Abstract
The oncogene amplified in breast cancer 1 (AIB1) is a nuclear receptor coactivator that plays a major role in the progression of various cancers. We previously identified a splice variant of AIB1 called AIB1-Δ4 that is overexpressed in breast cancer. Using mass spectrometry, we define the translation initiation of AIB1-Δ4 at Met224 of the full-length AIB1 sequence and have raised an antibody to a peptide representing the acetylated N terminus. We show that AIB1-Δ4 is predominantly localized in the cytoplasm, although leptomycin B nuclear export inhibition demonstrates that AIB1-Δ4 can enter and traffic through the nucleus. Our data indicate an import mechanism enhanced by other coactivators such as p300/CBP. We report that the endogenously and exogenously expressed AIB1-Δ4 is recruited as efficiently as full-length AIB1 to estrogen-response elements of genes, and it enhances estrogen-dependent transcription more effectively than AIB1. Expression of an N-terminal AIB1 protein fragment, which is lost in the AIB1-Δ4 isoform, potentiates AIB1 as a coactivator. This suggests a model whereby the transcriptional activity of AIB1 is squelched by a repressive mechanism utilizing the N-terminal domain and that the increased coactivator function of AIB1-Δ4 is due to the loss of this inhibitory domain. Finally, we show, using Scorpion primer technology, that AIB1-Δ4 expression is correlated with metastatic capability of human cancer cell lines.
Keywords: Breast Cancer, Coactivator Transcription, Estrogen, Growth Factors, Transcription Coactivators
Introduction
Gene transcription in eukaryotes is a complex and highly regulated process. One of the major controls of gene transcription is exerted by the coregulator family of proteins. These include both corepressors, which dampen transcription, and coactivators, which potentiate transcription. A subgroup of coactivators has been shown to be critical for the malignant progression of cancer and is known as the p160 steroid receptor coactivators (1). One member in particular was identified to be amplified in breast cancer. Amplified in breast cancer 1 (AIB1, SRC-3, NCOA3, ACTR, TRAM-1, pCIP, and RAC3) has been shown to be a gene amplified in breast cancer (2) and is also overexpressed at the mRNA and protein level in various cancers (1, 3, 4). Its role in tumorigenesis is attributed to its ability to coactivate both steroid hormone- and growth factor-dependent transcription (3, 5–7). In several oncogene-driven mouse models (8–11), reduction of AIB1 levels leads to a decrease in tumorigenesis, and overexpression of AIB1 leads to the formation of various tumors (12). Clinically, AIB1 expression in breast cancer cases is correlated with high HER2 levels, larger tumor size, higher tumor grade, and shorter disease-free survival (13–15). Also, high levels of AIB1 in conjunction with high HER2 levels coincide with reduced disease-free survival in patients treated with tamoxifen, suggesting a role for AIB1 in tamoxifen resistance (16).
We had previously identified a splice variant of AIB1, where exon 3 was spliced from the mature mRNA and the resulting protein named AIB1-Δ3 (17). More recently, an additional 5′ exon 81,164 bases upstream of the known 5′UTR was identified. Thus, the deleted exon is now exon 4, and we now refer to the splice variant as AIB1-Δ4. We had reported that AIB1-Δ4 mRNA results in an N-terminally truncated isoform of the AIB1 protein that was found to be a more potent coactivator of steroid-dependent transcription on a per mol basis when compared with the full-length AIB1 protein. AIB1-Δ4 mRNA expression was elevated in breast tumor tissue relative to normal breast tissue (17). It was also shown to increase the efficacy of estrogenic compounds and the agonist effects of the selective estrogen receptor modulator tamoxifen in breast and endometrial tumor cells (17, 18). Overexpression of AIB1-Δ4 in mice leads to ductal ectasia in the mammary gland with an increased expression of proliferative markers such as proliferating cell nuclear antigen, phospho-histone H3, and cyclin D1 (19) as well as increased cross-talk with ERα2 in epithelial and stromal responses (20). More recently, AIB1-Δ4 was shown to act as a molecular bridge between epidermal growth factor receptor (EGFR) and focal adhesion kinase (FAK) in the cytoplasm, and its overexpression increased the invasiveness of the MDA-MB-231 metastatic breast cancer cell line (21).
Because AIB1-Δ4 lacks a nuclear localization sequence (NLS), any function for this protein in cancer to date has been attributed predominantly to its role in the cytoplasm (21). We now show that AIB1-Δ4 can enter the nucleus by a noncanonical nuclear import mechanism. AIB1-Δ4 is recruited to estrogen-response elements of endogenous estrogen-regulated genes and increases their expression. We also determined that the N-terminal region absent from the AIB1-Δ4 protein contains an inhibitory domain. Through the use of Scorpion primer technology, we have created the first quantitative assay for the AIB1-Δ4 transcript and found a correlation between AIB1-Δ4 expression and the metastatic phenotype of human cancer cell lines. These data suggest that the nuclear activities of AIB1-Δ4 can contribute to its function in malignancy.
EXPERIMENTAL PROCEDURES
Plasmids
p300-HA and ERα constructs were provided by Dr. Maria L. Avantaggiati (Georgetown University) and from Dr. Pierre Chambon (INSERM, Strasbourg, France), respectively. AIB1, AIB1-Δ4, FLAG AIB1, and FLAG AIB1-Δ4 were described previously (17, 22). A C-terminal FLAG was added to the AIB1-Δ4 cDNA by deletion of the stop codon in AIB1-Δ4 and addition of FLAG peptide sequence by site-directed mutagenesis (Stratagene). HA AIB1 was generated by PCR amplification of the AIB1 sequence from pcDNA3-AIB1 and cloned into phCMV2. The AIB1 N-terminal construct was created by PCR amplification of the ACTR/AIB1 cDNA (184–777 bp) to add a new 5′ NotI site and 3′BglII site. The PCR product was then cloned into p3×FLAG-CMV-10 (Sigma).
Cell Lines and Transient Transfection
MDA231-BrM2 were kindly provided by Dr. Joan Massagué (Sloan Kettering Institute); 4175-TR and SCP2-TR cells by Yibin Kang (Princeton University); AIB1 KO/SRC-3−/− mouse embryonic fibroblasts by Dr. Jiangming Xu (Baylor College of Medicine); T47D A1-2 cells by Dr. Steve Nordeen (University of Colorado), and COLO SL and COLO PL cells by Dr. John M. Jessup (Georgetown University). Chinese hamster ovary (CHO) (23) and COLO 357 were purchased from ATCC. HEK293, HEK293T, COS-7, MCF-7, and MDA-MB-231 were obtained from the Tissue Culture Shared Resource at Georgetown University. The human mammary epithelial cells (HMEC) were purchased and cultured in commercially supplied medium (BulletKit, Lonza). HEK293T, COS-7, COLO 357, COLO SL, and COLO PL, MDA-MB-231, MDA231-BrM2 (brain), 4175-TR (lung), SCP2-TR (bone), and AIB1 KO mouse embryonic fibroblasts were grown in Dulbecco modified Eagle's medium (DMEM, Invitrogen) with 10% FBS. CHO cells were grown in DMEM F-12 (Invitrogen) with 10% FBS. HEK293 cells were grown in phenol red-free Iscove's modified Eagle's medium (IMEM, Invitrogen) with 10% charcoal-stripped serum (CCS). T47D A1-2 and MCF-7 cells were grown in phenol red-free IMEM + 5% CCS. HEK293, HEK293T, CHO, COS-7, and T47D A1-2 cells were transiently transfected with FuGENE 6 (Roche Applied Science).
