Abstract
Fungal allergens are associated with the development of asthma, and some have been characterized as proteases. Here, we established an animal model of allergic airway inflammation in response to continuous exposure to proteolytically active Pen c 13, a major allergen secreted by Penicillium citrinum. In functional analyses, Pen c 13 exposure led to increased airway hyperresponsiveness, significant inflammatory cell infiltration, mucus overproduction, and collagen deposition in the lung, dramatically elevated serum levels of total IgE and Pen c 13-specific IgE and IgG1, and increased production of the Th2 cytokines IL-4, IL-5, and IL-13 by splenocytes stimulated in vitro with Pen c 13. To examine the mechanisms involved in the regulation of allergenicity by Pen c 13, we performed two-dimensional fluorescence difference gel electrophoresis analysis combined with nano-LC-MS/MS, followed by bioinformatics analysis to identify potential targets that associated with allergic inflammation, which suggested that galectin-3 and laminin might be involved in novel pathogenic mechanisms. Finally, we focused on junctional proteins between cells, because, in addition to opening of the epithelial barrier by environmental proteases possibly being the initial step in the development of asthma, these proteins are also associated with actin rearrangement. Taken together, our findings indicate that Pen c 13 exposure causes junctional structure alterations and actin cytoskeletal rearrangements, resulting in increased permeability and airway structural changes. These effects probably change the lung microenvironment and foster the development of allergic sensitization.
Keywords: Cell Junctions, Cell Permeabilization, Epithelial Cell, Lung, Mouse, Proteomics, Serine Protease, Allergen, Allergic Airway Inflammation, Airway Remodeling
Introduction
The prevalence of asthma has been steadily increasing around the world (1, 2). Asthma is a chronic respiratory disease characterized by airway hyperresponsiveness (AHR)2 to allergens, together with eosinophilic inflammation, airway edema, mucus hypersecretion, airway remodeling, and airflow obstruction. In addition, a characteristic feature of allergic inflammation is the predominance of Th2 lymphocytes, and the cytokines they produce (e.g. IL-4, IL-5, and IL-13) play a pivotal role in recruiting effector inflammatory cells (mast cells, eosinophils, and lymphocytes) to the airway and in the production of IgE (3).
Airway remodeling in asthma refers to structural changes in the airways due to repeated cycles of airway injury and repair, causing abnormal deposition of extracellular matrix components, goblet cell hyperplasia, subepithelial fibrosis, and airway smooth muscle hyperplasia and hypertrophy (4, 5). Adherens junctions (AJs) and tight junctions (TJs) play a critical role in cell-cell adhesion and, although they are made up of different proteins, have similar specialized transmembrane proteins, which form an intact paracellular space, and intracellular scaffolding protein linked to the actin cytoskeleton. When protease allergens are inhaled, the protective barrier may be destroyed, potentially allowing access of atmospheric components and pathogens to the circulation and probably involving the reorganization of the actin cytoskeleton, causing airway remodeling (6–9).
Previous studies have shown that numerous allergens from various organisms, including the house dust mite, cockroach, Penicillium sp., and Aspergillus sp., have protease activity. The crude allergen extracts, which contain several components, are rich in proteases that have been shown to be major allergens. These are involved in the pathogenesis of allergic diseases through (i) the release of proinflammatory cytokines by the activation of protease-activated receptors (PARs), which are widely expressed on cells in blood vessels, connective tissue, leukocytes, epithelium, and many airway cells; (ii) the cleavage of CD23 from activated B cells and of CD25 from T cells to favor the development of Th2-type responses; (iii) the degradation of junctional proteins, thus increasing the permeability of the lung epithelium and leading to morphologic changes and cell desquamation in the lung epithelium; and (iv) the induction of an imbalance between proteases and antiproteases (10–14). Many proteases have been shown to be important mediators of inflammation in a murine model of asthma and in lung injury (15–17). The cysteine and serine protease allergens from the dust mite have been examined for their ability to stimulate allergic immune responses in animal models (18, 19), the cockroach serine protease Per a 10 induces allergic airway inflammation in a mouse model (20), and an alkaline serine protease from Aspergillus sp. has synergistic effects on the immune response induced by a major Aspergillus fumigatus allergen, Asp f 2, in mice (21). However, the connection between proteolytic allergens and the development of hypersensitivity reactions requires more investigation to understand the molecular mechanisms involved in allergic diseases.
In the present study, we focused our attention on Pen c 13, an immunodominant human allergen secreted by Penicillium citrinum identified as a 33-kDa alkaline serine protease (22), which we previously showed induces proinflammatory cytokine release in airway epithelial cells in vitro through PAR-1 and PAR-2 activation and increase in intracellular calcium levels (23). To investigate the effects of Pen c 13 in augmenting allergic airway inflammation in a murine model, BALB/c mice were exposed to active Pen c 13 or PBS for 10 consecutive days, and their immunological responses were assessed. Furthermore, we combined 2-D DIGE technology and nano-LC-MS/MS analyses to profile changes in protein expression in lungs from PBS-treated and native Pen c 13 (n-Pen c 13)-sensitized mice. Our studies demonstrated the possible pathogenic mechanisms by which Pen c 13 might directly contribute to asthma susceptibility in naïve subjects and increased severity in affected asthmatics.
EXPERIMENTAL PROCEDURES
Materials
Female BALB/c mice age 6–8 weeks were obtained from the Animal Center of the National Taiwan University College of Medicine and allowed to acclimatize to their new surroundings for a week prior to experimentation. Animal care and handling conformed to the Guide for the Care and Use of Laboratory Animals (75).
Cell Culture
The NCI-H441 cell line, derived from a human lung epithelial adenocarcinoma, was obtained from the American Type Culture Collection (ATCC HTB-174) and was grown in DMEM supplemented with 10% FBS (both from Invitrogen) and 100 units/ml penicillin/streptomycin at 37 °C in a humidified chamber in 5% CO2.
Purification of Native and His-tagged Recombinant Pen c 13
Because n-Pen c 13 is heat-labile, we used a recombinant Pen c 13, which is structurally misfolded and lacks enzymatic activity but is thermally stable and retains linear allergenic epitopes (denatured Pen c 13 (d-Pen c 13)). The two forms were purified as described previously (22). Briefly, n-Pen c 13 was purified from the culture broth of P. citrinum by a combination of anion-exchange (HiTrap DEAE-Sepharose Fast Flow) and cation-exchange (HiTrap SP-Sepharose Fast Flow) chromatography. To produce recombinant Pen c 13, cDNA encoding mature Pen c 13 was obtained from the P. citrinum cDNA library by PCR amplification, and the amplified fragment was ligated into the expression vector, pQE 30 (Qiagen, Chatsworth, CA) to generate a construct coding for the N-terminally fused His6-tagged protein (plasmid PQE-30/Pen c 13), which was used to transform expressing Escherichia coli M15 cells (Qiagen). The His-tagged recombinant Pen c 13 was bound to a Ni2+-chelate affinity column, and then endotoxin was removed by washing the resin in a centrifuge tube with binding buffer (20 mm Tris-HCl, pH 7.9, 0.5 m NaCl, 8 m urea, and 5 mm imidazole) containing 1% Triton X-114 (Sigma). The resin was loaded into a column and washed with binding buffer containing 0.1% Triton X-114 before elution of the recombinant protein with elution buffer (20 mm Tris-HCl, pH 7.9, 0.5 m NaCl, 8 m urea, and 60 mm imidazole) (24). The purified recombinant protein was denatured by heat at 95 °C and called d-Pen c 13. Protein concentrations were measured using the 2-D Quant kit (GE Healthcare).
Intratracheal Inoculation
Female BALB/c mice (n = 34) aged 6–8 weeks were divided into four experimental groups, the n-Pen c 13-sensitized group, the d-Pen c 13-sensitized group, the PBS-treated group (n = 10 in each group), and naïve animals (n = 4). The animal was anesthetized with an intraperitoneal injection of sodium pentobarbital (SCI Pharmtech (Taoyuan, Taiwan); 10 mg/ml solution, 0.005 ml/g body weight) and placed in dorsal recumbence on an inclined board with its mouth kept open by hooking the upper incisors onto a wire frame and the tongue extended to one side using forceps. Intratracheal administration of n-Pen c 13 or d-Pen c 13 resuspended at a concentration of 25 μg/ml in pyrogen-free PBS was performed using a number 23 steel gavage tube and a 1.0-ml microsyringe. The gavage tube was directed into the proximal trachea, and 40 μl of n-Pen c 13, d-Pen c 13, or PBS was dropped on the back of the tongue using the microsyringe, allowing the inoculum to be inhaled into the lungs by normal breathing. This method for intratracheal inoculation of mice was found to work well in preliminary tests using trypan blue dye and to allow the aspirated material to spread over the whole lung but not the esophagus or stomach. All mice were inoculated on 10 consecutive days, followed by 72 h of rest.
