Abstract
During the biosynthesis of heme d1, the essential cofactor of cytochrome cd1 nitrite reductase, the NirE protein catalyzes the methylation of uroporphyrinogen III to precorrin-2 using S-adenosyl-l-methionine (SAM) as the methyl group donor. The crystal structure of Pseudomonas aeruginosa NirE in complex with its substrate uroporphyrinogen III and the reaction by-product S-adenosyl-l-homocysteine (SAH) was solved to 2.0 Å resolution. This represents the first enzyme-substrate complex structure for a SAM-dependent uroporphyrinogen III methyltransferase. The large substrate binds on top of the SAH in a “puckered” conformation in which the two pyrrole rings facing each other point into the same direction either upward or downward. Three arginine residues, a histidine, and a methionine are involved in the coordination of uroporphyrinogen III. Through site-directed mutagenesis of the nirE gene and biochemical characterization of the corresponding NirE variants the amino acid residues Arg-111, Glu-114, and Arg-149 were identified to be involved in NirE catalysis. Based on our structural and biochemical findings, we propose a potential catalytic mechanism for NirE in which the methyl transfer reaction is initiated by an arginine catalyzed proton abstraction from the C-20 position of the substrate.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, Enzyme Structure, S-Adenosylmethionine (SAM), Site-directed Mutagenesis, Pseudomonas aeruginosa, SUMT, Heme d1, Tetrapyrrole, Uroporphyrinogen III
Introduction
Tetrapyrroles such as hemes, (bacterio)chlorophylls, siroheme, or cobalamin play essential roles in many fundamental biological processes, including respiration, photosynthesis, and numerous enzymatic reactions (1). The dioxo-isobacteriochlorin heme d1, another member of the tetrapyrrole family, serves as the catalytically essential prosthetic group in the cytochrome cd1 nitrite reductase that catalyzes the second step of the denitrification process in denitrifying bacteria such as Pseudomonas denitrificans, Paracoccus pantotrophus, or the human pathogen Pseudomonas aeruginosa (2).
All naturally occurring tetrapyrroles are synthesized along a branched multistep biosynthetic pathway and share uroporphyrinogen III (uro'gen III)3 as the last common precursor. Uro'gen III is either decarboxylated to yield coproporphyrinogen III for heme and (bacterio)chlorophyll formation or methylated for the biosynthesis of siroheme, cobalamin, coenzyme F430, and heme d1 (1, 3). The methylation of uro'gen III at positions C-2 and C-7 leads to the formation of precorrin-2 (Fig. 1). The two methyl groups are derived from S-adenosyl-l-methionine (SAM) and are transferred onto uro'gen III by SAM-dependent uro'gen III methyltransferases (SUMTs) (4). Up until now, several SUMTs from different organisms have been identified, and their biochemical functions in cobalamin (CobA), siroheme (CysG, SirA, UPM1, Met1p), and heme d1 (NirE) biosynthesis were characterized (5–13). Among these SUMTs three subclasses can be distinguished based on their size and functionalities. SUMTs like CobA, SirA, Met1p, and NirE are usually dimeric proteins with subunits of about 30 kDa and possess SUMT activity only (5–10, 13). In contrast, CysG is a dimeric protein of 50-kDa subunits, each of which contains two independent enzymatic modules. The C-terminal module (CysGA) of the enzyme is homologous to CobA, SirA, Met1p, and NirE and carries SUMT activity. The N-terminal module (CysGB) of CysG functions as a bifunctional dehydrogenase-ferrochelatase converting precorrin-2 into siroheme via the intermediate sirohydrochlorin. Thus, CysG is a trifunctional siroheme synthase (14, 15). Finally, in some organisms the cobA gene is fused to hemD resulting in the production of a fusion protein carrying uro'gen III synthase and SUMT activities (16, 17).
FIGURE 1.
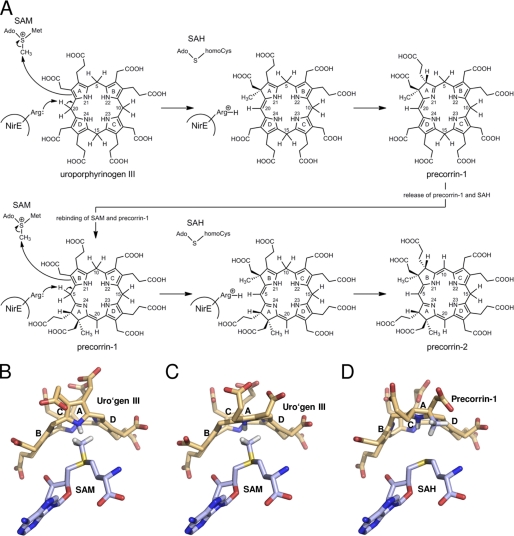
Reaction catalyzed by NirE. NirE and other SUMTs catalyze the bis-methylation of uroporphyrinogen III to precorrin-2. The two methyl groups that are attached to C-2 and C-7 are derived from SAM. SAH is produced as a reaction by-product.
The NirE protein was only recently characterized as a monofunctional SUMT specifically required for heme d1 biosynthesis. SUMT activity was demonstrated for NirE from P. aeruginosa and P. pantotrophus in vivo and in vitro (10, 13). Homodimeric NirE from P. aeruginosa showed inhibition by its substrate uro'gen III and the reaction by-product S-adenosyl-l-homocysteine (SAH) as previously observed for other SUMTs like CobA (5, 9). However, the homologous NirE protein from P. pantotrophus did not show these substrate inhibition phenomena (13).
Crystal structures for SUMT proteins were solved for CysG from Salmonella enterica (18), CobA from P. denitrificans (19), and the SUMT from Thermus thermophilus (ttSUMT) (20), all in complex with the reaction product SAH. As expected, the overall protein fold of the different SUMT proteins is conserved. Each subunit of the dimeric SUMTs consists of two domains that are connected by a single linker. Based on their overall structure SUMTs can be classified as class III methyltransferases (21). In all three available SUMT structures SAH was found to bind to the inner surface of the large active site pocket between the two domains.
Despite the previous biochemical and structural studies of SUMTs the current knowledge of the catalytic mechanism of these enzymes is limited. The two methyl groups are sequentially attached to uro'gen III with the C-2 position being methylated before the C-7 position (22, 23). After the first methylation at C-2 the resulting products precorrin-1 and SAH are thought to be released from the active site of the enzyme (5). For the second methylation at C-7 another SAM molecule and precorrin-1 have to bind to the active site. The methyl transfer reaction itself probably occurs through an SN2 mechanism as is the case for other SAM-dependent methyltransferases (24). After each methyl transfer a prototropic rearrangement of the macrocycle double bonds is presumed to take place (18, 25). Based on a model of the enzyme-substrate complex and site-directed mutagenesis several predictions were made which amino acids within the active site could be involved in catalysis. However, due to the lack of a structure for the enzyme-substrate complex the definite assignment of the precise functional roles for these amino acid residues was not possible.
Here, we report the crystal structure of NirE from P. aeruginosa in complex with its substrate uro'gen III and the reaction by-product SAH at a resolution of 2.0 Å. Based on the structure of this enzyme-substrate complex and supported by biochemical studies employing NirE variants, we identified several highly conserved amino acid residues most likely involved in catalysis. We propose a catalytic mechanism in which two arginines that are highly conserved in all SUMTs play essential roles for the methyl transfer reaction.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals, reagents, and antibiotics were purchased from Sigma-Aldrich or Merck. Restriction enzymes and other DNA-modifying enzymes were obtained from New England Biolabs. For purification of DNA fragments, the QIAquick PCR purification and gel extraction kits were purchased from Qiagen as well as all crystallization suites. Ni2+-SepharoseTM 6 Fast Flow and the HiLoadTM 26/60 SuperdexTM 75 preparation grade column were obtained from GE Healthcare. All primers were purchased from metabion international AG (Martinsried, Germany). Uroporphyrin III was obtained from Frontier Scientific Europe (Carnforth, UK).