Identification of N Terminus of AIB1-Δ4
HEK293T cells grown to 80% confluence were transiently transfected with 18 μg of C-terminal FLAG AIB1-Δ4 cDNA. 24 h later, whole cell lysates were prepared and subjected to immunoprecipitation using anti-FLAG M2 affinity gel (Sigma). After washing, AIB1-Δ4 protein was recovered by heating the affinity gel to 95 °C, and the sample was subjected to SDS-PAGE. A band corresponding to AIB1-Δ4 protein was isolated and trypsinized using a conventional in-gel digestion protocol where cysteines were reduced with DTT and alkylated by iodoacetamide. Extracted tryptic peptides were analyzed using the MIDAS-MS-based algorithm on an LC-electrospray ionization-MS 4000QTRAP instrument (AB SCIEX, Framingham, MA). In silico predicted peptides and corresponding collision energy settings were generated using recommended settings in MRMpilot software (AB SCIEX). The list of predicted precursors includes the potential variable modification of methionine oxidation and the fixed modification of cysteine alkylation. A final MS method was created for detection of the tryptic peptides produced from the full-length AIB1 protein. This approach allows detecting only tryptic peptides that overlaps with the spliced version of AIB1 and was tested on endogenous AIB1 protein isolated by immunoprecipitation followed by SDS-PAGE. In silico predictions for tryptic peptides with methionine as an initial amino acid residue were applied to data for identification of the N terminus. More detailed methods are provided in the supplemental material.
Generation of Affinity-purified AIB1-Δ4 Antibodies and Characterization
Rabbit polyclonal antibodies were generated against the identified N terminus using a N-acetylated peptide MQCFALSQPRK, and detection of AIB1-Δ4 reactive antibodies was monitored through ELISA by Abgent. Anti-AIB1-Δ4 serum was then subjected to positive and negative selection with N-acetylated AIB1-Δ4 N-terminal peptide and nonacetylated AIB1-Δ4 N-terminal peptide, respectively. The resultant affinity-purified antibodies reactive against N-acetylated AIB1-Δ4 were used in experiments.
Endogenous and transfected AIB1-Δ4 was detected after pulldown with AIB1-Δ4-specific antibody and detected with an AIB1 antibody that detects both full-length AIB1 and AIB1-Δ4 in MCF-7 and HEK293T cells, respectively. HEK293T cells were transfected with C-terminal FLAG-tagged AIB1-Δ4 used to determine the N terminus of AIB1-Δ4. For both experiments, AIB1-Δ4-containing lysates were immunoprecipitated with 10 μg of polyclonal antibody/1 mg of lysate overnight.
Peptide competition assays to validate the affinity-purified AIB1-Δ4 antibodies were performed by adding 1 μg of either N-acetylated or nonacetylated MQCFALSQPRK peptide to 1 mg of MCF-7 cell lysate with 10 μg of affinity-purified AIB1-Δ4 antibodies.
Western Blot Analysis and Immunoprecipitation (IP)
Western blotting was done with the following antibodies: AIB1 (5E11, Cell Signaling); FLAG M2 (Sigma); HA (Cell Signaling); ERα (Ab-10, Neomarkers); ERα (G-20, Santa Cruz Biotechnology); and human actin (Millipore). (i) For interaction of AIB1 with AIB1-Δ4, HEK293T cells were transfected with 6 μg of either FLAG AIB1, AIB1-Δ4, or FLAG AIB1 and AIB1-Δ4 together. After washing with cold 1× PBS, whole cell lysates were prepared by adding 1% Nonidet P-40 lysis buffer containing 1 mm NaO3VO4 and 1× Complete protease inhibitor tablet (Roche Applied Science). IP was performed with anti-FLAG M2 affinity gel as described previously (6), and samples were subjected to SDS-PAGE. (ii) For interaction of AIB1 and AIB1-Δ4 with p300-HA, HEK293T cells were transfected with either FLAG AIB1, FLAG AIB1-Δ4, or p300-HA. Whole cell lysates were prepared as indicated in (i). Equal amounts of FLAG AIB1 and FLAG AIB1-Δ4 were added to equal amounts of p300-HA lysate. After immunoprecipitation using HA antibody (Cell Signaling), the amounts of FLAG AIB1 or FLAG AIB1-Δ4 were detected with FLAG M2 antibody (Sigma). Densitometry was performed by using Adobe Photoshop 7.0 normalizing AIB1-Δ4 bands to AIB1 bands for both input and IP. (iii) For immunoprecipitation with AIB1-Δ4 antibody, 500 μg of lysate from MCF-7 or HEK293T transfected with C-terminal FLAG-tagged AIB1-Δ4 were subjected to IP with 5 μg of AIB1-Δ4 antibody. AIB1 or AIB1-Δ4 proteins were detected with AIB1 antibody (5E11, Cell Signaling).
Nuclear Cytoplasmic Fractionation
Fractionation of lysates was carried out as per the protocol recommended by the NE-PER nuclear and cytoplasmic extraction reagents (78833, Pierce). Endogenous or transfected AIB1-Δ4 was detected by either AIB1 (5E11) or FLAGM2 antibody. Controls used for nuclear and cytoplasmic fractions were HDAC1 (catalog no. 2062, Cell Signaling) and HSP90 (05-594, Upstate).
Immunofluorescence and Leptomycin B Treatment
CHO cells were plated on glass coverslips and transfected with 500 ng of either FLAG AIB1 or FLAG AIB1-Δ4. 24 h later, cells were fixed with 3.7% paraformaldehyde in PBS for 10 min at 25 °C. Cells were then washed three times with 1× PBS and permeabilized with 1× PBS containing 0.2% Triton X-100 for 5 min 25 °C. Cells were then washed with three times with 1× PBS. Coverslips with cells were then blocked for 30 min with 1% BSA in PBS. Cells were then incubated with FLAG M2 antibody (1:500, Sigma) and HA antibody (1:500, Abcam ab9110) for 20 min. After three 5-min washes with 1× PBS, cells were incubated with anti-mouse IgG AlexaFluor488 (1:1000, Invitrogen) and anti-rabbit AlexaFluor594 (1:1000, Invitrogen) for 20 min. Coverslips were then washed three times with 1× PBS and mounted with ProLong Gold antifade reagent with DAPI (Invitrogen) onto glass slides. 50 nm leptomycin B was added into the culture medium 4 h before fixation. 200 cells were counted, and the percentage of nuclear, nuclear/cytoplasmic, and cytoplasmic stained cells was quantified from three different experiments. Nuclear staining was defined as a protein-specific signal that overlaid with the DAPI signal only. Nuclear/cytoplasmic was defined as a protein-specific staining that overlaid with the DAPI signal but also showed staining in the cytoplasmic compartment. Cytoplasmic staining was defined as protein-specific staining that did not overlay with the DAPI staining of the nucleus. Imaging of stained CHO cells was performed on a Nikon E600 fluorescence digital microscope system and analyzed with Nuance multispectral imaging system and software (Cambridge Research and Instrumentation).
