Abstract
Various types of post-translational modifications of the histone tails have been revealed, but a few modifications have been found within the histone core sequences. Histone core post-translational modifications have the potential to modulate nucleosome structure and DNA accessibility. Here, we studied the histone H2B core domain and found that phosphorylation of H2B serine 32 occurs in normal cycling and mitogen-stimulated cells. Notably, this phosphorylation is elevated in skin cancer cell lines and tissues compared with normal counterparts. The JB6 Cl41 mouse skin epidermal cell line is a well established model for tumor promoter-induced cell transformation and was used to study the function of H2B during EGF-induced carcinogenesis. Remarkably, cells overexpressing a nonphosphorylatable H2BS32A mutant exhibited suppressed growth and EGF-induced cell transformation, possibly because of decreased activation of activator protein-1, compared with control cells overexpressing wild type H2B. We identified ribosomal S6 kinase 2 (RSK2) as the kinase responsible for H2BS32 phosphorylation. Serum-starved JB6 cells contain very little endogenous H2BS32 phosphorylation, and EGF treatment induced this phosphorylation. The phosphorylation was attenuated in RSK2 knock-out MEFs and RSK2 knockdown JB6 cells. Taken together, our results demonstrate a novel role for H2B phosphorylation in cell transformation and show that H2BS32 phosphorylation is critical for controlling activator protein-1 activity, which is a major driver in cell transformation.
Keywords: DNA, Histones, Mouse, Phosphorylase, Rsk
Introduction
The eukaryote genome is packaged into the highly organized chromatin. The fundamental repeating unit of chromatin is the nucleosome, which consists of 146 bp of DNA wrapped approximately two turns around a core histone octamer (two copies each of histone H2A, H2B, H3, and H4) (1). Generally speaking, chromatin is inaccessible to the outside environment because of the strong DNA/histone(s) and histone(s)/histone(s) interactions. Thus, the chromatin structure must be remodeled in different ways to facilitate any process that requires access to the DNA (2).
The chromatin structure is regulated by four main processes: 1) post-translational modifications of histone variants; 2) incorporation (or replacement) of histone variants; 3) DNA methylation; and 4) ATP-dependent chromatin remodeling (3). The post-translational modifications (e.g. acetylation, methylation, phosphorylation, ubiquitylation, sumoylation, or proline isomerization) of histone variants have drawn much attention and are a primary focus in the field of epigenetics (4). Some studies suggest that these modifications are performed by modifiers with multiple chromatin-binding domains to engage histone tails (5). Evidence also indicates that combinations of post-translational histone modifications cooperate to increase the specificity of the signal to be elicited or translated to distinct biological events (6).
Histone tails protrude from their own nucleosome, and they are able to make contact with adjacent nucleosomes. Thus, modifications of the histone N-terminal tails are likely to affect regional protein/DNA and protein/protein interaction(s) (7). This in turn has the potential to regulate the overall chromatin architecture. Over the past decade, histone tails have been the main focus for scientists to understand how post-translational modifications affect chromatin structure. However, less attention has been given to the potential post-translational modifications located close to or in the histone core. Here, we identified histone H2B serine 32 (H2BS32)3 as a novel H2B post-translational phosphorylation site. Phosphorylation of H2BS32 (H2BS32ph) was mitogen-inducible and associated with malignant transformation of mouse skin epidermal cells. Notably, we determined that ribosomal S6 kinase 2 (RSK2) was the kinase responsible for this modification, providing a new link between a histone core post-translational modification and cell transformation.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Chemical reagents, including Tris, NaCl, and SDS, for molecular biology and buffer preparation were purchased from Sigma-Aldrich. Restriction enzymes and some modifying enzymes were from New England Biolabs, Inc. (Ipswich, MA). The Taq DNA polymerase was obtained from Qiagen. The DNA ligation kit (version 2.0) was purchased from TAKARA Bio, Inc. (Madison, WI), and the pcDNA3.1(+) plasmid used for the construction of the expression vector was from Invitrogen. Cell culture medium and other supplements were purchased from Invitrogen. Antibodies for Western blot and immunofluorescence analyses were from Abcam (Cambridge, MA), Sigma-Aldrich, Invitrogen, Cell Signaling Technology, Inc. (Danvers, MA), Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and Upstate Biotechnology, Inc. (Lake Placid, NY), with the following dilutions: phospho-p44/42 ERK1/2 (Thr202/Tyr204) (9101, Cell Signaling), 1:2000; p44/42 ERK1/2 (9102; Cell Signaling), 1:2000; phospho-RSK (Ser380) (9341; Cell Signaling), 1:1000; phospho-RSK2 (Thr577) (sc-16407; Santa Cruz), 1:50 for immunofluorescence analysis; RSK2 (06-918; Millipore), 1:1000; phosphorylated H2BS32 (H2BS32ph) (ab10476; Abcam), 1:500 for Western blot analysis, 1:50 for immunofluorescence analysis; H2BS14ph (sc-31671; Santa Cruz), 1:1000; H2B (07-371; Upstate), 1:10000; FLAG (F3165, Sigma), 1:5000; Xpress (46-0528; Invitrogen), 1:5000; and H3 (9715; Cell Signaling), 1:5000. H2B peptides were purchased from Abcam: H2B (unmodified) (ab5489; Abcam), H2B (S14ph) (ab5496; Abcam), and H2B (S32ph) (ab18504; Abcam). The specificity of the H2BS32ph antibody was validated by peptide competition using an ELISA and peptide dot blots. Its specificity was also confirmed by Western blot of endogenous purified histones as well as immunofluorescence staining from EGF-stimulated JB6 Cl41 cells preincubated with a specific H2BS32ph or nonspecific unmodified H2B blocking peptide, respectively (8).
Histone Substrates
Pure histone proteins H2A, H2B, H3, and H4 were purchased from Upstate Biotechnology, Inc. The core histones and mononucleosomes were prepared according to the purification scheme described previously (9).
Construction of pGST-H2B, pFLAG-H2B, and the Corresponding Mutants
The Homo sapiens H2B gene coding sequence (NM_021058) was amplified from pHCE-H2B by PCR. After restriction digestion, the H2B sequence was ligated to the BamHI/XhoI site of pGEX-5X-1 to generate the pGST-H2B. For generation of pFLAG-H2B, the H2B coding sequence amplified from pGST-H2B was performed with the addition of three copies of FLAG tag at its 5′ end and subcloned into the BamHI/XbaI site of pcDNA3.1(+). To construct the point mutant of H2B (H2BS32A), the QuikChange® II site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used following the recommended protocols. The recombinant plasmids were confirmed by agarose gel electrophoresis and DNA sequencing by Genewiz (South Plainfield, NJ). The plasmid encoding RSK2 was described previously (10).