Construction of Vectors
The construction of the vector pET32a-nirE-Trx was described previously (10). For the construction of the vector pET32a_PreS2-4_nirE the nirE gene fused to a DNA fragment encoding a PreScissionTM cleavage site was PCR-amplified using the primers 54nirE_MscI_fw (GAATTGGCCATCTGGAAGTTCTGTTCCAGGG) and 14nirE_XhoI_rv (GACTCGAGGGCGCATGCGAC) containing restriction sites for MscI and XhoI (underlined), respectively, and the vector pGEX6p1-nirE as the template. The resulting DNA fragments were purified, digested with MscI and XhoI, and subsequently ligated into the similarly digested vector pET32a (Novagen) to generate the vector pET32a_PreS2–4_nirE. The template vector pGEX6p1-nirE was generated by cloning the PCR-amplified P. aeruginosa nirE gene into the vector pGEX6p1 (GE Healthcare) using the BamHI and NotI restriction sites. For the construction of expression vectors containing the nirE gene carrying different point mutations the overlap extension PCR technique was used (26). After the PCR amplification of two DNA fragments containing an overlapping region with the appropriate point mutation, a fusion PCR was performed to amplify the complete DNA fragment including the point mutation. The obtained DNA fragments were cloned into the vector pET32a_PreS2–4_nirE using BamHI and XhoI restriction sites to replace the wild-type (WT) nirE with the mutant nirE. After cloning, the correct nucleotide sequence of the nirE gene was verified by sequencing of all nirE constructs (GATC Biotech AG, Konstanz, Germany).
Protein Production and Purification
Production and purification of recombinant NirE from P. aeruginosa were performed as described previously with minor modifications (10). Buffer A was slightly modified and contained 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% (w/v) glycerol. The soluble protein fraction was treated three times for 10 s with ultrasound before being applied directly to a 1.5-ml Ni2+-SepharoseTM 6 Fast Flow column. After washing and elution steps the NirE-containing fractions were pooled, and dithiothreitol (DTT) was added to a final concentration of 5 mm. PreScissionTM protease or Tag off enterokinase (Novagen) was added to the NirE solution which was dialyzed overnight against buffer A. When cleaved with PreScissionTM protease the dialyzed protein solution was again applied to the Ni2+-SepharoseTM 6 Fast Flow column to remove the tag. For further purification of the now untagged NirE, a gel permeation chromatography was performed using buffer A. The NirE-containing fractions were pooled and concentrated by ultrafiltration (Amicon, Millipore GmbH, Eschborn, Germany). Finally, the buffer was exchanged in an anaerobic chamber (Coy Laboratories, Grass Lake, MI) against thoroughly degassed buffer A. The purified and concentrated NirE protein (10 mg/ml) was directly used for crystallization.
Reduction of Uroporphyrin III
Uroporphyrin III was chemically reduced to uro'gen III either with 3% sodium amalgam or with H2/palladium as described previously (27, 28). For all crystallization setups the H2/palladium-reduced uro'gen III was used, and for the determination of the specific activity of NirE WT and mutant enzymes the sodium amalgam-reduced uro'gen III was used.
SAM Binding Assay
The SAM binding assay was performed as described previously (10).
Determination of NirE Activity
The enzymatic activity of NirE WT and mutant enzymes was determined in an in vitro activity assay as described previously (10). Briefly, the activity assays were performed in an anaerobic chamber (Coy Laboratories) under anaerobic conditions. Uro'gen III was generated from 1 mm 5-aminolevulinic acid using the enzymes HemB from P. aeruginosa (0.14 μm), HemC (0.15 μm), and HemD (0.17 μm) both from Bacillus megaterium in a final volume of 1 ml of degassed buffer B (50 mm Tris-HCl, pH 8.0, 100 mm KCl, 5 mm MgCl2, 50 mm NaCl, 5 mm DTT). NirE WT and mutant enzymes were added to a final concentration of 1.5 μm, respectively. SirC from B. megaterium (1.5 μm) and NAD+ (100 μm) were added to convert precorrin-2 to sirohydrochlorin for quantification using the published extinction coefficient of sirohydrochlorin (ϵ376 nm = 2.4 × 105 m−1 cm−1) (29). 200 μm SAM was added to start the reaction. The assay mixture was incubated at 37 °C in the dark, and product formation was monitored using a V-550 or V-650 spectrophotometer (Jasco, Gross-Umstadt, Germany). The specific activity of NirE was determined as described previously (10) using chemically reduced uro'gen III at a final concentration of 17 μm.
Determination of Highly Conserved Amino Acid Residues
For the determination of conserved amino acid residues among SUMTs from different organisms an amino acid sequence alignment (ClustalW) was created with the sequences of more than 60 different SUMTs. Those amino acids that were identical in all SUMTs are termed “highly conserved” throughout this manuscript.
Crystallization
For crystallization of NirE without uro'gen III, the protein (9.2 mg/ml) was incubated with 2 mm SAM for 1 h prior to use. A crystallization robot (Mosquito; TTP abTech Ltd., Melbourne, UK) was used for preparing the sitting-drop vapor diffusion setups. 200 nl of reservoir buffer of the JCSG I-IV and ammonium sulfate screens (Qiagen) was mixed with 200 nl of NirE solution and incubated at 18 °C in the dark. Crystals grew within 3–6 days and were soaked with 20% glycerol prior to shock cooling in liquid nitrogen. For crystallization of NirE with uro'gen III, the protein (10 mg/ml) was crystallized by sitting-drop vapor diffusion at 17 °C in an anaerobic chamber. Prior to use NirE was incubated with an 82-fold molar excess of uro'gen III and 1 mm SAH for 1 h. For the crystallization drops, 1 μl of protein solution was mixed with 1 μl of reservoir solution of the JCSG+ Screen (Qiagen) and incubated in the dark. Crystals grew within 4–8 days and were soaked with 20% glycerol prior to shock cooling in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
X-ray diffraction data were collected in house at a Rigaku MicroMax 007HF rotating anode x-ray generator equipped with a Saturn 944+ charge-coupled device detector (Rigaku, Sevenoaks, UK) and at beamline 14.2 of the Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY, Berlin, Germany). Datasets of NirE in complex with SAH and NirE in complex with SAH and uro'gen III were processed with HKL2000 (30) and XDS (31, 32), respectively. Data were phased by molecular replacement with MOLREP (33) using the monomer of CobA (PDB ID code 1S4D) (19) as the search model. The MOLREP solution was manually corrected and rebuilt in COOT (34) and refined through rigid body, restrained, and TLS refinement in REFMAC5 (35). Atomic displacement factors were treated as being isotropic whereas anisotropic motion of the protein was accounted for by performing TLS refinement (36) using two or five TLS groups for dataset I and dataset II, respectively. The TLS groups were determined using the TLSMD server (37). Refinement was stopped after converging values of Rwork and Rfree were reached. Data collection and refinement statistics are summarized in Table 1. Figures were created with ESPript (38), LIGPLOT (39), and PyMOL (40).
TABLE 1.