Chromatin Immunoprecipitation Assays (ChIP)
HEK293 cells in a 10-cm dish were transfected with 5 μg of ERα and either 6 μg of FLAG AIB1 or 3 μg of FLAG AIB1-Δ4 in phenol red-free IMEM + 10% CCS. 24 h later, cells were treated with estrogen for 0, 15, 30, 45, or 60 min. Cells were fixed with formaldehyde fixation solution (3.7% formaldehyde, 100 mm NaCl, 50 mm Tris/HCl, pH 8.0, 1 mm EDTA, 0.5 mm EGTA) for 10 min at 37 °C and stopped with 0.125 m glycine in 1× PBS for 5 min at 25 °C. Cells were washed three times with 1× PBS and resuspended in SDS lysis buffer (50 mm Tris, pH 8.0, 10 mm EDTA, pH 8.0, 1% SDS). Cells were sonicated and resuspended in ChIP dilution buffer (20 mm Tris, pH 8.0, 2 mm EDTA, pH 8.0, 150 mm NaCl, 1% Triton X-100) and precleared with 30 μl of protein G-agarose/salmon sperm DNA (Millipore) for 1 h. 500 μg of total protein was immunoprecipitated with 2 μl of FLAG M2 antibody (Sigma) for 16 h and immunoprecipitated with 30 μl of protein G-agarose/salmon sperm DNA for 2 h. Agarose was washed with once with low salt buffer (20 mm Tris, pH 8.0, 2 mm EDTA, pH 8.0, NaCl 150 mm, 0.1% SDS, 1% Triton X-100), twice with high salt buffer (20 mm Tris, pH 8.0, 2 mm EDTA, pH 8.0, 500 mm NaCl, 0.1% SDS, 1% Triton X-100), once with LiCl salt buffer (10 mm Tris, pH 8.0, 1 mm EDTA, pH 8.0, 250 mm LiCl, 1% sodium deoxycholate, 1% Nonidet P-40), and twice with TE buffer (10 mm Tris, pH 8.0, 1 mm EDTA, pH 8.0). Samples were eluted with elution buffer (1% SDS, 0.1 m NaHCO3) for 15 min on a rotator and 10 min on a vortexer. Cross-links were removed with 200 mm NaCl for 6 h at 65 °C and proteins digested with 1 μg of proteinase K for 1 h at 45 °C. DNA was purified using GENECLEAN® Turbo kit (Qbiogene, Inc.). Samples were analyzed by real time PCR to examine the ERE recruitment of FLAG AIB1 or FLAG AIB1-Δ4 with the following primers: pS2 ERE S, 5′-GGCCATCTCTCACTATGAATCACTTC, and pS2 ERE AS, 5′-GGCAGGCTCTGTTTGCTTAAAGAGCG-3′; hC3 ERE S, 5′-GAGAAAGGTCTGTGTTCACCAGG-3′ hC3, and ERE AS, 5′-TGCAGGGTCAGAGGGACAGA-3′; and HER2 ERE S, 5′-GTTCCTCCCTCCTGTTCCTC-3′, and HER2 ERE AS, 5′-CCACAAACTGGTGGTCTCCT-3′. Cycling conditions for real time PCR using iCycler were 95 °C for 3 min followed by 40 cycles of 95 °C for 20 s, 57 °C for 30 s, and 72 °C for 40 s for hC3 ERE and pS2 ERE. For HER2, ERE cycling conditions were 95 °C for 3 min followed by 40 cycles of 95 °C for 20 s, 65 °C for 30 s, and 72 °C for 40 s. The percentage of the input for each time point is plotted on the graphs and normalized at time 0 for each transfection.
For ChIP assays with endogenous AIB1-Δ4, MCF-7 cells were plated in 15-cm dishes in phenol red-free IMEM + 5% CCS. Fresh IMEM + 5% CCS was added for 3 days. Cells were treated as the HEK293 cells except they were subjected to immunoprecipitation with either 4 μg of affinity-purified AIB1-Δ4 antibodies (Abgent), 2 μg of AIB1 (C-20, Santa Cruz Biotechnology), or 2 μg of ER antibody (HC-20, Santa Cruz Biotechnology). After immunoprecipitation, the ChIP procedure for HEK293 cells was then followed. All primers were synthesized by Integrated DNA Technologies (IDT).
Real Time PCR Analysis
HEK293 cells were transfected with FLAG AIB1 or FLAG AIB1-Δ4 at levels that give equal amounts of transfected protein in IMEM + 10% CCS. 16 h later, cells were stimulated with estrogen for 0, 4, 8, and 24 h. Total RNA was harvested using RNeasy mini kit (Qiagen) and reverse-transcribed with iScript cDNA synthesis kit (Bio-Rad) using 1 μg of total RNA. Samples were analyzed by real time PCR (iCycler, Bio-Rad) using the following conditions: 95 °C for 3 min, 40 cycles of 95 °C for 20 s, 56 °C for 30 s, and 72 °C for 40 s. Primer sequences used were as follows: pS2 sense, 5′-CCCCGTGAAAGACAGAATTGT-3′, and pS2 AS, 5′-GGTGTCGTCGAAACAGCAG-3′; hC3 S, 5′-CTGTCCACGACTTCCCAGG-3′, and hC3 AS, 5′-CCCTTTTCTGACTTGAACTCCC; HER2 S, 5′-AAAGGCCCAAGACTCTCTCC, and HER2 AS, 5′-CAAGTACTCGGGGTTCTCCA-3′; human actin S, 5′-CCTGGCACCCAGCACAAT-3′, and human actin as, 5′-GCCGATCCACACGGAGTACT-3′. All primers were synthesized by IDT. The expression level for each gene is normalized to actin expression and multiplied by either 1000 or 100,000 to obtain whole value numbers.
Quantitation of AIB1-Δ4 and AIB1 mRNA Levels Using Scorpion Primer-based Quantitative RT-PCR
A total of 2 × 106 cells were plated for each cell line. 24 h later, total RNA was extracted with RNeasy mini kit (Qiagen) and reverse-transcribed with iScript cDNA synthesis kit (Bio-Rad) using 1 μg of total RNA. Real time PCR was performed using IQ SYBR Green Supermix (Bio-Rad) with AIB1Δ4-Scorpion primer and human actin primers. Cycling conditions for the AIB1-Δ4-Scorpion primer consisted of an initial denaturing step at 94 °C (2 min) and 50 cycles (20 s at 94 °C, 15 s at 55.5 °C, and 20 s at 72 °C). Unlike SYBR Green real time PCR analysis where data are collected during the extension step, data for the Scorpion primer reactions were collected during the 55.5 °C annealing step (iCycler; Bio-Rad). Cycling conditions for the human actin primers include a denaturing step at 94 °C (2 min) and 45 cycles (20 s at 94 °C, 30 s at 58 °C, and 40 s at 72 °C). Primer sequences for the AIB1-Δ4-Scorpion reaction are as follows: 5′-FAMCCCGCGCTTGGAAATAGTTTTTCCCTTGTCCGCGCGGG-BHQ1HEG-CGCAAATTGCCATGTGATAC; for AIB1-Δ4 reverse primer, 5′-CCATCCAATGCCTGAAGTAA-3′. The expression level of AIB1-Δ4 is normalized to actin expression levels and multiplied by either 10,000 or 100,000 to obtain whole number values.
For AIB1-Scorpion primer detection, the same procedures were followed except the primer sequence of the AIB1-Scorpion primer was as follows: 5′-HEXCCCGCGCGTTTTTCACCACTGCAGGTAAGAGCGCGGG-BHQ1HEG-GCCATGTGATACTCCAGGA; for AIB1 reverse primer, 5′-ACGTATCTGTCTTACTGTTTCC-3′. The AIB1 and AIB1-Δ4-Scorpion primers were custom designed and synthesized by Sigma.
Luciferase Assay
25,000 COS-7 cells per well in a 24-well dish were transfected in DMEM without serum with 100 ng of MMTV luciferase, 25 ng of progesterone receptor, 5 ng of thymidine kinase Renilla luciferase, and either 500 ng of pcDNA3, 500 ng of FLAG AIB1, 500 ng of FLAG N terminus, or 125, 500, and 750 ng of FLAG N terminus with 500 ng of FLAG AIB1. 24 h later cells were treated with 10 nm R5020 or an equivalent volume of ethanol. 24 h after stimulation, cells were lysed, and luciferase values were measured using the Dual-Luciferase reporter assay system (Promega). Firefly luciferase values were normalized to Renilla luciferase values and averaged for each transfection condition plated in triplicate.
