Abstract
The 26 S proteasome comprises two multisubunit subcomplexes as follows: 20 S proteasome and PA700/19 S regulatory particle. The cellular mechanisms by which these subcomplexes assemble into 26 S proteasome and the molecular determinants that govern the assembly process are poorly defined. Here, we demonstrate the nonequivalent roles of the C termini of six AAA subunits (Rpt1–Rpt6) of PA700 in 26 S proteasome assembly in mammalian cells. The C-terminal HbYX motif (where Hb is a hydrophobic residue, Y is tyrosine, and X is any amino acid) of each of two subunits, Rpt3 and Rpt5, but not that of a third subunit Rpt2, was essential for assembly of 26 S proteasome. The C termini of none of the three non-HbYX motif Rpt subunits were essential for cellular 26 S proteasome assembly, although deletion of the last three residues of Rpt6 destabilized the 20 S-PA700 interaction. Rpt subunits defective for assembly into 26 S proteasome due to C-terminal truncations were incorporated into intact PA700. Moreover, intact PA700 accumulated as an isolated subcomplex when cellular 20 S proteasome content was reduced by RNAi. These results indicate that 20 S proteasome is not an obligatory template for assembly of PA700. Collectively, these results identify specific structural elements of two Rpt subunits required for 26 S proteasome assembly, demonstrate that PA700 can be assembled independently of the 20 S proteasome, and suggest that intact PA700 is a direct intermediate in the cellular pathway of 26 S proteasome assembly.
Keywords: ATPases, Protease, Proteasome, Protein Assembly, Protein Degradation, PA700, Rpt Subunits
Introduction
Cellular assembly of the 26 S proteasome, a 2,500,000-dalton protease complex responsible for the selective intracellular degradation of polyubiquitylated proteins, requires coordinated association of over 60 subunits representing the products of ∼35 genes (1, 2). These subunits are arranged as two structurally and functionally distinct subcomplexes as follows: a protease, termed 20 S proteasome or core particle, and an ATPase regulator, termed PA700 or 19 S regulatory particle (3). The 700,000-dalton 20 S proteasome consists of four axially stacked heptameric rings. Each of the two identical inner rings of eukaryotic 20 S proteasome contains seven different but homologous β-type subunits (β1–β7), and each of the two identical outer rings contains seven different but homologous α-type subunits (α1–α7) (4, 5). The resulting α7-β7-β7-α7 cylindrical complex features C2 symmetry about an axial plane through the β rings (6).
In eukaryotic cells, the 20 S proteasome is assembled with the aid of at least five dedicated assembly chaperones (7, 8). The process proceeds through multiple intermediate subassemblies and concludes when two immature half-proteasomes (α1–7, β1–7, and associated chaperones) combine. The final assembly step results in exposure of active site threonine residues by autolytic removal of pro-sequences from the N termini of three catalytic β subunits (β1, β2, and β5) and degradation of the associated chaperones. Formation of intact 20 S proteasome, however, seals the catalytic threonines in an interior chamber that physically sequesters them from protein substrates (5, 9). Two other structural features of the 20 S proteasome further limit its catalytic potential. First, the only route for substrates to reach the interior degradation chamber is through 13-Å pores in the center of the outer α rings (6). Second, the pores are reversibly gated by alternative conformations of the N termini of α subunits (10). Thus, substrate access to the central degradation chamber requires both an open gate conformation of the proteasome and unfolded polypeptide substrates capable of traversing the narrow access portal. As described below, the PA700 subcomplex functions to satisfy each of these requirements.
PA700 is composed of about 20 different subunits, including six homologous AAA (ATPase associated with various cellular activities) proteins (Rpt1–Rpt6) arranged as a heterohexameric ring that forms the binding interface of PA700 with either or both of the heteroheptameric α rings of the 20 S proteasome (3). Three additional subunits (Rpn1, Rpn2, Rpn13, and in many species a fourth, Uch37) associate with the ATPase ring to collectively constitute a PA700 substructure termed “base” (11). The remaining PA700 subunits (Rpn3, Rpn5–12, and Rpn15) constitute the “lid,” a substructure positioned distally to 20 S proteasome.
With the exception of peptide bond hydrolysis per se, all functions necessary for 26 S proteasome-catalyzed degradation of polyubiquitylated proteins are provided by PA700 (12). Thus, PA700 selects substrates by binding polyubiquitin chains (13), prepares the client protein for passage through the substrate access pore by unfolding it and detaching its ubiquitin chains (14–16), and translocates the unfolded deubiquitylated polypeptide to the catalytic chamber for hydrolysis (17). These processes appear to be mechanistically coupled to one another by PA700-catalyzed ATP hydrolysis, which is required for overall proteolysis of ubiquitylated substrates (18).
In addition to substrate binding and processing, PA700 also mediates 26 S proteasome function by inducting the open gate conformation of the substrate access pore (19). Recent work has revealed critical roles of the extreme C termini of certain Rpt subunits in this process. Three of the six Rpt subunits (Rpt2, Rpt3, and Rpt5) feature C-terminal residues that conform to an HbYX motif (where Hb is a hydrophobic residue; Y is tyrosine, and X is any amino acid) (20). These residues bind to pockets between specific α subunits of 20 S proteasome (21–24). Binding of HbYX residues of either of two Rpt subunits (Rpt2 and Rpt5) or of the HbYX residues of PAN, an Rpt subunit ortholog from archaea, induces gate opening, even when these residues are in the form of isolated short peptides corresponding to the C terminus of respective subunits (20, 24, 25).
Despite the documented physical interactions between HbYX residues of PA700 and the 20 S proteasome, the roles and contributions of these interactions to cellular 26 S proteasome assembly and to the structural stability of the assembled complex remain unclear and are the subject of conflicting results. Support for an important role of HbYX residues in these processes comes from both in vitro and cellular studies. For example, in vitro reconstitution of 26 S proteasome from purified 20 S proteasome and PA700 is abrogated when the HbYX residues of Rpt2 and Rpt5 are enzymatically removed (24). Moreover, Rpt3 lacking its HbYX residues is defective for incorporation into 26 S proteasome in mammalian cells (25). However, other results appear to contradict these findings and seem inconsistent with a model in which HbYX motifs are the principal binding elements for the PA700–20 S proteasome interaction. For example, 26 S proteasome assembly is not appreciably diminished in yeast when HbYX motif tyrosine residues are mutated in Rpt2, Rpt3, or Rpt5 (20), and deletion of the last residue of each Rpt subunit severely diminishes 26 S proteasome assembly for only the non-HbYX motif subunits Rpt4 or Rpt6 (26). Such apparent discrepancies may reflect differences in experimental design and detail among studies or reflect roles for Rpt C-terminal residues in cellular assembly processes other than binding per se to the proteasome (see below). In any case, these inconsistent results highlight the lack of understanding of the specific and relative roles of the C termini of Rpt subunits in proteasome assembly.
In contrast to the relatively advanced knowledge of 20 S proteasome assembly, much less is known about cellular mechanisms of PA700 assembly and the relationship of this process to 26 S proteasome assembly. Recent studies have provided important but incomplete and, in some instances, conflicting data regarding cellular assembly of PA700. Multiple studies in yeast and more limited studies in mammalian cells show that the six Rpt subunits initially associate as three separate but discrete subunit pairs in association with pair-specific chaperones (26–33). The three Rpt subassemblies subsequently combine to form a structure equivalent to the base. Conflicting data, however, have been obtained regarding the role of the 20 S proteasome in both this and subsequent steps of PA700 formation. These conflicts include a possible requirement for 20 S proteasome as a template for base assembly, the mechanism of lid assembly, and the physical and temporal relationship between assembly of the base and the lid (26, 34, 35). Thus, it remains uncertain whether PA700 is formed as a separate intact entity prior to its binding to 20 S proteasome or whether it is assembled sequentially from subassemblies using 20 S proteasome and perhaps other PA700 subassemblies as templates.
The purpose of this study was to determine the relative roles of the C termini of Rpt subunits for 26 S proteasome assembly in intact mammalian cells. We sought to test the hypothesis, based in part on previous in vitro biochemical studies, that intact HbYX residues of Rpt2, Rpt3, and Rpt5, but not corresponding non-HbYX residues of Rpt1, Rpt4, and Rpt6, would be essential for cellular 26 S proteasome assembly and to establish whether the HbYX motifs of different subunits played independent or cooperative roles in the assembly process (24, 25). We also sought to evaluate the physiologic significance of our in vitro reconstitution assay of 26 S proteasome from isolated 20 S proteasome and PA700 subcomplexes by determining whether PA700 is formed as an intact complex prior to its cellular assembly into 26 S proteasome. Our results reveal that features of cellular 26 S proteasome assembly mirror some but not all of those expected from biochemical studies and provide new insights to the mechanisms of this process.
EXPERIMENTAL PROCEDURES
DNA Constructs
cDNAs encoding each full-length (WT) human Rpt subunit and each subunit lacking three C-terminal amino acids (−C3) were subcloned into pIRESpuro3 expression vector (Clontech) featuring the N-terminal FLAG epitope. Each construct was confirmed by DNA sequencing.