Crystallographic data and refinement statistics
| Statistics | Dataset I: NirE + SAH | Dataset II: NirE + SAH + uro'gen III |
|---|---|---|
| Data collection | ||
| Beamline | Rigaku MircoMax 007HF | BESSY (BL14.2) |
| Wavelength (Å) | 1.541 | 0.91841 |
| Resolution (Å)a | 46.3-2.0 (2.05-2.00) | 35.0-2.0 (2.05-2.00) |
| Space group | C2221 | C2221 |
| Unit cell parameters | ||
| a, b, c (Å) | 60.9, 115.1, 76.8 | 60.9, 116.2, 77.1 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Unique reflectionsa | 19,360 (1,411) | 18,362 (1,418) |
| Multiplicitya | 4.4 (4.5) | 3.9 (4.0) |
| Completeness (%)a | 99.82 (99.86) | 92.6 (99.9) |
| I/σIa | 20.19 (2.58) | 22.8 (4.0) |
| Rmergea,b | 0.043 (0.586) | 0.083 (0.451) |
| Wilson B (Å2) | 35.1 | 34.4 |
| Solvent content (%) | 46.4 | 49.7 |
| Refinement | ||
| Rworkc | 0.2349 | 0.2169 |
| Rfreed | 0.2868 | 0.2526 |
| No. of atoms | ||
| Protein | 1,864 | 1,825 |
| Ligand | 26 | 86 |
| Water | 195 | 129 |
| Total | 2,085 | 2,040 |
| Atomic displacement factors B (Å2)e | ||
| Overall | 21.4 | 33.6 |
| Protein | 21.4 | 33.7 |
| SAH | 23.6 | 22.7 |
| Uro'gen III | 58.8 | |
| Water | 21.4 | 24.0 |
| Molecules per asymmetric unit | 1/1/0 | 1/1/1 |
| (NirE/SAH/uro'gen III) | ||
| Real space correlation coefficientsf | ||
| Total | 0.7676 | 0.7806 |
| Protein | ||
| Backbone | 0.8204 | 0.8546 |
| Side chains | 0.7848 | 0.8232 |
| SAH | 0.9314 | 0.9427 |
| Uro'gen III | 0.7769 | |
| Root mean square deviation from ideal | ||
| Bond lengths (Å) | 0.0239 | 0.0219 |
| Bond angles (°) | 2.167 | 1.935 |
| Ramachandran plot | ||
| Favored (%) | 97.1 | 97.0 |
| Allowed (%) | 2.9 | 3.0 |
| Disallowed (%) | 0.0 | 0.0 |
a Values in parentheses account for the highest resolution shell.
b Rmerge = Σ| Io−〈I〉|/ΣIo.
c Rwork = (Σ‖Fo|−|Fc‖/Σ|Fo|); |Fo|, structure factor magnitudes observed; |Fc|, structure factors calculated.
d Rfree is computed as Rwork but using 5% randomly assigned reflections excluded from refinement.
e These B factors constitute the total B factors representing isotropic (thermal) and anisotropic (domain) movement.
f Real space correlation coefficient (RSCC) = Σ|ρo−〈ρo〉|Σ|ρc − 〈ρc〉|/(Σ| ρo−〈ρo〉|2Σ|ρc−〈ρc〉2|)1/2; ρ, electron density values at grid points that cover the molecule in question; ρo, experimental electron density; ρc, model density.
Protein Data Bank Accession Numbers
The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank. The entry codes are 2YBO for NirE + SAH, and 2YBQ for NirE + SAH + uro'gen III.
RESULTS AND DISCUSSION
Crystallization of NirE and Structure Determination
Recombinant purified NirE from P. aeruginosa was crystallized either in the presence of added SAM or in the presence of added uro'gen III plus SAH by the sitting-drop vapor diffusion method. Crystals of NirE without uro'gen III were obtained in Tris, pH 8.0, 24% PEG 6000, and crystals of the NirE-substrate complex grew in 0.2 m lithium sulfate, 0.1 m Tris, pH 8.5, 1.26 m ammonium sulfate. NirE crystals were base-centered orthorhombic. Apart from minor deviations in unit cell dimensions, crystals of NirE with SAH and NirE with both SAH and uro'gen III were isomorphous (Table 1). The molecular replacement resulted in a single peak in the rotation function indicating the presence of one NirE molecule in the asymmetric unit. This was in agreement with Matthews coefficients (41) of 2.29 Å3 Da−1 (dataset I) and 2.44 Å3 Da−1 (dataset II). Electron density unexplained by the protein molecule indicated the presence of SAH and uro'gen III, each being present with one molecule in the asymmetric unit.
Overall Structure of NirE
The NirE monomer consists of two equally sized domains that are connected to each other by a short linker. Each of the two domains consists of a five-stranded β-sheet that is surrounded by helices (Fig. 2). In the N-terminal domain A (residues 1–129), the five β-strands are parallel and arranged in the order β5 β1 β4 β2 β3 (the numbering represents the sequential appearance of the β-strands in the NirE primary structure). Helices hA, hB, and hC are located on one face of the parallel β-sheet whereas hD and hE occupy the opposite face. The short linker that connects the two domains comprises amino acid residues 130–133. The β-strands of the mixed β-sheet of the C-terminal domain B (residues 134–279) are arranged in the order β6 β7 β10 β8 β9. Helices hF and hJ are located on the same side of the mixed β-sheet whereas helices hG, hH, and hI lie on the opposite side. The two domains of the NirE monomer show a similar arrangement of secondary structure elements but do not share the same topology (Fig. 2, A and B).
FIGURE 2.
Structure of NirE. A, topology diagram of NirE. Helices are depicted as cylinders and β-strands as arrows. The N-terminal and C-terminal domains are labeled with A and B, respectively. B, monomer of NirE (content of the asymmetric unit) in schematic representation with SAH (light blue) and uro'gen III (light orange) shown as sticks. Domains A and B are shown in lighter and darker shades of green, respectively, and the linker region is shown in gray. C, schematic representation of the physiological dimer of NirE created by a crystallographic symmetry operation (rotation axis is indicated by a dashed line). The two monomers are dark green and light green. SAH and uro'gen III are represented as in B. The two different views of dimeric NirE are related by a rotation about ∼90° around the 2-fold symmetry axis.
In the crystal structure, a homodimeric assembly of NirE is present which is created by a 2-fold crystallographic symmetry axis (Fig. 2C). This is in accordance with the dimeric organization of the protein in solution (10). In the dimer, the two five-stranded β-sheets of the domains B combine to form one large, slightly twisted, 10-stranded β-sheet. The total buried surface of the dimer is 5140 Å2. Interestingly, the long loop between hF and β6 of domain B of one monomer protrudes toward the active site cleft of the other monomer where it participates in substrate binding and catalysis (see below).
The overall fold and dimeric arrangement are highly similar for both NirE with and without substrate bound. Apparently, no major conformational changes of the NirE backbone occur upon uro'gen III binding. As expected, the NirE structure is very similar to the previously published SUMT structures, namely those of S. enterica CysG (PDB ID code 1PJQ) (18), P. denitrificans CobA (1S4D) (19), and T. thermophilus SUMT (1V9A) (20), which share 49, 41, and 40% amino acid sequence identity with P. aeruginosa NirE, respectively (Fig. 3). The root mean square deviations between common Cα positions are 0.776 Å for NirE and CysG, 0.603 Å for NirE and CobA, and 0.836 Å for NirE and ttSUMT. Thus, the highest degree of similarity is found between NirE and CobA. The structural comparison of all four SUMTs also reveals that there are two loops that are highly flexible (Fig. 3B). In the NirE structure, these loops are located between β3 and hD of domain A and between β6 and hG of domain B. In all SUMT structures, the amino acids of these loops were found disordered. Although the flexible loop of domain B does not contain highly conserved amino acid residues, the disordered loop of domain A contains a highly conserved lysine residue (Lys-72 in NirE) which was found to be essential for catalysis in CysG (Lys-270) through site-directed mutagenesis. It was proposed that this lysine could either be involved in the coordination of the substrate carboxylate groups or play an essential role for the prototropic rearrangements of the macrocycle (18). Surprisingly, in our NirE-uro'gen III complex structure the two flexible loops were still found disordered, and Lys-72 was not included in the final model. Thus, the exact function of this lysine remains unresolved.
