Abstract
Heparin, which has been used as an anticoagulant drug for decades, inhibits angiogenesis, whereas thrombin promotes tumor-associated angiogenesis. However, the mechanisms underlying the regulation of angiogenesis by heparin and thrombin are not well understood. Here, we show that microRNA-10b (miR-10b) is down-regulated by heparin and up-regulated by thrombin in human microvascular endothelial cells (HMEC-1). Overexpression of miR-10b induces HMEC-1 cell migration, tube formation, and angiogenesis, and down-regulates homeobox D10 (HoxD10) expression via direct binding of miR-10b to the putative 3′ UTR of HoxD10. In addition, HMEC-1 cell migration and tube formation are induced by HoxD10 knockdown, whereas angiogenesis is arrested when HoxD10 expression is increased after anti-miR-10b or heparin treatments. Furthermore, expression of miR-10b and its transcription factor Twist are up-regulated by thrombin, whereas HoxD10 expression is impaired by thrombin. Using quartz crystal microbalance analysis, we show that heparin binds to thrombin, thereby inhibiting thrombin-induced expression of Twist and miR-10b. However, the expression of miR-10b is not attenuated by heparin any more after thrombin expression is silenced by its siRNA. Interestingly, we find that heparin attenuates miR-10b expression and induces HoxD10 expression in vivo to inhibit angiogenesis and impair the growth of MDA-MB-231 tumor xenografts. These results provide insight into the molecular mechanism by which heparin and thrombin regulate angiogenesis.
Keywords: Cell Migration, Heparin, Heparin-binding Protein, MicroRNA, Thrombin, Tumor, HoxD10, Twist, Angiogenesis, miR-10b
Introduction
Thrombosis is considered an early clinical indication and frequent complication of cancer (1, 2). Malignant tumors often exhibit increased expression of tissue factor and cancer procoagulant, which can be followed by activation of cell surface protease receptors and fibrin generation (3). In addition, tumor cells can interact with blood cells, particularly monocytes, macrophages, and platelets, leading to the generation of thrombin and thrombosis through the clotting cascade or platelet activation (4). Moreover, aggressive antitumor therapies such as chemotherapy, radiation, and surgery also increase the risk of thrombosis.
Positive feedback signaling loops exist between tumor tissue and the coagulation system (5, 6). For instance, it is widely accepted that elements of the coagulation and fibrinolytic system may aid in cancer cell survival, proliferation, invasion, and metastasis, as well as tumor angiogenesis (7). Therefore, inhibition of the activation of coagulation could be a useful antitumor strategy. A number of studies have shown that anticoagulant drugs can extend survival in patients with certain types of cancer (8), and studies are currently ongoing to confirm the effects of anticoagulant therapy in a range of tumor types. However, the molecular mechanisms involved in the action of anticoagulants in this process are not well understood.
Heparins, in particular those of low molecular weight, are effective in the prevention and treatment of thromboembolic events in cancer patients (9, 10). As early as the 1930s, heparins were reported to interfere with several vital steps of tumor progression, including growth, motility, migration, invasion, metastasis, and angiogenesis. Angiogenesis is an important determinant of tumor growth and metastasis. Notably, heparin can interfere with the actions of many pro- and antiangiogenic endogenous factors, including basic fibroblast growth factor (bFGF), hepatocyte growth factor, vascular endothelial growth factor-A, and tissue factor pathway inhibitor. During tumor growth and metastasis, the tight regulatory balance that normally exists between pro- and antiangiogenic factors is disturbed. Heparins have been shown to inhibit capillary tube formation by human endothelial cells (EC)3 from the macrovascular bed (i.e. human umbilical vein endothelial cells, HMVECs), which can be induced by standard proangiogenic factors, such as bFGF and vascular endothelial growth factor (11). In addition, heparin can inhibit capillary tube formation by EC of microvascular origin (i.e. human microvascular endothelial cells (HMEC-1)), which is stimulated by tumor cell-derived factors (12). Animal studies have shown that heparins have antitumor activity and antiangiogenic activity, which is mediated in part through the inhibition of FGF-2 (13). In addition to inhibiting angiogenic factors, heparin may also modulate angiogenesis via anticoagulant action, inhibiting proteolytic enzymes, binding to extracellular matrix components, or via their effects on pericytes.
MicroRNAs (miRNAs), which are short, single-stranded endogenous RNAs transcribed from noncoding genes, regulate gene expression post-transcriptionally by either cleaving or binding the 3′-untranslated region (3′ UTR) of mRNA to inhibit translation. Recently, miRNAs have been implicated in the regulation of a variety of tumor processes. For instance, miR-10b is an miRNA that is associated with metastasis and/or invasiveness in various cancer types, including breast carcinoma (14), pancreatic adenocarcinomas (15), esophageal cancer (16), hepatocellular carcinomas (17), glioblastomas (18), and neurofibromatosis type 1 tumorigenesis (19). Recently, miRNAs have emerged as important modulators of angiogenesis (20). Specific endothelial miRNAs have been implicated in response to angiogenic stimuli, growth factor stimulation, and hypoxia, suggesting that miRNAs may be an integral component of angiogenic signal transduction pathways (21). Additionally, dynamic changes in miRNA expression in response to treatment with anticancer drugs have been observed (22). Therefore, the identification of angiogenic miRNAs via expression profiling in cultured endothelial cells treated with angiogenic factors may be a useful strategy. However, the regulatory pathways controlled by miRNAs and the utility of therapeutic manipulation of miRNA expression to control vascular formation in human disease states have not yet been fully elucidated.
Previous work suggested that heparan sulfate chains on heparan sulfate proteoglycans can act as co-receptors for FGF2 to facilitate tumor cell growth (23). In addition, genetic evidence suggests that cell membrane heparan sulfate proteoglycans are required for tumor angiogenesis (24). These results led to the discovery that heparan sulfate mimetics, such as PI-88, are useful as anti-angiogenesis and anti-cancer therapeutics (25). We previously described a heparan sulfate mimetic, sulfated glycan, WSS25, which could disrupt angiogenesis and inhibit tumor growth in vivo (26). Interestingly, genome-wide miRNA screening indicated that WSS25 could modify miRNA expression, implicating this factor in angiogenesis.4 Therefore, we hypothesize that heparin inhibits angiogenesis and tumor growth through the disruption of miRNA function. In this study, we describe the roles of miR-10b and HoxD10 in angiogenesis, and the relationship of these factors to the actions of thrombin and heparin.
EXPERIMENTAL PROCEDURES
Cell Culture
HMEC-1 were cultured in MCDB131 (Invitrogen) medium containing 15% fetal bovine serum (FBS, Sijiqing Co. Ltd., Hangzhou, China), 2 mm l-glutamine, 10 ng/ml of epidermal growth factor (EGF, Shanghai PrimeGene Bio-Tech Co., Ltd., China), 100 units/ml of penicillin, and 100 μg/ml of streptomycin (Invitrogen) at 37 °C with 5% CO2. MDB-MA-231, HepG-2, Bel7402, and HEK293 cells were obtained from the Cell Bank in the Type Culture Collection Center of the Chinese Academy of Sciences. HepG2 and Bel7402 cells were maintained in RPMI1640 medium (HyClone) supplemented with 10% FBS. MDB-MA-231 and HEK293 cells were incubated in Dulbecco's modified Eagle's medium (HyClone).
Plasmid Constructs and Transfections
A fragment containing human miR-10b was PCR amplified from normal genomic DNA using the following primers: sense, 5′-CCCATTAGGCTACCTGAACTGTCT-3′, and antisense, 5-TCCAAGGTAATAAAACAGAACGAG-3′, and subcloned into the pll3.7 vector. The fragments of 3′ UTR sequences of various genes containing the miR-10b target sequences were also subcloned into the psiCHECK-2 plasmid using the following primers: HoxD10 sense, 5′-CCGCTCGAGCATATGGTCAGAGGCCAGGATTGGAG-3, and antisense, 5′-TATATGTCATTTTTAAAGTACTGGATG-3′; neuropilin 2 (NRP2) sense, 5′-CCGCTCGAGCATATGCCCAACTCACTGCTGATCCTATTA-3′, and antisense, 5′-CACAGTTTGACATTGTTGTTTATTTTT-3′; and nuclear receptor subfamily 4 group A member 3 (NR4A3) sense, 5′-CCGCTCGAGCATATGGGGGTTATAGTTCATGAGGGTTTT-3′, and antisense, 5′-ACCTTGAGTAACTCTTCACCCTTC-3′. The sequences of the resultant vectors were confirmed by DNA sequencing. Anti-miR-10b and siRNA directed against HoxD10 were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). siRNA directed against thrombin were obtained from Guangzhou RiboBio Co., Ltd., China. The sequence of 2′-O-methyl anti-miR-10b was 5′-CACAAAUUCGGUUCUACAGGGUA-3′. Scrambled 2′-O-methyl modified RNA (5′-CAGUACUUUUGUGUAGUACAA-3′) was used as a negative control. The most effective siRNA sequence for HoxD10 knock down was 5′-CGAAUGAAACUCAAGAAGATT-3′. Transfections were carried out using Lipofectamine 2000 (Invitrogen) and pools of stable transfectants were selected using 0.2 μg/ml of puromycin according to a modification of the protocol described previously (27).