Preparation of HEK293 Cell Lines with Stable Expression of FLAG-tagged Rpt Subunits
HEK293 cell lines were cultured in Dulbecco's modified Eagle's medium (Invitrogen) containing high glucose and glutamine, supplemented with 10% fetal bovine serum in the presence of 5% CO2 at 37 °C. HEK293 cells were transfected at ∼60% confluence with respective vectors using FuGENE 6 (Roche Applied Science). Forty eight h after transfection, the media were replaced with media containing 5 μg/ml puromycin, and cells were incubated further for 4 weeks for selection of clones exhibiting stable expression of respective proteins. Each stable cell line was grown to ∼95% confluence, harvested, and washed with phosphate-buffered saline. Cells were disrupted in ice-cold buffer consisting of 50 mm Tris-HCl, pH 7.5, at 4 °C, 5 mm MgCl2, 1 mm β-mercaptoethanol, 1 mm ATP, and, depending on the experiment, protease inhibitor mixture (Roche Applied Science) by 15 passages through a 27-gauge needle. The lysates were centrifuged for 20 min to obtain a crude soluble fraction. Expression of Rpt proteins was determined by Western blot analysis using anti-FLAG M2 antibody (Sigma) and corresponding anti-Rpt antibodies.
Glycerol Density Gradient Centrifugation
Glycerol density gradient centrifugation was conducted as described previously using 10–40% linear glycerol gradients in buffer containing 50 mm Tris-HCl, pH 7.5, 20 mm NaCl, 1 mm β-mercaptoethanol, 1 mm ATP, and 5 mm MgCl2 (36) Control centrifugations established the sedimentation positions of purified bovine 26 S proteasome, PA700, and 20 S proteasome (18).
Affinity Purification of Proteasome Complexes
Cells were cultured, harvested, and lysed as described above. Approximately 20 mg of each cell extract normalized for the level of FLAG-Rpt protein was mixed gently for 2 h at 4 °C with 100 μl of anti-FLAG M2-agarose beads (Sigma) in 50 mm Tris-HCl, pH 7.5, at 4 °C, 100 mm NaCl, 1 mm β-mercaptoethanol, 1 mm ATP, 5 mm MgCl2, 10% glycerol, and 0.1% Nonidet P-40. The beads were harvested by centrifugation and washed three times with the same buffer. Bound proteins were eluted at 4 °C with 150 μl of elution buffer containing 200 μg/ml FLAG peptide (Sigma). Identical volumes of each affinity-purified protein sample (10 μl) were characterized by proteasome activity, SDS-PAGE, Western blotting, and native PAGE, as described in the text and individual figure legends.
Proteasome Activity
Proteasome activity was measured fluorescently by determining the rate of hydrolysis of AMC2 from the peptide Suc-Leu-Leu-Val-Tyr-AMC, as described previously (18). All values represent the mean of triplicate assays and are expressed as arbitrary fluorescent units (AFU) produced per min.
PA700 Activity
PA700 activity was determined by the ATP-dependent activation of 20 S proteasome activity (38). In brief, PA700 was preincubated with 100 ng of purified bovine 20 S proteasome, prior to addition of Suc-Leu-Leu-Val-Tyr-AMC and proteasome assay, as described above. Assays were performed in triplicate, and activities are expressed as AFU produced per min.
Native PAGE
Nondenaturing PAGE was conducted with 4% polyacrylamide gels, as described previously (18). Gels were either stained with silver, blotted for proteins of interest, or assayed for in-gel proteasome activity by overlaying a 50 μm solution of Suc-Leu-Leu-Val-Tyr-AMC. After incubation for 15–30 min at 37 °C, AMC production was visualized by exposure of the gel to fluorescent light.
RNA Interference (RNAi) with siRNA
RNAi of the β5 subunit of 20 S proteasome was conducted by transfection of double-stranded siRNA oligonucleotides (5′-GAAGGUGAUAGAGAUCAAC-3′, Dharmacon, Boulder, CO), as described previously (36). Control transfections used a proven nontargeting siRNA (D-001210-01-05) provided by Dharmacon. The HEK293 cells expressing FLAG-Rpt6 (WT) were plated, grown to 40–50% confluence, and transfected with 20 nm siRNA using Lipofectamine 2000 (Invitrogen). 72 h after transfection, cells were harvested, and cell lysates were prepared and analyzed by Western blotting. The remaining cell extracts were subjected to FLAG affinity purification and characterized as described in the text.
RESULTS
Exogenously Expressed Rpt Subunits of PA700 Are Incorporated into Intact 26 S Proteasome in Mammalian Cells
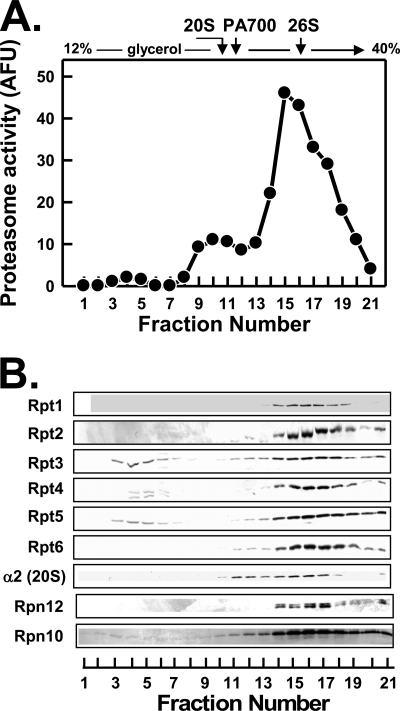
Structural and functional states of proteasome complexes in cultured cells can be analyzed simply and reproducibly by glycerol density gradient centrifugation of corresponding cell-free extracts. Normal HEK293 cells, like most other commonly studied mammalian cells, feature a bimodal distribution of proteasome activity in gradient fractions. Most proteasome activity sediments at a position characteristic of 26 S proteasome, whereas a smaller amount of the slower sedimenting activity is found in a position characteristic of 20 S proteasome (Fig. 1, panel A). Western blotting of gradient fractions reveals a bimodal distribution of 20 S proteasome subunits corresponding to the proteasome activity profiles (Fig. 1, panel B). Western blotting also showed that the majority of each Rpt subunit was present in 26 S proteasome. Thus, these subunits co-sedimented with one another, with other subunits of the 26 S proteasome, and with 26 S proteasome activity (Fig. 1). Lesser amounts of the Rpt subunits sedimented at a position characteristic of free intact PA700. Small but detectable amounts of several subunits, including Rpt3, Rpt4, and Rpt5, sedimented significantly slower than intact PA700 and probably represent subassemblies of PA700 described previously (29, 30, 32).
FIGURE 1.
Most endogenous Rpt proteins are present in 26 S proteasome. Soluble extracts of HEK293 cells were prepared and fractionated by glycerol density gradient centrifugation as described under “Experimental Procedures.” Panel A, gradient fractions were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC as substrate. The sedimentation positions of purified 26 S proteasome and PA700 were determined in independent experiments and are indicated by arrows. Panel B, gradient fractions were subjected to Western blotting with the antibodies against the indicated subunits of 26 S proteasome. AFU, arbitrary fluorescent units.
To analyze the relative roles of the C termini of Rpt subunits in 26 S proteasome assembly, we created a series of HEK293 cell lines that stably express N-terminally FLAG-tagged versions of a given Rpt subunit either as a wild-type (WT) protein or as a protein whose three C-terminal residues are deleted (−C3). We utilized the epitope tag to monitor the steady-state incorporation of these proteins into 26 S proteasome and/or other protein complexes by glycerol density gradient centrifugation, as described above for endogenous proteins. We also exploited the epitope tag to affinity-purify and characterize protein complexes into which these proteins were assembled. In this manner, we could determine the influence of the C terminus on assembly of a given Rpt subunit into 26 S proteasome, compare the relative roles of C termini of different Rpt subunits for this process, and determine the fate of Rpt subunits that were defective for 26 S proteasome assembly. To avoid potential complications of protein overexpression in this analysis, we initially selected cell lines that expressed the FLAG-tagged proteins at levels approximately equal to or less than those of their respective endogenous counterparts.
With the exception of Rpt2 (see below), each wild-type FLAG-tagged Rpt protein was incorporated into intact 26 S proteasomes as judged by the distribution of FLAG-Rpt protein in fractions from the glycerol density gradient centrifugation of cell extracts (panel B in Figs. 2, 3, and 5–7). Additional structural and functional analysis of protein complexes purified with anti-FLAG beads confirmed this conclusion. For example, SDS-PAGE of the affinity-purified proteins revealed a set of stained proteins characteristic of purified 26 S proteasome subunits. Western blotting with antibodies against selected 26 S proteasome subunits revealed the presence of subunits for both the base and lid subassemblies of PA700 as well as for 20 S proteasome (panel C in Figs. 2, 3, and 5–7). The affinity-purified proteins also featured proteasome-specific peptidase activity (panel D in Figs. 2, 3, and 5–7) and electrophoretic migration positions characteristic of singly and doubly capped 26 S proteasome on native polyacrylamide gels (panel E in Figs. 2, 3, and 5–7). The latter were detected by protein staining, Western blotting with anti-FLAG antibodies, and proteasome activity using an in-gel peptidase overlay assay.