FIGURE 3.
Comparison of NirE with its homologs. A, structure-based amino acid sequence alignment of NirE from P. aeruginosa (NirE_PA) with the SUMT domain of CysG from S. enterica (CysGA_SE) (18), a SUMT from T. thermophilus (SUMT_TT) (20), and CobA from P. denitrificans (CobA_PD) (19). Helices and β-structures as found in NirE are indicated. Red boxes highlight identical amino acids among these four SUMTs; black stars indicate highly conserved amino acids identified through an alignment of 60 SUMTs from different organisms (see “Experimental Procedures”). Dashed green lines indicate flexible regions whose amino acids were not included in the final NirE structure. Blue triangles highlight the amino acid residues that were subjected to site-directed mutagenesis in this study. B, superposition of NirE (green) with the structures of CobA (salmon), the SUMT domain of CysG (blue), and ttSUMT (gray). The structural models are shown in ribbon representation, and the SAH molecules are shown as ball and stick models. The beginnings and ends of flexible loops are indicated by a star and a triangle, respectively.
SAH Binds Tightly in the Active Site of NirE
The active site pocket of NirE is located between the two domains of each monomer, as previously observed for CysG, CobA, and ttSUMT (18–20). In both NirE structures, i.e. without and with bound uro'gen III, SAH was bound deeply in the interior of the pocket (Fig. 2 and see Fig. 5). The presence of SAH in the NirE structure without uro'gen III indicates that SAH was copurified with NirE and was not exchanged against SAM, which had been added in 7-fold molar excess to the crystallization setups. In both NirE structures the SAH molecule adopts the same bent conformation as in the previously reported SUMT structures. Additionally, the localization and coordination of the SAH are identical in both NirE structures independent of the presence or absence of uro'gen III. Polar contacts between SAH and the protein mainly involve the amide groups of the protein backbone (see Fig. 5). Hydrophobic interactions of the SAH with the protein involve Met-186 and Leu-52, which face each other on opposite sites of the molecule with the SAH sulfur atom sitting between them at distances of about 4 Å, respectively. Pro-242 and Ala-134 contribute to the hydrophobic environment of the adenine ring of SAH whereas Tyr-185 stacks along the aliphatic part of the homocysteine moiety. The coordination of the SAH in the active site pocket of NirE is highly similar to that previously observed in CysG which, among the three other crystallized SUMTs, also shows the highest degree of amino acid sequence identity to NirE.
FIGURE 5.
Active site of NirE. Stereo top view (A) and stereo side view (B) of the NirE active site are shown. The protein chain is represented schematically; important amino acids are shown as sticks. The chain of monomer A is dark green and that of monomer B light green. The hydrogen bonds and salt bridges are drawn as black broken lines.
Uro'gen III Binds in a Twisted Conformation in the Active Site of NirE
The main goal of this study was to obtain the structure of NirE in complex with its substrate uro'gen III. For this purpose, NirE was incubated with an 82-fold molar excess of uro'gen III that had been prepared by chemical reduction of uroporphyrin III with H2/palladium. Additionally, SAH was added to the NirE-uro'gen III mixture instead of SAM to prevent a catalytic reaction and to trap the substrate in the active site of the enzyme. From these crystallization setups, we obtained NirE crystals with uro'gen III bound in the active site pocket.
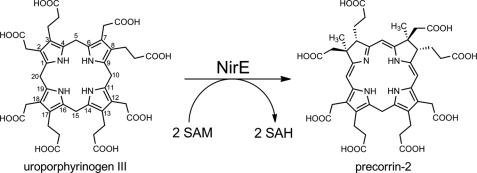
In the refined NirE-uro'gen III complex structure, the additional electron density clearly defined the localization, orientation, and conformation of the substrate. The large uro'gen III molecule sits above the SAH and is partially in contact with the solvent (Figs. 4 and 5). In the omit map, initial electron density was clearly visible for the tetrapyrrole macrocycle as well as for the acetate and propionate substituents of two opposite pyrrole rings (Fig. 4A). From the size and shape of the density for these substituents it was possible to unambiguously distinguish between acetate and propionate groups and therefore to assign the respective pyrrole rings as being either B or D. This left us with two possible orientations of the uro'gen III which could be converted into each other by flipping the molecule 180° overhead keeping the same positions for rings A and C. However, because the stereochemistry of the methylation reaction is known and the SAH is located beneath one face of the tetrapyrrole, one of the two possible orientations of the substrate could be excluded. Thus, in our NirE-uro'gen III complex structure the substrate orientation shows rings A–D arranged in a clockwise manner when viewed from the top (Fig. 5A).
FIGURE 4.
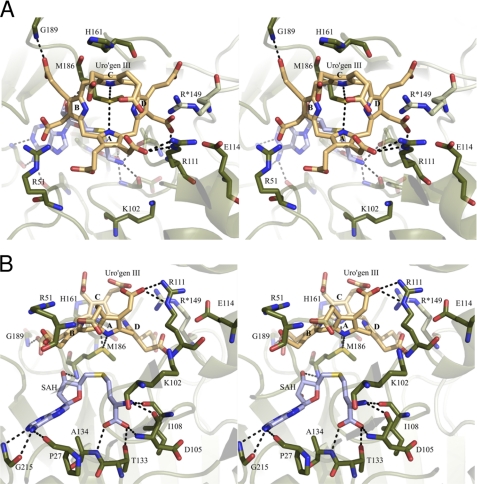
Substrate binding site of NirE. A, stereo view of the omit maps of SAH and uro'gen III calculated in the absence of the substrates from the final structure model. The 2Fo − Fc map is contoured to 1 σ and represented by a black mesh. The Fo − Fc map is contoured to 3 σ and represented by a green mesh. B, stereo view of the 2Fo − Fc map of SAH and uro'gen III calculated in the presence of the substrates shown as black mesh contoured to 1 σ. C, schematic representation of interactions between NirE and uro'gen III. Polar interactions are drawn as green broken lines with their lengths given in angstroms. Residues/atoms involved in hydrophobic contacts are surrounded by a red corona.