RESULTS
Identification of the N Terminus of AIB1-Δ4
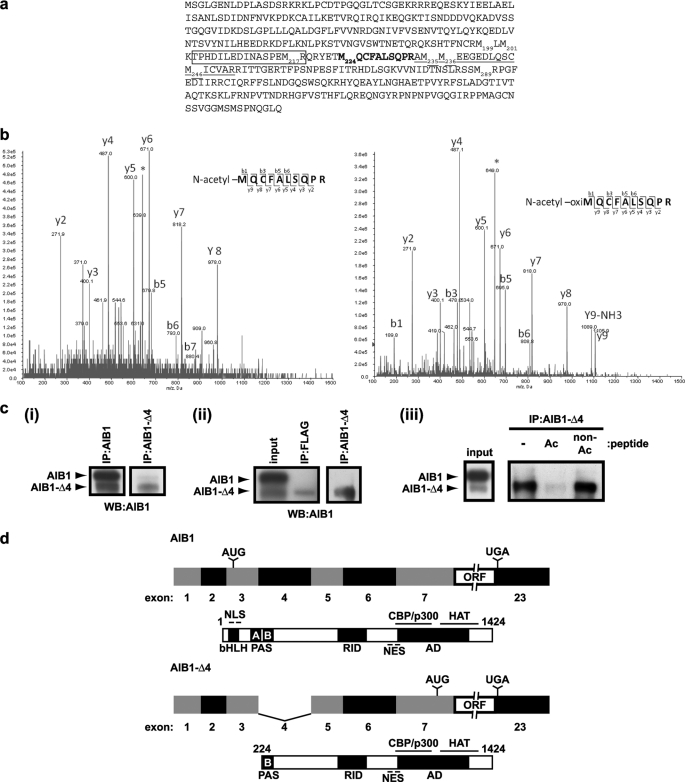
As we have identified previously, AIB1-Δ4 is a splice variant of the nuclear receptor coactivator AIB1 (17), which results in the translation of an N-terminally truncated isoform of the full-length AIB1 protein with a molecular mass of ∼130 kDa. The translation start site of AIB1-Δ4 was predicted to be at the methionine at position 199 in the full-length AIB1 protein because this was the next in-frame methionine residue; however, the translation start site was not identified experimentally. There is a cluster of methionines at positions 199, 201, 217, 224, 235, 236, 246, and 289 of the full-length AIB1 sequence from which initiation of translation could result in an ∼130-kDa protein (Fig. 1a). To determine which of these 8 methionines is the translation start site of the AIB1-Δ4 protein, we used mass spectrometry. We isolated AIB1-Δ4 protein from HEK293T cells transfected with a C-terminal FLAG AIB1-Δ4 cDNA construct. After immunoprecipitation with FLAG antibody, we isolated AIB1-Δ4 from a Coomassie-stained SDS-polyacrylamide gel and subjected the tryptic peptides to mass spectrometric analysis. Through an initial protein identification, we detected an AIB1-related protein based on the initial analysis of the peptides. A search for the peptide TPHDILEDINASPEM217R (boxed in Fig. 1a) was employed because it is an easily detectable fragment present in the full-length AIB1 protein in previous studies (22). This peptide was not found in the analysis of the tryptic fragments from AIB1-Δ4. This eliminated the possibility of the translation start site at Met199 and Met201. The most N-terminal peptide identified from the AIB1-Δ4 protein was AM235M236EEGEDLQSCM246ICVAR (underlined in Fig. 1a). This ruled out the Met235, Met236, Met246, and Met289 as possible translation start sites and limited the possibilities to the Met217 and Met224 residues. As a next step, we tested these methionines using in silico predictions for tryptic peptides with an initial methionine as the first amino acid residue. We also included predicted precursors that could appear as a result of cotranslational modifications such as N-terminal acetylation. In eukaryotes, 80% of all proteins have been described with an acetyl moiety added to the N terminus (24, 25). Interestingly, we observed only one predicted peptide 224MQCFALSQPR that retained an initiator methionine, which was Met224. This peptide harbored N-terminal acetylation, and the double charge state of this peptide defines an m/z value of 640.3 (Fig. 1b). Because of the chemical oxidation of methionines, the precursor ions are split between the m/z ratio of 640.3 and an m/z shift of +8 atomic mass units to 648.3. Fragmentation analysis of both precursors matched the corresponding peptides with different oxidation states of methionine and with Mascot scores assigned of 45 and 65, respectively, where scores above 23 indicate peptide identity. Therefore, we define the N terminus of AIB1-Δ4 to be at the Met224 residue of full-length AIB1. To further verify that this was the correct N terminus of AIB1-Δ4, we used the N-terminal acetylated sequence 224MQCFALSQPR determined by mass spectrometry to generate polyclonal antibodies that detect the AIB1-Δ4 with little or no detectable cross-reaction with full-length AIB1. Immunoprecipitations with the affinity-purified antibodies for AIB1-Δ4 detected both endogenous and transfected AIB1-Δ4 in MCF-7 and HEK293T cells, respectively (Fig. 1c, panels i and ii). The immunoprecipitated AIB1-Δ4 is competed by the N-acetylated peptide but not by the nonacetylated peptide, which also suggests that the unique acetylation present at Met224 provides an epitope not present in full-length AIB1 (Fig. 1c, panel iii).
FIGURE 1.
Identification of the translation start site of AIB1-Δ4 and characterization of affinity-purified AIB1-Δ4 antibodies. a, after overexpression of a C-terminal FLAG AIB1-Δ4 construct in HEK293T cells, we determined by mass spectrometric analysis that the AIB1-Δ4 transcript initiates translation at the methionine at position 224 in the full-length AIB1 protein. The TPHDILEDINASPEMR peptide was not identified in AIB1-Δ4 (boxed), and the AMMEEGEDLQSCMICVAR peptide was identified (underlined) by mass spectrometric analysis. The MQCFALSQPR peptide identified through further mass spectrometry analysis containing the translation start site is shown in boldface. b, annotated fragmentation spectra of the MQCFALSQPR peptide are shown. This tryptic peptide is bearing acetylation of the initial methionine as a result of cotranslational modification. The left spectrum shows collision-induced dissociation fragmentation of the double charged peptide with a nonoxidized methionine and m/z ratio of 640.3. The right spectrum depicts fragmentation of the double charged peptide with m/z ratio of 648.3 due to a mass shift caused by oxidation of the initial methionine. Asterisks indicate the position of parental ions in the MS/MS spectra. Based on the analysis of the collision-induced dissociation fragmentation, acetylation was assigned to the initial methionine residue. c, panel i, IP of endogenous AIB1-Δ4 in MCF-7 cells using AIB1-Δ4-specific antibody and detection with AIB1 (5E11) antibody that detects both isoforms of AIB1. Panel ii, IP of transfected C-terminal FLAG AIB1-Δ4 in HEK293T cells. Panel iii, peptide competition assay during IP of AIB1-Δ4 from MCF-7 cells using both an N-acetylated and nonacetylated MQCFALSQPRK peptide. IP was performed as in panel i except 1 μg of peptides was added during immunoprecipitation. WB, Western blot. d, full-length human AIB1 transcript consists of 23 exons. This leads to the creation of a 155-kDa protein consisting of 1424 amino acids. The AIB1-Δ4 transcript lacks exon 4 due to alternative splicing and the resultant protein lacks the N-terminal 223 amino acids of the full-length protein. This results in a 130-kDa protein consisting of 1201 amino acids. NES, nuclear export sequence.
The full-length and alternatively spliced AIB1 mRNAs and proteins derived from these data are depicted schematically in Fig. 1d. The full-length human AIB1 transcript has 23 exons coding for a 1424-amino acid protein of 155 kDa. Translation of AIB1 is initiated in exon 3 and continues until the stop codon in exon 23. The AIB1-Δ4 transcript lacks exon 4 because of alternative splicing. Translation of the AIB1-Δ4 isoform is initiated in exon 7, and the resultant protein lacks the N-terminal 223 amino acids of the full-length AIB1 protein. Because of the loss of exon 4 in the AIB1-Δ4 transcript, the NLS, basic helix loop helix (bHLH), and PAS A domain are lost from the AIB1-Δ4 protein. Thus, we now define AIB1-Δ4 as a 130-kDa protein of 1201 amino acids.