Cell Culture and Transfections
JB6 Cl41 (CRL-2010) and HEK293 (CRL-1573) cells were purchased from the American Type Culture Collection (Manassas, VA) and maintained in Eagle's MEM with 5% FBS and DMEM with 10% FBS, respectively. RSK2+/+ and RSK2−/− MEFs (a generous gift from Dr. J. C. Bruning, Institute for Genetics, Center for Molecular Medicine Cologne, Cologne, Germany) were cultured with DMEM with 10% FBS. Transfection of the expression vectors was performed using Lipofectamine (Invitrogen) following the manufacturer's suggested protocol. Human keratinocyte (HaCaT) and human malignant melanoma cell lines (SK-MEL-28, SK-MEL-31, and RPMI-7951) were also from the American Type Culture Collection. These cell lines were cultured in MEM containing 10% FBS or 15% FBS for SK-MEL-31 cells, 2 mm l-glutamine, and 25 μg/ml gentamicin, 0.1 mm nonessential amino acids, and 1 mm sodium pyruvate (SK-MEL-31 also required 1.5 g/L sodium bicarbonate) at 37 °C in humidified air with 5% CO2. For generation of H2B stable cells, JB6 cells transfected with the DNA constructs were cultured in 400 μg/ml G418-containing medium for 2 weeks, and then stable clones were pooled together and expanded. For generation of RSK2 knockdown cells, lentiviral shRNA to RSK2 (Clone TRCN0000022728) was purchased (Thermo Scientific, Huntsville, AL), and viral particles were packaged according to the manufacturer's instructions. JB6 cells, grown to about 70% confluence, were infected with the above lentiviral shRNA in the presence of 8 μg/ml polybrene for 24 h. Uninfected cells were eliminated by exposure to 2 μg/ml puromycin for 4 days before use.
Cell Proliferation (MTS) Assay
To estimate proliferation, JB6-H2BWT and JB6-H2BS32A cells were seeded (2 × 103) in 96-well plates. After culturing for various times, 20 μl of the CellTiter 96 Aqueous One Solution (Promega) were added to each well, and cells were incubated for 1 h in a 37 °C, 5% CO2 incubator, and absorbance was measured at 492 nm.
Anchorage-independent Cell Transformation Assay
EGF-induced cell transformation was investigated in pFLAG-H2BWT or pFLAG-H2BS32A stably transfected cells. In brief, cells (8 × 103/ml) were exposed to EGF (10 ng/ml) in 1 ml of 0.3% basal medium Eagle agar containing 10% FBS. The cultures were maintained in a 37 °C, 5% CO2 incubator for 10 days, and the cell colonies were scored using a microscope and the Image-Pro PLUS (v.4) computer software program (Media Cybernetics, Bethesda, MD) as described previously (10).
Reporter Gene Assays
The AP-1-luciferase reporter plasmid construct containing the −73 to +63 collagenase promoter sequence was used as described previously (11). AP-1 activity was analyzed by transfection of the AP-1-luc reporter plasmid into H2B-overexpressing cells, which were established by stable transfection. The cells were disrupted with lysis buffer (dual-luciferase reporter assay system; Promega) at room temperature for 30 min by gentle shaking, and then firefly luciferase activity was measured. The AP-1-luciferase activity was normalized against Renilla luciferase activity (phRL-SV40).
In Vitro Kinase Assay
To detect histone phosphorylation, histone proteins (1 μg) were mixed with active RSK2 or active MST1 (Upstate Biotechnology, Inc.), 0.2 mm ATP, and 1× kinase buffer and incubated at 30 °C for 30 min. To detect 32P incorporation, histone proteins (1 μg) were mixed with active RSK2, 10 μmol/L unlabeled ATP, 1 μCi of [32P]ATP, and 1× kinase buffer and incubated at 30 °C for 30 min. The reaction was stopped by adding 5× SDS sample buffer. The samples were boiled and then separated by 15% SDS-PAGE and visualized by autoradiography, Western blotting, or Coomassie Blue staining.
Mononucleosome Immunoprecipitation
Oligonucleosome fragments were prepared from the nuclear pellets and digested with micrococcal nuclease (Sigma) as described (12). Nucleosomes containing FLAG-H2BWT or FLAG-H2BS32A were purified from the resulting material by immunoprecipitation with anti-FLAG-agarose beads.
Western Blotting
Samples containing equal amounts of protein were resolved by the appropriate percentage SDS-PAGE and transferred onto PVDF membranes. The membranes were incubated in blocking buffer and then probed with various specific primary antibodies and the appropriate anti-rabbit or anti-mouse HRP as the secondary antibody. The Western blots were visualized using an ECL detection system (Amersham Biosciences).
Extraction of Nuclear Proteins
To analyze H2BS32 phosphorylation, RSK2+/+, RSK2−/−, and JB6 and its corresponding shMock or shRSK2 knockdown cells were each seeded in 10-cm dishes and cultured to 90–95% confluence. The cells were serum-starved in culture medium with 0.1% FBS for 24 h and then stimulated with EGF. The cells were harvested, and nuclear fractions were obtained by the use of NE-PER nuclear and cytoplasmic extraction reagents (Pierce) following the manufacturer's suggested protocols. To obtain nuclear extracts containing histones, the nuclear fractions were treated with 250 units of benzonase (Sigma-Aldrich) for 30 min on ice. The samples were mixed for 15 s by vortex every 10 min, and the supernatant fraction was recovered by centrifugation (13,000 rpm, 5 min at 4 °C). The supernatant fraction was the nuclear fraction containing histone proteins. To detect H2BS32ph and total H2B protein, 2 μg of total nuclear fraction protein were used for Western blot analysis. To detect soluble and chromatin-bound histones, total soluble histones from histone stable cells were extracted using radioimmunoprecipitation buffer (Millipore). After centrifugation, the resulting pellets were washed extensively with buffer (37 mm EDTA, 10 mm Tris-Cl, pH 7.4). Pellets containing chromatin-bound histones were extracted with 0.4 n H2SO4. The histones were concentrated by acetone precipitation and redissolved in 4 m urea. Histones from soluble and chromatin-bound fractions were subjected to Western blot analysis using FLAG and H3 antibodies.
Immunofluorescence and Confocal Microscopy
JB6 cells were seeded in two-chamber culture slides. At the end of the experiment, they were fixed for 10 min in PBS containing 4% formalin, rinsed with PBS, and permeabilized for 20 min in PBS containing 0.1% (v/v) Triton X-100. After blocking with 5% BSA in PBS for 1 h, the cells were stained for 2 h with rabbit polyclonal antibody against phosphorylated H2B at Ser32 (H2BS32ph; 1:50) in 1% BSA in PBS. To show the specificity of this antibody, blocking peptide was preincubated for 30 min with antibody before adding to the cells. The cells were then washed with PBS and then stained with a 1:500 dilution of fluorescein-conjugated goat anti-rabbit IgG (Molecular Probes/Invitrogen) in 1% BSA in PBS for 1 h at room temperature in the dark. The cells were washed once with PBS and then incubated with 1 μg/ml of Hoechst 33258 in PBS for 5 min. The cells were finally washed four times with PBS, and the coverslips were mounted onto glass slides with Vectashield mounting medium (Vector Laboratories). The coverslips were fixed with nail varnish. The samples were analyzed by fluorescence microscope (magnification, ×200). Detection of RSK2T577ph was also performed by using the goat polyclonal antibody against phosphorylated RSK2 at Thr577 (RSK2T577ph; 1:50) and following the above procedures. The images were subjected to blind deconvolution using the Autoquant algorithm, and the degree of colocalization of signals was analyzed according to Manders' overlap coefficient. This coefficient varies from 0 to 1, the former corresponding to nonoverlapping images and the latter reflecting 100% colocalization between both images (13).
A human skin tissue array (SK804) was purchased from U.S. Biomax, Inc., deparaffinized in xylene, and rehydrated in alcohol, and the antigen was retrieved in 10 mmol/L sodium citrate buffer. The tissues were first blocked in 5% normal goat serum/PBS for 1 h at room temperature and then incubated overnight at 4 °C with a 1:50 dilution of the H2BS32ph antibody in PBS containing 1% BSA. The tissues were washed and then incubated in the dark with a 1:200 dilution of a Cy3-donkey anti-rabbit IgG antibody for 2 h at room temperature. Image stacks were captured using laser scanning confocal microscopy (NIKON C1si Confocal Spectral Imaging System, NIKON Instruments Co.) and analyzed with the Image J (version 1.37v) software program. The intensity score of fluorescence from each sample was measured, and the average intensity score from duplicate scores of each case was recorded as the H2BS32 phosphorylation level.