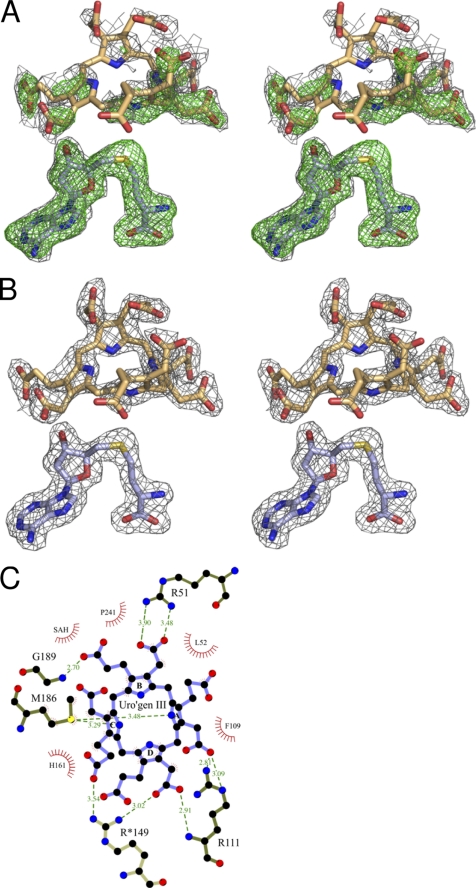
The uro'gen III was observed to bind to NirE in a twisted “two-up, two-down” conformation, i.e. the two pyrrole rings that face each other point in the same direction either upward or downward out of the tetrapyrrole plane (Figs. 4 and 5). In the NirE active site, pyrrole rings A and C point down toward the interior of the active site pocket and the bound SAH, whereas rings B and D point upward. Rings A and C are inclined by about 70° and 60° out of the tetrapyrrole plane, respectively, rings B and D both by about 45°. Such a “puckered” conformation was also observed for uro'gen III in the enzyme-product complex structure of uro'gen III synthase (28). Interestingly, Met-186 is located directly below the tetrapyrrole and stacks against pyrrole rings B and D with its sulfur atom being placed beneath the exact center of the macrocycle (Fig. 5). Met-186 seems to stabilize the central part of the substrate by providing a hydrophobic environment and forming hydrogen bonds (∼3.5 Å each) with the N-H groups of pyrrole rings A and C (Fig. 4C). The distances and angles between the A and C rings N-H groups and the Met-186 sulfur are in accordance with the presence of these unusual sulfur-containing H bonds (42). Therefore, Met-186 is important for the binding of both substrates, SAM (see above) and uro'gen III. Accordingly, a P. denitrificans CobA variant in which the corresponding methionine residue (Met-184) had been replaced by an alanine was unable to bind SAM and showed almost no SUMT activity (19). Besides the interaction between uro'gen III and Met-186, another hydrophobic contact between the substrate and the protein is provided by His-161. The imidazole ring of His-161 stacks against pyrrole ring C of uro'gen III (Fig. 5A). This histidine residue is part of the flexible loop of the C-terminal domain of NirE and moves slightly backward upon substrate binding to accommodate the tetrapyrrole (Fig. 6A).
FIGURE 6.
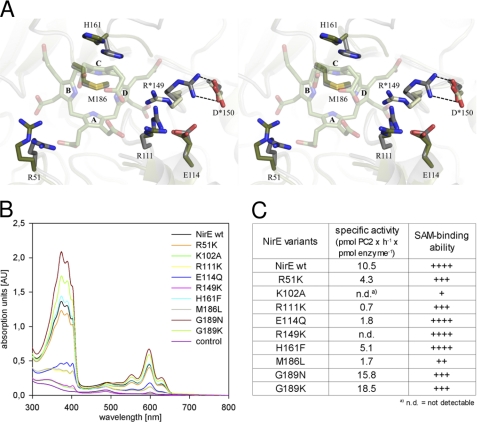
Active site of NirE and enzymatic activity. A, superposition of the NirE active site in the presence (green) and absence (gray) of uro'gen III. Amino acid residues Arg-51, Arg*-149, and His-161 clearly change their orientation upon substrate binding. B, UV-visible absorption spectra of sirohydrochlorin formed in NirE activity assay mixtures containing either WT NirE or variants thereof. The substrate uro'gen III was produced enzymatically as described under “Experimental Procedures,” and the NirE reaction product precorrin-2 was converted into sirohydrochlorin by the enzyme SirC. The assay mixtures were incubated overnight at 37 °C to allow for sirohydrochlorin formation before the spectra were recorded. The shown spectra represent the average of two independent activity assay setups. C, specific activity of WT NirE and NirE variants determined as described under “Experimental Procedures” using chemically produced uro'gen III as the substrate at a final concentration of 17 μm. In contrast to the overnight activity assay (B) the values for the specific activity represent the initial reaction rates. The given values represent the average of three independent measurements. The SAM binding ability of WT NirE and NirE variants was determined as described under “Experimental Procedures.” The strength of SAM binding is represented as identical to WT NirE (++++), slightly reduced (+++), reduced (++), and strongly reduced (+).
The eight carboxylate groups of the acetate and propionate substituents of uro'gen III are involved in polar contacts (hydrogen bonds or salt bridges) with three conserved arginine residues, the NirE protein backbone, and several water molecules (Figs. 4C and 5; to avoid too crowded figures, the water molecules are not shown). The acetate carboxylate of ring A forms a polar contact with the guanidinium group of Arg-111 and hydrogen bonds with three water molecules, whereas the propionate carboxylate of this ring makes a polar contact with the guanidinium group of Arg-51. This arginine residue is also involved in a polar contact with the carboxylate group of the acetate substituent of ring B. The propionate side chain of ring B is held in its position via hydrogen bonds to the backbone N-H group of Gly-189 and two water molecules. The ring C acetate carboxylate group forms hydrogen bonds with three water molecules. The propionate carboxylate of the same ring also makes a hydrogen bond with a water molecule. The highly conserved amino acid Arg*-149, which belongs to the polypeptide chain of the second monomer of dimeric NirE (indicated by an asterisk) and is located on the loop between hF and β6 of domain B (see above), is also involved in the binding of uro'gen III, although it does not directly form hydrogen bonds or salt bridges. Rather, Arg*-149 seems to compensate the negative charges of the ring C propionate and the ring D acetate carboxylates with its guanidinium group. Of the three arginine residues involved in the coordination of the carboxylate groups of the substrate, Arg-51 and Arg*-149 clearly change their orientation upon substrate binding (Fig. 6A). In the NirE structure without bound uro'gen III the guanidinium group of Arg-51 points upward and moves down in the presence of the substrate. This movement is even more pronounced when the NirE-substrate-bound structure is compared with the CysG structure. Here, the side chain of the corresponding arginine residue (Arg-249) would have to hinge down by about 90° to adopt the same conformation as Arg-51 of NirE in the presence of uro'gen III. Depending on the absence or presence of the substrate, the side chain of Arg*-149 also drastically changes its orientation. In the NirE structure without uro'gen III bound, this arginine residue forms a salt bridge with the adjacent Asp*-150. The presence of the substrate induces a flipping of the guanidinium group which then points toward the opposite direction and participates in the coordination of the ring C and D carboxylate groups (Fig. 6A).
Overall, the acetate and propionate substituents of uro'gen III are not very tightly fixed in their positions by the surrounding amino acid residues. Rather, residues Arg-111, Arg-51, and Arg*-149 and possibly also the highly conserved Lys-72 lying on the disordered loop of domain A provide a positively charged environment to accommodate the carboxylate groups of the substrate. This loose coordination of the substrate carboxylate groups is reflected in a high temperature factor (58.8 Å2) and a weaker electron density for the substrate compared with that of the bound SAH (Fig. 4A). The observed relatively loose coordination of the uro'gen III side chains is in line with the fact that the enzyme has to bind the monomethylated precorrin-1 in the same active site pocket to perform the second methylation at C-7. Additionally, it also explains the observation that SUMTs are able to accept uroporphyrinogen isomers other than uro'gen III, such as uro'gens I, II, or IV, as substrates (23, 43).
Generation of NirE Variants Carrying Amino Acid Exchanges of Residues Potentially Involved in Catalysis
Our NirE-uro'gen III complex structure revealed for the first time the direct amino acid environment of the substrate uro'gen III in the active site of a SUMT. The amino acids surrounding uro'gen III are involved in the coordination of the substrate and/or catalysis. To identify those amino acid residues that are directly involved in catalysis we performed site-directed mutagenesis. Previously, it was proposed that the SAM-dependent methyl transfer reaction could be initiated through a proton abstraction from uro'gen III carried out by a basic amino acid residue (18). This H+ abstraction can occur from either a methylene bridge between the pyrrole rings (most likely the C-20 position for the first methylation) or a pyrrole N-H group. Subsequent to the methyl group transfer from SAM onto uro'gen III, a bond rearrangement within the tetrapyrrole macrocycle is presumed to take place (18, 25). Possibly, amino acid residues of the NirE active site could promote this prototropic rearrangement through acid/base catalysis.