Site-directed Mutagenesis
3′ UTR of HoxD10 mutation was generated by site-directed mutagenesis using the overlap extension PCR method. Briefly, PCR amplification of two oligonucleotides were performed first using the following primers: forward, CAGGAGGACGCTCAGATGAA and mutant reverse, AGTCTGGGTGCATTACTTTGAAAAAATAATAAATTACACGTGC; reverse, GCGAGGTCCGAAGACTCATT and mutant forward, ATTATTTTTTCAAAGTAATGCACCCAGACTATTATTGCGCATT. PCR amplification conditions were denaturation for 30 s at 94 °C, annealing for 30 s at 60 °C, and extension for 45 s at 72 °C. PCR products were precipitated by ethanol and resuspended in 10–20 μl of water. Those fragments were then extended using forward and reverse primers. The products were subcloned into psiCHECK-2 after the fragments were digested by XhoI and SacI. The mutants were verified by DNA sequencing. The results indicated that the direct binding region of miR-10b, TCGTAATGCAGGGTAAC, had been changed to AAGTAATGCACCCAGAC.
RNA Isolation, RT-PCR, and Quantitative Real-time PCR
Following transfection or reagent treatment, total RNA was extracted from frozen primary tumors and/or cell lines using TRIzol (Invitrogen). RNA (2.0 μg) was used to synthesize cDNA via Moloney murine leukemia virus reverse transcriptase (TaKaRa, Japan) according to the manufacturer's instructions. Detection of mature miRNAs was performed using the miRNA Primer (Guangzhou RiboBio Co., Ltd., China) according to the manufacturer's instructions. Semi-quantitative RT-PCR was performed to evaluate HoxD10 and Twist mRNA levels as previously described (14). The primers used were: HoxD10, 5′-ATAAGCGCAACAAACTCATTTCG-3′ (sense) and 5′-CCTTCGGGGCTATTATTGTACTC-3′ (antisense); Twist, 5′-GTCCGCAGTCTTACGAGGAG-3′ (sense) and 5′-GCTTGAGGGTCTGAATCTTGCT-3′ (antisense); thrombin, 5′-CTTGTGAGACAGCGAGGAC-3′ (sense) and 5′-AGGATGGGTAGTGGAGTTGA-3′ (antisense). PCR was conducted using the following conditions: denaturation for 30 s at 94 °C, annealing for 30 s at 60 °C, and extension for 30 s at 72 °C. Quantitative real-time PCRs were performed using an Applied Biosystems 7500 Fast Real-time PCR system with SYBR Green Premix Ex Taq kit (TaKaRa). Real-time PCR cycle conditions included the following steps: denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 20 s, and extension at 72 °C for 25 s. Each sample was run in triplicate and Ct was determined for the target transcripts. Twist and HoxD10 levels were normalized using the ΔΔCt method.
Western Blotting Analysis
Proteins were isolated from cells treated under different conditions as described previously (26). After separation by electrophoresis using 10% SDS-PAGE gels, proteins were transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with Tris-buffered saline (TBS) plus 5% nonfat dry milk and 0.1% Tween 20 before incubation with antibody against β-actin (Sigma), HoxD10 (Santa Cruz), Twist (Santa Cruz), and thrombin (Boster Biological Technology, Ltd., Wuhan, China) at 4 °C overnight. After incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) for 1 h, ECL Western blot substrate (Pierce) was used for the detection.
Quartz Crystal Microbalance (QCM) Analysis
QCM analysis was performed as previously described (26). The biosensor experiments were carried out using an Attana A100 QCM instrument (Attana AB, Stockholm, Sweden). Thrombin (200 μg/ml, 50 μl) (Sigma) dissolved in running buffer was injected on the heparin biosensor and streptavidin surfaces (as a reference), respectively. A continuous flow (25 μl/min) of running buffer was used throughout the experiment. The frequency responses produced from the interactions were monitored by frequency logging with Attester 1.1, with mass changes of bound or released ligands recorded as the resulting frequency shifts (Δf).
Dual Luciferase Reporter Assays
Luciferase reporter assays were performed using the psiCHECK2–3′UTR vector. Cells were grown to ∼70% confluence in 48-well plates and co-transfected with psiCHECK2–3′UTR plus pll3.7-miR-10b or empty vector as described above and previously (28). Cells were incubated with a transfection reagent-DNA complex for 36 h followed by luciferase reporter assay using the Dual Luciferase Assay System (Promega). Renilla luciferase activity was normalized to firefly luciferase activity. Cell lysates were subjected to luciferase activity measurement according to the manufacturer's instructions.
Scratch Wound Healing Assay
The scratch wound migration assay was performed as previously described (29). Briefly, HMEC-1 cells (5 × 105) were cultured in 6-well plates for 24 h. Confluent cell monolayers were scraped with a yellow pipette tip to generate a wound and rinsed twice with growth medium. The cells were photographed immediately after the scratch (t = 0 h) and 24 h later (t = 24 h) with an Olympus IX51 digital camera microscope (Olympus). The width of the wound area was measured and calculated by Image J to determine cell migration distance: relative migration rate = (distancet=0 h − Distancet=24 h)/Distancet=0 h × 100%.
Tube Formation Assay
Endothelial cell tube formation was assessed using a modification of previous methods (12). Briefly, HMEC-1 cells were transfected with miR-10b vector or anti-miR-10b siRNA. After 24 h, 3 × 104 cells were plated on 96-well plates coated with 50 μl of Matrigel (BD Biosciences) and incubated for 8 h with or without heparin. Cells were photographed using the Olympus digital camera. Five randomly selected fields of view were photographed in each well, and averaged to analyze for total capillary structure length using Image J software.
Cell Proliferation Assay
To examine the cell proliferation ability, cells were seeded at the density of 2,000/well and cultured with or without heparin at concentrations of 100 and 200 ng/ml. Cell growth status at each time point was evaluated with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Briefly, 3-(4,5-methylthiazol-2-yl)-2,5-diphenyltetrazolium bromide was added (100 μg/well) to each well of the 96-well plate and incubated in 37 °C for 4 h. Formazan products were solubilized with DMSO, and the optical density was measured at 490 nm.
Implanted Matrigel Plug Model
A modified method was employed to determine whether miR-10b directly impacts the ability of HMEC-1 to form vessels in vivo (27, 30). Briefly, 1 × 106 HMEC-1 cells (as control) or miR-10b-transfected HMEC-1 cells (100 μl) were mixed with 400 μl Matrigel, and subcutaneously injected into the midventral abdominal region of 4–6-week-old nude mice. After 12 days, mice were sacrificed. The implants were retrieved, photographed, fixed in 4% buffered formaldehyde for histologic analysis, and probed with anti-human CD34 antibody (Boster Biological Technology, Ltd., China). Vessels were defined as those structures possessing a patent lumen and positive endothelial nuclei (30).
Tumor Xenograft Growth Assay
All animal experiments were approved by the Institutional Animal Care and Use Committee and Local Ethical Board. 1 × 107 MDA-MB-231 cells were subcutaneously injected into the mammary fat pads of 4–6-week-old nude female mice. Tumor volume was determined according to the equation: V = (L × W2) × 0.5, where V is volume, L is length, and W is width. When tumor volume reached around 100 mm3, the mice were randomly assigned into control and treatment groups. The vehicle (normal saline) or 20 mg/kg of heparin was administrated subcutaneously every other day. At the 22nd day after injection, mice were sacrificed and tumors were harvested, weighed, and photographed. Half of the tumor tissue was fixed in 4% neutral buffered formaldehyde for immunohistochemistry, and the rest was lysed for mRNA and protein detection.