Measurement of AHR
AHR was assessed as described previously (25) by measuring changes in airway resistance (RL, cm H2O/ml/s) after a challenge with aerosolized methacholine using the FlexiVent system (SCIREQ, Montreal, Canada). On day 12, mice were weighed and anesthetized with an injection of chloralose (100 mg/kg, intraperitoneal; Sigma) and urethane (500 mg/kg, intraperitoneal; Sigma). A tracheostomy was performed, and a cannula was inserted into the Y-adaptor of a ventilator. After initiating mechanical ventilation, the mouse was paralyzed with a pancuronium bromide (1 mg/kg, intraperitoneal; Sigma) and subjected to a deep lung inflation (slow inflation to a pressure of 30 cm H2O held for 3 s) before the plethysmograph was sealed for the rest of the experiment. Mice were ventilated at a tidal volume of 10 ml/kg, a respiratory rate of 150 breaths/min, and an inspiratory/expiratory ratio of 3:2. Positive end-expiratory pressure was maintained between 2 and 3 cm H2O. Doubling doses of methacholine chloride (Sigma) were inhaled (0.75–24 mg/ml), and resistance was measured.
Bronchoalveolar Lavage Fluid Cytology
At 72 h after the last challenge, blood samples were taken (see below), and then the mice were killed and the lungs were immediately lavaged three times via the tracheal cannula with 1 ml of Hanks' balanced salt solution (HBSS), free of ionized calcium and magnesium (Sigma), and the bronchoalveolar lavage fluid samples were pooled and stored on ice. Total cell numbers were determined by counting in a hemocytometer, and viability was assessed by trypan blue exclusion. Cytospin preparations of 100–150 μl of bronchoalveolar lavage fluid were made by centrifugation onto glass slides (Shandon Cytospin: 200 rpm, 15 min) and stained with Liu's stain for different cell counts. A minimum of 200 cells were counted and classified as eosinophils, lymphocytes, or neutrophils based on standard morphological criteria.
Measurement of Antibody Levels in Serum
At 72 h after the last challenge, the mice were bled from the retro-orbital venous plexus, and serum samples were prepared and stored at −80 °C. The serum samples were initially titrated to determine the optimal dilution (1:50) for testing for antibodies. Total IgE in serum was measured in a sandwich ELISA using a monoclonal anti-mouse IgE antibody (clone R35-72, Pharmingen) as the capture antibody and a second biotinylated monoclonal anti-mouse IgE antibody (clone R35-118, Pharmingen) as the detection antibody according to the manufacturer's instructions by comparison with a standard curve for a purified mouse IgE standard (clone C38-2, Pharmingen) included on each microtiter plate. To measure levels of Pen c 13-specific IgE, IgG1, and IgG2a, serum samples were added to ELISA plates coated with a 10 μg/ml solution of d-Pen c 13, and then, after overnight incubation at 4 °C, the plates were washed and incubated with biotinylated rat monoclonal antibody against mouse IgE (R35-118), IgG1 (A85-1), or IgG2a (R19-15) (2 μg/ml; Pharmingen) in 1% BSA in PBS (PBSB) for 1 h at room temperature, followed by washes with 0.05% Tween 20 in PBS and incubation with streptavidin-HRP conjugate (1:5000 dilution in PBSB) (Sigma) for 30 min at room temperature. The plates were washed and developed with the TMB microwell peroxidase substrate system, and the absorbance was measured at 450 nm.
Spleen Cell Culture and Cytokine Analyses
To measure cytokine levels in splenocyte culture supernatants, the spleen was harvested and crushed into a single cell suspension, and red blood cells were lysed with ACK lysis buffer (0.15 m NH4Cl, 10 mm KHCO3, 0.1 mm Na2EDTA, pH 7.2), and then the cells were washed with HBSS and resuspended in RPMI 1640 medium (Invitrogen) containing 10% FBS, 100 units/ml penicillin/streptomycin, and 2 mm l-glutamine at a concentration of 5 × 106 cells/ml and cultured in medium alone or medium supplemented with d-Pen c 13. After 48 h of culture, supernatants were collected, and IL-4, IL-5, IL-13, and IFN-γ were measured by ELISA according to the manufacturer's recommended protocols (R&D Systems, Minneapolis, MN).
Lung Histology
After the lavage was collected, the lungs were immediately removed and washed with PBS until all blood was removed. The left lung was fixed in 10% formalin and embedded in OCT (tissue freezing medium). Tissue sections (5 μm) were stained with H&E, periodic acid-Schiff (PAS), and Masson's trichrome. H&E staining was used for morphological comparison of airway epithelial damage and allergic inflammatory cell infiltration. Mucus hypersecretion of goblet cells was visualized by PAS staining, and collagen deposition was assessed by Masson's trichrome staining.
Hydroxyproline Assay
Total lung collagen levels were determined using a previously described assay (26). Briefly, a left lobe of each lung tissue was hydrolyzed with 6 n HCl (10 μl/mg tissue) for 16 h at 120 °C. After brief cooling at room temperature, the lost volume by evaporation was replenished by water. The samples were centrifuged at 8000 × g for 10 min at room temperature, and supernatant was transferred to a fresh microcentrifuge tube. An equal volume of 6 n NaOH was added, and the solution was adjusted to pH 6–8 using litmus paper. Aliquots of neutralized samples and standard hydroxyproline were oxidized with a buffered chloramine-T reagent. The chromophore was developed with the addition of Ehrlich's reagent and was incubated for 35 min in the dark at 60 °C. Absorbance of each sample was read at 540 nm using a spectrophotometer and was plotted against the concentration of standard hydroxyproline.
Protein Extraction
For protein extraction, the right lung was stored at −80 °C until use. It was then placed in a clean mortar containing liquid nitrogen and finely ground, and then 1 ml of lysis buffer (30 mm Tris, 2 m thiourea, 7 m urea, 4% CHAPS, adjusted to pH 8.8 at 4 °C with HCl) was added, and the sample was mixed thoroughly and then sonicated for 30 min at 4 °C and centrifuged at 20,000 × g for 20 min at 4 °C. Then proteins in the supernatant were precipitated using the 2-D Clean-Up Kit (GE Healthcare) and resuspended in lysis buffer, with pH adjustment to 8.5 at 4 °C. Protein concentration was measured using the 2-D Quant kit (GE Healthcare).
2-D DIGE
The lung lysates were labeled according to the manufacturer's instructions (GE Healthcare). Briefly, 50 μg of lysate was mixed with 400 pmol of CyDyeTM (Cy2, Cy3, and Cy5; GE Healthcare) by vortexing, and the mixture was incubated on ice in the dark for 30 min. The lung lysates from n-Pen c 13-sensitized and PBS-treated mice were labeled with Cy3 and Cy5, respectively, mixed with Cy2-labeled internal pooled standard, and run in the same gel. Analysis was repeated with reciprocal labeling, and each labeling was repeated two times. The internal pooled standard sample was prepared by pooling 50 μg of protein from each of the eight lung lysates. The labeling reaction was stopped by the addition of 1 μl of 10 mm l-lysine solution (Sigma), and the sample was left on ice for 10 min. The combined proteins from a Cy3-labeled lysate, a Cy5-labeled lysate, and the Cy2-labeled pool (150 μg/gel) were mixed and denatured in 2-D sample buffer (8 m urea, 2% CHAPS, 2% immobilized pH gradient (IPG) 3–10 buffer, 60 mm DTT), and then the samples were actively rehydrated into 24-cm pH 3–10 immobilized pH gradient strips (ImmobilineTM DryStrip, GE Healthcare) for 16 h at 20 °C in a strip holder for first dimension isoelectric focusing, which was performed on an Ettan IPGphor II horizontal electrophoresis system (GE Healthcare). The electrophoresis conditions were 500 V for 4 h, 1000 V for 2 h, 8000 V for 10 h, and 8000 V up to 40,000 V-h. After focusing, the IPG strips were equilibrated for 15 min with gentle shaking in 10 ml of 50 mm Tris-HCl (pH 8.8), 6 m urea, 50% (w/v) glycerol, 2% (w/v) SDS, and 0.01% (w/v) bromphenol blue, and then second dimension electrophoresis was performed on a 12.5% SDS-polyacrylamide gel in an Ettan Daltsix electrophoresis system (GE Healthcare) at 2 watts/gel for 2 h and then at 17 watts/gel until the bromphenol blue front reached the end of the gel. For preparative gels for MS analysis, 300 μg of internal pooled standard proteins was used, and the gel was silver-stained according to the manufacturer's instructions (GE Healthcare). Cy2-, Cy3-, and Cy5-labeled images were acquired using a Typhoon 9400TM scanner (GE Healthcare). Intragel spot detection and intergel matching were carried out using, respectively, the Differential In-gel Analysis (DIA) module and Biological Variation Analysis (BVA) module of the DeCyder software (GE Healthcare). After manual inspection of this automatic matching, protein spots differentially expressed between n-Pen c 13-sensitized and PBS-treated mice were identified by Student's t test (p < 0.05).