Upon inspection of the NirE active site, we selected the conserved basic residues Arg-51, Lys-102, Arg-111, Arg-149, and His-161 for site-directed mutagenesis. Arg-51 is located near the methylene bridge C-5 between rings A and B, the residues Arg-111 and Arg*-149 are close to the C-20 position between rings A and D, and finally His-161 is located in the direct neighborhood of methylene bridge positions C-10 and C-15 as well as the N-H group of ring C. To knock out the putative function of His-161 as catalytically active base but to retain its hydrophobic stacking interaction with pyrrole ring C, it was replaced by a phenylalanine. Although arginine is usually considered to be a poor base, there are several examples reported in the literature showing the involvement of arginine residues in proton abstractions (44). In these cases, the arginine residues are solvent-accessible and adjacent to carboxylate groups of glutamate or aspartate side chains. In the NirE active site, an acidic environment is provided by the highly conserved residues Glu-114 and Glu-115 as well as the carboxylate groups of the substrate, which are close to the conserved residues Arg-51, Arg-111, and Arg*-149. Therefore, all three arginines were chosen for site-directed mutagenesis and were replaced by lysines. Additionally, Glu-114 was subjected to site-directed mutagenesis as well and was exchanged against a glutamine.
Further, to investigate the role of the highly conserved Met-186 that is strikingly located in the center of the active site and participates in both SAH and substrate coordination, we exchanged this residue against a leucine. This substitution is more conservative than the previously reported methionine to alanine exchange in CobA from P. denitrificans which resulted in a complete loss of enzyme activity (19). Finally, Gly-189 was selected for replacement by an asparagine or a lysine based on the superposition of NirE with the structures of CysG and CobA in which it corresponds to Asn-385 and Lys-187, respectively. The superposition revealed that Asn-385 and Lys-187 could form polar contacts to the propionate carboxylate of the substrate ring B, and we aimed to study the effect of their presence at this particular position in NirE.
All NirE variants were produced in Escherichia coli, purified, and analyzed for their SAM binding ability and catalytic SUMT activity. A summary of the obtained results is shown in Fig. 6, B and C.
Exchange of Amino Acid Residues Arg-51, Arg-111, Glu-114, Arg-149, His-161, and Gly-189 Does Not Disturb SAM Binding
In our binding assay, the SAM binding ability of the NirE variants E114Q, R149K, and H161F was identical to that of the WT protein. For the NirE variants R51K, R111K, G189N, and G189K a slightly reduced (about 75% of WT) SAM binding was measured. In contrast, the protein variant M186L was more impaired in SAM binding (about 40% of WT), and the K102A variant exhibited a strongly reduced ability to bind the cofactor (about 20% of WT). Overall, these results are in accordance with the localization of the respective amino acid residues in the NirE active site. Residues Arg-51, Arg-111, Glu-114, Arg*-149, His-161, and Gly-189 are obviously not crucial for SAM binding, and therefore, any defects in SUMT activity caused by their substitution must be due to their contribution to uro'gen III coordination or NirE catalysis. The reduced SAM binding of the M186L variant was not unexpected because the NirE structure and the structures of CysG and CobA had revealed the hydrophobic interaction between this methionine residue and the cofactor. The M186L variant, however, still exhibited SAM binding strong enough to allow for performing SUMT activity assays and investigating the role of Met-186 in uro'gen III binding and NirE catalysis (see below). The strongly reduced SAM binding capacity of K102A was surprising because it is not located in the direct neighborhood of the cofactor. However, this lysine residue participates in an extended network of hydrogen bonds also involving the residues Glu-115, Gly-110, Arg-100, Gln-81, and Asp-50. All of these amino acid residues except Arg-100 are highly conserved in SUMTs from different organisms. Therefore, Lys-102 together with the mentioned residues is essential for the overall structural integrity of the active site.
Amino Acids Arg-51, His-161, and Met-186 Are Involved in Uro'gen III Coordination
The SUMT activity of NirE WT and variants was measured using two slightly different assay systems (Fig. 6, B and C). In one of them, the overnight production of sirohydrochlorin was determined (Fig. 6B), and in the second the specific activity (i.e. the initial reaction rates) was measured (Fig. 6C). For NirE variants carrying exchanges of amino acid residues solely involved in substrate coordination and not directly involved in catalysis a reduced specific activity, but an equal overnight activity compared with the WT NirE was expected. Michaelis-Menten kinetics cannot be performed with NirE due to the substrate inhibition by uro'gen III and product inhibition by SAH (10).
Compared with the WT, all analyzed NirE variants showed reduced SUMT activity except for G189N and G189K, which exhibited a higher activity in both assay systems. The G189K variant catalyzed the methylation of uro'gen III almost twice as fast as the WT protein (specific activity). The active sites of the NirE variants G189N and G189K resemble those of CysG and CobA, respectively (see above). Interestingly, in previous studies these two enzymes also showed a higher specific activity than NirE (18, 19). The increased activity of enzymes carrying an asparagine or lysine at this specific position might be due to a more precise and specific binding of uro'gen III and precorrin-1 through polar contacts between the asparagine or lysine side chains with the carboxylate groups of the substrates.
Both NirE variants K102A and M186L showed reduced SAM binding (see above). Additionally, the exchange of Lys-102 to an alanine residue resulted in the complete loss of SUMT activity. Therefore, the replacement of the lysine by an alanine probably disrupted the active site architecture interfering with both SAM and uro'gen III binding. For the M186L variant which still exhibited a SAM binding ability of about 40% compared with WT NirE, only residual SUMT activity was observed. This dramatic decrease of the catalytic activity clearly shows that Met-186 is critically involved in both SAM and uro'gen III binding as suggested by its pronounced localization in the active site. Even the rather conservative exchange of methionine against a leucine led to the almost complete loss of SUMT activity. This indicates that the sulfur atom of the methionine might help to position the substrate correctly through hydrogen bonds with the N-H groups of pyrrole rings A and C.
NirE variants R51K and H161F were able to bind SAM but showed reduced specific SUMT activities. However, both variants were still clearly able to catalyze the methyl transfer reaction. The amount of precorrin-2 produced by them in the overnight incubation assay even equaled that of WT NirE. Both residues, Arg-51 and His-161, were found to coordinate the substrate uro'gen III in our NirE-uro'gen III complex structure and hence to be directly involved in the binding of the substrate. As mentioned above, these amino acids were initially also considered suitable to act as base for proton abstraction from uro'gen III to initiate the methyl transfer reaction. However, because R51K and H161F retained the ability to convert uro'gen III into precorrin-2, it can be ruled out that they are directly involved in catalysis. Rather, they solely assist in the coordination of uro'gen III as their substitution leads to reduced initial reaction rates that result from reduced substrate affinities.
Amino Acids Arg-111, Glu-114, and Arg*-149 Are Involved in NirE Catalysis
Like R51K and H161F, the NirE mutants R111K and R149K were able to bind SAM but showed reduced SUMT activity. However, whereas the former were still able to form precorrin-2 (see above), both R111K and R149K showed drastically reduced SUMT activity. Strikingly, both arginines, Arg-111 and Arg*-149, are located in close proximity to the uro'gen III C-20 position where the proton abstraction could take place. Further, both arginines are in close contact to several carboxylate groups either originating from the substrate or from the protein. Therefore, they are both situated in an environment that could enhance their ability to act as base for proton abstraction (44). Arg-111 is in close contact to the strictly conserved Glu-114 which helps to position it near the substrate. To support this hypothesis we also modified the conserved Glu-114 by changing it to glutamine. The resulting E114Q mutant was able to bind SAM like the WT NirE but showed likewise strongly reduced SUMT activity. This result underlines the important role of Glu-114 for the correct functioning of Arg-111. In fact, Arg-111 is the only proteinogenic polar contact partner for the carboxylate group of Glu-114, which is otherwise only in contact with three water molecules.