Immunohistochemistry
Tumor tissues were excised, fixed in 4% neutral paraformaldehyde, embedded in paraffin, and sectioned for immunohistochemical analysis as previously described (26). Identification of endothelial cells was performed by immunostaining using a monoclonal antibody against CD34 at a 1:100 dilution at 4 °C overnight. For HoxD10 expression detection in vivo, the sections were stained using anti-HoxD10 antibody at 1:50 dilution at 4 °C overnight. To evaluate the protein expression, semi-quantitative image analysis on the immunohistochemical section was employed to measure the integrated optical density using Image Pro Plus software (Media Cybernetics, Silver Spring, MD).
Statistical Analysis
Results are described as mean ± S.D. In all experiments, statistical analysis was performed using Student's t test and p < 0.05 was considered statistically significant (*, p < 0.05; **, p < 0.01).
RESULTS
Heparin Down-regulates miR-10b Expression in Vitro
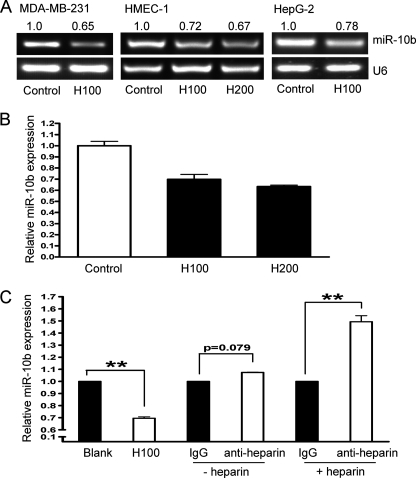
Heparin inhibits metastasis (31, 32), likely through the modulation of genes involved in the metastatic process, such as P-selectin, L-selectin, chemokine (C-X-C motif) ligand 12 (CXCL12), CXC chemokine receptor 4 (CXCR4), and heparanase. Recent studies have demonstrated that miRNAs have a role in cancer metastasis (14, 33, 34). In fact, miR-10b was highly expressed in metastatic breast cancers (14) and malignant glioma (18), and promoted cell migration and invasion. Therefore, heparin-dependent inhibition of metastasis might be linked to miR-10b. Indeed, we found that heparin treatment (100 ng/ml) greatly reduced miR-10b levels in MDA-MB-231 breast cancer cells. In addition, expression of miR-10b was also impaired by heparin in HepG-2, Bel7402, HMEC-1, and HEK293 cells (Fig. 1A and supplemental Fig. S1). To confirm this down-regulation effect, quantitative real-time PCR was used to determine the miR-10b transcription after heparin treatment in HMEC-1 (Fig. 1B). This effect was confirmed with anti-heparin antibody (Millipore) treatment, which reversed heparin-induced miR-10b inhibition (Fig. 1C). These results suggest that heparin can inhibit miR-10b expression in multiple cell types.
FIGURE 1.
Heparin inhibits miR-10b expression. A, breast cancer MDA-MB-231, human hepatoma HepG2, and HMEC-1 cells were treated with 100 (H100) or 200 ng/ml (H200) of heparin for 24 h. RT-PCR analysis of miR-10b expression was performed using U6 as control. Representative results of three experiments were shown. Normalized band densities were shown above the figures. B, HMEC-1 cells were treated with 100 (H100) or 200 ng/ml (H200) of heparin for 24 h. Quantitative real-time PCR was used to determine the miR-10b expression using U6 as a control. C, HMEC-1 cells were treated with or without 4 μg/ml of anti-heparin antibody or IgG (Abcam Inc.) as control for 1 h, and then cultured in the presence or absence of 100 ng/ml of heparin for 18 h. miR-10b expression was measured by RT-PCR in each condition and normalized using U6 as control. **, p < 0.01.
miR-10b Promotes HMEC-1 Cell Migration, Tube Formation, and Angiogenesis
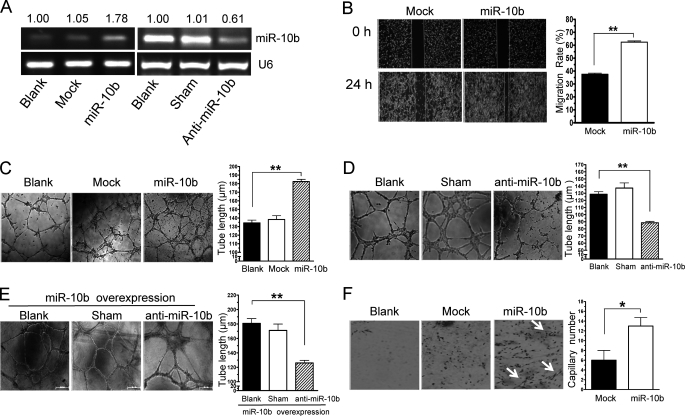
Endothelial cell migration and metastasis are essential for angiogenesis. Therefore, we sought to determine the effect of heparin-mediated inhibition of miR-10b expression on angiogenesis. A vector expressing miR-10b and green fluorescent protein (GFP) was used to stably transfect HMEC-1 cells, and miR-10b expression levels were evaluated by RT-PCR. We found that miR-10b was highly expressed in HMEC-1 cells stably transfected with miR-10b (Fig. 2A). Although cell growth was not influenced by miR-10b overexpression (supplemental Fig. S2), wound healing assays showed that HMEC-1 cell migration was increased significantly by miR-10b overexpression (Fig. 2B). Then we further evaluated the impact of miR-10b expression on angiogenesis by assessing HMEC-1 cell tube formation on Matrigel (26). The length of capillary networks formed by HMEC-1 cells stably transfected with miR-10b was greater than that of control cells (Fig. 2C). To confirm this data, 2-O-methyl-modified oligo RNA (anti-miR-10b), which is complimentary and can anneal to miR-10b, was employed. miR-10b expression was nearly abolished by anti-miR-10b in HMEC-1 cells (Fig. 2A), whereas migration and tube formation were obviously impaired after anti-miR-10b treatment in these cells (Fig. 2D and supplemental Fig. S3). To confirm that the tube formation and angiogenesis inducing phenotype were caused by overexpression of miR-10b, a genetic gain-loss-of-function experiment was performed. Down-regulating miR-10b by anti-miR-10b in the miR-10b-overexpressed HMEC-1 cells completely reversed the promotion on tube formation by miR-10b to the background level (Fig. 2, C and E, and supplemental Fig. S4). To test whether miR-10b overexpression would induce angiogenesis in vivo, we mixed untransfected cells (as control) or miR-10b-transfected HMEC-1 cells with Matrigel and subcutaneously injected these mixtures into nude mice. After 12 days Matrigel plugs were harvested followed by the immunostaining test. We observed that control HMEC-1 cells formed complete functional vessels containing red blood cells, as determined by staining probed with human-specific CD34 (Fig. 2F). In stark contrast, HMEC-1 cells overexpressing miR-10b formed more microvessels. Hence, miR-10b might induce angiogenesis in vitro and in vivo.
FIGURE 2.
HMEC-1 cell migration, tube formation, and angiogenesis are induced by miR-10b. A, miR-10b expression was measured by RT-PCR in HMEC-1 cells, mock transfected cells, and miR-10b stably transfected HMEC-1 cells (miR-10b), and transfected without or with scrambled 2-O-methyl-modified RNA (sham), or 2-O-methyl-modified oligo RNA against miR-10b (anti-miR-10b) (right). Normalized band densities are shown above the figures. B, HMEC-1 cells were stably transfected with mock transfected (control) or miR-10b (miR-10b). Confluent monolayers were scraped to generate a wound (t = 0 h), and photographed 24 h later (t = 24 h). Overexpression of miR-10b resulted in a significant increase in cell migration. The widths of the scratches were quantified (right). C, HMEC-1 cells (blank) transfected without or with miR-10b were seeded onto Matrigel-coated plates and tube formations were assessed. D, HMEC-1 cells (Blank) were treated with vehicle (sham) or transfected with 100 nm anti-miR-10b followed by tube formation assay. E, miR-10b stable overexpressed HMEC-1 cells were transfected without (blank) or with vehicle (sham), or anti-miR-10b, and then tube formation was analyzed. The representative photomicrographs (×40) and statistical analysis of tube length are shown in C–E (right), respectively. F, Matrigel plug assays were performed using Matrigel alone (blank), HMEC-1 cells transfected with control vector, or stably transfected with miR-10b (miR-10b) and assayed as described under “Experimental Procedures.” Plug sections were evaluated for CD34-positive vessels (arrows). Quantization of mean capillary number in implanted Matrigel plugs were shown (right). *, p < 0.05; **, p < 0.01.