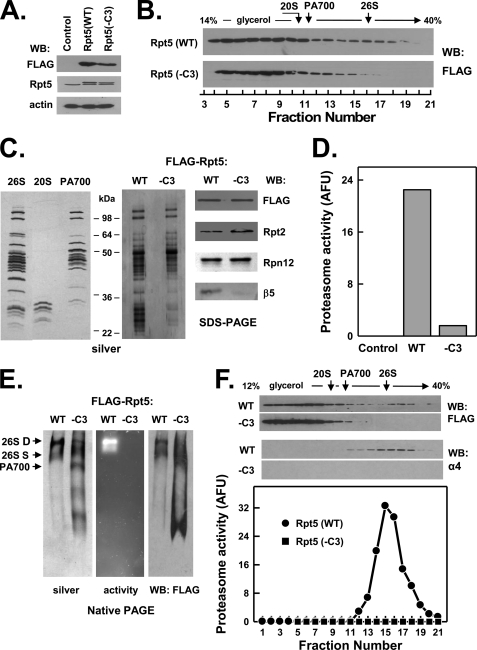
FIGURE 2.
C-terminal HbYX motif of Rpt5 is essential for assembly of the 26 S proteasome. HEK293 cells stably expressing FLAG-tagged wild-type Rpt5 (FLAG-Rpt5-WT) or FLAG-tagged Rpt5 lacking the last three C-terminal residues (FLAG-Rpt5−C3) were prepared as described under “Experimental Procedures.” Panel A, cell extracts of nontransfected (Control) and each Rpt-expressing cell line were prepared and subjected to Western blotting (WB) with antibodies against FLAG epitope, Rpt5 protein, and actin. Panel B, extracts from cell lines expressing indicated proteins were subjected to glycerol density gradient centrifugation. Gradient fractions were subjected to Western blotting with anti-FLAG antibodies. Arrows indicate the known sedimentation positions of purified bovine 20 S proteasome, PA700, and 26 S proteasome. Panels C–F, affinity purification of FLAG proteins from extracts of indicated cell lines was conducted as described under “Experimental Procedures.” Panel C, affinity-purified samples were subjected to SDS-PAGE and stained with silver (center panel) or Western-blotted with antibodies against indicated proteins (right panel). For reference, purified bovine 26 S proteasome, 20 S proteasome, and PA700 were subjected to SDS-PAGE and stained with silver (right panel). Panel D, affinity-purified samples were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate. WT and −C3 samples contained equal amounts of FLAG protein. Control represents a sample of nontransfected cell extracts subjected to the affinity purification procedure. Panel E, affinity-purified samples were subjected to native PAGE and stained with silver (left panel), analyzed for in-gel proteasome activity peptide substrate overlay (center panel), or Western-blotted with antibodies against FLAG (right panel). Arrows indicate the known migration positions of purified bovine 26 S proteasome containing two (D) or one (S) PA700 complex or PA700. Panel F, affinity-purified samples from indicated cell lines were subjected to glycerol density gradient centrifugation. Fractions were Western-blotted with antibodies against FLAG or the α4 subunit of 20 S proteasome (upper panel). Fractions from WT extracts were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate (lower panel). Similar results were obtained in three independent cell preparations. AFU, arbitrary fluorescent units.
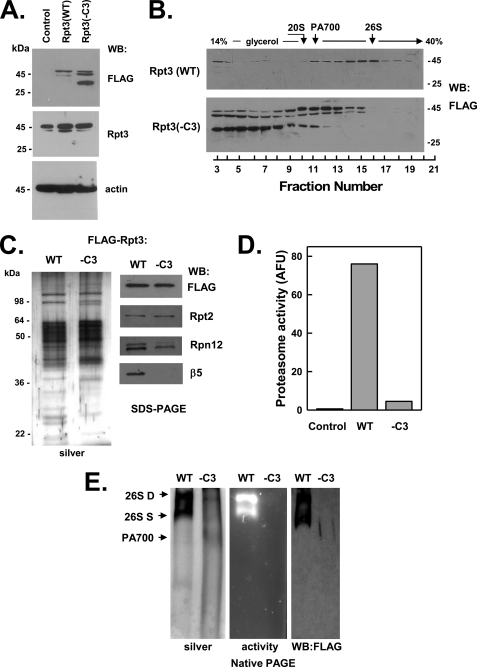
FIGURE 3.
C terminus of Rpt3 is essential for assembly of the 26 S proteasome. HEK293 cells stably expressing FLAG-tagged wild-type Rpt3 (FLAG-Rpt3-WT) or FLAG-tagged Rpt3 lacking the last three C-terminal residues (FLAG-Rpt3−C3) were prepared as described under “Experimental Procedures.” Panel A, cell extracts of nontransfected (Control) and each Rpt-expressing cell line were prepared and subjected to Western blotting (WB) with antibodies against FLAG epitope, Rpt3 protein, and actin. Panel B, extracts from cell lines expressing indicated proteins were subjected to glycerol density gradient centrifugation. Gradient fractions were subjected to Western blotting with anti-FLAG antibodies. Arrows indicate the known sedimentation positions of purified bovine 20 S proteasome, PA700, and 26 S proteasome. Panels C–E, affinity purification of FLAG proteins from extracts of indicated cell lines was conducted as described under “Experimental Procedures.” Panel C, affinity-purified samples were subjected to SDS-PAGE and stained with silver (left panel) or Western-blotted with antibodies against indicated proteins (right panel). Panel D, affinity-purified samples were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate. WT and −C3 samples contained equal amounts of FLAG protein. Control represents a sample of nontransfected cell extract subjected to the affinity purification procedure. Panel E, affinity-purified samples were subjected to native PAGE and stained with silver (left panel), analyzed for in-gel proteasome activity peptide substrate overlay (center panel), or Western-blotted with antibodies against FLAG (right). Arrows indicate the known migration positions of purified bovine 26 S proteasome containing two (D) or one (S) PA700 complex or PA700. Similar results were obtained in three independent cell preparations. AFU, arbitrary fluorescent units.
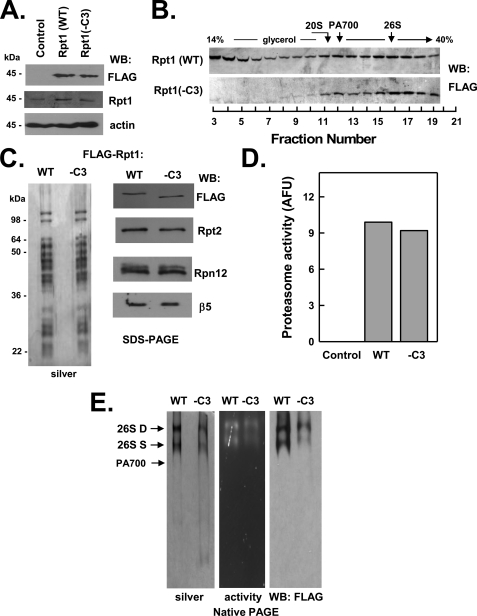
FIGURE 5.
C terminus of Rpt1 is not essential for assembly of the 26 S proteasome. HEK293 cells stably expressing FLAG-tagged wild-type Rpt1 (FLAG-Rpt1-WT) or FLAG-tagged Rpt1 lacking the last three C-terminal residues (FLAG-Rpt1−C3) were prepared as described under “Experimental Procedures.” Panel A, cell extracts of nontransfected (Control) and each Rpt-expressing cell line were prepared and subjected to Western blotting (WB) with antibodies against FLAG epitope, Rpt1 protein, and actin. Panel B, extracts from cell lines expressing indicated proteins were subjected to glycerol density gradient centrifugation. Gradient fractions were subjected to Western blotting with anti-FLAG antibodies. Arrows indicate the known sedimentation positions of purified bovine 20 S proteasome, PA700, and 26 S proteasome. Panels C–E, affinity purification of FLAG proteins from extracts of indicated cell lines was conducted as described under “Experimental Procedures.” Panel C, affinity-purified samples were subjected to SDS-PAGE and stained with silver (left panel) or Western-blotted with antibodies against indicated proteins (right panel). Panel D, affinity-purified samples were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate. WT and −C3 samples contained equal amounts of FLAG protein. Control represents a sample of nontransfected cell extracts subjected to the affinity purification procedure. Panel E, affinity-purified samples were subjected to native PAGE and stained with silver (left panel), analyzed for in-gel proteasome activity peptide substrate overlay (center panel), or Western-blotted with antibodies against FLAG (right panel). Arrows indicate the known migration positions of purified bovine 26 S proteasome containing two (D) or one (S) PA700 complex or PA700. Similar results were obtained in three independent cell preparations. AFU, arbitrary fluorescent units.