AIB1-Δ4 Is a Predominantly Cytoplasmic Protein That Enters the Nucleus
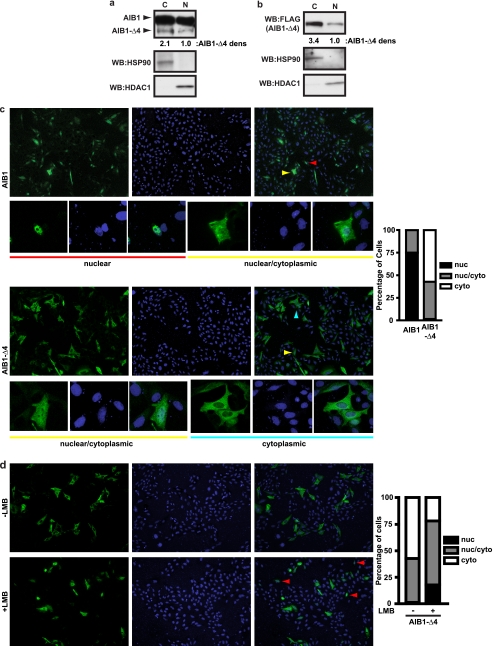
It has not been determined if the phenotypic effects of AIB1-Δ4 are only due to its cytoplasmic function or if there are additional direct transcriptional effects of this protein in the nucleus. Others have published the presence of a bipartite NLS in the N terminus of the AIB1 protein at amino acids 16–19 and 35–38 (26–28) indicating that AIB1-Δ4 lacks this NLS sequence. Initially, we examined the subcellular distribution of endogenous AIB1-Δ4 in fractionated MCF-7 cell extracts. Using an AIB1 antibody that detects both full-length AIB1 and AIB1-Δ4, we determined that both AIB1 isoforms were detected in the cytoplasmic and nuclear extracts from MCF-7 cells (Fig. 2a). AIB1-Δ4 is present in the nuclear extracts, although at levels considerably less than in the cytoplasm (68% cytoplasmic versus 32% nuclear).
FIGURE 2.
AIB1-Δ4 is found predominantly in the cytoplasm but can be detected in the nucleus. a, distribution of AIB1-Δ4 in nuclear (N) and cytoplasmic (C) fractions from MCF-7 cells was determined with AIB1 (5E11) antibody. HSP90 was used as a cytoplasmic fraction control and HDAC1 as a nuclear fraction control. b, FLAG AIB1-Δ4 localization in the HEK293T cells was determined as in a using FLAG M2 antibody for Western blotting (WB). c, Chinese hamster ovary cells were grown in DMEM F-12 + 10% FBS and transfected with FLAG AIB1 or FLAG AIB1-Δ4. Cells were fixed and stained for FLAG peptide and DAPI and visualized by indirect immunofluorescence. FLAG staining is shown in the left panels (green), DAPI staining DNA in the nucleus is shown in the middle panels (blue), and merge images in the right panels. Typical nuclear, nuclear/cytoplasmic, and cytoplasmic staining is shown in the smaller panels below the larger images. Red, yellow, and blue arrowheads note the nuclear, nuclear/cytoplasmic, and cytoplasmically stained cells shown in the insets below the large field images. The percentage of nuclear, nuclear/cytoplasmic, and cytoplasmic staining cells is graphed. 200 cells were counted per transfection, and cell compartment staining was quantified for three separate experiments. Nuclear (nuc), nuclear/cytoplasmic, and cytoplasmically (cyto) stained cells are represented by the black, gray, and white bars, respectively. d, CHO cells were transfected with FLAG AIB1-Δ4 and treated as in c. 24 h after transfection, either the carrier EtOH or 50 nm leptomycin B (LMB in EtOH) was added to the culture media for 4 h before fixing and staining cells. The percentage of nuclear, nuclear/cytoplasmic, and cytoplasmically staining cells is shown in the absence and presence of leptomycin B. Cells were quantified as in c. Nuclear, nuclear/cytoplasmic, and cytoplasmically stained cells are represented by the black, gray, and white bars, respectively. Red arrowheads note the nuclear stained cells in the field of FLAG AIB1-Δ4-transfected cells.
We next examined the dynamics of the subcellular distribution of AIB1-Δ4 by immunofluorescence. We utilized a transfected FLAG-tagged construct of AIB1-Δ4 (22) to determine its cellular localization because the AIB1-Δ4 affinity-purified antibodies signal in immunofluorescence for endogenous protein is very weak and not quantifiable. This transfected FLAG AIB1-Δ4 protein was detected in both the nucleus and cytoplasm after subcellular fractionation, and the distribution was 77% cytoplasmic versus 23% nuclear (Fig. 2b). Chinese hamster ovary cells (CHO) were transfected with either FLAG AIB1 or FLAG AIB1-Δ4 in media with 10% FBS, and the number of nuclear, nuclear/cytoplasmic, and cytoplasmically stained cells were quantified. CHO cells were chosen because their endogenous AIB1 is not detected by available AIB1 antibodies (17). We confirmed the fractionation result that the majority of AIB1-Δ4 resides in the cytoplasm (Fig. 2c). CHO cells transfected with AIB1 showed nuclear and nuclear/cytoplasmic staining in 75 and 25% of the cells, respectively. Cells transfected with AIB1-Δ4 showed nuclear, nuclear/cytoplasmic, and cytoplasmic staining in 1.5, 41.3, and 57.2% of cells, respectively. Large field images of staining for AIB1 and AIB1-Δ4 as well as typical staining for nuclear, nuclear/cytoplasmic, and cytoplasmically stained cells are shown.
Because we detected some nuclear and nuclear/cytoplasmic staining for AIB1-Δ4, we wanted to determine whether AIB1-Δ4 traffics through the nucleus. We took advantage of the nuclear export inhibitor leptomycin B, which inhibits CRM1-dependent nuclear export of proteins that contain a nuclear export sequence. Both AIB1 and AIB1-Δ4 contain the nuclear export sequence that is in the C-terminal half of the proteins (Fig. 1d) (27), so we hypothesized that if we blocked nuclear export then the AIB1-Δ4 protein would accumulate in the nucleus if it is imported into the nucleus. CHO cells were transfected with FLAG AIB1-Δ4 in media with 10% FBS and were either treated with vehicle or leptomycin B. Cells were fixed and stained, and the localization was quantified as before (Fig. 2d). We observed the same distribution of staining in the transfected CHO cells treated with vehicle; however, we saw an increase in the number of cells showing nuclear staining for AIB1-Δ4. CHO cells transfected with AIB1-Δ4 and treated with leptomycin B showed staining in the nuclear, nuclear/cytoplasmic, and the cytoplasmic compartments in 18, 60, and 22% of cells, respectively. The increase in the number of nuclear stained cells after leptomycin B treatment suggests that AIB1-Δ4 is imported into the nucleus; however, lack of a canonical NLS and the present data suggest that nuclear import of AIB1-Δ4 is an inefficient process, which contributes to the lower steady state levels of nuclear staining of the protein.
Nuclear Import of AIB1-Δ4 Can Be Facilitated by Other NLS-containing Proteins
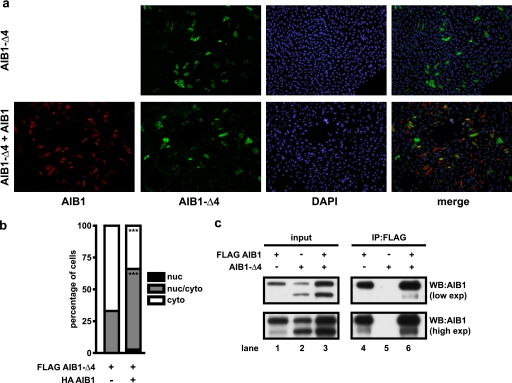
Various proteins involved in signaling have been shown to be imported into the nucleus in an NLS-independent manner (29–34). To study if the localization of AIB1-Δ4 could be altered by other NLS-containing proteins, CHO cells were transfected with FLAG AIB1-Δ4 in the presence of HA AIB1 (Fig. 3a). We observed a statistically significant increase in nuclear staining of AIB1-Δ4 after transfection with HA AIB1. CHO cells transfected with AIB1-Δ4 alone showed 67 ± 3.6 and 33 ± 3.6% of cells staining in cytoplasm and nuclear/cytoplasmic compartments, respectively. After cotransfection of AIB1-Δ4 with HA AIB1, cells stained the nuclear, nuclear/cytoplasmic, and cytoplasmic staining patterns in 2.7 ± 1.5, 63.3 ± 1.5, and 34 ± 1% of cells. To determine that this effect was not cell line-specific, we confirmed that AIB1 expression increased AIB1-Δ4 nuclear levels in mouse embryonic fibroblasts derived from AIB1 knock-out mice (supplemental Fig. 1). The pattern of staining of AIB1-Δ4 that was cytoplasmic, nuclear/cytoplasmic, or nuclear was 86 ± 1, 14 ± 1, and 0%, respectively, and shifted to 70.3 ± 3.5, 29.3 ± 4, and 0.3 ± 0.6% with expression of full-length AIB1.