ChIP Assay
Nuclear factors associated with chromatin in JB6 cells were cross-linked to DNA by using formaldehyde (1%). The cells were harvested, and cross-linked chromatin was sheared by sonication. DNA fragments were <1 kb and averaged 450 bp as verified by agarose gel electrophoresis. Immunoprecipitation was performed with 100 μg (DNA content) of chromatin extracts diluted in ChIP dilution buffer (1.1% Triton X-100, 0.01% SDS, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, and 167 mm NaCl). The samples were precleared with salmon sperm DNA/protein A-agarose beads (Upstate) for 30 min and then incubated overnight (16 h) with 4 μg of control IgG or anti-RSK2, respectively, at 4 °C. DNA present in the immunoprecipitated chromatin was isolated after reversed cross-link and proteinase K digestion. The DNAs were extracted with phenol-chloroform and chloroform and precipitated with ethanol and water. Input DNAs were quantified by UV spectrophotometry. Purified DNA was used as a template for 30 cycles of PCR amplification using the designated primers for c-jun (−733 to −572), c-fos (−131 to +93), and β-globin (+261 to +439) sequences (numbers are with respect to the transcription start sites) as previously described (14). The PCR products were then analyzed by agarose gel electrophoresis and visualized using ethidium bromide staining.
RESULTS
Confirmation of Antibody Specifically against H2BS32ph
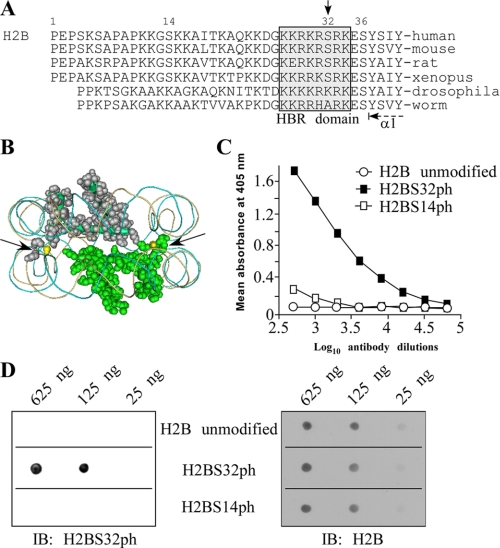
Our group has been investigating novel functions of histone modifications by histone writers (15, 16). In the present study, we focused on phosphorylation sites of histone H2B located close to or in the histone core. By primary sequence alignment of the H2B N-terminal “tails” from different species, we found that only vertebrates have a well conserved Ser32 (Fig. 1A), whereas Ser36 is conserved from humans to worms. Interestingly, Ser32 is located within the histone H2B repression domain (HBR). The crystal structure of the nucleosome indicates that H2BS32 lies in a structurally important region of the nucleosome, positioned adjacent to the extreme N terminus of the first helix of H2B (α1 helix) and between the grooves of the DNA double helix (Fig. 1, A and B, arrows). This region of H2B and phosphorylation of Ser32 very likely play an important role in nucleosome structure/stability. To study the phosphorylation of H2BS32, we used polyclonal antisera to detect H2B peptides encompassing a phosphorylated Ser32 residue. The specificity of the antisera determined by ELISA assay confirmed that the resulting IgG population specifically binds an H2BS32ph peptide, but not the nonphosphorylated version of the peptide (Fig. 1C). Furthermore, peptide competition analyses using ELISA and peptide dot blots indicated that the antibody preparation did not cross-react to other sites of histone phosphorylation, such as Ser14 (Fig. 1, C and D).
FIGURE 1.
Confirmation of antibody to specifically detect H2BS32ph. A, comparison of the primary sequences of histone H2B N-terminal “tails” from different species shows that only vertebrates have a well conserved Ser32 or Ser14, whereas a highly conserved Ser36 is evident in the H2B tails of most species from human to worm. Ser32 (downward arrow) is the only serine located within the HBR domain (gray box). B, location of Ser32 residues (yellow) in the nucleosome crystal structure (Protein Data Bank code 2CV5). Shown are two molecules of H2B present in the nucleosome (image drawn using Cn3D version 4.1). The arrows indicate the location of the pair of H2BS32 residues, each at the extreme end of the N-terminal tail of H2B and in between the DNA grooves. C, peptide competition analyses confirm the specificity of the H2BS32ph antibody. The specificity of the antibody was validated by peptide competition using ELISA. The antibody, which was raised against an H2BS32ph peptide, specifically recognizes only this peptide and fails to detect the identical but unmodified H2B peptide. Similarly, the antibody does not recognize other phosphoserine sites like H2BS14 previously shown to be modified on H2B. D, the indicated amounts of various phosphorylated histone peptides were spotted onto a PVDF membrane and probed with the H2BS32ph antibody. This antibody only recognizes the peptide containing phosphorylated Ser32 (left panel). The same blot was stripped and reprobed with the H2B antibody as a loading control (right panel). IB, immunoblot.
Phosphorylation of H2BS32 Is a Marker of Living Cells
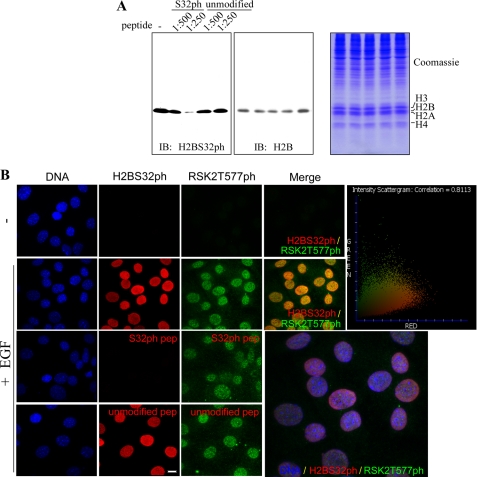
To determine the physiological relevance of phosphorylation of H2BS32, immunofluorescence microscopy was performed using the H2BS32ph antibody and JB6 cells. First, we tested the specificity of this antibody by immunoblot analyses with total soluble cellular protein extracts from asynchronously growing JB6 cells. The H2BS32ph antibody reacted with H2B from JB6 cells (Fig. 2A, first lane). To exclude the possibility that the H2BS32ph antibody simply cross-reacted with H2B, we performed the immune reactions in the presence of a competing peptide encompassing the unmodified or phosphorylated Ser32. In this case, the reaction was blocked in a concentration-dependent manner by the H2BS32ph peptide (Fig. 2A, second and third lanes) but not by the unmodified peptide (Fig. 2A, fourth and fifth lanes). Thus, this antibody specifically recognized H2BS32ph. We then stimulated JB6 cells with EGF and observed the cells with immunofluorescent microscopy. The results indicated that serum-starved JB6 cells contain very little H2BS32ph, whereas EGF treatment induced H2BS32 phosphorylation as reflected by the increased fluorescent signal (Fig. 2B). Similarly, incubation of the specific blocking peptide (H2BS32ph) but not the nonspecific blocking peptide (H2B unmodified) with the antibody before addition to the cells abolished the fluorescent signal, indicating that this antibody specifically recognizes H2BS32ph, which is present in the chromatin (Fig. 2B). These data suggest that H2BS32ph is detectable in normal cycling cells as well as mitogen-treated JB6 cells. Notably, cells positive for H2BS32ph colocalized with phosphorylated RSK2. This did not occur in the Hoechst densely stained region, indicating that the colocalization occurs in euchromatin, where active gene transcription takes place (Fig. 2B, lower right panel). The degree of colocalization of signals was analyzed according to Manders' overlap coefficient. The intensity scattergram (Fig. 2B, upper right panel) showed a value of 0.8113, indicating a very strong correlation (overlapping) between the two images (H2BS32ph and phosphorylated RSK2 exist in JB6 cells treated with EGF). Considered together, these data indicate an association between H2BS32 phosphorylation and an EGF-induced proliferation marker, activated RSK2, a relatively early event in the process of the response to EGF stimulation. Our results clearly demonstrate an induction of H2BS32 phosphorylation in proliferating cells following the administration of EGF.