In-gel Digestion and Protein Identification by Nano-LC-MS/MS
Proteins of interest were manually excised from a silver-stained preparative gel and destained with a 1:1 solution of 30 mm potassium ferricyanide and 100 mm sodium thiosulfate to remove silver staining and then dried in a SpeedVac concentrator. The protein was digested overnight at 37 °C with sequencing grade trypsin (Promega, Madison, WI) in 25 mm ammonium bicarbonate, pH 7.8. The resulting peptides were extracted sequentially with 50 μl of 1% TFA and 0.1% TFA, 60% acetonitrile, and the combined extracts were lyophilized and analyzed using a QSTARTM XL mass spectrometer (Applied Biosystems) coupled to an UltiMateTM nano-LC system (Dionex/LC Packings, Amsterdam, Netherlands). Peak lists of MS/MS spectra were created using the mascot.dll script version 1.6b13 in the Analyst QS program version 1.1 (Applied Biosystems) and uploaded to the Mascot MS/MS Ions Search program (Mascot version 2.2) on the Matrix Science public Web site, and protein identification was performed against the National Center for Biotechnology Information non-redundant (NCBInr) data base (NCBInr_20090627 (9,184,702 sequences; 3,146,847,487 residues)) with the taxonomy limited to Mus musculus (house mouse) (143,340 sequences). The data base searches were performed with the following variable modifications: cysteine carbamidomethylation and methionine oxidation with 0.3 Da precursor ion mass tolerance and 0.5 Da mass tolerance for fragment ions. Enzyme specificity for tryptic digests was selected to semitrypsin with two missed cleavages. MH22+ and MH33+ were selected as the precursor peptide charge states in the search. Ions scores greater than 41 indicate identity or extensive homology (p < 0.05); the individual score for the MS/MS spectrum of each peptide was more than 20.
Immunoblotting Analysis
To validate the 2-D DIGE and bioinformatics results, lung tissue lysates were prepared from n-Pen c 13-sensitized and PBS-treated mice, and then the proteins (30 μg of protein/lane) were separated by 12.5% SDS-PAGE and transferred to PVDF membranes (Millipore, Bedford, MA). The membranes were blocked with blocking buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 5% skim milk) for 1 h at room temperature with gentle shaking and then were washed with TNT buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% Tween 20) and incubated overnight at 4 °C with rabbit antibodies against Grp94 (glucose-regulated protein 94) (diluted 1:250), Grp78 (diluted 1:1000), moesin (diluted 1:500), or CFL1 (cofilin-1) (diluted 1:500) (all from Bioworld Technology) or mouse anti-CORO1A (coronin actin-binding protein 1A) antibodies (diluted 1:200) or goat anti-CAPZA1 (F-actin capping protein subunit α-1) antibodies (diluted 1:100) (both from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) (all dilutions in TNT buffer containing 1% skim milk). The membranes were then incubated for 1 h at room temperature with HRP-conjugated anti-mouse IgG antibody (BD Biosciences Pharmingen), anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), or anti-goat IgG antibody (Sigma) diluted 1:2000 in TNT buffer containing 1% skim milk. Bound antibodies were visualized using ECL reagents (PerkinElmer Life Sciences). β-Actin was used as an internal control of protein loading. The relative amounts of protein expression were analyzed using ImageQuantTM TL software (GE Healthcare) and normalized to the amounts of β-actin. To analyze the effect of n-Pen c 13 on junctional proteins, the lung lysates were subjected to immunoblot analysis using rabbit antibodies against occludin (diluted 1:2000), ZO-1 (diluted 1:3000), or E-cadherin (diluted 1:1000) (all from Zymed Laboratories Inc.. San Francisco, CA) and HRP-conjugated anti-rabbit IgG antibody (1:2000, Jackson ImmunoResearch Laboratories, Inc.). For a detailed description of immunoblot analysis, please see above.
In Vitro Assay
To further evaluate the effect of n-Pen c 13 on airway epithelium cells, NCI-H441 cells (3–5 × 105 cells) were grown on 6-well plates and cultured for 5–7 days until confluence and then were incubated for the indicated time with n-Pen c 13 (30 nm). Whole-cell extracts were prepared by washing the cells twice with PBS containing a protease inhibitor mixture (Calbiochem). Sample buffer (50 mm Tris-HCl, pH 6.8, 4% SDS, 12% glycerol, 2% β-mercaptoethanol, and a trace of bromphenol blue) was added to the cells, and the lysates were scraped into a microcentrifuge tube and boiled for 10 min at 95 °C. After SDS-PAGE, the proteins were subjected to immunoblot analysis as above using antibodies against occludin, ZO-1, and E-cadherin.
Immunocytochemistry
For immunocytochemical studies, NCI-H441 cells (5 × 104 cells/cm2) were grown on 24-well plates on glass coverslips coated with collagen. After attaining confluence, the cells were incubated in serum-free DMEM with either vehicle (PBS) or n-Pen c 13 (30 nm) for 0.5 h. All subsequent procedures were performed at room temperature. The cells were fixed in 4% paraformaldehyde in PBS for 15 min, washed with PBS, and permeabilized by incubation with 0.1% Triton X-100 in PBS for 15 min. They were then incubated with PBSB for 30 min and incubated for 1 h with a 1:50 dilution of rabbit antibodies against occludin, ZO-1, or E-cadherin (Zymed Laboratories Inc.. San Francisco, CA) in PBSB, washed, and incubated for 1 h with a 1:100 dilution of Alexa Fluor-488-conjugated goat anti-rabbit IgG antibodies in PBSB. Following a final wash, the cells were mounted with mounting medium and examined using a Leica TCS SP2 confocal laser-scanning microscope.
Transepithelial Electrical Resistance (TEER) Measurements
NCI-H441 cells (5 × 104 cells/cm2) were grown in a 24-well Transwell until confluence before use in the TEER study. The NCI-H441 cell monolayers were equilibrated in prewarmed HBSS at 37 °C before the experiment, and then the HBSS on the apical side was replaced with prewarmed HBSS containing 10 nm n-Pen c 13, and the resistance across the cell monolayer was measured at the indicated times to determine the extent to which tight junctions were opened.
Data Analysis
The data for AHR, serum samples, splenocyte culture supernatants, and hydroxyproline level were analyzed by one-way ANOVA, followed by the Newman-Keuls multiple comparison test. Western blotting and TEER measurement results were analyzed using the paired Student's t test. p < 0.05 was considered statistically significant. Values for all measurements were expressed as the mean ± S.E.