HoxD10 Is a Functional Target of miR-10b in HMEC-1 Cells
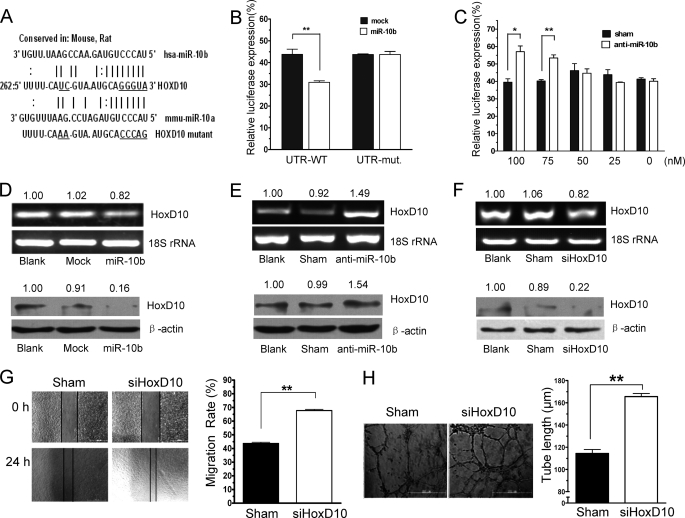
To determine the mechanisms by which miR-10b induces angiogenesis, we evaluated several potential targets of miR-10b computationally predicted using public algorithms. Among the predicted miR-10b targets, priority was given to pro- and anti-angiogenic proteins. HoxD10, NRP2, and NR4A3 (Fig. 3A and supplemental Fig. S5) were predicted to be miR-10b targets by both Miranda and Target Scan (35). To determine whether miR-10b directly bound to the 3′ UTRs of the mRNA, we constructed luciferase reporter vectors that encoded the complete 3′ UTR of each gene, respectively. The vector were then co-transfected with miR-10b into HEK293 cells. We found that miR-10b reduced the activity of a luciferase reporter gene fused to the wild-type 3′ UTR of HoxD10 (Fig. 3B), but had no effect on the 3′ UTR of NRP2 or NR4A3 (supplemental Fig. S5). To test the binding specificity of miR-10b, specific base pair mutations on the 3′ UTR of HoxD10 mRNA was performed using the site-directed mutagenesis method followed by co-transfection with miR-10b. As expected, miR-10b had no effect on the luciferase activity of the reporter, which contained the mutated 3′ UTR of HoxD10. This suggested that miR-10b specifically targeted the 3′ UTR of HoxD10 mRNA (Fig. 3B). In addition, we observed significant expression of the luciferase reporter gene fused to the 3′ UTR of HoxD10 after anti-miR-10b treatment (Fig. 3C). To determine whether miR-10b could disrupt endogenous HoxD10 expression in endothelial cells, cell lysates from miR-10b-transfected or vector control HMEC-1 cells were analyzed. Although miR-10b slightly inhibited mRNA expression of HoxD10 in HMEC-1 cells, it might impair HoxD10 expression through translational inhibition. Indeed, expression of the HoxD10 protein was significantly reduced in cells that overexpressed miR-10b (Fig. 3D). This result was in agreement with the data published by Ma et al. (14). They claimed that miR-10b did not cause degradation of HoxD10, but reduced protein expression. Furthermore, HoxD10 mRNA and protein expression were increased after anti-miR-10b treatment (Fig. 3E). It was reported that HoxD10 represses expression of genes that are involved in cell migration and angiogenesis (27). We next ascertained whether reduction of HoxD10 levels might provide an explanation for the induction of cell migration and angiogenesis observed following miR-10b overexpression. HMEC-1 cells (3 × 105) were transfected with siRNA directed against HoxD10 or control, scrambled siRNA, and assessed using a wound healing assay. A marked reduction in HoxD10 expression was observed in HMEC-1 cells after siRNA transfection compared with scrambled siRNA transfection and control (Fig. 3F). Cell migration rates were significantly increased after HoxD10 knockdown (Fig. 3G). In addition, tube formation was also increased after HoxD10 knockdown in HMEC-1 cells (Fig. 3H). These results suggest that HoxD10 is indeed an important functional target of miR-10b in HMEC-1.
FIGURE 3.
HoxD10 is a functional target of miR-10b in HMEC-1 cells. A, alignment of potential miR-10b-binding sites in the 3′ UTR of the HoxD10 mRNA of different species. The schematic shows the sequences of mature miR-10b and the miR-10b seed region. The seed sequence of mature miR-10b is underlined. B, HEK293 cells were transfected with miR-10b or vehicle plus with psicheck2-HoxD10-3′UTR or mutant 3′ UTR reporter gene. Luciferase activities were measured after the transfection for 36 h. C, HEK293 cells were co-transfected with psicheck2-HoxD10–3′UTR and anti-miR-10b at concentrations of 0, 25, 50, 75, or 100 nm for 36 h, followed by luciferase assay. D, HMEC-1 cells were either transfected without (Blank) or with vector control (Mock) or miR-10b for 36 h. The expression of HoxD10 was measured by RT-PCR and Western blot analysis using 18S rRNA and β-actin as loading control, respectively. E, HMEC-1 cells were either left untransfected (Blank) or were transfected with modified RNA control (Sham) or anti-miR-10b. HoxD10 expression was analyzed 36 h later via RT-PCR and Western blot. F, HMEC-1 cells were transfected with sham control or siRNA of HoxD10. HoxD10 expression was analyzed 36 h later via RT-PCR and Western blot. Normalized band densities are indicated above the figures in D–F. G, migration assay was performed 24 h later. Tube formation (H) was also measured as described under “Experimental Procedures” after the siRNA of HoxD10 transfection as in F. Representative photomicrographs (×100) and statistical analysis of tube length are shown (right). *, p < 0.05; **, p < 0.01.
Heparin Inhibits HMEC-1 Cells Migration and Tube Formation and Induces HoxD10 Expression
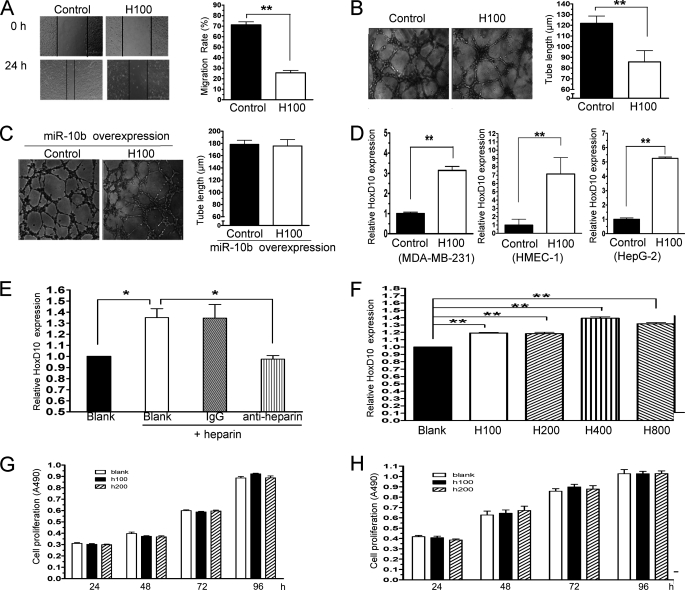
The above results suggested that heparin treatment reduced miR-10b expression, whereas miR-10b might induce angiogenesis in vitro and in vivo. To understand whether heparin inhibits HMEC-1 migration, tube formation, and via the down-regulation of miR-10b, HMEC-1 cells were incubated with or without heparin (100 ng/ml), followed by cell migration and tube formation assays. We found that heparin inhibited cell migration (Fig. 4A) and disrupted tube formation (Fig. 4B). Moreover, overexpression of miR-10b rescue these responses from inhibition by heparin (Fig. 4C). Heparin treatment reduced miR-10b expression, whereas miR-10b down-regulated HoxD10 expression. Therefore, HoxD10 expression might be induced by heparin. To test this hypothesis, MDB-MA-231, HepG2, and HMEC-1 cells were treated with 100 ng/ml of heparin for 24 h followed by mRNA expression of HoxD10 by real-time PCR. Indeed, HoxD10 mRNA expression increased significantly after heparin treatment (Fig. 4D). However, treatment with an anti-heparin antibody reduced HoxD10 expression levels in HMEC-1 (Fig. 4E). Furthermore, HoxD10 protein expression was also up-regulated by heparin under different concentrations (Fig. 4F). To exclude the possibility that inhibition of cell migration and tube formation are due to the cell growth arresting, HMEC-1 cells were treated with heparin for 24, 48, 72, and 96 h, respectively, followed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. The results showed heparin could not arrest HMEC-1 growth at least at concentrations of 100 or 200 ng/ml (Fig. 4G). In addition, heparin also could not inhibit MDA-MB-231 growth under these conditions (Fig. 4H).