FIGURE 2.
Phosphorylation of H2BS32 is a marker of living cells. H2BS32ph is detectable in normal cycling JB6 cells, and the level is increased by mitogen stimulation in serum-starved JB6 cells. A, the specificity of the H2BS32ph antibody was assessed by Western blot using whole cell lysates with the corresponding specific H2BS32ph and nonspecific unmodified H2B blocking peptide. JB6 cells were disrupted in 1× SDS sample buffer and boiled for 10 min at 95 °C. Total soluble proteins were subjected to immunoblot (IB) analysis using specific antibodies to detect H2BS32ph and total H2B. Incubation of lysates with primary antibodies was performed without (−) or with the competing peptide (shown are peptide dilutions) phosphorylated at Ser32 or the unmodified peptide. Extracts identical to those loaded onto the gel for immunoblot (left panel) have also been probed with the H2B antibody (middle panel) as well as stained with Coomassie Blue (right panel) to assure that identical amounts of proteins were loaded in each lane. B, JB6 cells were seeded into two-chamber slides and cultured overnight. After serum starvation for 24 h, the cells were or were not stimulated with 10 ng/ml EGF for 30 min. Expression and subcellular localization of H2BS32ph were determined by immunofluorescence microscopy. JB6 cells were washed, fixed, and incubated with H2BS32ph and RSK2T577ph antibodies or with antibodies preincubated with 5 μg of the H2BS32ph or unmodified H2B blocking peptide. The nuclei were stained with Hoechst before viewing by immunofluorescence microscopy. Colocalization of H2BS32ph and RSK2T577ph occurs in the nucleus of EGF-stimulated JB6 cells. Staining was not apparent in the densely Hoechst-stained region, indicating that colocalization occurs in the euchromatin, where active gene transcription takes place. An enlarged merged image of all three signals from EGF-stimulated JB6 cells is shown (lower right panel). Scale bar, 10 μm. The degree of colocalization of signals was analyzed according to Manders' overlap coefficient. The intensity scattergram (upper right panel) showed a value of 0.8113, indicating a very strong correlation (overlap) between the two images (H2BS32ph and phosphorylated RSK2 in EGF-treated JB6 cells).
H2BS32ph Is Highly Abundant in Skin Cancer Tissues and Skin Cancer Cell Lines Compared with Normal Counterparts
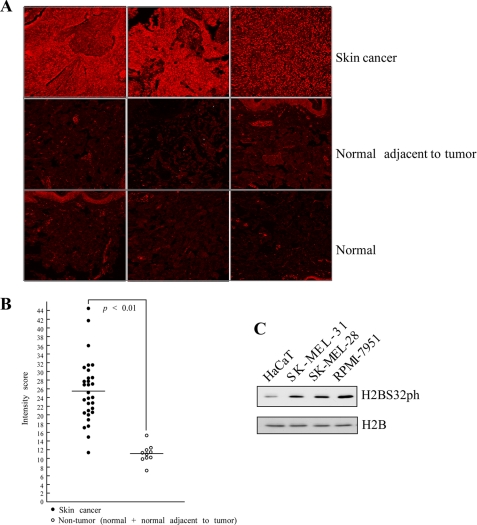
Our group reported that RSK2, an ERK downstream serine/threonine protein kinase, is regulated by tumor promoters such as EGF or 12-O-tetradecanoylphorbol-13-acetate and RSK2 is highly expressed in human skin cancer tissues and cancer cell lines (10, 17). EGF-induced activation of ERKs and RSK2 is a well known cellular event, and thus we wondered whether H2BS32ph might also be highly expressed in skin cancer compared with normal tissues. To determine the expression of H2BS32ph in human skin cancer, we examined a human skin cancer tissue array that contained duplicate core samples of 9 cases of squamous cell carcinoma, 10 cases of basal cell carcinoma, 11 cases of malignant melanoma, and 5 each of adjacent normal tissue and normal tissue (US Biomax, Inc.). We found that H2BS32ph was significantly higher in skin cancer tissues (Fig. 3, A and B), with a median expression level of 25.49 relative intensity units, compared with expression in nontumor tissues, which had a median score of 11.07 (Fig. 3B). Similarly, the levels of H2BS32ph were assessed in normal skin cells and several different human melanoma cell lines. The results showed that H2BS32ph was elevated in all three melanoma cell lines compared with the human keratinocyte (HaCaT) cell line (Fig. 3C). These results confirmed that like RSK2, the level of H2BS32ph is higher in human skin cancer. Based on results thus far, we hypothesized that RSK2 might be an upstream kinase to phosphorylate H2BS32.
FIGURE 3.
H2BS32ph is highly abundant in skin cancer tissues and skin cancer cell lines compared with normal counterparts. A, immunofluorescent staining and confocal microscopy were used to detect the levels of H2BS32ph in tissues from duplicate core samples of 9 cases of squamous cell carcinoma, 10 cases of basal cell carcinoma, 11 cases of malignant melanoma, and 5 each of adjacent normal tissue and normal tissue. H2BS32ph was detected by immunofluorescent staining with the H2BS32ph antibody as the primary antibody and Cy3-conjugated donkey anti-rabbit as the secondary antibody. Photographs from representative cases are shown. B, the intensity score of fluorescence from each sample was determined using Image J (National Institutes of Health); horizontal lines indicate the median fluorescence scores of skin cancer or noncancerous tissue samples; p < 0.01, Mann-Whitney U test. C, the level of H2BS32ph in several skin cell lines. Human keratinocytes (HaCaT) and several different human melanoma cell lines were screened by Western blot to determine the level of H2BS32ph. Total H2B protein was used to verify equal protein loading.