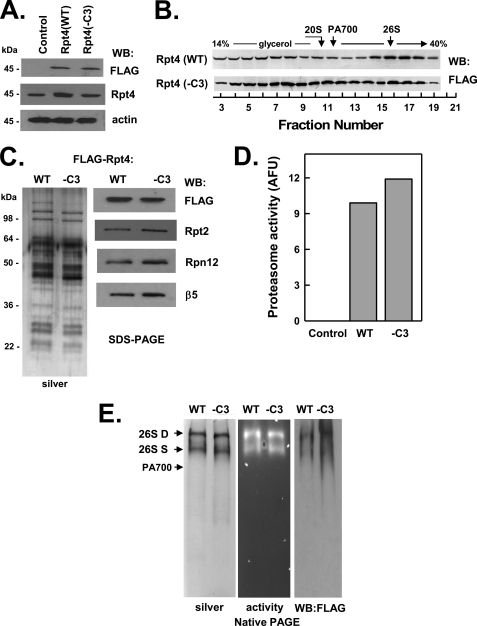
FIGURE 6.
C terminus of Rpt4 is not essential for assembly of the 26 S proteasome. HEK293 cells stably expressing FLAG-tagged wild-type Rpt4 (FLAG-Rpt4-WT) or FLAG-tagged Rpt4 lacking the last three C-terminal residues (FLAG-Rpt4−C3) were prepared as described under “Experimental Procedures.” Panel A, cell extracts of nontransfected (Control) and each Rpt-expressing cell line were prepared and subjected to Western blotting (WB) with antibodies against FLAG epitope, Rpt4 protein, and actin. Panel B, extracts from cell lines expressing indicated proteins were subjected to glycerol density gradient centrifugation. Gradient fractions were subjected to Western blotting with anti-FLAG antibodies. Arrows indicate the known sedimentation positions of purified bovine 20 S proteasome, PA700, and 26 S proteasome. Panels C–E, affinity purification of FLAG proteins from extracts of indicated cell lines was conducted as described under “Experimental Procedures.” Panel C, affinity-purified samples were subjected to SDS-PAGE and stained with silver (left panel) or Western-blotted with antibodies against indicated proteins (right panel). Panel D, affinity-purified samples were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate. WT and −C3 samples contained equal amounts of FLAG protein. Control represents a sample of nontransfected cell extracts subjected to the affinity purification procedure. Panel E, affinity-purified samples were subjected to native PAGE and stained with silver (left panel), analyzed for in-gel proteasome activity peptide substrate overlay (center panel), or Western-blotted with antibodies against FLAG (right panel). Arrows indicate the known migration positions of purified bovine 26 S proteasome containing two (D) or one (S) PA700 complex or PA700. Similar results were obtained in three independent cell preparations. AFU, arbitrary fluorescent units.
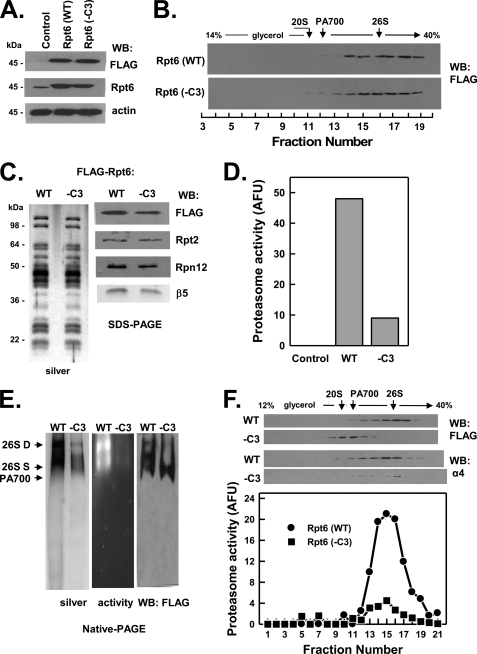
FIGURE 7.
C terminus of Rpt6 is not essential for assembly of the 26 S proteasome but reduces 26 S proteasome stability. HEK293 cells stably expressing FLAG-tagged wild-type Rpt6 (FLAG-Rpt6-WT) or FLAG-tagged Rpt6 lacking the last three C-terminal residues (FLAG-Rpt6−C3) were prepared as described under “Experimental Procedures.” Panel A, cell extracts of nontransfected (Control) and each Rpt-expressing cell line were prepared and subjected to Western blotting (WB) with antibodies against FLAG epitope, Rpt6 protein, and actin. Panel B, extracts from cell lines expressing indicated proteins were subjected to glycerol density gradient centrifugation. Gradient fractions were subjected to Western blotting with anti-FLAG antibodies. Arrows indicate the known sedimentation positions of purified bovine 20 S proteasome, PA700, and 26 S proteasome. Panels C–F, affinity purification of FLAG proteins from extracts of indicated cell lines was conducted as described under “Experimental Procedures.” Panel C, affinity-purified samples were subjected to SDS-PAGE and stained with silver (left panel) or Western-blotted with antibodies against indicated proteins (right panel). Panel D, affinity-purified samples were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate. WT and −C3 samples contained equal amounts of FLAG protein. Control represents a sample of nontransfected cell extract subjected to the affinity purification procedure. Panel E, affinity-purified FLAG proteins were subjected to native PAGE and stained with silver (left panel), analyzed for in-gel proteasome activity peptide substrate overlay (center panel), or Western-blotted with antibodies against FLAG (right panel). Arrows indicate the known migration positions of purified bovine 26 S proteasome containing two (D) or one (S) PA700 complex or PA700. Panel F, affinity-purified FLAG proteins from indicated cell lines were subjected to glycerol density gradient centrifugation. Fractions were Western-blotted with antibodies against FLAG or the α4 subunit of 20 S proteasome (upper panel). Fractions from WT and −C3 extracts were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate (lower panel). Similar results were obtained in three independent cell preparations. AFU, arbitrary fluorescent units.
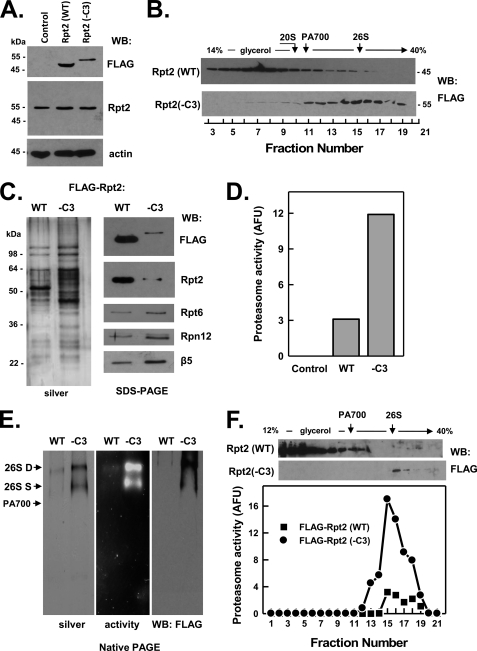
In contrast to other FLAG-Rpt wild-type proteins, FLAG-Rpt2(WT) appeared to undergo C-terminal proteolytic processing, as judged by its increased mobility on SDS-PAGE when compared with endogenous Rpt2 (Fig. 4, panel A). The processing, which decreased the apparent size of the protein by about 10 kDa, most likely occurred in intact cells rather than during preparation and analysis of the extract and also was observed when the protein was expressed transiently (data not shown). Most FLAG-Rpt2(WT) protein sedimented slowly during glycerol density gradient centrifugation, indicating that the processed protein was present mainly in low molecular weight complexes rather than in 26 S proteasome or intact PA700 complexes (Fig. 4, panel B). Affinity purification of FLAG-Rpt2(WT) complexes displayed low levels of both 26 S proteasome activity and intact 26 S proteasome protein (Fig. 4, panels C–E). Additional characterization of FLAG-Rpt2(WT) protein is described below.
FIGURE 4.
C terminus of Rpt2 is not critical for assembly of the 26 S proteasome. HEK293 cells stably expressing FLAG-tagged wild-type Rpt2 (FLAG-Rpt2-WT) or FLAG-tagged Rpt2 lacking the last three C-terminal residues (FLAG-Rpt2−C3) were prepared as described under “Experimental Procedures.” Panel A, cell extracts of nontransfected (Control) and each Rpt-expressing cell line were prepared and subjected to Western blotting (WB) with antibodies against FLAG epitope, Rpt2 protein, and actin. Panel B, extracts from cell lines expressing indicated proteins were subjected to glycerol density gradient centrifugation. Gradient fractions were subjected to Western blotting with anti-FLAG antibodies. Arrows indicate the known sedimentation positions of purified bovine 20 S proteasome, PA700, and 26 S proteasome. Panels C–F, affinity purification of FLAG proteins from extracts of indicated cell lines was conducted as described under “Experimental Procedures.” Panel C, affinity-purified samples were subjected to SDS-PAGE and stained with silver (left panel) or Western-blotted with antibodies against indicated proteins (right panel). Panel D, affinity-purified samples were assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate. WT and −C3 samples contained equal amounts of FLAG protein. Control represents a sample of nontransfected cell extract subjected to the affinity purification procedure. Panel E, affinity-purified samples were subjected to native PAGE and stained with silver (left panel), analyzed for in-gel proteasome activity peptide substrate overlay (center panel), or Western-blotted with antibodies against FLAG (right panel). Arrows indicate the known migration positions of purified bovine 26 S proteasome containing two (D) or one (S) PA700 complex or PA700. Panel F, affinity-purified proteins from WT and −C3 extracts were subjected to glycerol density gradient centrifugation. Gradient fractions were blotted for FLAG (WB: FLAG, upper panel) or assayed for proteasome activity (lower panel). Similar results were obtained in three independent cell preparations. AFU, arbitrary fluorescent units.