FIGURE 4.
Heparin arrests cell migration and tube formation and augments HoxD10 expression in vitro. A, HMEC-1 cells were incubated with or without 100 ng/ml of heparin (H100) and wound healing assays were performed. Width alteration of the scratch after wound healing is quantified (right). B, HMEC-1 cells were cultured in the presence or absence of 100 ng/ml of heparin (H100) followed by tube formation assay. Statistical analysis of tube length was shown (right). C, miR-10b stable overexpressed HMEC-1 cells were treated without (Control) or with 100 ng/ml of heparin (H100) followed by tube formation analysis. D, MDA-MB-231, HMEC-1, and HepG-2 cells were treated with 100 ng/ml of heparin (H100) for 24 h. mRNA expression of HoxD10 was measured by real-time PCR. E, anti-heparin antibody reversed the effect of heparin on HoxD10 expression. HMEC-1 cells were treated with heparin and anti-heparin antibody or control IgG, followed by RT-PCR analysis of HoxD10 expression using 18S rRNA as control. Normalized band densities are shown. F, HoxD10 protein expression is augmented by heparin. HMEC-1 cells were treated with 100 (H100), 200 (H200), 400 (H400), or 800 (H800) ng/ml of heparin for 24 h followed by Western blot detection. Band intensities were normalized to a loading control. Each of the duplicate determinations are shown. Cells proliferations were tested in the presence of heparin. HMEC-1 (G) and MDA-MB-231 (H) were treated without or with 100 (H100) and 200 (H200) ng/ml of heparin for 24, 48, 72, and 96 h. Cells proliferations were evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. *, p < 0.05; **, p < 0.01.
Heparin Impairs Angiogenesis and Attenuates the Growth of Xenografted Breast Cancer Cells in Vivo
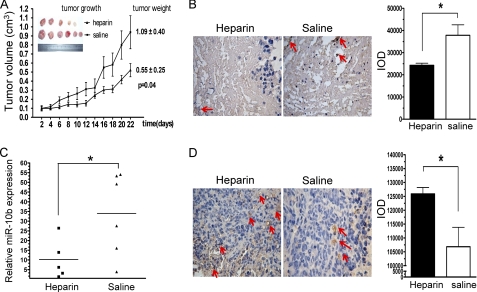
Molecules that prevent angiogenesis can efficiently hinder tumor growth. We found that heparin treatment inhibited miR-10b expression and stimulated HoxD10 expression in HMEC-1 cells, leading to a reduction in cell migration and tube formation. To determine whether heparin utilizes these anti-angiogenic mechanisms in vivo, nude mice were inoculated with MDA-MB-231 cells. After the xenografts became visible and attained a diameter of 100 mm3, heparin (20 mg/kg/days) or saline were administered subcutaneously every other day and tumor size was measured. Heparin treatment significantly inhibited the growth of MDA-MB-231 xenografts (Fig. 5A). There was no significant change in mouse body weight in this experiment (data not shown). To determine whether heparin could affect angiogenesis in primary tumors, average microvessel density was measured via immunohistochemical staining for CD34. We found fewer blood vessels in tumors of the heparin-treated group compared with controls (Fig. 5B). These results confirm previous reports that heparin can inhibit angiogenesis in vivo (36, 37). Quantitative real-time PCR analysis of tumor tissues revealed that heparin treatment significantly reduced levels of miR-10b expression when compared with tumors from control animals (Fig. 5C). In addition, we found that HoxD10 expression levels were significantly higher in tissues from the heparin-treated group compared with control group (Fig. 5D). Taken together, these observations indicated that heparin could disrupt tumor angiogenesis in vivo. The molecular mechanisms by which heparin may interfere with angiogenesis has been an area of great focus (38–40). Our results showed that inhibition of miR-10b expression and up-regulation of HoxD10 expression were implicated in this process.
FIGURE 5.
Heparin inhibits tumor angiogenesis in vivo and reduces miR-10b expression. A, MDA-MB-231 cells were subcutaneously injected into the mammary fat pads of nude female mice. After the tumor volume grew to 100 mm3, mice were injected subcutaneously every other day with either 20 mg/kg of heparin (Δ, 5 mice/group) or saline (■, 6 mice/group). Mice were sacrificed after 22 days of treatment. Each data point represents the mean ± S.D. of 5 or 6 mice. Primary tumors removed from mice are shown on the left. B, immunohistochemical analysis was performed on tumor sections probed with CD34 antibody. The left panel shows representative tissue sections stained for CD34, whereas the right panel demonstrates mean integrated optical density of staining intensity for CD34 protein expression. Arrows indicate CD34-positive vessels. Compared with animals treated with saline, there was a significant reduction in the number CD34 expressing cells in the tumors of animals treated with heparin. C, miR-10b expression in xenografts was measured by real-time PCR and normalized to U6 expression. miR-10b expression in tumor tissue was significantly inhibited by heparin (■) compared with saline (▴) treatment. D, primary mammary xenografts were stained with anti-HoxD10 antibody. Representative tissue sections stained for HoxD10 and mean integrated optical density for HoxD10 protein expression (right) are shown. Arrows indicate HoxD10-positive staining. *, p < 0.05; **, p < 0.01.
Heparin Impedes miR-10b Expression through Thrombin
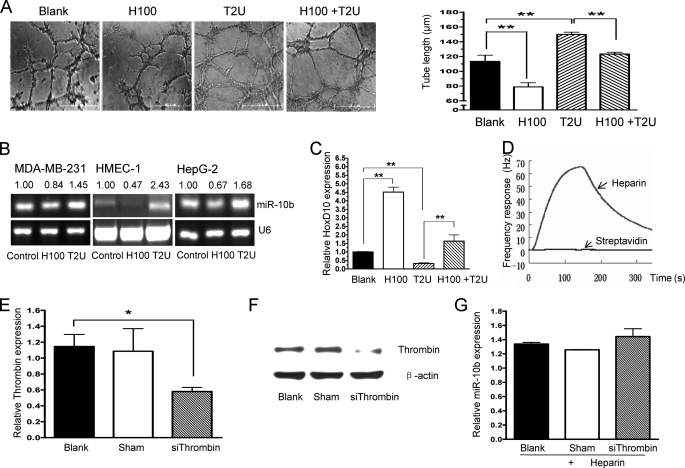
Heparin may interact with a variety of vascular growth factors released from the endothelium and/or tumor cells to inhibit angiogenesis. To date, anti-coagulant therapy is the main clinical application of heparins. However, the anti-tumor effect of heparin also appears to be related to its anti-coagulant activity (8, 36, 41). Thrombin is the primary heparin-response factor in the coagulant system. Moreover, thrombin is a potent stimulator of tumor growth, metastasis, and angiogenesis. To determine whether the anti-angiogenesis activities of heparin are linked to its anti-coagulant effect, the relationships between thrombin, miR-10b, and HoxD10 function were explored. We found that thrombin has a significant stimulatory effect on HMEC-1 cell tube formation, however, heparin could negate the impact of thrombin on the induction of the cell tube formation (Fig. 6A). We assessed levels of miR-10b and HoxD10 expression after thrombin treatment using real-time PCR. As shown in Fig. 6B, miR-10b expression was induced by thrombin in HMEC-1, MDA-MB-231, and HepG2 cells. Furthermore, HoxD10 expression levels were reduced by thrombin treatment (Fig. 6C). When cells were treated with both heparin and thrombin, however, no reduction in HoxD10 expression was observed (Fig. 6C). Although the thrombin-heparin interaction has been characterized and supported by crystal structure analysis (42), we confirmed that heparin strongly bound thrombin by QCM analysis (Fig. 6D). To confirm the effects of heparin on miR-10b through thrombin, leading to inhibition of thrombin-mediated responses in HMEC-1 cells, thrombin was silenced using its siRNA followed by heparin treatment. Indeed, both mRNA and protein expression of thrombin were reduced after siRNA transfection in HMEC-1 cells (Fig. 6, E and F). After being transfected with siRNA of thrombin or vehicle, HMEC-1 cells were treated with 100 ng/ml (H100) of heparin for 24 h followed by miR-10b expression measurement using quantitative real-time PCR. As shown in Fig. 6G, heparin did not inhibit miR-10b expression after the knockdown of thrombin in HMEC-1 cells. These results support that thrombin plays an important role in the effects of heparin on miR-10b function.
