
- 50. Choi B. H., Choi M., Jeon H. Y., Rho H. M. (2001) DNA Cell Biol. 20, 75–80 [DOI] [PubMed] [Google Scholar]
- 51. Harrington E. A., Bruce J. L., Harlow E., Dyson N. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11945–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Knudsen K. E., Booth D., Naderi S., Sever-Chroneos Z., Fribourg A. F., Hunton I. C., Feramisco J. R., Wang J. Y., Knudsen E. S. (2000) Mol. Cell. Biol. 20, 7751–7763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 54. Viale A., De Franco F., Orleth A., Cambiaghi V., Giuliani V., Bossi D., Ronchini C., Ronzoni S., Muradore I., Monestiroli S., Gobbi A., Alcalay M., Minucci S., Pelicci P. G. (2009) Nature 457, 51–56 [DOI] [PubMed] [Google Scholar]
- 55. Zhu Y., Alvarez C., Doll R., Kurata H., Schebye X. M., Parry D., Lees E. (2004) Mol. Cell. Biol. 24, 6268–6277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao H., Piwnica-Worms H. (2001) Mol. Cell. Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xiao Z., Chen Z., Gunasekera A. H., Sowin T. J., Rosenberg S. H., Fesik S., Zhang H. (2003) J. Biol. Chem. 278, 21767–21773 [DOI] [PubMed] [Google Scholar]
- 58. Hu B., Zhou X. Y., Wang X., Zeng Z. C., Iliakis G., Wang Y. (2001) J. Biol. Chem. 276, 17693–17698 [DOI] [PubMed] [Google Scholar]
- 59. Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 60. Jirmanova L., Bulavin D. V., Fornace A. J., Jr. (2005) Cell Cycle 4, 1428–1434 [DOI] [PubMed] [Google Scholar]
- 61. Shimada M., Niida H., Zineldeen D. H., Tagami H., Tanaka M., Saito H., Nakanishi M. (2008) Cell 132, 221–232 [DOI] [PubMed] [Google Scholar]
- 62. Naruyama H., Shimada M., Niida H., Zineldeen D. H., Hashimoto Y., Kohri K., Nakanishi M. (2008) Biochem. Biophys. Res. Commun. 374, 79–83 [DOI] [PubMed] [Google Scholar]
- 63. Ariumi Y., Kuroki M., Dansako H., Abe K., Ikeda M., Wakita T., Kato N. (2008) J. Virol. 82, 9639–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kitay-Cohen Y., Amiel A., Hilzenrat N., Buskila D., Ashur Y., Fejgin M., Gaber E., Safadi R., Tur-Kaspa R., Lishner M. (2000) Blood 96, 2910–2912 [PubMed] [Google Scholar]
- 65. Machida K., Cheng K. T., Sung V. M., Shimodaira S., Lindsay K. L., Levine A. M., Lai M. Y., Lai M. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101, 4262–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Machida K., Cheng K. T., Sung V. M., Lee K. J., Levine A. M., Lai M. M. (2004) J. Virol. 78, 8835–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Machida K., Cheng K. T., Lai C. K., Jeng K. S., Sung V. M., Lai M. M. (2006) J. Virol. 80, 7199–7207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baek K. H., Park H. Y., Kang C. M., Kim S. J., Jeong S. J., Hong E. K., Park J. W., Sung Y. C., Suzuki T., Kim C. M., Lee C. W. (2006) J. Mol. Biol. 359, 22–34 [DOI] [PubMed] [Google Scholar]
- 69. Lai C. K., Jeng K. S., Machida K., Cheng Y. S., Lai M. M. C. (2008) Virology 370, 295–309 [DOI] [PubMed] [Google Scholar]
- 70. Machida K., McNamara G., Cheng K. T., Huang J., Wang C. H., Comai L., Ou J. H., Lai M. M. (2010) J. Immunol. 185, 6985–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hirano M., Kaneko S., Yamashita T., Luo H., Qin W., Shirota Y., Nomura T., Kobayashi K., Murakami S. (2003) J. Biol. Chem. 278, 5109–5115 [DOI] [PubMed] [Google Scholar]
- 72. Shimakami T., Honda M., Kusakawa T., Murata T., Shimotohno K., Kaneko S., Murakami S. (2006) J. Virol. 80, 3332–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kerkvliet J., Papke L., Rodriguez M. (2011) J. Virol. 85, 621–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. von dem Bussche A., Machida R., Li K., Loevinsohn G., Khander A., Wang J., Wakita T., Wands J. R., Li J. (2010) J. Hepatol. 53, 797–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.