The FLAG-Rpt(WT) proteins that successfully assembled into 26 S proteasome featured distribution profiles on glycerol density gradients that differed from one another and, in some cases, from their respective endogenous counterparts. For example, the distributions of FLAG-Rpt3 and FLAG-Rpt6 closely mirrored those of their respective endogenous counterparts such that most of these proteins were found in 26 S proteasome. In contrast, significant proportions of FLAG-Rpt5, -Rpt1, and -Rpt4 were present in fractions that sedimented slower than 26 S proteasome. Most FLAG-Rpt5 not associated with 26 S proteasome protein sedimented in fractions characteristic of intact PA700 (Fig. 1, panel B), whereas significant amounts of non-26 S proteasome-associated FLAG-Rpt1 and FLAG-Rpt4 sedimented both in fractions characteristic of PA700 and in gradient fractions smaller than PA700 (Figs. 5, panel B, and 6, panel B, respectively). These results suggest that different FLAG-Rpt proteins were incorporated into 26 S proteasome with different efficiencies. Despite these differences, the incorporation of wild-type FLAG-tagged Rpt subunits into 26 S proteasome indicates that these proteins can be used to monitor features of the assembly process.
C-terminal HbYX Motifs of Two Rpt Subunits, Rpt5 and Rpt3, Are Essential for Assembly of 26 S Proteasome
Previous work by us and others has established an obligatory role for the C-terminal HbYX motifs of eukaryotic Rpt2, Rpt3, and Rpt5 subunits and the related archaeal PAN in binding of these proteins to the 20 S proteasome. This analysis was conducted principally in vitro using isolated peptides corresponding to the C termini of the proteins or with various homomeric protein complexes (20, 21, 23–25). Therefore, to test the relative roles of the three HbYX motifs of the Rpt subunits in cellular assembly of mammalian 26 S proteasome, we repeated the analysis described above with FLAG-Rpt proteins whose HbYX motifs were deleted. FLAG-Rpt5 and FLAG-Rpt3 lacking their HbYX motifs failed to be incorporated to appreciable extents into 26 S proteasome, as judged by their greatly reduced presence in glycerol gradient fractions characteristic of 26 S proteasome (Figs. 2, panel B, and 3, panel B). Analysis of the corresponding affinity-purified protein complexes confirmed this conclusion and demonstrated clear distinctions between the respective wild-type and mutant proteins. Thus, affinity-purified proteins from neither FLAG-Rpt3(−C3) nor FLAG-Rpt5(−C3) cell extracts displayed significant proteasome activity or featured complexes characteristic of 26 S proteasome after native PAGE (Figs. 2, panels C–E, and 3, panels C–E, respectively). Instead, these affinity-purified protein complexes featured several smaller catalytically inactive complexes, including increased amounts of PA700. SDS-PAGE followed by protein staining or Western blotting for 26 S proteasome subunits revealed that the main difference between the wild-type and mutant proteins was the absence of 20 S proteasome subunits in complexes of the latter. To confirm the exclusion of the mutant proteins from 26 S proteasome complexes, we also subjected affinity-purified complexes from FLAG-Rpt5(WT) and FLAG-Rpt5(−C3) cell extracts to glycerol density gradient centrifugation. Only complexes purified from extracts expressing wild-type proteins displayed 26 S proteasome activity that co-sedimented with proteins detected with antibodies against FLAG and 20 S proteasome subunits (Fig. 2, panel F, and data not shown).
Unlike either its endogenous or FLAG-tagged wild-type counterparts, mutant Rpt3 appeared to undergo C-terminal proteolytic processing. Thus, in addition to the intact mutant protein (∼45 kDa), two truncated FLAG-Rpt3 proteins (∼40 and ∼30 kDa) also were detected by anti-FLAG Western blotting (Fig. 3, panels A and B). The 30-kDa protein differed from the others by sedimenting more slowly during glycerol density gradient centrifugation and not assembling significantly into PA700 (Fig. 3, panel B). Although the reason for the susceptibility of mutant Rpt3 to proteolysis may be a direct consequence of the HbYX deletion, we note that the same mutant protein was not processed when expressed transiently at about the same level in HEK293 cells (25). Nevertheless, the features of unprocessed mutant Rpt3 demonstrate that the loss of only the HbYX residues was sufficient to prevent incorporation of this subunit into 26 S proteasome but not into intact PA700.
C-terminal HbYX Motif of Rpt2 Is Not Essential for Assembly of 26 S Proteasome but Promotes Proteasome Gating
In contrast to Rpt3 and Rpt5, deletion of the HbYX motif from FLAG-Rpt2 did not prevent its assembly into 26 S proteasome. This result was readily apparent from the distribution profile of the mutant protein after glycerol density gradient centrifugation (Fig. 4, panel B), which closely resembled the distribution profile of endogenous Rpt2, and was confirmed by isolation of intact, catalytically active 26 S proteasome by anti-FLAG affinity purification (Fig. 4, panel C). Thus, unlike the HbYX motifs of Rpt3 and Rpt5, the HbYX motif of Rpt2 does not appear to be essential for assembly of the protein into 26 S proteasome. As described above, the proteolytic processing of FLAG-Rpt2(WT) greatly reduced the efficiency of incorporation of the protein into 26 S proteasome, thereby preventing a direct comparison of the deletion mutant with its wild-type counterpart. It is unclear why FLAG-Rpt2(WT) protein, but neither FLAG-Rpt2(−C3) nor endogenous Rpt2, is susceptible to modification.
Previous work has established the selective roles of the HbYX residues of Rpt2 and Rpt5 in proteasome activation by gating (20, 24, 25). Purification of 26 S proteasome containing Rpt2 lacking its HbYX motif allowed us to determine the contribution of Rpt2 to this process by comparing peptidase activities of the mutant and wild-type proteins. Because of the proteolytic modification of FLAG-Rpt2(WT), we utilized other wild-type 26 S proteasomes, including both FLAG-tagged 26 S proteasomes affinity-purified from HEK293 cells and 26 S proteasome purified from bovine red blood cells for this comparison. 26 S proteasome containing FLAG-Rpt2(−C3) displayed ∼20% of the specific peptidase activity of various wild-type 26 S proteasomes (supplemental Fig. S1). This result indicates that the HbYX motif of Rpt2 plays a significant role in proteasome gating and distinguishes this effect from its nonessential role in proteasome assembly.
Collectively, these results demonstrate differential effects of the three HbYX-containing Rpt subunits on their incorporation into 26 S proteasome. Remarkably, for each of the two subunits rendered assembly-defective by this deletion, the presence of five other wild-type Rpt subunits is insufficient to overcome this defect. Moreover, the two mutant subunits incapable of incorporation into 26 S proteasome are nevertheless competent for assembly into intact PA700.
C-terminal Residues of None of the Three Non-HbYX Motif Rpt Subunits Are Essential for Assembly of 26 S Proteasome
C-terminal peptides of the three non-HbYX motif Rpt subunits (Rpt1, Rpt4, and Rpt6), unlike those of subunits containing the HbYX motif, do not bind detectably to the 20 S proteasome in vitro (20, 25). This feature, however, does not exclude these residues from critical roles in cellular assembly of 26 S proteasome because their action may require cooperation with other PA700 components or may be exerted in assembly processes other than direct binding to 20 S proteasome. Nevertheless, as shown in Figs. 5–7, FLAG-Rpt1(−C3), FLAG-Rpt4(−C3), and FLAG-Rpt6(−C3) were assembled into intact 26 S proteasome similarly to their wild-type counterparts. Affinity purification of FLAG proteins from extracts of cells expressing mutant Rpt1 and Rpt 4 confirmed the presence of structurally intact and functionally active 26 S proteasome (Figs. 5, panels C–E, and 6, panels C–E). In contrast, much reduced active 26 S proteasome was detected in affinity-purified samples from extracts of cells expressing mutant Rpt6 (Fig. 7, panels C–F). Instead, most of the affinity-purified Rpt6(−C3) protein was in the form of PA700 and variable amounts of dissociated 20 S proteasome. We interpret these results to indicate that FLAG-Rpt6(−C3) is assembled normally into 26 S proteasome in cells but destabilizes the resulting complex. Thus, the high salt conditions used during affinity purification likely promote either loss of 20 S proteasome during the binding and washing phases of the purification or dissociation of any remaining holoenzyme during and after elution from the affinity matrix. To illustrate this feature, we repeated the initial glycerol density gradient centrifugation of cell extracts in the presence of the same salt concentrations used during affinity purification. Increased salt concentrations had no effect on the distribution profile of the FLAG-Rpt6(WT) protein but significantly shifted FLAG-Rpt6(−C3) to a position characteristic of PA700 (supplemental Fig. S2).