FIGURE 3.
Overexpression of AIB1 localizes AIB1-Δ4 to the nucleus. a, CHO cells were transfected with FLAG AIB1-Δ4 alone or with an equal amount of HA-AIB1 and plated on glass coverslips in DMEM F-12 + 10% FBS. Cells were fixed and permeabilized 24 h after plating and stained for FLAG-containing proteins (green), HA-tagged proteins (red), and nuclei were stained with DAPI (blue). Cells were then analyzed by indirect immunofluorescence. b, number of nuclear (nuc), nuclear/cytoplasmic, and cytoplasmic (cyto) staining cells was quantified for three experiments as in Fig. 2c. Data were analyzed by t test. ***, p < 0.001 when compared with AIB1-Δ4 transfection alone. c, HEK293T cells grown in DMEM + 10% FBS were transfected with either FLAG AIB1 alone, AIB1-Δ4 alone, or FLAG AIB1 and AIB1-Δ4 together, and lysates were immunoprecipitated using FLAG antibody to pull down FLAG AIB1 and any interacting proteins. WB, Western blot; exp, exposure.
We next investigated possible mechanisms whereby AIB1 could enhance AIB1-Δ4 nuclear localization. It has been shown that the PAS B domain is sufficient for heterodimerization and homodimerization of p160 SRC family members (35). Because AIB1 and AIB1-Δ4 both contain PAS B domains, we set out to determine whether AIB1-Δ4 could interact with full-length AIB1 and piggyback into the nucleus with AIB1. HEK293T cells were transfected with FLAG AIB1 alone, AIB1-Δ4 alone, or FLAG AIB1 and AIB1-Δ4 together. After immunoprecipitation with FLAG antibody to pull down AIB1, Western blotting with a pan-AIB1 antibody showed that AIB1-Δ4 immunoprecipitates with AIB1 (Fig. 3c, lane 6, top panel). There is also a detectable amount of endogenous AIB1-Δ4 protein in HEK293T cells, and immunoprecipitation with FLAG AIB1 is able to pull down endogenous AIB1-Δ4 protein as well (Fig. 3c, lane 4, bottom panel). When no FLAG AIB1 is present, we do not detect any AIB1-Δ4 present after immunoprecipitation (Fig. 3c, lane 5, top and bottom panels). However, it should be noted that we have been unable to detect full-length AIB1 in AIB1-Δ4 immunoprecipitates with a variety of tags (not shown) or with immunoprecipitation using AIB1-Δ4 affinity-purified antibodies (Fig. 1d). Thus, the AIB1 interaction with AIB1-Δ4 is most likely weak, and we conclude that most of the AIB1 and AIB1-Δ4 proteins are not heterodimerized with each other in the cells we have examined. This suggests that AIB1 may increase nuclear import of AIB1-Δ4 through a nondimerization-mediated mechanism.
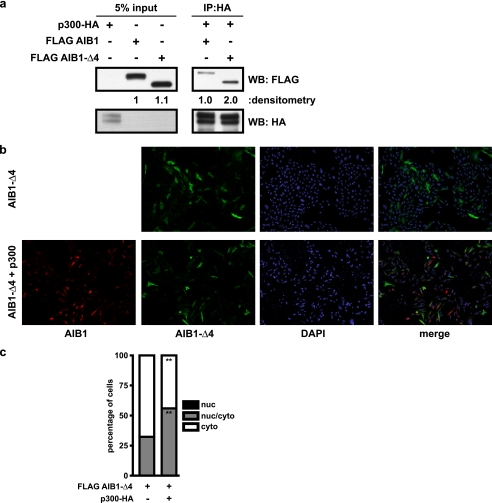
The interaction of AIB1 with other proteins involved in transcription is well characterized (7). One of these proteins is p300/CBP (36), and we show that AIB1-Δ4 interacts with p300 by immunoprecipitation (Fig. 4a). HEK293T cells were transfected with either p300-HA, FLAG AIB1, or FLAG AIB1-Δ4, and cell lysates were harvested. After normalization for protein content, equal amounts of either FLAG AIB1 or FLAG AIB1-Δ4 were added to the p300-HA cell lysate. The amounts of FLAG AIB1 and FLAG AIB1-Δ4 proteins were observed after immunoprecipitation with p300-HA. We found that both AIB1 and AIB1-Δ4 are associated with p300. However, with equal amounts of FLAG AIB1 or FLAG AIB1-Δ4 in the lysate, we found twice as much AIB1-Δ4 associated with p300 as compared with AIB1. This suggests that AIB1-Δ4 may be able to preferentially complex with p300 in active transcriptional complexes.
FIGURE 4.
Overexpression of p300 localizes AIB1-Δ4 to the nucleus. a, HEK293T cells were transfected with either p300-HA, FLAG AIB1, or FLAG AIB1-Δ4. Equal amounts of FLAG AIB1 or FLAG AIB1-Δ4 protein were incubated with equal amounts of p300-HA. After immunoprecipitation with HA antibody to pull down p300 and associated proteins, a FLAG Western blot (WB) was performed to determine how much AIB1 and AIB1-Δ4 immunoprecipitated with p300. b, CHO cells were transfected with FLAG AIB1-Δ4 alone or with an equal amount of p300-HA and plated on glass coverslips in DMEM F-12 + 10% FBS. Cells were fixed and permeabilized 24 h after plating and stained for DAPI (blue)-, FLAG (green)-, and HA (red)-containing proteins as in Fig. 3b. Cells were then analyzed by indirect immunofluorescence. c, number of nuclear (nuc), nuclear/cytoplasmic, and cytoplasmic (cyto) staining cells was quantified as in Fig. 2a. The percentage of nuclear, nuclear/cytoplasmic, and cytoplasmically stained cells is shown in the black, gray, and white bars, respectively, for three experiments is shown. Data were analyzed by t test. **, p < 0.01 when compared with AIB1-Δ4 transfection alone.
Consistent with these data, we see in CHO cells transfected with AIB1-Δ4 and p300-HA an increase in AIB1-Δ4 staining in the nucleus relative to cells transfected with AIB1-Δ4 alone (Fig. 4b). CHO cells transfected with FLAG AIB1-Δ4 alone showed staining in the nuclear/cytoplasmic and cytoplasmic compartments in 32.3 ± 1.5 and 67.7 ± 1.5%, respectively, which shifted to 56 ± 5.3 and 44 ± 5.3% (Fig. 4c). The induction of AIB1-Δ4 nuclear transport by p300 was not cell line-specific as we also replicated this finding in AIB1 KO mouse embryonic fibroblasts transfected with FLAG AIB1-Δ4 and p300-HA. The pattern of staining of AIB1-Δ4 that was nuclear, nuclear/cytoplasmic, or cytoplasmic was 0, 14 ± 1, and 86 ± 1%, respectively, and shifted to 13.3 ± 2.3, 63 ± 4.6, and 23.7 ± 2.5% (supplemental Fig. 2). Overall, the data suggest that AIB1-Δ4 may interact with other NLS-containing proteins to traffic into the nucleus.