RESULTS
Exposure of Mice to Intratracheal Pen c 13 Induces Pulmonary Allergic Responses
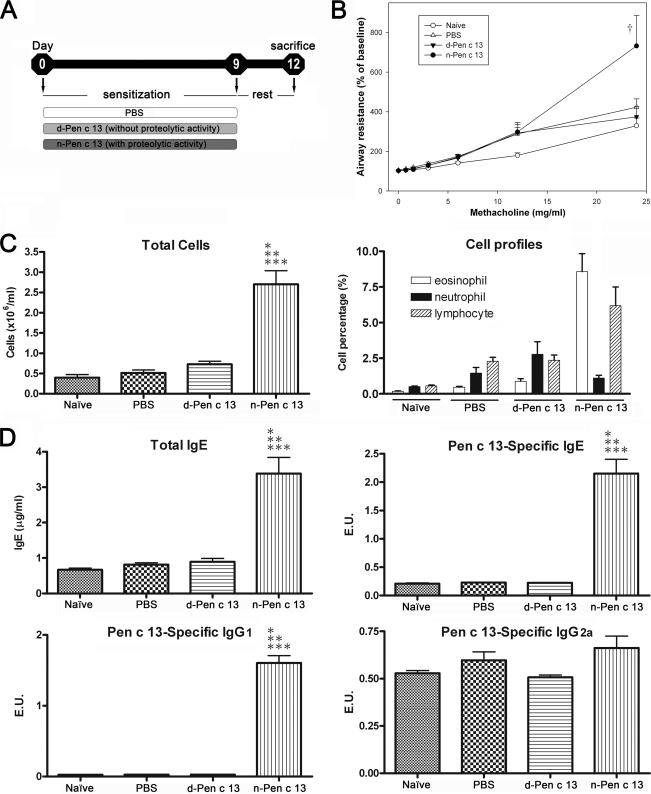
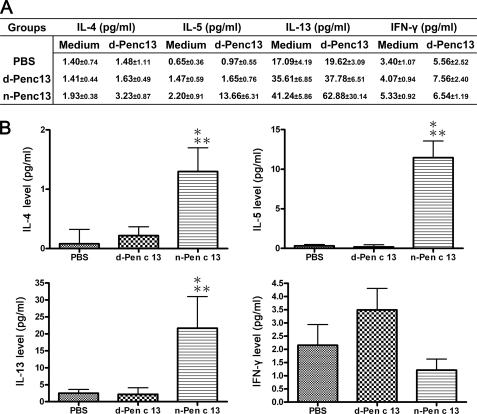
To investigate the effects of Pen c 13 protease in a murine model of induced allergic pulmonary inflammation, BALB/c mice were intratracheally inoculated with 1 μg of n-Pen c 13, d-Pen c 13, or PBS for 10 consecutive days (days 0–9) and were sacrificed 72 h after the last challenge (day 12) (Fig. 1A). There was no significant difference in body weight gain or loss between the groups throughout the period of intratracheal administration (supplemental Fig. S1). We further evaluated changes in airway resistance in response to increasing doses of methacholine. As seen in Fig. 1B, compared with d-Pen c 13-sensitized, PBS-treated, or naïve mice, the n-Pen c 13-sensitized mice showed significantly greater AHR at the highest dose (25 mg/ml) of methacholine (p < 0.05). In addition, significant numbers of leukocytes were increased in the BAL fluid (p < 0.001), of which 7.3–10% were eosinophils (Fig. 1C). A significant increase in levels of total IgE, Pen c 13-specific IgE, and Th2-associated Pen c 13-specific IgG1 was seen in the sera from the n-Pen c 13-sensitized mice compared with naïve or control animals (p < 0.001), but there was no significant difference in Th1-associated Pen c 13-specific IgG2a levels (Fig. 1D). To investigate the pattern of Th2-associated cytokines responses in this model, the d-Pen c 13-elicited secretion of cytokines in splenocyte cultures was determined as shown in Fig. 2A. The background level was measured in wells containing splenocytes in medium only. The increased levels of cytokine secretion were obtained by subtracting the background. The results are shown in Fig. 2B; a significant increase in the level of the Th2-associated cytokines IL-4, IL-5, and IL-13 was produced by splenocytes isolated from n-Pen c 13-sensitized mice compared with those from d-Pen c 13-sensitized mice or PBS-treated mice after in vitro stimulation with d-Pen c 13 (p < 0.05). However, there was no significant difference in Pen c 13-induced production of the Th1-associated cytokine IFN-γ between the n-Pen c 13-sensitized, d-Pen c 13-sensitized, and PBS-treated groups. In addition, we have also characterized this model regarding the levels of the gene expression of various Th2 cytokines, Th2 chemokines, and proinflammatory mediators in the lung to provide more information on the immunological aspect of n-Pen c 13-induced lung allergic inflammation. The n-Pen c 13 challenge caused a marked increase in the mRNA levels for the Th2 cytokines (IL-4, IL-5, and IL-13), Th2 chemokines (CCL17 and CCL22), and proinflammatory mediators (TNF-α and IL-10) (supplemental Fig. S2). These results suggest that the active protease Pen c 13 is a potent inducer of Th2 and IgE responses.
FIGURE 1.
Functional analyses of the ability of Pen c 13 to facilitate allergic sensitization and airway inflammation. A, schematic representation of the mouse model for inducing allergic inflammation by consecutive intratracheal administration of allergen. B, effect of n-Pen c 13 on methacholine-induced airway resistance in mice. AHR against aerosolized methacholine was determined by evaluation of percentage changes from the base-line level of airway resistance (n = 6–9/group). †, significant difference from control (naïve, PBS, and d-Pen c 13), p < 0.05. C, total cell count (left) and differential cell count (right) in the bronchoalveolar lavage. D, serum levels of total IgE (top left), specific IgE (top right), specific IgG1 (bottom left), and specific IgG2a (bottom right). Sera were collected from naïve mice (untreated) and mice intratracheally treated with PBS, d-Pen c 13, or n-Pen c 13. n = 4–10/group, p < 0.001 compared with naïve (*), PBS-treated (**), or d-Pen c 13-sensitized mice (***), respectively. The data are expressed as the mean ± S.E. Differences between experimental groups were assessed by one-way analysis of variance followed by the Newman-Keuls multiple comparison test. The levels of the specific IgE, IgG1, and IgG2a were expressed as ELISA unit (E.U.). The E.U. is calculated as the absorbance of sample subtracted by the blank (Asample − Ablank) divided by the absorbance of positive control subtracted by the blank (Apositive − Ablank).
FIGURE 2.
Th2-associated cytokine production after in vitro stimulation of splenocytes with d-Pen c 13. A, production of IL-4, IL-5, IL-13, and IFN-γ by splenocytes after in vitro stimulation with d-Pen c 13 or medium alone. n = 4–11/group; data are expressed as means ± S.E. B, the increased levels of cytokine secretion were determined by subtracting background activity of cells incubated with medium only without antigen. The data are expressed as the mean ± S.E. (error bars). p < 0.05 compared with PBS-treated (*) or d-Pen c 13-sensitized mice (**), respectively. Statistical analysis was performed using one-way analysis of variance followed by the Newman-Keuls multiple comparison test.
Histological Evaluation of Lung Pathology
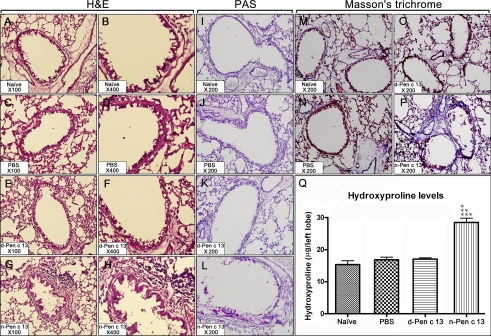
Histopathologic evaluation of the lavaged lung tissues of mice repeatedly challenged with n-Pen c 13 revealed extensive inflammatory infiltrates in both the peribronchiolar and perivascular connective tissues compared with d-Pen c 13- or PBS-challenged mice (Fig. 3, A–H). In addition, the PAS staining of the lung sections revealed markedly goblet cell hyperplasia and mucus hypersecretion in the bronchi of n-Pen c 13-sensitized mice but not d-Pen c 13-sensitized or PBS-treated mice (Fig. 3, I–L). Because peribronchial collagen deposition is one of the cardinal structural alterations associated with remodeling in asthma, lung tissue sections from four groups were stained with Masson trichrome to determine the relative amount and distribution of collagen deposition within the lung. The n-Pen c 13-sensitized mice showed a marked increase in collagen deposition compared with control mice (Fig. 3, M–P). Consistent with this morphometric analysis, the content of lung hydroxyproline in the n-Pen c 13-sensitized (p < 0.001) mice was markedly elevated compared with naïve or control mice (Fig. 3Q). Overall, these findings suggest that n-Pen c 13 can promote inflammatory cell infiltration, mucus overproduction, and collagen deposition leading to airway morphological changes.
FIGURE 3.
Histological evidence of induced inflammatory cell recruitment, mucus hypersecretion, and collagen deposition. Histopathologic analysis of a lung from a naïve mouse (A, B, I, and M) or a PBS-treated (C, D, J, and N), d-Pen c 13-sensitized (E, F, K, and O), or n-Pen c 13-sensitized (G, H, L, and P) mouse is shown. Lung sections were stained with H&E staining (magnification: ×100 (left) and ×400 (right)) for assessment of inflammatory changes, with PAS staining (magnification: ×200) for visualization of mucus-secreting cells in the intrapulmonary conducting airways (purple-red staining), and with Masson's trichrome staining (magnification: ×200) for showing collagen deposition (blue staining). The results shown are representative of those from at least four experiments. Q, the left lobe of the mice was harvested and assayed for hydroxyproline analysis. Values are expressed as the means ± S.E. (error bars) (n = 6) for each group. p < 0.001 compared with naïve (*), PBS-treated (**), or d-Pen c 13-sensitized mice (***), respectively. Differences between experimental groups were assessed by one-way analysis of variance followed by the Newman-Keuls multiple comparison test.
2-D DIGE Analysis of Lung Tissues from PBS-treated or n-Pen c 13-sensitized Mice
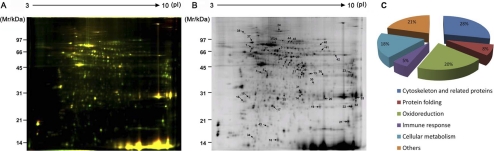
To explore the inflammatory mechanism induced by n-Pen c 13, its downstream regulatory effectors were analyzed. Using 2-D DIGE, we examined protein expression in tissue lysates of PBS-treated and n-Pen c 13-sensitized mice using a pH range of 3–10 and 12.5% SDS gels. The arrangement of samples on the four gels used is shown in supplemental Table 2, including the switching of samples between Cy3 and Cy5 to avoid dye labeling bias arising from the distinct fluorescence features of gels at different wavelengths. Fig. 4A shows representative images of a 2-D DIGE gel containing two test samples, one labeled with Cy3 (total protein in n-Pen c 13-sensitized tissue lysates) and the other with Cy5 (total protein in PBS-treated tissue lysates), and an internal standard pool labeled with Cy2 (combined total protein from all of the lungs of the four pairs of mice). The data were analyzed automatically using DeCyder image analysis software without user interference, and 4536 protein spots were detected on the master gel (gel 1). The other three gels were then matched to the master two-dimensional fluorescence map to detect protein spots showing consistent differences across all of the gels. Gel 2 contained 4524 spots and 3007 matches; gel 3 contained 3851 spots and 2695 matches; and gel 4 contained 3957 spots and 2487 matches. Statistical analysis using DeCyder software to apply a confidence level to each difference resulted in 45 significant reproducible spots induced by n-Pen c 13 (p < 0.05, >1.2-fold change). The location of these 45 protein spots on a representative gel is shown in Fig. 4B. The expression of 19 spots was decreased, and that of 26 spots was increased by n-Pen c 13 sensitization.