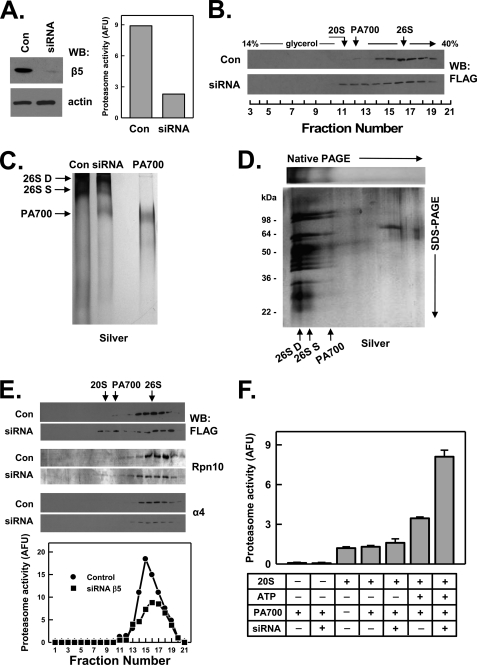
20 S Proteasome Is Not Required for Assembly of PA700
The results presented above indicate that intact C termini of certain Rpt subunits are required for assembly of those subunits into 26 S proteasome. In each case, however, the assembly defective subunit was present in a complex structurally indistinguishable from intact PA700. This finding is consistent with a model in which cellular 26 S proteasome assembly occurs by binding of intact PA700 to the 20 S proteasome. Such a model, however, differs from one presented by others, in which 26 S proteasome is formed by the sequential binding of PA700 subassemblies to the 20 S proteasome (26, 34). The mechanisms of these alternative models should result in different fates of Rpt subunits in the absence of 20 S proteasome. Thus, if the 20 S proteasome is a required template for PA700 assembly, Rpt subunits should accumulate as subassemblies in the absence of 20 S proteasome. In contrast, if 20 S proteasome is not an obligatory template for PA700 assembly, intact PA700 should accumulate in its absence. To distinguish between these models, we reduced 20 S proteasome content in FLAG-Rpt6(WT) cells using siRNA against the β5 subunit. RNAi reduced β5 content by more than 90% and proteasome activity in cell extracts by ∼75% (Fig. 8, panel A). Glycerol density gradient centrifugation of extracts from control and siRNA-treated cells confirmed that RNAi of β5 significantly reduced 26 S proteasome content and activity and promoted a relative redistribution of the FLAG-Rpt6 subunit from fractions characteristic of 26 S proteasome to those characteristic of PA700 (Fig. 8, panel B). These results indicate that intact PA700 accumulated in cells when the level of 20 S proteasome was reduced. To confirm the nature of the FLAG-Rpt6-containing complexes present in extracts of siRNA-treated cells, we purified these complexes on anti-FLAG beads and analyzed them by native PAGE and glycerol density gradient centrifugation. FLAG-Rpt6 purified from control extracts was present almost exclusively as a subunit of 26 S proteasome, as demonstrated by both methods (Fig. 8, panels C and E). In contrast, FLAG-Rpt6 purified from extracts of siRNA-treated cells was present both in the remaining but significantly reduced 26 S proteasome and in PA700 (Fig. 8, panels C and E). We also subjected the proteins separated on native PAGE to second dimension SDS-PAGE followed by silver staining. The band derived from the siRNA-treated cells that accumulated in the native gel with a migration position characteristic of PA700 featured a second dimension subunit pattern characteristic of purified PA700 and differed from the 26 S proteasome only by the absence of 20 S proteasome subunits (Fig. 8, panel D). A similar subunit pattern was observed when siRNA glycerol gradient fractions corresponding to the position of PA700 were subjected to SDS-PAGE and silver staining (supplemental Fig. S3). Interestingly, after siRNA-mediated knockdown of the β5 subunit, residual 26 S proteasome was enriched in the doubly capped form, as detected by both a slower migrating band on native PAGE (Fig. 8, panel C) and a faster sedimenting species during glycerol density gradient centrifugation (Fig. 8, panel E). This feature is a likely consequence of increased binding of accumulated PA700 to both ends of the remaining 20 S proteasomes as the number of total available binding sites on 20 S proteasomes declines during RNAi. A similar effect has been noted with yeast mutants defective in 20 S proteasome assembly (35). Finally, we tested whether PA700 that accumulated in siRNA-treated cells featured PA700 activity, defined as ATP-dependent activation of 20 S proteasome-catalyzed peptidase activity. Fractions 8–11 of glycerol density gradient centrifugation of affinity-purified proteins from control and siRNA-treated cells contained no significant proteasome activity themselves in the presence or absence of ATP nor did they stimulate the activity of exogenous purified 20 S proteasome in the absence of ATP (Fig. 8, panel F). However, in the presence of ATP, fractions from the siRNA gradients significantly stimulated 20 S proteasome activity. Notably, this activity was about 3-fold greater in gradient fractions derived from siRNA-treated cells compared with those from control cells (Fig. 8, panel F). These results confirm the increased presence of intact PA700 in cells subjected to RNAi of 20 S proteasome. In sum, these results are consistent with a model in which PA700 is assembled as an intact complex that then binds to 20 S proteasome to form 26 S proteasome (Fig. 9, lower panel, upper pathway).
FIGURE 8.
Assembly of PA700 is independent of 20 S proteasome. HEK293 cells stably expressing FLAG-Rpt6 WT were transfected with siRNAs against the β5 subunit of 20 S proteasome (siRNA) or a nonexistent mRNA (Control). Panel A, cell extracts from control and siRNA cells were subjected to Western blotting with antibodies against β5 or actin (left panel) or assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrates (right panel). Panel B, extracts from control and siRNA cells were subjected to glycerol density gradient centrifugation. Gradient fractions were Western-blotted with anti-FLAG antibodies. Arrows indicate the known sedimentation positions of purified bovine 20 S proteasome, PA700, and 26 S proteasome. Panels C–F, affinity purification of FLAG proteins from extracts of control and siRNA cells was conducted as described under “Experimental Procedures.” Panel C, affinity-purified FLAG proteins from control and siRNA cell extracts were subjected to native PAGE and stained with silver. Purified bovine PA700 is presented as a standard. Panel D, affinity-purified FLAG proteins from siRNA cell extracts were subjected to native PAGE. Identical lanes were either stained with silver (top panel) or subjected to second dimension SDS-PAGE and then stained with silver (bottom panel). Panel E, affinity-purified FLAG proteins from control and siRNA cell extracts were subjected to glycerol density gradient centrifugation. Gradient fractions were Western-blotted with antibodies against FLAG, Rpn10 subunit of PA700, or α4 subunit of the 20 S proteasome (upper panels) or assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC substrate (lower panels). Panel F, PA700 activity was determined in gradient factions from control and siRNA extracts. Fractions 8–11 of each gradient from panel E were combined (denoted PA700) and assayed for ATP-dependent activation of purified bovine 20 S proteasome as described under “Experimental Procedures.” “ + ” and “−” indicate the presence or absence, respectively, of PA700, ATP, 20 S proteasome, respectively, in fractions from control and siRNA gradients. Similar results were obtained for data in all panels in three independent cell preparations. AFU, arbitrary fluorescent units.
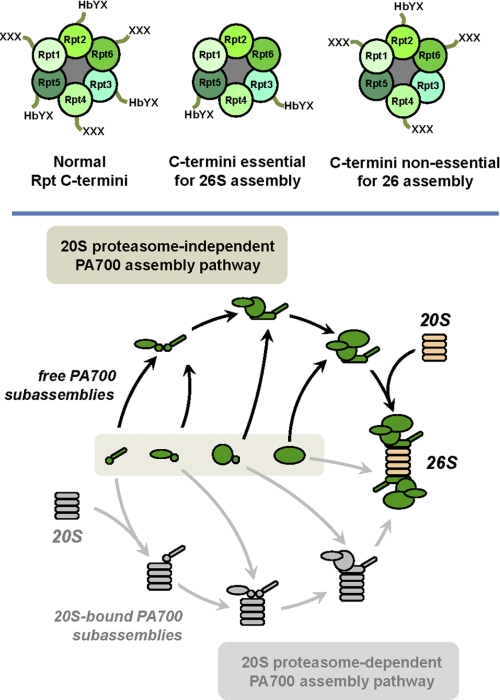
FIGURE 9.