AIB1-Δ4 Is Recruited to Active Transcriptional Complexes and Coactivates Estrogen-responsive Genes
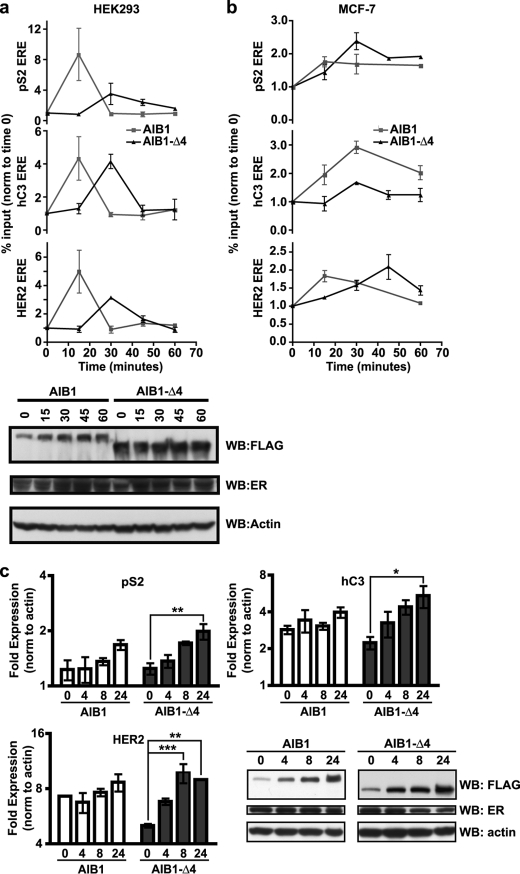
As it is known that both AIB1 and AIB1-Δ4 coactivate estrogen-dependent transcription, we decided to examine whether or not AIB1-Δ4 is recruited to known transcriptional targets of estrogen receptor. To examine the recruitment of AIB1-Δ4 to ERE, we transfected HEK293 cells with ER and either FLAG AIB1 or FLAG AIB1-Δ4 and performed ChIP analysis (Fig. 5a). Cells were treated with estrogen for 15, 30, 45, and 60 min to observe the recruitment of AIB1 or AIB1-Δ4 over time. Both AIB1 and AIB1-Δ4 were recruited to the three different estrogen-response elements in pS2, hC3, and HER2 (23, 37, 38), known to be binding sites for ERα (confirmed in supplemental Fig. 3a). Maximal recruitment of FLAG AIB1 was at 15 min for 8.7, 4.3, and 5.0% of the EREs in the pS2, hC3, and HER2 genes, respectively. In contrast, maximal recruitment of FLAG AIB1-Δ4 was at 30 min for 3.5, 4.2, and 3.1% of the EREs in the pS2, hC3, and HER2 genes, respectively. Thus, both AIB1 and AIB1-Δ4 are recruited to these EREs confirming that AIB1-Δ4 is found in the nucleus and found at active sites of transcription. The corresponding protein levels are shown by Western blot for all the different times of estrogen stimulation. Interestingly, the kinetics of AIB1-Δ4 recruitment is delayed, which is possibly due to inefficient or altered nuclear transport because of the lack of the NLS. Alternatively, this time delay could be an artifact generated by driving transcription with transiently transfected proteins. To determine whether this was the case, we also examined the dynamics of endogenous AIB1-Δ4 on estrogen-responsive genes using IP with the affinity-purified AIB1-Δ4 antibodies in MCF-7 cells (Fig. 5b). ChIP assays with this antibody confirmed that AIB1-Δ4 binds to ERE in estrogen-responsive genes, and consistent with the transfected AIB1-Δ4 protein, the peak of binding for AIB1-Δ4 was delayed relative to the peak binding for full-length AIB1 for the ERE in pS2 and HER2. Of note is that recruitment of AIB1-Δ4 is not exclusive to estrogen-regulated genes because we also observed its recruitment to a progesterone-response element in T47D A1-2 cells that have a stable integration of the MMTV luciferase construct (supplemental Fig. 3b) (39).
FIGURE 5.
AIB1-Δ4, like AIB1, is recruited to ERE in the nucleus. a, HEK293 cells grown in phenol red-free IMEM + 10% charcoal-stripped serum were transfected with either FLAG AIB1 or FLAG AIB1-Δ4 and ERα. 24 h later, cells were stimulated with estrogen and harvested at 0, 15, 30, 45, and 60 min after estrogen stimulation. These lysates were subjected to a quantitative ChIP analysis using a FLAG antibody. The percentage of the input recovered after immunoprecipitation for each ERE in pS2, hC3, or HER2 was determined. Data are representative of three independent experiments. The relative amount of transfected proteins and actin after transfection for each time point is shown by Western blot (WB). b, MCF-7 cells were grown in phenol red-free IMEM + 10% charcoal-stripped serum for 3 days before stimulation with 10 nm estrogen. Quantitative ChIP analysis was performed as in a except the immunoprecipitation was carried out with AIB1 antibody or AIB1-Δ4 affinity purified antibodies. Data are representative of two independent experiments. c, HEK293 cells were grown as in a, and total RNA was harvested from cells at 0, 4, 8, and 24 h after estrogen treatment to determine the relative gene expression for pS2, hC3, and HER2. Data were analyzed by two-way ANOVA with Bonferroni post-test where *, p < 0.05; **, p < 0.01; ***, p < 0.001 relative to time 0. None of the time points for AIB1 were significantly different. The relative amount of proteins after transfection for each time point is shown by the Western blot.
To confirm that recruitment of AIB1-Δ4 leads to an increase in these estrogen-regulated genes, we looked at the levels of expression of the pS2, hC3, and HER2 mRNA after estrogen stimulation. HEK293 cells were transfected with ER and either FLAG AIB1 or FLAG AIB1-Δ4 and stimulated with estrogen for 4, 8, and 24 h. RNA was harvested from the cells, and the gene expression was examined by real time PCR. We found maximal gene expression in AIB1-Δ4-transfected cells at 8 h for HER2 and at 24 h for pS2 and hC3 mRNA. Interestingly, significant differences in gene expression were only observed in the AIB1-Δ4 transfected cells suggesting that estrogen-responsive genes are more sensitive to changes in AIB1-Δ4 levels than full-length AIB1.
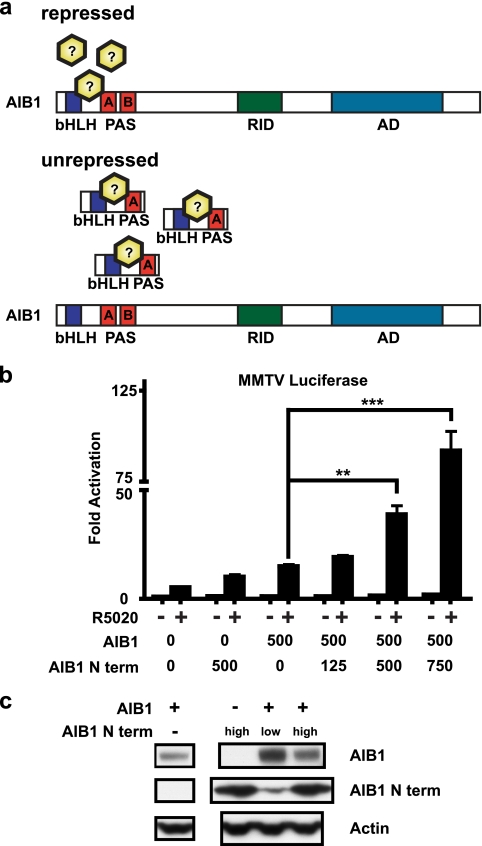
Region Deleted from AIB1-Δ4 Contains an Inhibitory Domain
A possible hypothesis explaining the more potent coactivator function of AIB1-Δ4 could be its lack of repression by an endogenous factor that is able to bind to the full-length AIB1 protein (Fig. 6a). One prediction of this model is that expression of an N-terminal fragment that bound this repressor could relieve repression of full-length AIB1. To test this hypothesis, we created a FLAG-tagged N-terminal AIB1 construct (AIB1 N term) that contains the bHLH and PAS A domains not present in the AIB1-Δ4 protein (Fig. 1c). This construct should have no coactivator function because it lacks the C-terminal activation domain responsible for binding to nuclear receptors and molecules that affect transcription such as p300/CBP and CARM1. For this experiment, we used progesterone-induced transcription in COS-7 cells as a readout of AIB1 activity as this has consistently given the strongest dose-response coactivator readout for AIB1 (17). We transfected COS-7 cells with progesterone receptor and AIB1 with increasing amounts of AIB1 N term and monitored the coactivator function of AIB1 on a progesterone-inducible MMTV luciferase reporter construct (Fig. 6b). As expected, AIB1 is able to coactivate transcription of the luciferase reporter relative to empty vector-transfected cells (AIB1 0 versus 500). Cotransfection of AIB1 with AIB1 N term significantly increased the amount of transcription from the luciferase reporter. This effect was increased with the amount of AIB1 N term cotransfected with AIB1 (AIB1 cotransfection with AIB1 N term 125, 500, 750). A relief of repression on endogenous AIB1 coactivator activity because of coexpression of the AIB1 N term construct alone was also apparent (AIB1 N term 0 versus 500). The levels of AIB1 and AIB1 N term protein are shown by Western blot, and there is no increase in the AIB1 protein with cotransfection of higher levels of AIB1 N term (Fig. 6b). These data suggest a suppressor role of the N-terminal region of AIB1 in the regulation of the coactivator function of AIB1. This region is the most highly conserved region among steroid receptor coactivator proteins and has been suggested to contain an inhibitory function for AIB1 also in the cytoplasm (21).