RSK2 Phosphorylates H2B at Serine 32
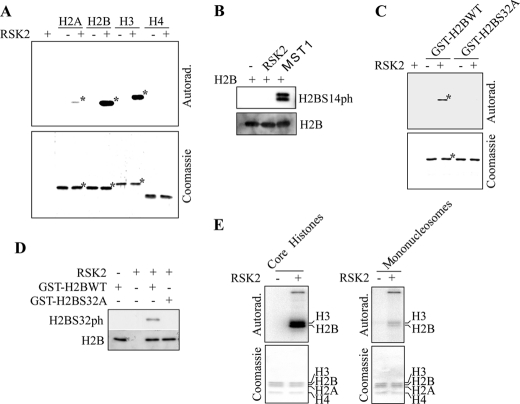
Results thus far showed that phosphorylation of H2BS32 is inducible by EGF and H2B is very likely to be phosphorylated by kinases that occupy the EGF signaling pathway. To determine which kinase is responsible for this modification, we used recombinant pure histone H2B as substrate to screen a library of active serine/threonine kinases by in vitro kinase assay. Interestingly, RSK2 was identified as one of the top candidates to phosphorylate H2B, and the phosphorylation was confirmed using 32P (Fig. 4A). We also confirmed that, in addition to H2B, RSK2 could also phosphorylate histone H3 as reported (18) (Fig. 4A). RSK2 also weakly phosphorylated histone H2A but did not phosphorylate histone H4 (Fig. 4A). Phosphorylation of mammalian H2BS14 by MST1 (mammalian sterile twenty kinase 1) was reported to be related to apoptosis (19). RSK2 is known to be an important mediator of cell survival and tumor promoter-induced cell transformation (10, 20). This suggests that RSK2 is not likely to phosphorylate H2BS14, and this was confirmed by Western blot analysis (Fig. 4B). Therefore, to determine the histone H2B site that is phosphorylated by RSK2, we focused on the substrate motif of RSK2, which is RXX(S/T) or RX(S/T). We found that Ser32 (i.e. RKRS32) is located within this motif. The crystal structure of the nucleosome indicates that Ser32 is located in the histone fold of H2B, between the grooves of the DNA double helix (Fig. 1B). By constructing a point mutant of histone H2B (H2BS32A) and performing a kinase assay using active RSK2, we determined that RSK2 could phosphorylate H2BS32. Importantly, the ability to phosphorylate H2B was severely abrogated when Ser32 is converted to alanine (Fig. 4C, third lane versus fifth lane). Thus, H2BS32 is a primary site within H2B that is phosphorylated by RSK2 in vitro. We also performed similar in vitro kinase assays to those described above but with nonradioactive ATP. In this case, the reaction products were detected by Western blot using the H2BS32ph antibody (Fig. 4D). The antibody detects phosphorylation of wild type H2B but not mutant H2BS32A. To determine whether RSK2 can phosphorylate the histone H2B present in the nucleosome, we performed a kinase assay using core histones or mononucleosomes. Our results showed that RSK2 phosphorylates histone H2B that is present in both configurations (Fig. 4E). We have previously observed that some histone kinases show a higher level of activity on free histones compared with nucleosomal substrates. The higher level of activity of RSK2 on free and core histones compared with mononucleosomes might be due to the presence of DNA in mononucleosomes. The strong DNA/histone(s) interactions would probably decrease the accessibility of the H2BS32 site to be phosphorylated by RSK2 in vitro. This could explain the weaker signal intensity of the phosphorylated histones in Fig. 4E (right panel). Collectively, our data suggest that RSK2 can phosphorylate Ser32 of free H2B and H2B present in the H3/H4/H2A/H2B core histones and mononucleosomes.
FIGURE 4.
RSK2 phosphorylates H2BS32. A, an in vitro kinase assay using active RSK2 and recombinant histone H2A, H2B, H3, or H4 as substrate in the presence of 32P was performed and results visualized by autoradiography (top panel). B, Western blot analysis of H2B phosphorylation after a kinase assay using active RSK2 or MST1 and recombinant H2B. A phospho-specific antibody was used to detect phosphorylation of H2BS14 (top panel). C, mapping of the H2B site phosphorylated by RSK2. A GST-H2B wild type or GST-H2BS32A mutant protein was used as substrate for active RSK2 in an in vitro kinase assay in the presence of 32P and visualized by autoradiography (top). D, Western blot analysis of H2B phosphorylation after a kinase assay using active RSK2 and GST-H2BWT or GST-H2BS32A mutant protein. The H2BS32ph antibody was used to detect phosphorylation of H2BS32 (top panel). For B and D, the same blot was also reprobed with an antibody against total H2B to monitor equal protein loading (bottom panels). E, RSK2 phosphorylates nucleosomal H2B. RSK2 phosphorylates H2B present in core histones (left panel) as well as H2B present as a component of the mononucleosomes (right panel). The experiments were performed as described under “Experimental Procedures,” except that different histone substrates were used for the kinase assays. Each asterisk indicates the respective protein band. For A, C, and E, the corresponding gels are stained with Coomassie Blue to monitor equal protein loading (bottom panels of each).
RSK2 Interacts with H2B under in Vitro and ex Vivo Conditions
To determine whether RSK2 interacts directly with histone H2B, we performed a GST pulldown assay using GST-H2B coupled with GSH-Sepharose beads and RSK2 overexpressed in HEK293 cells. This result showed that RSK2 does interact with histone H2B, and the interaction is specific because no interaction was observed in the pull-down using GST alone (Fig. 5A). To determine whether RSK2 and H2B interact ex vivo, plasmids encoding FLAG-H2B and Xpress-RSK2 were cotransfected into HEK293 cells. H2B proteins were immunoprecipitated with anti-FLAG agarose beads and subjected to Western blot using anti-Xpress. RSK2 was confirmed to interact with H2B (Fig. 5B). Likewise, we transfected plasmids encoding Xpress-RSK2 into HEK293 cells. RSK2 was immunoprecipitated with anti-Xpress and protein A/G-agarose beads and subjected to Western blot using anti-H2B. H2B was confirmed to interact with RSK2 (Fig. 5C).
FIGURE 5.

RSK2 interacts with histone H2B in vitro and associates with nucleosomes ex vivo. A, GST or GST-H2B beads were incubated with 300 μg of total lysates prepared from HEK293 cells transfected with pcDNA4-HisMaxA-RSK2 (2 μg), followed by immunoblotting (IB) with anti-Xpress. The input indicates the amount of cell lysate loaded. B and C, RSK2 binds with nucleosomal H2B ex vivo. pFLAG-H2B and pcDNA4-HisMaxA-RSK2 were transiently transfected into HEK293 cells, and 40 h later H2B was immunoprecipitated (IP) with anti-FLAG agarose beads followed by immunoblotting with anti-Xpress. RSK2 was coimmunoprecipitated with histone H2B. Similarly, pcDNA4-HisMaxA-RSK2 was or was not transiently transfected into HEK293 cells, and 40 h later RSK2 was immunoprecipitated with anti-Xpress and protein A/G-agarose beads followed by immunoblotting with anti-H2B. H2B was coimmunoprecipitated with RSK2.