Relative roles of C termini of Rpt subunits for 26 S proteasome assembly and model of cellular assembly pathway of PA700. Upper panel, relative arrangement of Rpt subunits and their C termini (left panel); C-terminal Rpt residues essential for (center panel) or nonessential for (right panel) respective subunit incorporation into 26 S proteasome. Lower panel, alternative models for cellular assembly of PA700 and its incorporation into 26 S proteasome. Each model involves assembly of PA700 from multiple PA700 subassemblies, here depicted arbitrarily in number and without defined composition. Our data are compatible with and favor the model depicted in the upper pathway (colorized) in which these PA700 subassemblies form intact PA700 prior to binding with intact 20 S proteasome to form 26 S proteasome. In contrast, our data are inconsistent with a model depicted in the lower pathway (gray) in which PA700 subassemblies use 20 S proteasome as a template for assembly PA700 and 26 S proteasome.
DISCUSSION
To investigate the roles of the C termini of Rpt subunits in cellular 26 S proteasome assembly, we utilized HEK293 cells stably expressing epitope-tagged forms of wild-type and C-terminally truncated Rpt proteins in a background of endogenous wild-type proteins. The successful incorporation of FLAG-Rpt(WT) proteins into the 26 S proteasome validates the strategy of using these proteins as monitors of the assembly process. Not surprisingly, varying proportions of some exogenously expressed Rpt proteins also existed in complexes smaller than 26 S proteasome to a greater extent than that observed for the corresponding endogenous proteins. This could reflect accumulation of the FLAG protein in normal assembly pathway intermediates that become stalled because of an excess of that protein over endogenous interacting partners required for continued progress along the assembly pathway. In fact, our initial characterization of selected low molecular weight complexes indicates that they likely represent authentic assembly intermediates (see below).
The C-terminal HbYX motifs of PA700 Rpt subunits have been studied previously for their roles in proteasome gating. The structural basis of this process has been advanced significantly by analogous studies of the homohexameric PAN and the monomeric PA200/Blm10 proteasome regulators, whose HbYX residues also likely provide the main determinants for proteasome binding and subsequent complex stability (20–23, 40, 41). However, unlike PAN and PA200/Blm10, the six Rpt subunits are structurally distinct proteins that feature both HbYX and non-HbYX motifs at their C termini. Thus, the relative roles Rpt of C termini among individual subunits and between the HbYX and non-HbYX subunits in various functions have been unclear and the source of conflicting results.
Our results demonstrate that deletion of the HbYX residues from either Rpt3 or Rpt5 was sufficient to prevent incorporation of the truncated subunit into 26 S proteasome and indicate that in each case the contributions of other intact binding elements of PA700 were not able to overcome the loss of a single critical binding interaction. Because assembly of intact PA700 was not affected by these deletions, it seems unlikely that the defects in 26 S proteasome assembly were a consequence of disrupted early steps of PA700 assembly. In fact, because of C-terminal proteolytic processing of a fraction of the FLAG-Rpt3(−C3) protein, we could directly discriminate between the effect of deletion of the HbYX residues and the effect of more extensive C-terminal deletions on PA700 assembly. Thus, unlike the unprocessed Rpt3(−C3), larger C-terminal truncations inhibited PA700 assembly per se, perhaps as a consequence of lost interactions between Rpt3 and cognate PA700 assembly chaperones, such as p28 (42). The exact reason for the lack of processing of wild-type Rpt3 compared with mutant Rpt3 is uncertain but seems unlikely to be related to an inherent structural role of the deleted residues because no processing occurred when the same mutant protein was expressed transiently (25).
In contrast to the essential roles of the HbYX residues for assembly of Rpt3 and Rpt5 into 26 S proteasome, deletion of HbYX residues from Rpt2 had no effect on incorporation of this subunit. This effect was clearly evident in the case of genetically truncated Rpt2, but direct comparison to the assembly of FLAG-Rpt2(WT) was complicated by apparent C-terminal processing of the latter protein. Processing of wild-type Rpt2 appreciably decreased the extent and efficiency of incorporation of the protein into 26 S proteasome. As with the FLAG-Rpt3(−C3), C-terminal residues of Rpt2 exclusive of the HbYX motif probably are involved in protein-protein interactions required in early steps of the assembly pathway. Nevertheless, despite the common in vitro proteasome binding capacities of C-terminal peptides of the HbYX-Rpt subunits, HbYX residues of Rpt2 are not essential for cellular incorporation of this subunit into the 26 S proteasome. 26 S proteasome with Rpt2 lacking an HbYX displayed significantly lower peptidase activity than wild-type 26 S proteasome, demonstrating an important role of this motif in proteasome gating. An important role for Rpt2 in proteasome gating was demonstrated previously using yeast Rpt2 mutants defective in ATP binding and hydrolysis (43, 44).
No peptide corresponding to the C terminus of any non-HbYX Rpt subunits binds detectably to 20 S proteasome (25). Nevertheless, it seems likely that these residues must interact with proteasome α subunits when present in assembled 26 S proteasomes. In support of this assertion, we found that deletion of non-HbYX residues from Rpt6 had no effect on cellular proteasome assembly but destabilized the structural integrity of 26 S proteasome by promoting dissociation of the holoenzyme into 20 S proteasome and PA700 subcomplexes. The former result, but not the latter, was observed when analogous deletions were made in other non-HbYX subunits. Thus, C-terminal non-HbYX residues of Rpt6 appear crucial for structural stability of the assembled 26 S proteasome.
Various aspects of the effects of Rpt subunit C termini on 26 S proteasome assembly reported here are both compatible and in apparent conflict with analogous but not identically designed studies in yeast. For example, yeast expressing Rpt1, Rpt2, Rpt3, or Rpt5 lacking a single C-terminal residue displayed normal or near-normal 26 S proteasome levels, whereas yeast expressing Rpt4 or Rpt6 with this deletion showed severely decreased levels (26). Similarly, intact 26 S proteasomes were affinity-purified from yeast strains in which the tyrosine of the HbYX motifs of Rpt2, Rpt3, or Rpt5 was mutated (20). Other than the obvious differences in experimental details of the studies, these apparent discrepancies have unclear bases.
Recently, Goldberg and co-workers (45) used features of ATP binding by PAN and Rpt subunits to propose a model that divides functions of HbYX and non-HbYX residues between 20 S gating and 20 S binding, respectively. This model posits that the fixed order arrangement of alternating HbYX and non-HbYX Rpt subunits (Rpt1, Rpt2, Rpt6, Rpt3, Rpt4, and Rpt5) (46) ensures simultaneous interactions of at least one para-positioned pair of gating and binding Rpt subunits with the 20 S proteasome at all times during ATP-dependent hydrolysis of proteins. Such an arrangement could reflect the need to maintain overall structural stability of the 26 S proteasome while allowing dynamic 20 S-Rpt subunit interactions that likely occur during ordered cycles of nucleotide binding, hydrolysis, and release by different subunit pairs. An appealing aspect of this model is its explanation for the symmetry mismatch of the opposing hexameric Rpt subunit and heptameric α subunit rings. This mismatch may also be related to various imaging results that reveal asymmetric contacts between the rings (47–50). We note that the important determinants of proteasome assembly and stability identified here are found in subunits oriented asymmetrically within the Rpt ring (Fig. 9, upper panel).
In addition to defining the relative roles of C termini of Rpt subunits for 26 S proteasome assembly, our results provide new information about intermediates of the assembly pathway. Each of the two Rpt subunits (Rpt5 and Rpt3) not assembled into 26 S protease due to C-terminal truncations was assembled normally into intact PA700. Furthermore, reduction of cellular 20 S proteasome content by RNAi promoted accumulation of PA700, indicating that intact PA700 was an immediate precursor to the 26 S proteasome. Inhibition of proteasome function by RNAi of 20 S proteasome subunits can potentially cause compensatory up-regulated expression of other proteasome subunits (51, 52). However, this effect was minimal under the conditions employed here and cannot account for the observed increase in PA700 content. The incorporation of mutant Rpt subunits into intact PA700 demonstrated here indicates that their failure to assemble into 26 S proteasome is not a consequence of a defect in an early stage of the assembly process. For example, previous work has documented the roles of specific assembly chaperones in the formation of multiple Rpt subassemblies and in the subsequent combination of these subassemblies to form the base (27, 28). Although our experiments have not systematically evaluated the effect of C-terminal mutations on the formation of Rpt subassemblies, assembly of these mutant Rpt proteins into intact PA700 argues that the last three residues are not critical for early stages of the assembly process and instead exert their effects at the level of 20 S proteasome binding. Our initial analysis of the low molecular weight complexes observed for FLAG-Rpt5 and FLAG-Rpt1 indicates that they represent complexes similar to those previously identified as assembly intermediates (supplemental Figs. S4 and S5) (24, 30, 32).