FIGURE 6.
Heparin interferes with thrombin to impede miR-10b expression and angiogenesis. A, HMEC-1 cells were cultured without (Blank) or with 100 ng/ml of heparin (H100), 2 units/ml of thrombin (T2U), or both heparin and thrombin (H100 + T2U), followed by cell tube formation assays. The statistical analysis of tube length is shown (right). B, thrombin treatment increased miR-10b expression. MDA-MB-231, HMEC-1, and HepG2 cells were cultured in the absence (control) or presence of 2 units/ml of thrombin (T2U) or 100 ng/ml of heparin (H100), and miR-10b expression was assayed by RT-PCR. Normalized band densities are shown above the figures. C, HMEC-1 cells were treated without (Blank) or with H100, T2U, or H100 + T2U for 24 h. HoxD10 expression was detected by real-time PCR. D, heparin binds thrombin, as assayed by QCM. The heparin biosensor surface was employed to measure the carbohydrate-protein interactions. Thrombin (200 μg/ml, 50 μl) was injected directly onto to the streptavidin surface as a reference, as well as the heparin biosensor surface, and the resulting frequency shifts (Δf) were recorded. E, HMEC-1 cells were transfected without (Blank) or with sham control or siRNA of thrombin for 36 h followed by thrombin expression detection using quantitative real-time PCR. 18S rRNA was employed as a control. Data are mean ± S.D. F, HMEC-1 cells were treated as in E followed by thrombin protein detection using Western blot analysis. β-Actin was used as a loading control. G, after transfection with siRNA of thrombin or vehicle for 36 h, HMEC-1 cells were treated with 100 ng/ml (H100) of heparin for 24 h. Then quantitative real-time PCR was used to determine the miR-10b expression. U6 was used as a control. *, p < 0.05; **, p < 0.01.
Twist Is a Transcription Factor of miR-10b, Promotes Angiogenesis, and Is Regulated by Thrombin and Heparin
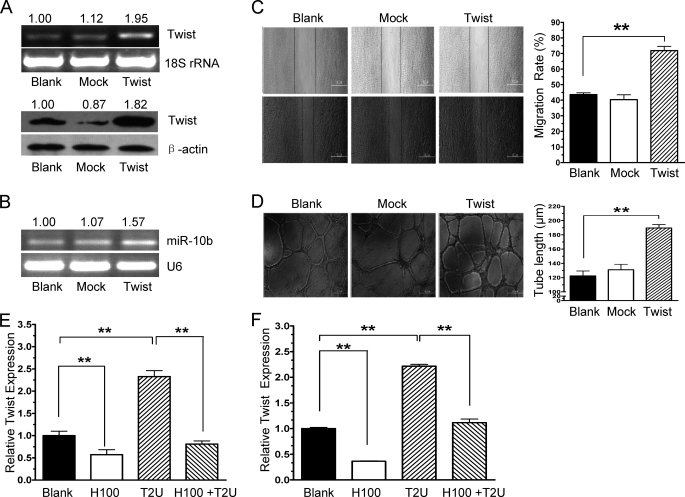
Twist is a transcription factor and specifically binds to the miR-10b promoter to activate expression in breast cancer (14). To further understand how miR-10b function is mediated by heparin and thrombin, the relationship between miR-10b, Twist, and their function under heparin and thrombin treatments in HMEC-1 cells was explored. Indeed, overexpression of Twist (Fig. 7A) up-regulate miR-10 expression in HMEC-1 cells (Fig. 7B). In accord with a previous report, the migration and tube formation were significantly induced by Twist (Fig. 7, C and D) (43). Interestingly, the expression of Twist was augmented by thrombin and attenuated by heparin at mRNA (Fig. 7E) and protein levels (Fig. 7F).
FIGURE 7.
Twist, a transcription factor of miR-10b, promotes angiogenesis and is regulated by thrombin and heparin. A, HMEC-1 cells were transfected without (Blank) or with vector control (Mock) or pcmv-Twist (TWIST) followed by Twist mRNA (top panel) or protein detection (bottom panel). Normalized band densities are shown above the figures. B, HMEC-1 cells were treated as in A. The expression of miR-10b was measured by RT-PCR. Normalized band densities are shown above the figures. C, cell migration was induced by overexpression of Twist in vitro. HMEC-1 cells were transfected without (Blank) or with Mock or Twist as described in A followed by wound healing assay. The widths of the scratches were quantified. D, cell tube formation was induced by overexpression of Twist in vitro. HMEC-1 cells (3 × 104) that were transfected without or with a control vector (Mock) or pcmv-Twist (TWIST) were seeded onto Matrigel-coated plates followed by tube formation assay. The statistical analysis of tube length is shown on the right. E and F, HMEC-1 cells were treated with 100 ng/ml of heparin (H100), no factors (control), 2 units/ml of thrombin (T2U) or heparin and thrombin (H100 + T2U) for 24 h, followed by real-time PCR analysis and Western blot of Twist expression. Heparin treatment inhibited Twist expression, whereas thrombin treatment increased Twist expression at mRNA level (E) and protein level (F). Band intensities normalized to β-actin for each of the duplicate determinations are shown. **, p < 0.01.
DISCUSSION
Heparins have been validated as antithrombotic agents and used in the prevention and treatment of thromboembolic diseases in cancer patients. Heparin inhibits angiogenesis, whereas thrombin induces angiogenesis. However, the mechanisms by which heparin and thrombin mediate their effects on angiogenesis remain uncertain. Previous work suggests that the antiangiogenic effects of heparin are unrelated to its anticoagulant actions (37, 44, 45). However, this is still in dispute. Our results argue that heparin and thrombin play conflicting roles in angiogenesis through their regulation of Twist, a transcription factor of miR-10b to further influence the expression of miR-10b, whereas miR-10b induces angiogenesis by decreasing the levels of a functional target, Hoxd10, which impairs angiogenesis. Indeed, we find that thrombin treatment increases Twist expression levels and leads to increased expression of miR-10b and decreased expression of HoxD10 in HMEC-1 cells. In addition, we demonstrate that heparin interferes with thrombin to reverse the stimulatory effect of thrombin on angiogenesis (Fig. 6, D and G). These results provide a new mechanism by which heparin and thrombin interact during angiogenesis. However, as shown in Fig. 1A, miR-10b is expressed endogenously in those cells. The effects of heparin on miR-10b are modest. This suggests that other possible mechanisms may also be involved in this process.
Although we focused on the role of heparin in the regulation of miR-10b, Twist, and HoxD10 expression in HMEC-1 cells and xenografted breast cancer cells, the phenomenon demonstrated here likely has wider relevance. First, thrombin was highly expressed in HMEC-1, MDA-MB-231, and HepG2 cells (supplemental Fig. S6). Furthermore, we observed miR-10b expression inhibition by heparin and its expression up-regulation by thrombin not only in HMEC-1 cells, but also in Bel7402, HepG2, and MDA-MB-231 cancer cells, and HEK293 cells (Figs. 1A and 6B and supplemental Fig. S1). Second, Twist is induced by thrombin in human umbilical vascular endothelial cells, human prostate DU145, breast MCF7, murine melanoma B16F10, and undifferentiated mouse UMCL cells (43). Third, miR-10b has previously been shown to be induced by Twist and inhibit translation of HoxD10 both in vitro and in vivo (14, 46). In addition, Twist is required for thrombin-induced tumor angiogenesis and growth, and tumor invasion and metastasis can be triggered by miR-10b (18, 46, 47). Indeed, angiogenesis is induced by miR-10b (Fig. 2F). Moreover miR-10b levels are higher in cancer patient serum (48). Thrombin, which is frequently present in excess in cancer patient serum (49), also increases the miR-10b expression level. However, the anti-angiogenic effect of heparin on HMEC-1 cells was reversed after stable transfection with miR-10b (Fig. 4C). This suggests that the effect of heparin on miR-10b expression occurs through an indirect pathway. In fact, our QCM binding experiment results show that heparin does not bind to miR-10b (data not shown).