FIGURE 4.
2-D DIGE analysis of lung proteome alterations in a mouse model. A, overlaid two-dimensional gel images of tissue lysates from n-Pen c 13-sensitized mice labeled with Cy3 (green), PBS-treated mice labeled with Cy5 (red), protein spot patterns. B, representative two-dimensional electrophoresis profiles of lung samples using the pooled standard proteins showing altered expression after n-Pen c 13 treatment and the spots identified by nano-LC-MS/MS spectrometry, which are numbered and shown in supplemental Table 3. C, classification of identified proteins based on their functional annotations using gene ontology of the UniProt Knowledgebase and Rat Genome Database (available on the World Wide Web).
Identification of the Differentially Expressed Proteins
To identify the proteins, each gel was silver-stained, and then the protein spots of interest were excised manually, digested in-gel with trypsin, and identified by nano-LC-MS/MS spectrometry, followed by a Mascot data base search. Thirty-nine different proteins were identified from the 45 spots that showed a significant difference in expression levels between the PBS-treated and n-Pen c 13-sensitized mouse groups (supplemental Table 3). The ontology analysis of the identified proteins indicated the relevance and diversity of molecular functions, including cytoskeleton and related proteins (28%), protein folding (8%), oxidoreduction (20%), immune response (5%), cellular metabolism (18%), and others (21%), which may provide clues for understanding the molecular pathogenesis of lung disease (Fig. 4C).
Validation of the Protein Results by Western Blotting
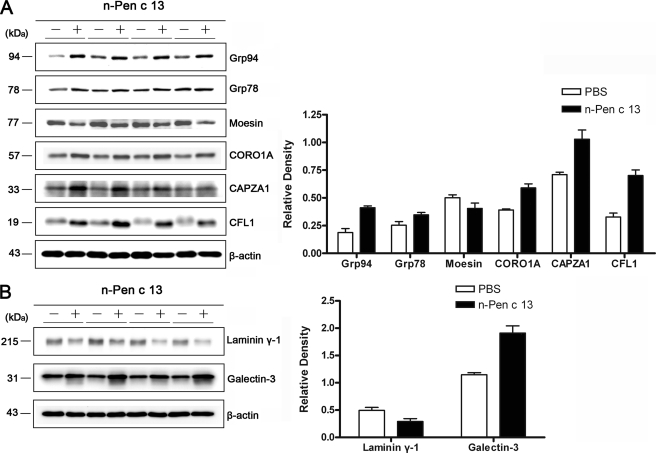
Bioinformatics analysis and a literature review for Pen c 13-regulated proteins identified in the present study showed that many of these proteins were involved in actin cytoskeleton signaling. To verify the 2-D DIGE results, we performed an immunoblotting analysis on six selected protein targets (Grp94, Grp78, moesin, CORO1A, CAPZA1, and CFL1) that have been suggested to be involved in actin cytoskeletal rearrangements. As shown in Fig. 5A, these results were consistent with the 2-D DIGE results. Five of the six proteins (Grp94, Grp78, CORO1A, CAPZA1, and CFL1) were up-regulated in lung tissues from n-Pen c 13-sensitized mice, whereas one protein, moesin, was down-regulated. We also performed immunoblotting analysis on two additional proteins (galectin-3 and laminin-γ1) identified by bioinformatics analysis (see supplemental material) but not by our proteomics experiments. Fig. 5B shows that galectin-3 expression in the lung was increased, whereas laminin-γ1 expression was decreased by n-Pen c 13. These data suggest that the allergic protease Pen c 13 might induce actin polymerization and cytoskeletal remodeling, thus allowing cells to migrate, which is closely linked to cellular and structural changes within the airway.
FIGURE 5.
Protein validation by Western blotting. A, confirmation for selected proteins that were identified as differentially expressed by 2-D DIGE. B, confirmation of the two proteins, laminin γ-1 and galectin-3, hypothetically identified by bioinformatics analysis. Left panels, representative immunoblots of lung lysates obtained from n-Pen c 13-sensitized mice (+) and PBS-treated mice (−). β-Actin was used as the loading control. Right panels, quantitative analysis of results of four Western blots performed using ImageQuantTM TL software. The data are shown as the mean ± S.E. (error bars).
Evaluation of Intercellular Barrier Disruption by n-Pen c 13
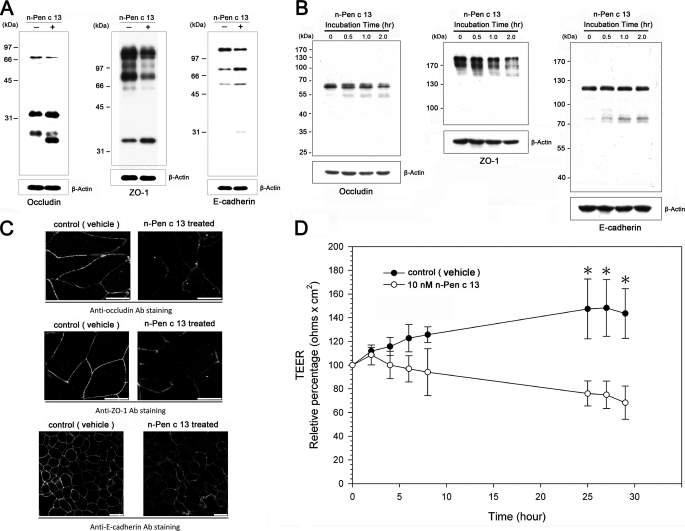
In order to understand the effects of proteolytic activity on epithelial function in models of n-Pen c 13-induced allergic airway inflammation, we examined lung tissue extracts from PBS-treated and n-Pen c 13-sensitized mice by Western blotting to ascertain whether loss of junctional proteins was caused by n-Pen c 13 and found that TJ proteins (occludin and ZO-1) and an AJ protein (E-cadherin) were markedly degraded in n-Pen c 13-sensitized mice (Fig. 6A). The effect of n-Pen c 13 on pulmonary epithelial barrier integrity in vivo was mimicked using an in vitro NCI-H441 cell model. Fig. 6B shows that n-Pen c 13 treatment of NCI-H441 cells resulted in time-dependent cleavage of occludin and E-cadherin into two major immunoreactive products. In addition, although ZO-1 is a cytoplasmic protein, it was also markedly reduced in the n-Pen c 13-treated cells, suggesting that ZO-1 may serve as a link between the transmembrane protein and the actin cytoskeleton, which is indirectly disrupted by n-Pen c 13. Similar results were obtained by immunofluorescence staining and confocal microscopy (Fig. 6C), which showed that n-Pen c 13 treatment of NCI-H441 cells resulted in less intense staining and some areas of discontinuous staining compared with the control, implying that disruption of TJ integrity had occurred. In addition, permeability measurements by TEER were performed to determine whether the effects of n-Pen c 13 on junctional proteins were associated with a diminished epithelial barrier. As shown in Fig. 6D, confluent NCI-H441 cells exhibited a mean TEER of 100 ohms cm2, and the TEER decreased in a time-dependent manner following n-Pen c 13 treatment compared with treatment with vehicle. These results suggest that breakdown of AJ/TJ proteins and increased permeability are associated with loss of pulmonary epithelium integrity in n-Pen c 13-sensitized mice.
FIGURE 6.
Disruption of junctional proteins by Pen c 13 increases epithelial paracellular permeability. A, immunoblots of junctional proteins from the lung tissues of PBS-treated or n-Pen c 13-sensitized mice. The results shown are representative of those from four separate experiments. B, immunoblots of junctional proteins from NCI-H441 cell monolayers under control conditions and after treatment for 0.5, 1, or 2 h with n-Pen c 13 (30 nm). The results shown are representative of those from at least four separate experiments. C, representative photomicrographs obtained by confocal microscopy after immunofluorescence staining for occludin, ZO-1, and E-cadherin in NCI-H441 cells exposed to PBS or n-Pen c 13 (30 nm) for 0.5 h. The results shown are representative of those from at least four separate experiments. Bar, 25 μm. D, time course of the decrease in the TEER of NCI-H441 cell monolayers after applying n-Pen c 13 (10 nm). Values are expressed as the means ± S.E. (error bars) (n = 3) for each measurement time point. p < 0.05 versus control.