Ectopic Expression of a Nonphosphorylatable H2BS32A Mutant Suppresses Cell Proliferation and EGF-induced Anchorage-independent Cell Transformation
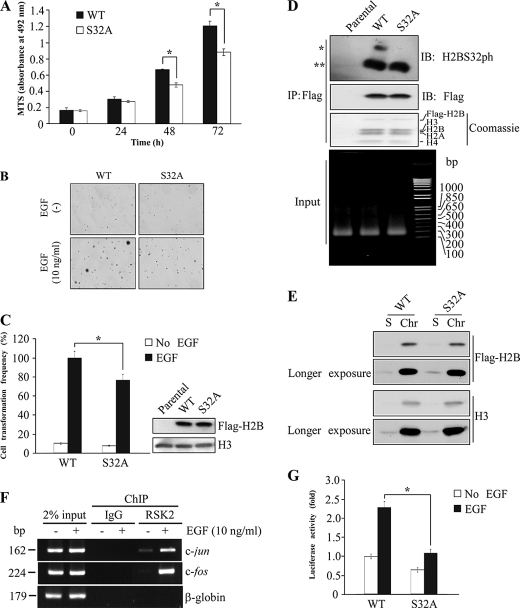
To investigate the role of H2BS32ph in skin cancer, we transfected plasmids encoding FLAG-H2BWT and FLAG-H2BS32A into JB6 cells, and stable transfectants were pooled and expanded. JB6-H2BWT and JB6-H2BS32A cells have similar expression levels of FLAG-H2BWT and FLAG-H2BS32A proteins (Fig. 6C, right panel). The results of the MTS cell proliferation assay indicated that JB6-H2BS32A mutant cells grow apparently slower than JB6-H2BWT cells (Fig. 6A). Because EGF-induced RSK2 activation is associated with cell transformation, we investigated the effect of EGF on cell transformation by soft agar assay in these histone-transfected cells. Cells overexpressing mutant H2BS32A suppressed the number of colonies compared with H2BWT-transfected cells (Fig. 6, B and C). We also performed a mononucleosome IP assay, and the result showed that similar levels of exogenous H2B had been incorporated into the chromatin of these cells and that FLAG-H2BWT was phosphorylated at S32 (Fig. 6D), which also confirmed the specificity of the H2BS32ph antibody ex vivo. Moreover, examination of histones in soluble versus chromatin-bound fractions indicated that the majority of exogenous histone is chromatin-bound (Fig. 6E). This suggests that exogenous H2B most likely exerts its effect in a chromatin-associated fashion, in which the exogenous FLAG-H2BWT and FLAG-H2BS32A are competing with endogenous H2B and are gradually incorporated into chromatin and exerting their functions. By ChIP assay, we showed that endogenous RSK2 is associated with the c-jun and c-fos promoters with EGF stimulation, indicating that endogenous RSK2 locates to genes induced by EGF (Fig. 6F). By AP-1-luciferase assay, we observed an attenuation of AP-1 activity in FLAG-H2BS32A cells as compared with FLAG-H2BWT cells, indicating that genes induced by EGF are altered by H2BS32ph (Fig. 6G). We showed that endogenous RSK2 phosphorylates H2BS32 and observed EGF-induced H2BS32ph and RSK2T577ph colocalization, as well as the enrichment of RSK2 in c-jun and c-fos promoters. These results clearly suggest that H2BS32 is phosphorylated by RSK2, which mediates the function of RSK2 in gene activation. Thus far, these data indicated that H2BS32 is involved in cell proliferation and transformation potential through the ERK/RSK/H2B signaling axis and requires the phosphorylation of H2BS32, which leads to the transcriptional activation of genes encoding for products in the AP-1 complex.
FIGURE 6.
Cell proliferation and EGF-induced anchorage-independent cell transformation are suppressed after serine 32 of H2B was mutated to alanine. A, JB6-H2BS32A cells grow slower than JB6-H2BWT cells. Proliferation was measured by MTS assay. B, ectopic expression of H2BS32A, but not H2BWT, suppresses neoplastic transformation of JB6 cells. JB6 cells stably transfected with pFLAG-H2BWT or pFLAG-H2BS32A were seeded in a six-well plate and exposed to EGF (10 ng/ml) in 1 ml of 0.3% basal medium Eagle agar containing 10% FBS. The cultures were maintained in a 37 °C, 5% CO2 incubator for 10 days and then photographed. Representative images of anchorage-independent colonies are shown. C, the colonies were counted using a microscope and the Image-Pro PLUS (v. 4) computer software program. The relative difference in average colony number is shown in the graph. Similar expression levels of FLAG-H2BWT and FLAG-H2BS32A in JB6 cells were also confirmed by Western blot using anti-FLAG as shown in the right panel. Total histone H3 was used to verify equal protein loading. Significant differences were evaluated using the Student's t test. The asterisk indicates a significant difference (p < 0.05). D, the incorporation of exogenous H2BWT and H2BS32A in the chromatin was determined by mononucleosome immunoprecipitation (IP). Oligonucleosome fragments were prepared from the nuclear pellets of JB6-parental, H2BWT, and H2BS32A cells and digested with micrococcal nuclease. The fragments were then used for immunoprecipitation with anti-FLAG-agarose beads, followed by immunoblotting (IB) with antibodies against H2BS32ph or FLAG. FLAG-H2BWT and FLAG-H2BS32A proteins are incorporated into the nucleosomes, and FLAG-H2BWT is phosphorylated at Ser32 in JB6 cells (*, FLAG-H2B; **, endogenous H2B). Coomassie Blue staining of nucleosomes precipitated by mononucleosome IP showed the presence of equal quantities of all the core histones and exogenous H2B proteins in both WT and mutant S32A cells. The input indicates that the preparation after micrococcal nuclease digestion was composed mainly of mononuclesomes, as reflected by the DNA size of ∼150 bp. DNA and histone proteins in mononuclesomes were separated by SDS and NaCl and then extracted with phenol-chloroform. The supernatant fraction containing the DNA was analyzed by 1% agarose gel electrophoresis. E, exogenous H2BWT and mutant H2BS32A proteins are mostly chromatin-bound in histone-expressing stable cells. Total soluble (S) and chromatin-bound (Chr) histones were extracted and subjected to Western blot analysis using antibodies against FLAG or histone H3. H3 served as a reference for endogenous histone level. Signals of longer exposure time from the same blots are also shown. The data are representative of three individual experiments. F, chromatin immunoprecipitation assay of endogenous RSK2. JB6 cells were seeded in 10-cm dishes and cultured overnight. The cells were subsequently cultured in 0.1% FBS-MEM for 24 h and then treated or not treated with 10 ng/ml EGF. After 15 min, the cells were harvested and subjected to ChIP assay using control IgG or an RSK2 antibody. RSK2 is enriched in c-jun and c-fos promoters upon EGF treatment but was not located in the β-globin gene. Results similar to those depicted here were obtained in at least two other independent sets of ChIP experiments. G, AP-1-luciferase assay using JB6 FLAG-H2BWT and FLAG-H2BS32A cells treated with EGF. Stable FLAG-H2BWT- and FLAG-H2BS32A-expressing JB6 cells were transiently transfected with the AP-1-luciferase reporter plasmid. At 36 h after transfection, the cells were starved for 24 h by incubating in serum-deprived MEM and then treated with 10 ng/ml of EGF. After 12 h, AP-1-luciferase activity was analyzed by disrupting the cells with lysis buffer (dual-luciferase reporter assay system; Promega) at room temperature for 30 min by gentle shaking, and then firefly luciferase activity was measured. The AP-1-luciferase activity was normalized against Renilla luciferase activity (phRL-SV40). Luciferase activity was presented as a fold increase (set at 1 for FLAG-H2BWT cells before EGF stimulation). The data are shown as the means ± S.D., and the asterisk indicates a significant difference (p < 0.05).
RSK2 Is an Important H2BS32 Kinase in the EGF-stimulated Signal Transduction Pathway
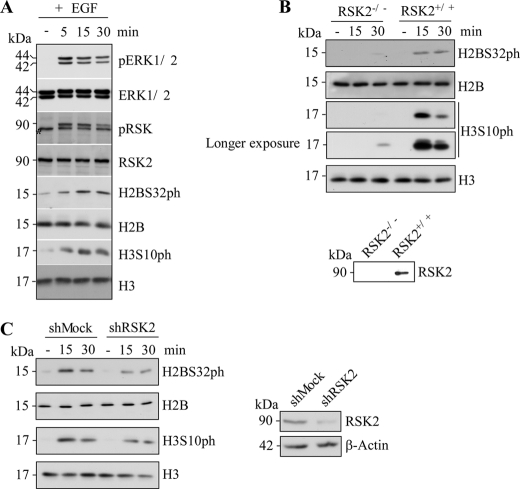
To determine the relationship of EGF-induced proliferation markers (10) and phosphorylation of H2BS32 in mouse epidermal JB6 cells, Western blot analysis was conducted. Serum-starved JB6 cells contain very little endogenous H2BS32ph, but EGF treatment induced phosphorylation at this site in a time-dependent manner (Fig. 7A). Notably, the kinetics of EGF-induced ERKs and RSK2 phosphorylation and activation closely parallel the induction of H2BS32 phosphorylation. H3S10 phosphorylation is also induced with EGF treatment, which is consistent with a previous report (Fig. 7A) (21).