In the experiments, anti-heparin antibody was employed to eliminate the effect of heparin. As shown in Figs. 1C and 4E, anti-heparin antibody not only blocked heparin to reverse the miR-10b expression inhibition induced by heparin, but also took the role of others in play. The antibody has been characterized previously to interact with both heparin and cell surface heparan sulfates, whereas heparan sulfates may also contribute to the antithrombotic properties (50). Thrombin activity in plasma is inhibited primarily by antithrombin III, which is accelerated by heparin and some similar glucosaminoglycans that are covalently linked to a core protein to form heparan sulfate proteoglycans (51–53). In addition, this antibody may cause releasing of thrombin from the extracellular matrix (54). Theoretically, the releasing thrombin may induce miR-10b expression. Indeed, it has been shown that this antibody could promote a procoagulant state by the blockade of heparan sulfate binding to antithrombin III, inhibiting the accelerated formation of thrombin-antithrombin III complexes (55).
In this study, the expression levels of miR-10b were decreased by heparin in solid tumors of mice (Fig. 5C). We speculated that one reason was that heparin might interfere with thrombin (Fig. 6C) in blood and blocked miR-10b function induced by thrombin in angiogenesis, invasion, and metastasis at the early stage of tumor development when tumor cells were in the blood circulation system. However, as heparins enter circulation, they also bind to, modify, and release a multitude of circulating, cell-bound or extracellular matrix-bound pro- and anti-angiogenic factors, growth factors, enzymes, proteins, and receptors that may influence the induction and progression of angiogenesis (36, 37). The molecular mechanisms involved in modulating angiogenesis by heparins are complex and not fully understood. This study just provides a new insight into the molecular mechanism by which heparin and thrombin regulate angiogenesis.
The significance of miRNAs in the regulation of angiogenesis was first investigated in mice mutant for Dicer and Drosha, two key enzymes in miRNA biogenesis. Dicer mutant mice die between embryonic day 12.5 and 14.5, exhibiting impaired blood vessel and yolk sac formation (56). Profiling of miRNAs highly expressed in endothelial cells showed that miR-10b is significantly down-regulated by Drosha and Dicer siRNA (21, 57). Furthermore, the miR-17–92 cluster, miR-126, miR-378, and miR-296 have been shown to promote tumor angiogenesis in vivo (20, 21). In addition, miR-21, miR-31, miR-130, miR-210, miR-296, let-7f, miR-221/222, and miR-27b have all been shown to mediate angiogenesis in vitro (20, 21).
In this work, we find that miR-10b induces HMEC-1 cell migration and tube formation. However, miR-10b had no significant effect on proliferation or viability in our experiments (supplemental Fig. S2). Therefore, the effect of miR-10b on angiogenesis is more likely due to its role in endothelial cell differentiation and reorganization than proliferation. These results are in agreement with those published by Ma et al. (14), in which ectopic expression of miR-10b had no effect on the proliferation of immortalized breast cancer cells, HMECs, or SUM149 cells in vitro. Here we show that changes in miR-10b levels have proportional effects on the induction of angiogenesis. However, miR-10b likely also plays important roles in physiological processes unrelated to angiogenesis through the regulation of multiple targets. These targets include HoxD10, human T-lymphoma invasion and metastasis (Tiam1), and Kruppel-like factor 4 (KLF4) in tumors. Here, we report that miR-10b may down-regulate HoxD10 expression via direct binding to sites within the HoxD10 3′ UTR in HMEC-1 cells. A previous study showed that HoxD10 maintains a quiescent, differentiated phenotype in endothelial cells by suppressing expression of genes involved in remodeling the extracellular matrix and cell migration (27). Indeed, HoxD10 blocked migration and angiogenesis in endothelial cells. We also show that HMEC-1 cell migration and tube formation are induced by HoxD10 gene knockdown, whereas angiogenesis was arrested by HoxD10 up-regulation induced by anti-miR-10b. In this study, the effects of anti-miR-10b and miR-10b on angiogenesis are modest but significant (>30% in Fig. 2, C and D). It is possible that miR-10b regulates additional targets that are involved in proangiogenesis cascade steps. According to the results predicted by Miranda, Pictar, and Target Scan programs online, some other targets are involved with angiogenesis, such as vascular endothelial growth factor receptor 1 (Flt1), N-methyl-d-asparatate receptor-regulated protein 1 (NARG1), chondroitin sulfate proteoglycan 4 precursor (CSPG4), and oxidoreductase HTATIP2. Although those targets were not predicted simultaneously by both Miranda and Target Scan programs, they could not be excluded from those targets of miR-10b. It is important and challenging work to identify all targets of miR-10b to understand fully the function of this miRNA in angiogenesis.
Although this study supports a new role for miR-10b in angiogenesis induced by thrombin, the details of the signaling pathway mediating miR-10b expression have yet to be discovered. Previous study has shown that multiple miRNAs can alter fibrinogen production in Huh7 cells (58). However, the roles of miR-10b in relationship to heparin and thrombin during the process of coagulation is not clear. In addition, the details involved in the anti-angiogenic and coagulation functions of HoxD10 also require further investigation. Furthermore, future studies will be needed to identify additional miRNAs and their targets to determine the contributions of miRNAs to angiogenesis and coagulation.
Supplementary Material
This work was supported National Natural Science Foundation of China Grant 30770484, “100 Talents Project” of Chinese Academy of Sciences, China (to K. D.), and National Science and Technology Major Project “Key New Drug Creation and Manufacturing Program” Grants 2009ZX09301-001, 2009ZX09501-011, and 2009ZX09103-071.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
H. Qiu and K. Ding, unpublished data.
- EC
- endothelial cell
- miR
- microRNA
- QCM
- quartz crystal microbalance
- HoxD10
- homeobox D10
- HMEC-1
- human microvascular endothelial cells
- NRP2
- neuropilin-2
- NR4A3
- nuclear receptor subfamily 4, group A, member 3.
REFERENCES
- 1. Noble S., Pasi J. (2010) Br. J. Cancer 102, Suppl. 1, S2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodrigues C. A., Ferrarotto R., Kalil Filho R., Novis Y. A., Hoff P. M. (2010) J. Thromb. Thrombolysis 30, 67–78 [DOI] [PubMed] [Google Scholar]
- 3. Agorogiannis E. I., Agorogiannis G. I. (2002) Lancet 359, 1440 [DOI] [PubMed] [Google Scholar]
- 4. Bick R. L. (2003) N. Engl. J. Med. 349, 109–111 [DOI] [PubMed] [Google Scholar]
- 5. Daly M. E., Makris A., Reed M., Lewis C. E. (2003) J. Natl. Cancer Inst. 95, 1660–1673 [DOI] [PubMed] [Google Scholar]
- 6. Khorana A. A. (2010) Thromb. Res. 125, 490–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Snyder K. M., Kessler C. M. (2008) Semin. Thromb. Hemost. 34, 734–741 [DOI] [PubMed] [Google Scholar]
- 8. Zacharski L. R. (2002) Cancer Lett. 186, 1–9 [DOI] [PubMed] [Google Scholar]