DISCUSSION
Pen c 13, a major allergen secreted from P. citrinum, has been identified as an alkaline serine protease (22). We developed an animal model of allergic airway inflammation in response to continuous exposure to Pen c 13. Our results showed that n-Pen c 13 can induce cellular and immunologic responses in BALB/c mice. These include increased numbers of infiltrating cells, specifically eosinophils, increased production of Th2 cytokines IL-4, IL-5, and IL-13 by splenocytes from n-Pen c 13-sensitized mice after in vitro antigen-specific activation and significantly elevated serum levels of total IgE, Pen c 13-specific IgE, and the anti-Pen c 13 IgG1/IgG2a antibody ratio, leading to structural changes in the airways. Conversely, administration of d-Pen c 13, which lacks proteolytic activity, did not result in allergic airway inflammation and structural alterations.
In this study, we used a proteomic approach to compare the lung protein profile in n-Pen c 13-sensitized and PBS-treated mice. A total of 39 differentially expressed proteins were identified, and their potential biological connections with development of allergic airway inflammation were explored by bioinformatics analysis (supplemental Table 4 and Fig. S3). The canonical pathways identified were (i) acute phase response and leukocyte extravasation signaling; (ii) NRF2 (NF-E2-related factor 2)-mediated oxidative stress response; (iii) actin cytoskeleton, integrin, and focal adhesion kinase signaling; and (iv) tight junction signaling.
Acute Phase Response and Leukocyte Extravasation Signaling
During acute lung injury, the lung releases acute-phase proteins, such as pentraxin 3, causing leukocyte recruitment out of blood vessels into the airways, which contributes to the severity of asthma (27).
NRF2-mediated Oxidative Stress Response
Protease-induced inflammation in the respiratory tract may be associated with abnormal iron metabolism characterized by impaired iron release from tissues. However, excess iron is potentially harmful because it can catalyze the formation of toxic reactive oxygen species. The observed increase in ferritin (spot 17 in Fig. 4B) is probably responsible for the diversion of labile iron into ferritin stores and reduction of reactive oxygen species accumulation (28, 29). This suggests that the lung has a particularly efficient antioxidant system to protect it from damage by reactive oxygen species (30). Moreover, levels of superoxide dismutase (spot 19) and peroxiredoxin 1 (spot 22) were also increased in our proteomic data. Recent studies have revealed that both play a critical role in protecting the lungs against oxidative damage and are involved in the development of asthma (31, 32).
Actin Cytoskeleton, Integrin, and Focal Adhesion Kinase Signaling
Actin cytoskeletal signaling participates in many fundamental processes, including the regulation of cell shape, migration, and adhesion. Moreover, the actin cytoskeleton also plays a critical role in the activation of T cells (33, 34). In this study, we identified several proteins categorized as involved in actin cytoskeleton organization or related proteins, including F-actin capping protein (spot 4 in Fig. 4B), coronin (spot 6), macrophage capping protein (spot 11), CFL1 (spot 21), vinculin (spot 24), Arp2/3 complex (spot 43), and moesin (spot 45) (35–39). These proteins may be potential novel therapeutic targets, particularly in preventing tissue remodeling by proteases.
Tight Junction Signaling
The 2-D DIGE analysis revealed that expression of vinculin, a ubiquitously expressed actin-binding protein, was decreased in the n-Pen c 13-sensitized mice. This protein is believed to strengthen the mechanical links between adhesion complexes and the actin cytoskeleton, which are required for TJ organization. In vinculin-null cells, apical junctional organization is impaired, and this phenotype is rescued by re-expression of vinculin (40). In addition, a recent study also identified vinculin as a novel regulator of E-cadherin function and as providing dynamic regulation to reinforce the adhesion junctions (41). These results indicate that vinculin might be involved in the assembly of the apical junctional complex in epithelia. This suggests that Pen c 13 might disrupt AJ and TJ structure and barrier function. This agrees with our finding that AJ and TJ proteins were obviously degraded by n-Pen c 13 treatment in both in vitro and in vivo studies. We conclude that the changes in vinculin protein levels and actin organization are linked to disruption of cellular adhesion molecules to allow easier penetration of allergens. Furthermore, repeated damage and repair of epithelial cells may result in mucous metaplasia and airway remodeling.
Previous studies have indicated that three important intrinsic features might render proteins allergenic; these are (i) protease activity, (ii) surface features, and (iii) glycosylation patterns (12, 42–47). Several proteases from different sources, such as P. citrinum (Pen c 13), Penicillium chrysogenum (Pen ch 13), A. fumigatus (Asp f 13), Epicoccum purpurascens (Epi p 1), Curvularia lunata (Cur l 1), Dermatophagoides pteronyssinus (Der p 1, Der p 3, and Der p 9), Dermatophagoides farinae (Der f1 and Der f 3), and Periplaneta americana (Per a 10), have been reported as important allergens with proteolytic activity (19, 48–54). Many reports have revealed that allergic inflammation induced by proteases may be due to activation of PARs (10, 55, 56). Our previous study showed that Pen c 13 induces proinflammatory cytokine release in airway epithelial cells through PARs (23). Among the known proteases, thrombin has similar effects on lung epithelial cells (57). Furthermore, thrombin also induces connective tissue growth factor expression in lung fibroblasts by activating PAR1 (58). In fact, connective tissue growth factor, apart from its regulation of wound repair and fibrosis in the lung, mediates the expression of both matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) (59). In addition, fibrogenesis is strongly linked to the development of a Th2 cell response, which is characterized by an increase in the MMP/TIMP ratio (60–62). Interestingly, we observed that TIMP-2 expression was significantly increased in lung tissue after n-Pen c 13 inoculation (supplemental Fig. S4). Based on the above evidence, we hypothesized that MMPs and their inhibitors, TIMPs, may contribute to the development of Pen c 13-induced pulmonary fibrosis.
In the absence of d-Pen c 13 stimuli, the basal levels of IL-4, IL-5, IL-13, and IFN-γ in cultures of splenocytes from antigen-sensitized mice were higher than in PBS-treated mice. A possible explanation for these results might be that the antigen-presenting cells process antigen and migrate to drive the splenocyte activation, but without protease activity, the antigen is insufficient to induce the allergic response. So far, the role of protease in antigen processing and presentation related to Th2-type inflammation in vivo is still unclear. However, the dendritic cells and basophils are most probably to prime a Th2 response due to their ability to take up antigen and migrate to draining lymph nodes to initiate adaptive immune responses. Reports have indicated that the migration of dendritic cells and basophils to the draining lymph node as well as the expression of thymic stromal lymphopoietin by airway epithelial cells and basophils are further increased under the influence of protease, providing a powerful amplification mechanism of Th2 differentiation (63–65). Furthermore, a recent study has demonstrated that the occurrence of protease-induced Th2 responses requires dendritic cell-basophil cooperation via reactive oxygen species-mediated signaling (66), but the specific role of Pen c 13 in dendritic cells and basophils remains to be elucidated.
Among a number of candidate inflammatory mediators, galectin-3, which belongs to a family of β-galactoside-binding animal lectins, has been shown to play a pivotal role in activated cells involved in allergic airway inflammation (67–69). Recent studies also suggest that galectin-3 contributes to allergic inflammation by promoting polarization toward a Th2 immune response (70, 71). The role of galectin-3 in airway remodeling was investigated in a murine model of chronic allergic airway inflammation recently. This study demonstrated that galectin-3 is an important lectin that promotes airway remodeling via airway recruitment of inflammatory cells, specifically eosinophils, and the development of a Th2 phenotype as well as increased expression of eosinophil-specific chemokines and profibrogenic and angiogenic mediators (70). On the basis of the dramatic increase in galectin-3 expression in n-Pen c 13-sensitized mice, we hypothesize that galectin-3 might play a role in the development of allergic airway inflammation and remodeling. Inhibitors of this lectin may prove useful in the treatment of allergic disease. Another candidate inflammatory mediator, laminin, is an abundant basement membrane glycoprotein that participates in the maintenance of airway architecture. Previous studies have shown an increased thickness in layers expressing laminin in patients with asthma (72, 73), but our results showed that laminin was degraded by n-Pen c 13. This suggests thickened laminin expression in the airways, possibly as a result of continual damage and recovery from airway inflammation in asthma.
In conclusion, we have established a mouse model of allergic airway inflammation and changes in epithelial barrier integrity and function in response to continuous exposure to a proteolytically active fungal allergen. Our data underline the important role of the active protease Pen c 13 in driving Th2-associated inflammation in the lungs. To gain insights into the mechanisms involved in the regulation of allergenicity, we used a 2-D DIGE approach to identify differentially expressed proteins and also adopted bioinformatics analysis software to explore the immunological and pathological development of asthma. Taken together, these data demonstrate that Pen c 13 may contribute to tissue damage associated with allergic airway inflammatory diseases, such as asthma (Fig. 7). Thus, we show that prolonged inhalation of Pen c 13 without adjuvant may represent an optimal condition for developing experimental animal models of allergic airway inflammation. In addition, our studies may lead to the treatment of allergy using serine protease inhibitors, as suggested previously (74).