FIGURE 7.
RSK2 is an important H2BS32 kinase in the EGF-stimulated signal transduction pathway. A, H2BS32 phosphorylation occurs in cells treated with tumor promoters such as EGF. JB6 cells were seeded in 10-cm dishes and cultured overnight. The cells were subsequently cultured in 0.1% FBS-MEM for 24 h and then treated or not treated with EGF (10 ng/ml) and harvested at various times (5, 15, and 30 min). The proteins were extracted, and Western blotting was conducted using specific antibodies as indicated. Equal protein loading was confirmed using antibodies to detect total ERK1/2, RSK2, histone H2B, or H3. The asterisk indicates a nonspecific band signal B, RSK2 deficiency blocks EGF-induced H2BS32ph. RSK2 wild type (RSK2+/+) and RSK2-deficient (RSK2−/−) MEFs (1 × 106) were each seeded in 10-cm dishes and grown to 90–95% confluence and subsequently cultured in 0.1% FBS-DMEM for 24 h. The cells were stimulated with EGF (10 ng/ml) and harvested 15 or 30 min later. Histone proteins were extracted, and Western blotting was conducted using specific antibodies as indicated. The RSK2 expression levels in RSK2 wild type (RSK2+/+) and RSK2-deficient (RSK2−/−) MEFs were also confirmed by Western blot (bottom). Signals of longer exposure time from the same blots are also shown for H3S10ph. C, knockdown of RSK2 in JB6 cells clearly attenuates the induction of H2BS32 phosphorylation by EGF. RSK2 expression levels in JB6 shMock and shRSK2 cells were first confirmed by Western blot (right). JB6 shMock and shRSK2 cells (1 × 106) were seeded in 10-cm dishes and grown to 90–95% confluence and then cultured in 0.1% FBS-MEM 24 h. The cells were stimulated with EGF (10 ng/ml) and harvested 15 or 30 min later. The histone proteins were extracted and visualized by Western blot. Equal protein loading was confirmed using antibodies to detect β-actin, histone H2B, or H3. Phosphorylation of H3S10 served as a positive marker for EGF stimulation.
Next we sought to determine whether RSK2 deficiency could block EGF-induced H2BS32 phosphorylation in the cell. We stimulated wild type RSK2 (RSK2+/+) or RSK2-deficient (RSK2−/−) MEFs with EGF, and the results showed that H2BS32ph was attenuated in RSK2−/− MEFs compared with wild type RSK2 MEFs in a 30-min time course period of EGF treatment (Fig. 7B). As a positive control, we also detected phosphorylation of H3S10 in these cells after EGF treatment. In agreement with the work of others and our findings, RSK2 is required for EGF-activated phosphorylation of H3S10, and H3S10ph was also severely attenuated in RSK2−/− MEFs compared with wild type RSK2 MEFs (Fig. 7B) (10, 21). Likewise, stable JB6 cells expressing shRNA knockdown of RSK2 clearly showed an attenuation of the induction of H2BS32 phosphorylation by EGF treatment (Fig. 7C). These data indicated that the ERK/RSK signaling pathway lies directly upstream of H2BS32 phosphorylation and that RSK2 is a bona fide H2BS32 kinase in the chromatin of cultured mammalian cells. Together, our results provide evidence suggesting that RSK2 phosphorylates H2BS32 in skin cells. Therefore, aberrantly increased expression of RSK2, as in skin cancer (17), might very likely cause hyperphosphorylation of H2BS32, an event linked to cell transformation and carcinogenesis.
DISCUSSION
Although histones have been studied for decades, the phosphorylation of serine, threonine, or tyrosine residues on the histone proteins and the subsequent biological meaning are still not completely understood, and this is especially true for histone H2B (22). So far, H2B serine phosphorylation has been reported in three different species. The mammalian H2BS14 or the yeast H2BS10 is phosphorylated by the mammalian sterile 20 kinase 1 (MST1), and the phosphorylation is associated with apoptosis (19, 23). The Drosophila H2BS33 is phosphorylated by the C-terminal kinase domain of the Drosophila TFIID subunit TAF1 and is essential for transcriptional activation events that promote cell cycle progression and development (24).
Genetic studies revealed the presence of a well conserved HBR, located at residues 30–37 of the H2B N-terminal tail (25). The HBR domain consists of a serine residue flanked by a series of lysine and arginine basic residues and is very important for transcriptional repression (25). Deletion of the yeast H2B tail segment, containing the HBR, was reported to decrease plasmid superhelical density, which probably reflects a release of DNA from the constraints of the nucleosome (26). Therefore, post-translational modification(s) within the HBR, such as phosphorylation, might result in relatively large increases in mass, which can induce structural changes of the nucleosome that open up or loosen DNA/histone interactions that affect gene regulation. Although these yeast studies provided some idea as to the function of this HBR domain, whether this domain has functions in higher eukaryotes is unclear. In this study, we focused on phosphorylation of residues in this HBR domain and identified H2BS32ph as new modification in mammalian cells. Results using the H2BS32ph antibody indicate that this modification mark exists in “normal” cycling cells in culture and that the phosphorylation occurs very rapidly (<5 min) following the addition of EGF. These observations raise the interesting question of whether a functional link exists between the turnover of this mark in normal cycling cells and its robust appearance in the early phases of EGF stimulation. During the immediate-early response of mammalian cells to mitogen stimulation, histone H3S10 is rapidly and transiently phosphorylated by one or more unidentified kinases (27). A recent report indicated that H3 phosphorylation at Thr45 is increased during apoptosis (28), and H2BS14 phosphorylation is suggested as a hallmark of apoptosis. Herein we showed that phosphorylation of H2BS32 is unrelated to apoptosis, suggesting that phosphorylation on the same histone molecule at different sites seems to result in very different biological functions.
Our results indicate that RSK2 is the kinase responsible for phosphorylation of H2BS32 during the early stages of EGF stimulation in skin cells. H2BS32 occupies a structurally interesting position within the nucleosome because it is positioned adjacent to the extreme N terminus of the first H2B α-helix and is an important determinant for the exit of the H2B N-terminal tail from the nucleosome. Perhaps its modification could have a role in nucleosome structure and stability.