- 9. Lindahl U. (2007) Thromb. Haemost. 98, 109–115 [PubMed] [Google Scholar]
- 10. Lee A. Y. (2007) Thromb. Res. 120, Suppl. 2, S121–127 [DOI] [PubMed] [Google Scholar]
- 11. Collen A., Smorenburg S. M., Peters E., Lupu F., Koolwijk P., Van Noorden C., van Hinsbergh V. W. (2000) Cancer Res. 60, 6196–6200 [PubMed] [Google Scholar]
- 12. Marchetti M., Vignoli A., Russo L., Balducci D., Pagnoncelli M., Barbui T., Falanga A. (2008) Thromb. Res. 121, 637–645 [DOI] [PubMed] [Google Scholar]
- 13. Castelli R., Porro F., Tarsia P. (2004) Vasc. Med. 9, 205–213 [DOI] [PubMed] [Google Scholar]
- 14. Ma L., Teruya-Feldstein J., Weinberg R. A. (2007) Nature 449, 682–688 [DOI] [PubMed] [Google Scholar]
- 15. Bloomston M., Frankel W. L., Petrocca F., Volinia S., Alder H., Hagan J. P., Liu C. G., Bhatt D., Taccioli C., Croce C. M. (2007) JAMA 297, 1901–1908 [DOI] [PubMed] [Google Scholar]
- 16. Tian Y., Luo A., Cai Y., Su Q., Ding F., Chen H., Liu Z. (2010) J. Biol. Chem. 285, 7986–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan H. X., Wang Q., Chen L. Z., Huang X. H., Chen J. S., Fu X. H., Cao L. Q., Chen X. L., Li W., Zhang L. J. (2010) Med. Oncol. 27, 654–660 [DOI] [PubMed] [Google Scholar]
- 18. Sasayama T., Nishihara M., Kondoh T., Hosoda K., Kohmura E. (2009) Int. J. Cancer 125, 1407–1413 [DOI] [PubMed] [Google Scholar]
- 19. Chai G., Liu N., Ma J., Li H., Oblinger J. L., Prahalad A. K., Gong M., Chang L. S., Wallace M., Muir D., Guha A., Phipps R. J., Hock J. M., Yu X. (2010) Cancer Sci. 101, 1997–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang S., Olson E. N. (2009) Curr. Opin. Genet. Dev. 19, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fish J. E., Srivastava D. (2009) Sci. Signal. 2, pe1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blower P. E., Chung J. H., Verducci J. S., Lin S., Park J. K., Dai Z., Liu C. G., Schmittgen T. D., Reinhold W. C., Croce C. M., Weinstein J. N., Sadee W. (2008) Mol. Cancer Ther. 7, 1–9 [DOI] [PubMed] [Google Scholar]
- 23. Ding K., Lopez-Burks M., Sánchez-Duran J. A., Korc M., Lander A. D. (2005) J. Cell Biol. 171, 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iozzo R. V., San Antonio J. D. (2001) J. Clin. Invest. 108, 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ferro V., Dredge K., Liu L., Hammond E., Bytheway I., Li C., Johnstone K., Karoli T., Davis K., Copeman E., Gautam A. (2007) Semin. Thromb. Hemost. 33, 557–568 [DOI] [PubMed] [Google Scholar]
- 26. Qiu H., Yang B., Pei Z. C., Zhang Z., Ding K. (2010) J. Biol. Chem. 285, 32638–32646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Myers C., Charboneau A., Cheung I., Hanks D., Boudreau N. (2002) Am. J. Pathol. 161, 2099–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Y., Gorski D. H. (2008) Blood 111, 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Würdinger T., Tannous B. A., Saydam O., Skog J., Grau S., Soutschek J., Weissleder R., Breakefield X. O., Krichevsky A. M. (2008) Cancer Cell 14, 382–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miao R. Q., Agata J., Chao L., Chao J. (2002) Blood 100, 3245–3252 [DOI] [PubMed] [Google Scholar]
- 31. Smorenburg S. M., Van Noorden C. J. (2001) Pharmacol. Rev. 53, 93–105 [PubMed] [Google Scholar]
- 32. Laubli H., Borsig L. (2009) Cancer Invest. 27, 474–481 [DOI] [PubMed] [Google Scholar]
- 33. Budhu A., Jia H. L., Forgues M., Liu C. G., Goldstein D., Lam A., Zanetti K. A., Ye Q. H., Qin L. X., Croce C. M., Tang Z. Y., Wang X. W. (2008) Hepatology 47, 897–907 [DOI] [PubMed] [Google Scholar]
- 34. Rosenfeld N., Aharonov R., Meiri E., Rosenwald S., Spector Y., Zepeniuk M., Benjamin H., Shabes N., Tabak S., Levy A., Lebanony D., Goren Y., Silberschein E., Targan N., Ben-Ari A., Gilad S., Sion-Vardy N., Tobar A., Feinmesser M., Kharenko O., Nativ O., Nass D., Perelman M., Yosepovich A., Shalmon B., Polak-Charcon S., Fridman E., Avniel A., Bentwich I., Bentwich Z., Cohen D., Chajut A., Barshack I. (2008) Nat. Biotechnol. 26, 462–469 [DOI] [PubMed] [Google Scholar]
- 35. John B., Sander C., Marks D. S. (2006) Methods Mol. Biol. 342, 101–113 [DOI] [PubMed] [Google Scholar]
- 36. Hasan J., Shnyder S. D., Clamp A. R., McGown A. T., Bicknell R., Presta M., Bibby M., Double J., Craig S., Leeming D., Stevenson K., Gallagher J. T., Jayson G. C. (2005) Clin. Cancer Res. 11, 8172–8179 [DOI] [PubMed] [Google Scholar]
- 37. Norrby K. (2006) APMIS 114, 79–102 [DOI] [PubMed] [Google Scholar]
- 38. Ashikari-Hada S., Habuchi H., Kariya Y., Kimata K. (2005) J. Biol. Chem. 280, 31508–31515 [DOI] [PubMed] [Google Scholar]
- 39. Soncin F., Strydom D. J., Shapiro R. (1997) J. Biol. Chem. 272, 9818–9824 [DOI] [PubMed] [Google Scholar]
- 40. Vlodavsky I., Ilan N., Nadir Y., Brenner B., Katz B. Z., Naggi A., Torri G., Casu B., Sasisekharan R. (2007) Thromb. Res. 120, Suppl. 2, S112–120 [DOI] [PubMed] [Google Scholar]
- 41. Solari V., Jesudason E. C., Turnbull J. E., Yates E. A. (2010) Br. J. Cancer 103, 593–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olson S. T., Halvorson H. R., Björk I. (1991) J. Biol. Chem. 266, 6342–6352 [PubMed] [Google Scholar]
- 43. Hu L., Roth J. M., Brooks P., Ibrahim S., Karpatkin S. (2008) Cancer Res. 68, 4296–4302 [DOI] [PubMed] [Google Scholar]
- 44. Mousa S. A. (2004) Cardiovasc. Drug Rev. 22, 121–134 [DOI] [PubMed] [Google Scholar]
- 45. Fu Y., Chen Y., Luo X., Liang Y., Shi H., Gao L., Zhan S., Zhou D., Luo Y. (2009) Biochemistry 48, 11655–11663 [DOI] [PubMed] [Google Scholar]
- 46. Ma L., Reinhardt F., Pan E., Soutschek J., Bhat B., Marcusson E. G., Teruya-Feldstein J., Bell G. W., Weinberg R. A. (2010) Nat. Biotechnol. 28, 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li G., Wu Z., Peng Y., Liu X., Lu J., Wang L., Pan Q., He M. L., Li X. P. (2010) Cancer Lett. 299, 29–36 [DOI] [PubMed] [Google Scholar]
- 48. Heneghan H. M., Miller N., Lowery A. J., Sweeney K. J., Newell J., Kerin M. J. (2010) Ann. Surg. 251, 499–505 [DOI] [PubMed] [Google Scholar]
- 49. Costantini V., De Monte P., Cazzato A. O., Stabile A. M., Deveglia R., Frezzato E., Paolucci M. C. (1998) Blood Coagul. Fibrinolysis 9, 79–84 [DOI] [PubMed] [Google Scholar]
- 50. Mertens G., Cassiman J. J., Van den Berghe H., Vermylen J., David G. (1992) J. Biol. Chem. 267, 20435–20443 [PubMed] [Google Scholar]
- 51. Zhang W., Swanson R., Xiong Y., Richard B., Olson S. T. (2006) J. Biol. Chem. 281, 37302–37310 [DOI] [PubMed] [Google Scholar]
- 52. Girardin E. P., Hajmohammadi S., Birmele B., Helisch A., Shworak N. W., de Agostini A. I. (2005) J. Biol. Chem. 280, 38059–38070 [DOI] [PubMed] [Google Scholar]
- 53. de Agostini A. I., Watkins S. C., Slayter H. S., Youssoufian H., Rosenberg R. D. (1990) J. Cell Biol. 111, 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bar-Shavit R., Eldor A., Vlodavsky I. (1989) J. Clin. Invest. 84, 1096–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shibata S., Harpel P., Bona C., Fillit H. (1993) Clin. Immunol. Immunopathol. 67, 264–272 [DOI] [PubMed] [Google Scholar]
- 56. Yang W. J., Yang D. D., Na S., Sandusky G. E., Zhang Q., Zhao G. (2005) J. Biol. Chem. 280, 9330–9335 [DOI] [PubMed] [Google Scholar]
- 57. Kuehbacher A., Urbich C., Zeiher A. M., Dimmeler S. (2007) Circ. Res. 101, 59–68 [DOI] [PubMed] [Google Scholar]
- 58. Fort A., Borel C., Migliavacca E., Antonarakis S. E., Fish R. J., Neerman-Arbez M. (2010) Blood 116, 2608–2615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.