FIGURE 7.
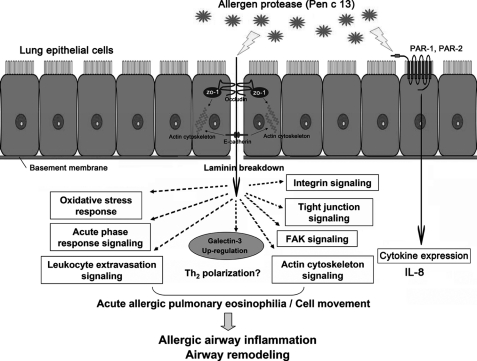
Schematic overview of possible signaling pathways in Pen c 13 allergen-induced allergic airway inflammation. Potential mechanisms by Pen c 13 might concomitantly disrupt the epithelial barrier and release IL-8. The epithelium damage might bring active Pen c 13 into contact with target cells and influence Th2 and IgE regulation via several signaling pathways.
Supplementary Material
Acknowledgment
We thank Hsin-Ying Huang (Graduate Institute of Immunology, College of Medicine, National Taiwan University) for essential technical support.
This work was supported in part by the Program for Excellence Research Teams from the Ministry of Education and Republic of China National Science Council Grant NSC 99-2320-B-002-012.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Results, Tables 1–4, and Figs. S1–S7.
- AHR
- airway hyperresponsiveness
- 2-D DIGE
- two-dimensional fluorescence difference gel electrophoresis
- AJ
- adherens junction
- HBSS
- Hanks' balanced salt solution
- MMP
- matrix metalloproteinase
- PAR
- protease-activated receptor
- PAS
- periodic acid-Schiff
- TEER
- transepithelial electrical resistance
- TIMP
- tissue inhibitor of metalloproteinase
- TJ
- tight junction
- n- and d-Pen c 13
- native and denatured Pen c 13, respectively.
REFERENCES
- 1. Lai C. K., De Guia T. S., Kim Y. Y., Kuo S. H., Mukhopadhyay A., Soriano J. B., Trung P. L., Zhong N. S., Zainudin N., Zainudin B. M. (2003) J. Allergy Clin. Immunol. 111, 263–268 [DOI] [PubMed] [Google Scholar]
- 2. Rabe K. F., Adachi M., Lai C. K., Soriano J. B., Vermeire P. A., Weiss K. B., Weiss S. T. (2004) J. Allergy Clin. Immunol. 114, 40–47 [DOI] [PubMed] [Google Scholar]
- 3. Gould H. J., Sutton B. J. (2008) Nat. Rev. Immunol. 8, 205–217 [DOI] [PubMed] [Google Scholar]
- 4. Elias J. A., Zhu Z., Chupp G., Homer R. J. (1999) J. Clin. Invest. 104, 1001–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Folli C., Descalzi D., Scordamaglia F., Riccio A. M., Gamalero C., Canonica G. W. (2008) Curr. Opin. Allergy Clin. Immunol. 8, 367–375 [DOI] [PubMed] [Google Scholar]
- 6. Wan H., Winton H. L., Soeller C., Gruenert D. C., Thompson P. J., Cannell M. B., Stewart G. A., Garrod D. R., Robinson C. (2000) Clin. Exp. Allergy 30, 685–698 [DOI] [PubMed] [Google Scholar]
- 7. Schneeberger E. E., Lynch R. D. (2004) Am. J. Physiol. Cell Physiol. 286, C1213–C1228 [DOI] [PubMed] [Google Scholar]
- 8. Lai C. H., Kuo K. H., Leo J. M. (2005) Brain Res. Brain Res. Rev. 50, 7–13 [DOI] [PubMed] [Google Scholar]
- 9. Hartsock A., Nelson W. J. (2008) Biochim. Biophys. Acta 1778, 660–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reed C. E., Kita H. (2004) J. Allergy Clin. Immunol. 114, 997–1008; quiz 1009 [DOI] [PubMed] [Google Scholar]
- 11. Wan H., Winton H. L., Soeller C., Tovey E. R., Gruenert D. C., Thompson P. J., Stewart G. A., Taylor G. W., Garrod D. R., Cannell M. B., Robinson C. (1999) J. Clin. Invest. 104, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shakib F., Ghaemmaghami A. M., Sewell H. F. (2008) Trends Immunol. 29, 633–642 [DOI] [PubMed] [Google Scholar]
- 13. Kauffman H. F., Tomee J. F., van de Riet M. A., Timmerman A. J., Borger P. (2000) J. Allergy Clin. Immunol. 105, 1185–1193 [DOI] [PubMed] [Google Scholar]
- 14. Johnson J. R., Wiley R. E., Fattouh R., Swirski F. K., Gajewska B. U., Coyle A. J., Gutierrez-Ramos J. C., Ellis R., Inman M. D., Jordana M. (2004) Am. J. Respir. Crit. Care Med. 169, 378–385 [DOI] [PubMed] [Google Scholar]
- 15. Chen C. L., Lee C. T., Liu Y. C., Wang J. Y., Lei H. Y., Yu C. K. (2003) J. Immunol. 170, 528–536 [DOI] [PubMed] [Google Scholar]
- 16. Fattouh R., Pouladi M. A., Alvarez D., Johnson J. R., Walker T. D., Goncharova S., Inman M. D., Jordana M. (2005) Am. J. Respir. Crit. Care Med. 172, 314–321 [DOI] [PubMed] [Google Scholar]
- 17. Cates E. C., Fattouh R., Wattie J., Inman M. D., Goncharova S., Coyle A. J., Gutierrez-Ramos J. C., Jordana M. (2004) J. Immunol. 173, 6384–6392 [DOI] [PubMed] [Google Scholar]
- 18. Yu C. K., Shieh C. M., Lei H. Y. (1999) J. Allergy Clin. Immunol. 104, 228–236 [DOI] [PubMed] [Google Scholar]
- 19. Gough L., Campbell E., Bayley D., Van Heeke G., Shakib F. (2003) Clin. Exp. Allergy 33, 1159–1163 [DOI] [PubMed] [Google Scholar]
- 20. Sudha V. T., Arora N., Singh B. P. (2009) Eur. J. Clin. Invest. 39, 507–516 [DOI] [PubMed] [Google Scholar]
- 21. Kurup V. P., Xia J. Q., Shen H. D., Rickaby D. A., Henderson J. D., Jr., Fink J. N., Chou H., Kelly K. J., Dawson C. A. (2002) Int. Arch. Allergy Immunol. 129, 129–137 [DOI] [PubMed] [Google Scholar]
- 22. Su N. Y., Yu C. J., Shen H. D., Pan F. M., Chow L. P. (1999) Eur. J. Biochem. 261, 115–123 [DOI] [PubMed] [Google Scholar]
- 23. Chiu L. L., Perng D. W., Yu C. H., Su S. N., Chow L. P. (2007) J. Immunol. 178, 5237–5244 [DOI] [PubMed] [Google Scholar]
- 24. Jensen L. B., Torp A. M., Andersen S. B., Skov P. S., Poulsen L. K., Knol E. F., van Hoffen E. (2008) J. Immunol. Methods 335, 116–120 [DOI] [PubMed] [Google Scholar]
- 25. Vanoirbeek J. A., Rinaldi M., De Vooght V., Haenen S., Bobic S., Gayan-Ramirez G., Hoet P. H., Verbeken E., Decramer M., Nemery B., Janssens W. (2010) Am. J. Respir. Cell Mol. Biol. 42, 96–104 [DOI] [PubMed] [Google Scholar]
- 26. Reddy G. K., Enwemeka C. S. (1996) Clin. Biochem. 29, 225–229 [DOI] [PubMed] [Google Scholar]
- 27. Deban L., Russo R. C., Sironi M., Moalli F., Scanziani M., Zambelli V., Cuccovillo I., Bastone A., Gobbi M., Valentino S., Doni A., Garlanda C., Danese S., Salvatori G., Sassano M., Evangelista V., Rossi B., Zenaro E., Constantin G., Laudanna C., Bottazzi B., Mantovani A. (2010) Nat. Immunol. 11, 328–334 [DOI] [PubMed] [Google Scholar]
- 28. Ryan T. P., Krzesicki R. F., Blakeman D. P., Chin J. E., Griffin R. L., Richards I. M., Aust S. D., Petry T. W. (1997) Free Radic. Biol. Med. 22, 901–908 [DOI] [PubMed] [Google Scholar]
- 29. Orino K., Lehman L., Tsuji Y., Ayaki H., Torti S. V., Torti F. M. (2001) Biochem. J. 357, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen X. L., Kunsch C. (2004) Curr. Pharm. Des. 10, 879–891 [DOI] [PubMed] [Google Scholar]
- 31. Comhair S. A., Ricci K. S., Arroliga M., Lara A. R., Dweik R. A., Song W., Hazen S. L., Bleecker E. R., Busse W. W., Chung K. F., Gaston B., Hastie A., Hew M., Jarjour N., Moore W., Peters S., Teague W. G., Wenzel S. E., Erzurum S. C. (2005) Am. J. Respir. Crit. Care Med. 172, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Inoue K., Takano H., Koike E., Warabi E., Yanagawa T., Yanagisawa R., Ishii T. (2009) Int. Immunopharmacol. 9, 1281–1288 [DOI] [PubMed] [Google Scholar]