Our cellular data also support multiple types of in vitro results consistent with the view that cellular 26 S proteasome is assembled by the binding of 20 S proteasome with intact PA700 via essential Rpt C-terminal residues. First, intact PA700 exists in vitro and can be purified from a variety of cells and tissues (53). Although it is possible that free PA700 is derived artifactually by dissociation of 26 S proteasome during the purification process, free intact PA700 also exists in crude extracts of mammalian cells lysed under gentle conditions that favor maintenance of intact 26 S proteasome. Second, 26 S proteasome can be reconstituted in vitro from purified PA700 and 20 S proteasome, suggesting that this process mirrors one that occurs in intact cells (37, 53, 54). Significantly, this ATP-dependent in vitro reconstitution requires intact C termini of HbYX residues shown here to mediate cellular 26 S proteasome assembly (24, 25). Finally, PA700 can be reconstituted in vitro from purified subassemblies that may represent cellular assembly intermediates (39). Reconstitution of PA700 from subassemblies requires neither intact C termini of Rpt subunits nor 20 S proteasome, and reconstituted PA700, like independently purified PA700, can subsequently bind to 20 S proteasome to form 26 S proteasome. Thus, our collective data support a model in which intact PA700 is formed independently of 20 S proteasome and is an immediate precursor to 26 S proteasome (Fig. 9, lower panel, upper pathway).
Despite these cellular and biochemical data, evidence for an alternative model for PA700 and 26 S proteasome formation has been presented previously in both yeast and mammalian cells. These studies show that Rpt-containing subassemblies of PA700 associate with the 20 S proteasome prior to formation of the intact PA700 and indicate that the 20 S proteasome provides a template function for PA700 formation (Fig. 9, lower panel, lower pathway) (26, 34). Moreover, subassemblies of PA700 accumulate in yeast genetically defective for normal 20 S proteasome assembly (35). The basis for discrepancies among these experiments is not readily apparent. However, these alternative assembly models need not be mutually exclusive. Different assembly pathways may be preferentially favored by different cells or by given cells under different physiologic states. Nevertheless, the data presented here for mammalian cells under normal growth conditions favor a model in which PA700 can be assembled independently of the 20 S proteasome and suggest that intact PA700 is a direct intermediate in the pathway of 26 S proteasome assembly (Fig. 9, lower panel, upper pathway).
Supplementary Material
Acknowledgments
We thank Dr. Thomas Gillette and Dr. Brajesh Kumar for helpful discussions and David Thompson and Xiaohua Li for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK46181. This work was also supported by Welch Foundation Grant I-500 (to G. N. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- AMC
- 7-amino-4-methylcoumarin
- Suc
- succinyl.
REFERENCES
- 1. Voges D., Zwickl P., Baumeister W. (1999) Annu. Rev. Biochem. 68, 1015–1068 [DOI] [PubMed] [Google Scholar]
- 2. Bedford L., Paine S., Sheppard P. W., Mayer R. J., Roelofs J. (2010) Trends Cell Biol. 20, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Demartino G. N., Gillette T. G. (2007) Cell 129, 659–662 [DOI] [PubMed] [Google Scholar]
- 4. Groll M., Bochtler M., Brandstetter H., Clausen T., Huber R. (2005) ChemBioChem. 6, 222–256 [DOI] [PubMed] [Google Scholar]
- 5. Baumeister W., Walz J., Zühl F., Seemüller E. (1998) Cell 92, 367–380 [DOI] [PubMed] [Google Scholar]
- 6. Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 7. Kusmierczyk A. R., Hochstrasser M. (2008) Biol. Chem. 389, 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murata S., Yashiroda H., Tanaka K. (2009) Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 9. Bochtler M., Ditzel L., Groll M., Hartmann C., Huber R. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 295–317 [DOI] [PubMed] [Google Scholar]
- 10. Groll M., Bajorek M., Kohler A., Moroder L., Rubin D. M., Huber R., Glickman M. N., Finley D. (2001) Nat. Struct. Biol. 11, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 11. Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) Cell 94, 615–623 [DOI] [PubMed] [Google Scholar]
- 12. Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee C., Schwartz M. P., Prakash S., Iwakura M., Matouschek A. (2001) Mol. Cell 7, 627–737 [DOI] [PubMed] [Google Scholar]
- 15. Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 16. Yao T., Cohen R. E. (2002) Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 17. Smith D. M., Benaroudj N., Goldberg A. (2006) J. Struct. Biol. 156, 72–83 [DOI] [PubMed] [Google Scholar]
- 18. Liu C. W., Li X., Thompson D., Wooding K., Chang T. L., Tang Z., Yu H., Thomas P. J., DeMartino G. N. (2006) Mol. Cell 24, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stadtmueller B. M., Hill C. P. (2011) Mol. Cell 41, 8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stadtmueller B. M., Ferrell K., Whitby F. G., Heroux A., Robinson H., Myszka D. G., Hill C. P. (2010) J. Biol. Chem. 285, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., Cheng Y. (2008) Mol. Cell 30, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu Y., Smith D. M., Kim H. M., Rodriguez V., Goldberg A. L., Cheng Y. (2010) EMBO J. 29, 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gillette T. G., Kumar B., Thompson D., Slaughter C. A., DeMartino G. N. (2008) J. Biol. Chem. 283, 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar B., Kim Y. C., DeMartino G. N. (2010) J. Biol. Chem. 285, 39523–39535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park S., Roelofs J., Kim W., Robert J., Schmidt M., Gygi S. P., Finley D. (2009) Nature 459, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park S., Tian G., Roelofs J., Finley D. (2010) Biochem. Soc. Trans. 38, 6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Besche H. C., Peth A., Goldberg A. L. (2009) Cell 138, 25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaneko T., Hamazaki J., Iemura S., Sasaki K., Furuyama K., Natsume T., Tanaka K., Murata S. (2009) Cell 137, 914–925 [DOI] [PubMed] [Google Scholar]
- 31. Saeki Y., Toh-E A., Kudo T., Kawamura H., Tanaka K. (2009) Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 32. Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Le Tallec B., Barrault M. B., Guérois R., Carré T., Peyroche A. (2009) Mol. Cell 33, 389–399 [DOI] [PubMed] [Google Scholar]
- 34. Hendil K. B., Kriegenburg F., Tanaka K., Murata S., Lauridsen A. M., Johnsen A. H., Hartmann-Petersen R. (2009) J. Mol. Biol. 394, 320–328 [DOI] [PubMed] [Google Scholar]
- 35. Kusmierczyk A. R., Kunjappu M. J., Funakoshi M., Hochstrasser M. (2008) Nat. Struct. Mol. Biol. 15, 237–244 [DOI] [PubMed] [Google Scholar]
- 36. Koulich E., Li X., DeMartino G. N. (2008) Mol. Biol. Cell 19, 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adams G. M., Falke S., Goldberg A. L., Slaughter C. A., DeMartino G. N., Gogol E. P. (1997) J. Mol. Biol. 273, 646–657 [DOI] [PubMed] [Google Scholar]
- 38. Chu-Ping M., Vu J. H., Proske R. J., Slaughter C. A., DeMartino G. N. (1994) J. Biol. Chem. 269, 3539–3547 [PubMed] [Google Scholar]
- 39. Thompson D., Hakala K., DeMartino G. N. (2009) J. Biol. Chem. 284, 24891–24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Förster A., Masters E. I., Whitby F. G., Robinson H., Hill C. P. (2005) Mol. Cell 18, 589–599 [DOI] [PubMed] [Google Scholar]
- 41. Sadre-Bazzaz K., Whitby F. G., Robinson H., Formosa T., Hill C. P. (2010) Mol. Cell 37, 728–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakamura Y., Nakano K., Umehara T., Kimura M., Hayashizaki Y., Tanaka A., Horikoshi M., Padmanabhan B., Yokoyama S. (2007) Structure 15, 179–189 [DOI] [PubMed] [Google Scholar]
- 43. Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. (1998) EMBO J. 17, 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Köhler A., Cascio P., Leggett D. S., Woo K. M., Goldberg A. L., Finley D. (2001) Mol. Cell 7, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 45. Smith D. M., Fraga H., Reis C., Kafri G., Goldberg A. L. (2011) Cell 144, 526–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tomko R. J., Jr., Funakoshi M., Schneider K., Wang J., Hochstrasser M. (2010) Mol. Cell 38, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. da Fonseca P. C., Morris E. P. (2008) J. Biol. Chem. 283, 23305–23314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bohn S., Beck F., Sakata E., Walzthoeni T., Beck M., Aebersold R., Förster F., Baumeister W., Nickell S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 20992–20997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Förster F., Lasker K., Beck F., Nickell S., Sali A., Baumeister W. (2009) Biochem. Biophys. Res. Commun. 388, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nickell S., Beck F., Scheres S. H., Korinek A., Förster F., Lasker K., Mihalache O., Sun N., Nagy I., Sali A., Plitzko J. M., Carazo J. M., Mann M., Baumeister W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11943–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wójcik C., DeMartino G. N. (2002) J. Biol. Chem. 277, 6188–6197 [DOI] [PubMed] [Google Scholar]
- 52. Radhakrishnan S. K., Lee C. S., Young P., Beskow A., Chan J. Y., Deshaies R. J. (2010) Mol. Cell 38, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. DeMartino G. N. (2005) Methods Enzymol. 398, 295–306 [DOI] [PubMed] [Google Scholar]
- 54. Adams G. M., Crotchett B., Slaughter C. A., DeMartino G. N., Gogol E. P. (1998) Biochemistry 37, 12927–12932 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.