In summary, the described biochemical results for the NirE variants R111K, E114Q, and R149K strongly suggest that these residues are not only involved in the coordination of the substrate carboxylate side chains but also participate directly in catalysis. Based on our results, we cannot definitely state which one of the two arginines, Arg-111 or Arg*-149, acts as the base that initiates the methyl transfer reaction. On one hand, both arginines are involved in polar contacts with two carboxylate groups. These are the carboxylates of Glu-114 and the substrate ring A acetate side chain for Arg-111 and those of the substrate ring D acetate side chain and ring C propionate substituent for Arg*-149. Such a carboxylate-rich environment was proposed to be a common motif for some proteins that use arginine residues as the catalytically essential base (44). It was argued that the carboxylate groups in the neighborhood of the arginine could facilitate the rapid exchange of protons with the solvent. Accordingly, another common feature of this group of enzymes was the solvent accessibility of their active sites. Indeed, in our NirE-uro'gen III complex structure the active site is also exposed to the solvent, although the two unresolved flexible loops mentioned above might cover the active site to a certain extend and shield it from the surrounding medium. Considering the distances of the guanidinium groups of Arg-111 and Arg*-149 from the substrate C-20 position the side chain of Arg*-149 is positioned in a slightly more favorable location than that of Arg-111. For Arg*-149 the guanidinium NH2 group located closest to C-20 is found at a distance (N → C) of 4.66 Å. For Arg-111 the ϵ-NH group is located closest to C-20 at a distance of 4.75 Å, whereas one of the terminal NH2 groups of this residue is positioned at a distance of 6.15 Å. However, the C-20 position of the substrate might even move closer to both arginine residues when the true cosubstrate SAM is present in the active site. Because in our NirE-substrate complex the product SAH was bound, that does not carry the methyl group, the uro'gen III might in fact be positioned deeper in the active site than it would be in the presence of SAM.
Another striking observation that might explain the need of both Arg-111 and Arg*-149 for catalysis is their direct neighborhood to each other. In fact, the two guanidinium groups are arranged almost in parallel with their planes at a distance of about 3.5 Å. It is possible that the proximity of the positive charge of one of the guanidinium groups favors the deprotonated state of the second which would then be available for proton abstraction from C-20. Interestingly, the side chain of Arg*-149 forms a salt bridge with Asp*-150 in the absence of uro'gen III (see above). Possibly, the binding of the substrate to the active site causes the side chain of Arg*-149 to move away from Asp*-150 toward the active site where it forms polar contacts with two of the substrate carboxylate side chains. However, in this new position Arg*-149 is now in close proximity to positively charged Arg-111 which facilitates deprotonation of the former. Subsequently, uncharged Arg*-149 can abstract a proton from C-20, thus initiating the methyl transfer reaction. Of course, positively charged Arg*-149 could also promote the deprotonation of Arg-111, which then in turn might act as the catalytically essential base.
Implications for the Catalytic Mechanism of NirE and Other SUMTs
Based on the crystal structure of NirE in complex with its substrate uro'gen III, reported in this study, it was possible to identify several amino acid residues potentially involved in the NirE catalytic mechanism. Further biochemical analyses of the influence of these amino acid residues on NirE enzyme activity showed that arginines Arg-111 and Arg*-149 as well as glutamate 114 are probably directly involved in NirE catalysis. All three amino acid residues are highly conserved among different SUMTs from diverse organisms. Based on the observed localization of Arg-111, Arg*-149, and Glu-114 with respect to the substrate uro'gen III we propose a potential mechanism for the NirE reaction that might be valid for all members of the SUMT family (Fig. 7A). In previous studies it was proposed that the methyl transfer reaction catalyzed by SUMTs could be initiated by a proton abstraction from the substrate uro'gen III. Based on our structural and biochemical observations described herein, we propose that either Arg-111 or Arg*-149 acts as the catalytically active base which abstracts a proton from C-20 of uro'gen III. The resulting movement of electrons facilitates the nucleophilic attack of C-2 at the methyl group of SAM.
FIGURE 7.
Potential reaction mechanism and model of SAM in the NirE active site. A, proposed catalytic mechanism of NirE (see “Results and Discussion” for detailed explanation). B–D, proposed conformation of bound uro'gen III in the presence of SAM in the NirE active site instead of SAH. The molecules are shown in stick representation. SAM/SAH is shown in light blue, uro'gen III/precorrin-1 in light orange. B, clash between the methyl group of modeled SAM and the N-H group of ring A in case that uro'gen III adopts its two-up, two-down conformation. C, proposed conformation of uro'gen III in the presence of SAM. D, proposed conformation of precorrin-1 after the methyl transfer reaction and prototropic rearrangements.
In our NirE structure uro'gen III binds in a two-up, two-down conformation that brings the N-H group of ring A close to the sulfur atom of SAH and leaves the C-2 atom in a relatively large distance to the SAH sulfur (6.15 Å). To investigate whether this uro'gen III conformation is compatible with the presence of SAM in the active site, we modeled SAM in place of SAH under the assumption that SAM would adopt an identical conformation. This assumption was based on x-ray structures of published SAM complexes (e.g. PDB ID 3GX3 and 3GX6) (45). In the presence of SAM, the ring A N-H group clashes with the methyl group of the cosubstrate in case that uro'gen III adopts its two-up, two-down conformation (Fig. 7B). Thus, we assume that in the presence of SAM the uro'gen III actually binds in a conformation in which the ring A is flipped down to avoid this clash (Fig. 7C). This conformation of uro'gen III with a planar arrangement of ring A potentially resembles that of the reaction transition state and might facilitate the formation of the new double bond between C-20 and C-1 upon proton abstraction from C-20. Furthermore, with the ring A flipped down the C-2 atom is positioned close enough to the SAM methyl group to perform the nucleophilic attack.
After the methyl transfer a rearrangement of double bonds within the tetrapyrrole macrocycle was proposed to take place. Accordingly, it was reported that the double bond that is located between C-3 and C-4 in uro'gen III is found either between C-4 and C-5 or between C-4 and N-21 in precorrin-1 (22). Possibly, the highly conserved Lys-72 of NirE might be involved in this tautomerization of the tetrapyrrole as previously proposed for the corresponding Lys-270 of S. enterica CysG (18). Unfortunately, this lysine residue is located on the flexible loop of domain A which was not resolved in our NirE structure. However, the K270I variant of CysG was hardly active indicating the involvement of this lysine in catalysis (18). Additionally, the water molecules that were found around uro'gen III might also be involved in the tautomerization of precorrin-1. After the first methylation at C-2 the newly formed precorrin-1 (Fig. 7D) and the reaction side product SAH are released from the active site. To facilitate the release of the reaction products Arg*-149 potentially reforms its salt bridge with Asp*-150 that was observed in the NirE structure without bound substrate. For the second methylation at C-7 a new SAM molecule and precorrin-1 have to bind to the active site. We propose that precorrin-1 binds to the enzyme in an orientation representing a 90° counterclockwise rotation with respect to the orientation of the initial substrate uro'gen III. This rotation would place the methylene bridge C-5 in a position similar to that of the C-20 during the first methylation. Thus, to initiate the second methylation reaction we propose a proton abstraction from C-5 again catalyzed by the arginine pair Arg-111/Arg*-149 (Fig. 7A). Finally, to yield precorrin-2 another rearrangement of double bonds has to occur.