- 33. Dustin M. L., Cooper J. A. (2000) Nat. Immunol. 1, 23–29 [DOI] [PubMed] [Google Scholar]
- 34. Föger N., Rangell L., Danilenko D. M., Chan A. C. (2006) Science 313, 839–842 [DOI] [PubMed] [Google Scholar]
- 35. Cooper J. A., Schafer D. A. (2000) Curr. Opin. Cell Biol. 12, 97–103 [DOI] [PubMed] [Google Scholar]
- 36. de Hostos E. L. (1999) Trends Cell Biol. 9, 345–350 [DOI] [PubMed] [Google Scholar]
- 37. Hashimoto S., Amaya F., Matsuyama H., Ueno H., Kikuchi S., Tanaka M., Watanabe Y., Ebina M., Ishizaka A., Tsukita S., Hashimoto S. (2008) Am. J. Physiol. Lung Cell Mol. Physiol. 295, L566–L574 [DOI] [PubMed] [Google Scholar]
- 38. Young C. L., Feierstein A., Southwick F. S. (1994) J. Biol. Chem. 269, 13997–14002 [PubMed] [Google Scholar]
- 39. Rüdiger M. (1998) BioEssays 20, 733–740 [DOI] [PubMed] [Google Scholar]
- 40. Watabe-Uchida M., Uchida N., Imamura Y., Nagafuchi A., Fujimoto K., Uemura T., Vermeulen S., van Roy F., Adamson E. D., Takeichi M. (1998) J. Cell Biol. 142, 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peng X., Cuff L. E., Lawton C. D., DeMali K. A. (2010) J. Cell Sci. 123, 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ghaemmaghami A. M., Shakib F. (2002) Clin. Exp. Allergy 32, 728–732 [DOI] [PubMed] [Google Scholar]
- 43. Furmonaviciene R., Shakib F. (2001) Mol. Pathol. 54, 155–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aalberse R. C. (2000) J. Allergy Clin. Immunol. 106, 228–238 [DOI] [PubMed] [Google Scholar]
- 45. Furmonaviciene R., Sutton B. J., Glaser F., Laughton C. A., Jones N., Sewell H. F., Shakib F. (2005) Bioinformatics 21, 4201–4204 [DOI] [PubMed] [Google Scholar]
- 46. Deslée G., Charbonnier A. S., Hammad H., Angyalosi G., Tillie-Leblond I., Mantovani A., Tonnel A. B., Pestel J. (2002) J. Allergy Clin. Immunol. 110, 763–770 [DOI] [PubMed] [Google Scholar]
- 47. Shreffler W. G., Castro R. R., Kucuk Z. Y., Charlop-Powers Z., Grishina G., Yoo S., Burks A. W., Sampson H. A. (2006) J. Immunol. 177, 3677–3685 [DOI] [PubMed] [Google Scholar]
- 48. Shen H. D., Tam M. F., Tang R. B., Chou H. (2007) Curr. Allergy Asthma Rep. 7, 351–356 [DOI] [PubMed] [Google Scholar]
- 49. Tai H. Y., Tam M. F., Chou H., Peng H. J., Su S. N., Perng D. W., Shen H. D. (2006) Allergy 61, 382–388 [DOI] [PubMed] [Google Scholar]
- 50. Bisht V., Arora N., Singh B. P., Pasha S., Gaur S. N., Sridhara S. (2004) FEMS Immunol. Med. Microbiol. 42, 205–211 [DOI] [PubMed] [Google Scholar]
- 51. Tripathi P., Kukreja N., Singh B. P., Arora N. (2009) J. Clin. Immunol. 29, 292–302 [DOI] [PubMed] [Google Scholar]
- 52. Gough L., Schulz O., Sewell H. F., Shakib F. (1999) J. Exp. Med. 190, 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sun G., Stacey M. A., Schmidt M., Mori L., Mattoli S. (2001) J. Immunol. 167, 1014–1021 [DOI] [PubMed] [Google Scholar]
- 54. Sudha V. T., Arora N., Gaur S. N., Pasha S., Singh B. P. (2008) Allergy 63, 768–776 [DOI] [PubMed] [Google Scholar]
- 55. Matsuwaki Y., Wada K., White T. A., Benson L. M., Charlesworth M. C., Checkel J. L., Inoue Y., Hotta K., Ponikau J. U., Lawrence C. B., Kita H. (2009) J. Immunol. 183, 6708–6716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shpacovitch V., Feld M., Bunnett N. W., Steinhoff M. (2007) Trends Immunol. 28, 541–550 [DOI] [PubMed] [Google Scholar]
- 57. Asokananthan N., Graham P. T., Fink J., Knight D. A., Bakker A. J., McWilliam A. S., Thompson P. J., Stewart G. A. (2002) J. Immunol. 168, 3577–3585 [DOI] [PubMed] [Google Scholar]
- 58. Yu C. C., Hsu M. J., Kuo M. L., Chen R. F., Chen M. C., Bai K. J., Yu M. C., Chen B. C., Lin C. H. (2009) J. Immunol. 182, 7916–7927 [DOI] [PubMed] [Google Scholar]
- 59. Fan W. H., Karnovsky M. J. (2002) J. Biol. Chem. 277, 9800–9805 [DOI] [PubMed] [Google Scholar]
- 60. Wynn T. A. (2004) Nat. Rev. Immunol. 4, 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wynn T. A., Cheever A. W., Jankovic D., Poindexter R. W., Caspar P., Lewis F. A., Sher A. (1995) Nature 376, 594–596 [DOI] [PubMed] [Google Scholar]
- 62. Sandler N. G., Mentink-Kane M. M., Cheever A. W., Wynn T. A. (2003) J. Immunol. 171, 3655–3667 [DOI] [PubMed] [Google Scholar]
- 63. Paul W. E., Zhu J. (2010) Nat. Rev. Immunol. 10, 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kouzaki H., O'Grady S. M., Lawrence C. B., Kita H. (2009) J. Immunol. 183, 1427–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sokol C. L., Barton G. M., Farr A. G., Medzhitov R. (2008) Nat. Immunol. 9, 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tang H., Cao W., Kasturi S. P., Ravindran R., Nakaya H. I., Kundu K., Murthy N., Kepler T. B., Malissen B., Pulendran B. (2010) Nat. Immunol. 11, 608–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen H. Y., Sharma B. B., Yu L., Zuberi R., Weng I. C., Kawakami Y., Kawakami T., Hsu D. K., Liu F. T. (2006) J. Immunol. 177, 4991–4997 [DOI] [PubMed] [Google Scholar]
- 68. Zuberi R. I., Hsu D. K., Kalayci O., Chen H. Y., Sheldon H. K., Yu L., Apgar J. R., Kawakami T., Lilly C. M., Liu F. T. (2004) Am. J. Pathol. 165, 2045–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. López E., del Pozo V., Miguel T., Sastre B., Seoane C., Civantos E., Llanes E., Baeza M. L., Palomino P., Cárdaba B., Gallardo S., Manzarbeitia F., Zubeldia J. M., Lahoz C. (2006) J. Immunol. 176, 1943–1950 [DOI] [PubMed] [Google Scholar]
- 70. Ge X. N., Bahaie N. S., Kang B. N., Hosseinkhani M. R., Ha S. G., Frenzel E. M., Liu F. T., Rao S. P., Sriramarao P. (2010) J. Immunol. 185, 1205–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Saegusa J., Hsu D. K., Chen H. Y., Yu L., Fermin A., Fung M. A., Liu F. T. (2009) Am. J. Pathol. 174, 922–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Altraja A., Laitinen A., Virtanen I., Kämpe M., Simonsson B. G., Karlsson S. E., Håkansson L., Venge P., Sillastu H., Laitinen L. A. (1996) Am. J. Respir. Cell Mol. Biol. 15, 482–488 [DOI] [PubMed] [Google Scholar]
- 73. Ward C., Pais M., Bish R., Reid D., Feltis B., Johns D., Walters E. H. (2002) Thorax 57, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Smith P. K., Harper J. I. (2006) Allergy 61, 1441–1447 [DOI] [PubMed] [Google Scholar]
- 75. National Research Council (1996) Guide for the Care and Use of Laboratory Animals, National Academy Press, Washington, D. C [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.