In an effort to gain insight into the functional role of H2BS32ph in the induction of genes by EGF stimulation, we performed chromatin immunoprecipitations. However, the H2BS32ph-associated DNA was not immunoprecipitated. This could be due to the fact that the H2BS32ph antibody immunoprecipitates nucleosomes very poorly, presumably because H2BS32 is located close to the DNA gyres. Consequently, recognition of the epitope might be sterically hindered after cross-linking in the ChIP assay. Nevertheless, our data indicate that RSK2 is the kinase responsible for phosphorylation of H2BS32 during the early phase of EGF stimulation. By ChIP assays using FLAG antibody, we also showed that FLAG-H2BWT and FLAG-H2BS32A are indeed incorporated in the c-jun and c-fos promoters (data not shown). In addition, when we examined the AP-1 activation of these cells under EGF treatment, AP-1 activity was attenuated in the FLAG-H2BS32A cells as compared with FLAG-H2BWT cells. This suggests that Ser32 phosphorylation is critical for the transcriptional activation of c-jun and c-fos expression. Because AP-1 activity is a major driver in cell transformation, this could also explain the decreased cell growth and cell transformation observed in H2BS32A cells. We suggest that phosphorylation of H2BS32, with the resulting introduction of a negative charge close to the DNA, would affect the binding energies of the nucleosome. Perhaps the altered nucleosomal/chromatin structure favors DNA processing in gene activation/transcription. Alternatively, phosphorylation of H2BS32 might provide a docking site for a protein in a manner similar to that of phosphorylated H3 (S10ph or S28ph) where the 14-3-3 protein binds (29).
Taken together, the data reveal that H2B is a novel physiological substrate of RSK2, and H2BS32ph is indispensable for mediating the effects of RSK2 and AP-1 activity. Notably, besides the well known function of H2BS14 phosphorylation in apoptosis, H2BS32 is a novel mitogen-responsive phosphorylation site that is associated with cell transformation. Importantly, because H2BS32ph is related to cell transformation and, similar to RSK2, is found at high levels in skin cancers, understanding the exact role of this histone modification in neoplastic transformation could have implications in the treatment/screening of certain human pathological conditions that are linked to skin cancer (30).
Acknowledgments
We thank Dr. Shigeyuki Yokoyama (RIKEN Genomic Sciences Center, 1-7-22 Suehiro-cho, Tsurumi, Yokohama 230-0045, Japan) for the pHCE-H2B plasmid and Dr. Edward H. Hinchcliffe from the Cellular Dynamics Section of the Hormel Institute for image deconvolution and analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants CA077646, R37CA081064, CA120388, and ES016548. This work was also supported by the Hormel Foundation.
- H2BS32
- histone H2B serine 32
- H2BS32ph
- phosphorylation of H2BS32 or phosphorylated H2BS32
- RSK2
- ribosomal S6 kinase 2
- HBR
- H2B repression domain
- AP-1
- activator protein-1
- MEM
- minimum essential medium
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Cheung P., Allis C. D., Sassone-Corsi P. (2000) Cell 103, 263–271 [DOI] [PubMed] [Google Scholar]
- 2. Wang G. G., Allis C. D., Chi P. (2007) Trends Mol. Med. 13, 363–372 [DOI] [PubMed] [Google Scholar]
- 3. Bönisch C., Nieratschker S. M., Orfanos N. K., Hake S. B. (2008) Expert Rev. Proteomics 5, 105–119 [DOI] [PubMed] [Google Scholar]
- 4. Delcuve G. P., Rastegar M., Davie J. R. (2009) J. Cell. Physiol. 219, 243–250 [DOI] [PubMed] [Google Scholar]
- 5. Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 7. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 8. Sarma K., Nishioka K., Reinberg D. (2004) Methods Enzymol. 376, 255–269 [DOI] [PubMed] [Google Scholar]
- 9. Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z. (2008) Cell 134, 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cho Y. Y., Yao K., Kim H. G., Kang B. S., Zheng D., Bode A. M., Dong Z. (2007) Cancer Res. 67, 8104–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu Y. M., Zhu F., Cho Y. Y., Carper A., Peng C., Zheng D., Yao K., Lau A. T., Zykova T. A., Kim H. G., Bode A. M., Dong Z. (2010) Cancer Res. 70, 3218–3227 [DOI] [PubMed] [Google Scholar]
- 12. Kanda T., Sullivan K. F., Wahl G. M. (1998) Curr. Biol. 8, 377–385 [DOI] [PubMed] [Google Scholar]
- 13. Manders E. M., Stap J., Brakenhoff G. J., van Driel R., Aten J. A. (1992) J. Cell Sci. 103, 857–862 [DOI] [PubMed] [Google Scholar]
- 14. Clayton A. L., Rose S., Barratt M. J., Mahadevan L. C. (2000) EMBO J. 19, 3714–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu C., Zhu F., Cho Y. Y., Tang F., Zykova T., Ma W. Y., Bode A. M., Dong Z. (2006) Mol. Cell 23, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee S. Y., Lau A. T., Jeong C. H., Shim J. H., Kim H. G., Kim J., Bode A. M., Dong Z. (2010) J. Biol. Chem. 285, 29525–29534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cho Y. Y., Yao K., Pugliese A., Malakhova M. L., Bode A. M., Dong Z. (2009) Cancer Res. 69, 4398–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho Y. Y., He Z., Zhang Y., Choi H. S., Zhu F., Choi B. Y., Kang B. S., Ma W. Y., Bode A. M., Dong Z. (2005) Cancer Res. 65, 3596–3603 [DOI] [PubMed] [Google Scholar]
- 19. Cheung W. L., Ajiro K., Samejima K., Kloc M., Cheung P., Mizzen C. A., Beeser A., Etkin L. D., Chernoff J., Earnshaw W. C., Allis C. D. (2003) Cell 113, 507–517 [DOI] [PubMed] [Google Scholar]
- 20. Clark D. E., Errington T. M., Smith J. A., Frierson H. F., Jr., Weber M. J., Lannigan D. A. (2005) Cancer Res. 65, 3108–3116 [DOI] [PubMed] [Google Scholar]
- 21. Sassone-Corsi P., Mizzen C. A., Cheung P., Crosio C., Monaco L., Jacquot S., Hanauer A., Allis C. D. (1999) Science 285, 886–891 [DOI] [PubMed] [Google Scholar]
- 22. Lall S. (2007) Nat. Struct. Mol. Biol. 14, 1110–1115 [DOI] [PubMed] [Google Scholar]
- 23. Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., Allis C. D. (2005) Cell 120, 25–36 [DOI] [PubMed] [Google Scholar]
- 24. Maile T., Kwoczynski S., Katzenberger R. J., Wassarman D. A., Sauer F. (2004) Science 304, 1010–1014 [DOI] [PubMed] [Google Scholar]
- 25. Parra M. A., Kerr D., Fahy D., Pouchnik D. J., Wyrick J. J. (2006) Mol. Cell. Biol. 26, 3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lenfant F., Mann R. K., Thomsen B., Ling X., Grunstein M. (1996) EMBO J. 15, 3974–3985 [PMC free article] [PubMed] [Google Scholar]
- 27. Bode A. M., Dong Z. (2005) Sci. STKE 2005, re4 [DOI] [PubMed] [Google Scholar]
- 28. Hurd P. J., Bannister A. J., Halls K., Dawson M. A., Vermeulen M., Olsen J. V., Ismail H., Somers J., Mann M., Owen-Hughes T., Gout I., Kouzarides T. (2009) J. Biol. Chem. 284, 16575–16583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Macdonald N., Welburn J. P., Noble M. E., Nguyen A., Yaffe M. B., Clynes D., Moggs J. G., Orphanides G., Thomson S., Edmunds J. W., Clayton A. L., Endicott J. A., Mahadevan L. C. (2005) Mol. Cell 20, 199–211 [DOI] [PubMed] [Google Scholar]
- 30. Bode A. M., Dong Z. (2009) Sci. Signal. 2, mr2 [DOI] [PMC free article] [PubMed] [Google Scholar]