Acknowledgments
We thank Dr. Bohdan Snovydovych (Institute of Organic Chemistry, Technische Universität (TU) Braunschweig) for technical help with the reduction of uroporphyrin III with H2/palladium, and Nick Quade (Department of Molecular Structural Biology, Helmholtz Centre for Infection Research, Braunschweig) for assistance with data collection. We would like to thank Dr. Jürgen Moser (Institute of Microbiology, TU Braunschweig) for helpful discussions and Prof. Dieter Jahn (Institute of Microbiology, TU Braunschweig) for continuous support.
This work was supported by grants from the Deutsche Forschungsgemeinschaft Emmy-Noether Program (to G. L.) and FOR 1220 (to D. W. H.) and by the Fonds der Chemischen Industrie (to G. L. and D. W. H.).
The atomic coordinates and structure factors (codes 2YBO and 2YBQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- uro'gen III
- uroporphyrinogen III
- CobA
- SUMT involved in cobalamin biosynthesis
- CysG
- SirA, UPM1, Met1p, SUMTs involved in siroheme biosynthesis
- NirE
- SUMT involved in heme d1 biosynthesis
- PDB
- Protein Data Base
- SAH
- S-adenosyl-l-homocysteine
- SAM
- S-adenosyl-l-methionine
- SUMT
- SAM-dependent uroporphyrinogen III methyltransferase.
REFERENCES
- 1. Battersby A. R. (2000) Nat. Prod. Rep. 17, 507–526 [DOI] [PubMed] [Google Scholar]
- 2. Zumft W. G. (1997) Microbiol. Mol. Biol. Rev. 61, 533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Layer G., Reichelt J., Jahn D., Heinz D. W. (2010) Protein Sci. 19, 1137–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leeper F. J. (1985) Nat. Prod. Rep. 2, 561–580 [DOI] [PubMed] [Google Scholar]
- 5. Blanche F., Debussche L., Thibaut D., Crouzet J., Cameron B. (1989) J. Bacteriol. 171, 4222–4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blanche F., Robin C., Couder M., Faucher D., Cauchois L., Cameron B., Crouzet J. (1991) J. Bacteriol. 173, 4637–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan J., Wang D., Liang Z., Guo M., Teng M., Niu L. (2006) Protein Expr. Purif. 46, 40–46 [DOI] [PubMed] [Google Scholar]
- 8. Leustek T., Smith M., Murillo M., Singh D. P., Smith A. G., Woodcock S. C., Awan S. J., Warren M. J. (1997) J. Biol. Chem. 272, 2744–2752 [DOI] [PubMed] [Google Scholar]
- 9. Robin C., Blanche F., Cauchois L., Cameron B., Couder M., Crouzet J. (1991) J. Bacteriol. 173, 4893–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Storbeck S., Walther J., Müller J., Parmar V., Schiebel H. M., Kemken D., Dülcks T., Warren M. J., Layer G. (2009) FEBS J. 276, 5973–5982 [DOI] [PubMed] [Google Scholar]
- 11. Warren M. J., Roessner C. A., Santander P. J., Scott A. I. (1990) Biochem. J. 265, 725–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woodcock S. C., Raux E., Levillayer F., Thermes C., Rambach A., Warren M. J. (1998) Biochem. J. 330, 121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zajicek R. S., Bali S., Arnold S., Brindley A. A., Warren M. J., Ferguson S. J. (2009) FEBS J. 276, 6399–6411 [DOI] [PubMed] [Google Scholar]
- 14. Spencer J. B., Stolowich N. J., Roessner C. A., Scott A. I. (1993) FEBS Lett. 335, 57–60 [DOI] [PubMed] [Google Scholar]
- 15. Warren M. J., Bolt E. L., Roessner C. A., Scott A. I., Spencer J. B., Woodcock S. C. (1994) Biochem. J. 302, 837–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson P. J., Entsch B., McKay D. B. (2001) Gene 281, 63–70 [DOI] [PubMed] [Google Scholar]
- 17. Lobo S. A., Brindley A., Warren M. J., Saraiva L. M. (2009) Biochem. J. 420, 317–325 [DOI] [PubMed] [Google Scholar]
- 18. Stroupe M. E., Leech H. K., Daniels D. S., Warren M. J., Getzoff E. D. (2003) Nat. Struct. Biol. 10, 1064–1073 [DOI] [PubMed] [Google Scholar]
- 19. Vévodová J., Graham R. M., Raux E., Schubert H. L., Roper D. I., Brindley A. A., Ian Scott A., Roessner C. A., Stamford N. P., Elizabeth Stroupe M., Getzoff E. D., Warren M. J., Wilson K. S. (2004) J. Mol. Biol. 344, 419–433 [DOI] [PubMed] [Google Scholar]
- 20. Rehse P. H., Kitao T., Tahirov T. H. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 913–919 [DOI] [PubMed] [Google Scholar]
- 21. Schubert H. L., Blumenthal R. M., Cheng X. (2003) Trends Biochem. Sci. 28, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brunt R. D., Leeper F. J., Grgurina I., Battersby A. R. (1989) J. Chem. Soc. Chem. Commun. 428–431 [Google Scholar]
- 23. Warren M. J., Gonzalez M. D., Williams H. J., Stolowich N. J., Scott A. I. (1990) J. Am. Chem. Soc. 112, 5343–5345 [Google Scholar]
- 24. Mascaro L., Jr., Horhammer R., Eisenstein S., Seller L. K., Mascaro K., Floss H. G. (1977) J. Am. Chem. Soc. 99, 273–274 [DOI] [PubMed] [Google Scholar]
- 25. Warren M. J., Bolt E., Woodcock S. C. (1994) Ciba Found. Symp. 180, 26–49 [DOI] [PubMed] [Google Scholar]
- 26. Aiyar A., Xiang Y., Leis J. (1996) Methods Mol. Biol. 57, 177–191 [DOI] [PubMed] [Google Scholar]
- 27. Layer G., Verfürth K., Mahlitz E., Jahn D. (2002) J. Biol. Chem. 277, 34136–34142 [DOI] [PubMed] [Google Scholar]
- 28. Schubert H. L., Phillips J. D., Heroux A., Hill C. P. (2008) Biochemistry 47, 8648–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leech H. K., Raux E., McLean K. J., Munro A. W., Robinson N. J., Borrelly G. P., Malten M., Jahn D., Rigby S. E., Heathcote P., Warren M. J. (2003) J. Biol. Chem. 278, 41900–41907 [DOI] [PubMed] [Google Scholar]
- 30. Minor W., Cymborowski M., Otwinowski Z. (2002) Acta Phys. Polonica 101, 613–619 [Google Scholar]
- 31. Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 34. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 36. Schomaker V., Trueblood K. N. (1968) Acta Crystallogr. B 24, 63–76 [Google Scholar]
- 37. Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 38. Gouet P., Robert X., Courcelle E. (2003) Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 40. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 41. Matthews B. W. (1968) J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 42. Zhou P., Tian F., Lv F., Shang Z. (2009) Proteins 76, 151–163 [DOI] [PubMed] [Google Scholar]
- 43. Scott A. I., Williams H. J., Stolowich N. J., Karuso P., Gonzalez M. D., Blanche F., Thibaut D., Müller G., Savvidis E., Hlineney K. (1989) J. Chem. Soc. Chem. Commun. 522–525 [Google Scholar]
- 44. Guillén Schlippe Y. V., Hedstrom L. (2005) Arch. Biochem. Biophys. 433, 266–278 [DOI] [PubMed] [Google Scholar]
- 45. Montange R. K., Mondragón E., van Tyne D., Garst A. D., Ceres P., Batey R. T. (2010) J. Mol. Biol. 396, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]