FIGURE 6.
N terminus of AIB1 contains an inhibitory domain that is lost in AIB1-Δ4. a, proposed mechanism of repression of the full-length AIB1 protein. An inhibitor that binds to the N terminus of AIB1 ordinarily represses its coactivator function. Overexpression of an N-terminal fragment of AIB1 lost in AIB1-Δ4 may bind to the squelching factor, which normally regulates the coactivator function of AIB1 thereby relieving repression on the full-length AIB1 protein. b, COS-7 cells were transfected with human progesterone receptor B (25 ng), MMTV luciferase (100 ng), and either pcDNA3 or FLAG AIB1 (500 ng) with increasing amounts of FLAG AIB1 N term (125, 500, and 750 ng). 24 h later, cells were treated with 10 nm R5020 for 24 h before reporter activity was determined. Luciferase values were normalized to thymidine kinase Renilla (10 ng) reporter activity. The assay was plated in triplicate, and a representative graph is shown from three separate experiments. Data were analyzed by one-way ANOVA with Tukey's multiple comparison post-test. **, p < 0.01; ***, p < 0.001 when compared with FLAG AIB1 transfection alone. c, relative amount of FLAG proteins in the COS-7 cells is shown by Western blot.
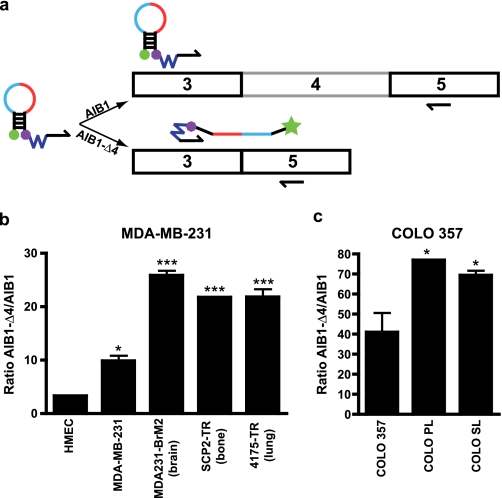
AIB1-Δ4 Expression Is Correlated with Metastatic Potential
We have shown previously that the mRNA expression of AIB1-Δ4 transcript was increased in breast tumor tissue relative to normal breast tissue and was higher in the MCF-7 breast cancer cell line relative to nontransformed breast cell lines (17). However, the only method we had previously to detect AIB1-Δ4 transcript was by semi-quantitative hybridization of a radioactive DNA probe to total RNA isolated from cells, thus making accurate quantification difficult. We therefore designed a quantitative assay to measure AIB1-Δ4 mRNA levels through the use of Scorpion primer technology (40, 41). We designed a Scorpion primer with a primer sequence complementary to a sequence in exon 3 and a probe sequence complementary to the unique sequence generated by the splicing of exon 3 and exon 5 (Fig. 7a). This Scorpion primer specifically recognizes the AIB1-Δ4 transcript and not the AIB1 transcript due to the fact that after amplification of AIB1 transcript, the probe sequence is not complementary to any sequence generated in the full-length AIB1 transcript. In addition, we designed an AIB1-specific Scorpion primer with a primer sequence in exon 3 and a probe sequence complementary to exon 4, which is not contained in the splice variant AIB1-Δ4 transcript. We subjected plasmids containing either AIB1 or AIB1-Δ4 cDNA to an analysis by real time PCR with the AIB1 and AIB1-Δ4 Scorpion primers (supplemental Fig. 4, a and b). We show that the AIB1-Δ4 Scorpion primer specifically identifies AIB1-Δ4 cDNA and not full-length AIB1 cDNA and vice versa for the AIB1-Scorpion primer. To confirm the increased expression of AIB1-Δ4 in breast cancer cell lines, total RNA was harvested from HMEC, parental MDA-MB-231, and three tissue-specific metastatic variants of MDA-MB-231 cells. The three tissue-specific variants homed either to the brain, bone, or lung after intravenous or intracardiac injection (42–44). After reverse transcription, cDNA generated from these RNAs was subjected to real time PCR analysis with Scorpion primers for AIB1 or AIB1-Δ4 (Fig. 7b). Expression of AIB1-Δ4 mRNA relative to AIB1 mRNA was higher in the parental and three tissue-specific metastatic variants of MDA-MB-231 cells relative to HMECs confirming increased expression of AIB1-Δ4 mRNA with malignant phenotype.
FIGURE 7.
AIB1-Δ4 expression is increased in metastatic cancer cell lines. a, the AIB1-Δ4 scorpion primer was designed to specifically recognize the unique splice junction of exon 3 and exon 5. The Scorpion primer consists of primer (black half-arrow), blocker (blue jagged line), quencher (purple octagon), probe (red, exon3; light blue, exon 5), stem (black attached lines), and reporter (green ball). The stem region will only be dissociated when a target sequence is created during the process of PCR. In the case of AIB1, no appropriate target is generated, so the preferred conformation is to remain in the stem loop with the reporter quenched. For the AIB1-Δ4 transcript, an appropriate target is generated, and the probe can then bind its target sequence allowing the reporter to fluoresce. Scorpion primers specific for AIB1-Δ4 transcript had a probe sequence complementary to the splice junction of exons 3 and 5. Scorpion primers for AIB1 had a probe sequence complementary to exon 4. b, Scorpion primers were used to quantitate the amount of AIB1 and AIB1-Δ4 transcript from the RNA of HMEC, parental MDA-MB-231 breast cancer cells, and three tissue-specific metastatic variants of MDA-MB-231 cells. The Ct values were normalized to actin expression as a control. The ratio of AIB1-Δ4 to AIB1 is shown. Data were analyzed by one-way ANOVA with Bonferroni post-test. *, p < 0.05; ***, p < 0.001 when compared with HMEC. c, Scorpion primers were used as in b to analyze RNA from parental COLO 357 pancreatic cancer cells and two metastatic variants that metastasized from pancreas to liver (COLO PL) or from spleen to liver (COLO SL). Ct values were normalized to actin expression as a control. The ratio of AIB1-Δ4 to AIB1 is shown. Data were analyzed by one-way ANOVA with Newman-Keuls post test. *, p < 0.05 when compared with parental COLO 357 cells.
We also examined if AIB1-Δ4 mRNA expression is also correlated with metastatic potential in another cancer metastasis model. We used parental COLO 357 pancreatic cancer cells and two metastatic derivative cell lines, which were selected in vivo to metastasize from the pancreas to liver or spleen to liver (COLO PL and COLO SL) (45). The expression of AIB1-Δ4 mRNA was found increased in the more metastatic variants of the COLO 357 cell line (Fig. 7c) suggesting a correlation between increased metastatic capability and AIB1-Δ4 expression.
DISCUSSION
We show in this study that AIB1-Δ4 enters the nucleus and has a nuclear function. In accordance with previous data that the NLS is contained in the N terminus of AIB1 (26–28), we found that the majority of AIB1-Δ4, which lacks an NLS, is predominantly in the cytoplasm at steady state levels. This is in contrast to the localization of AIB1, which resides mostly in the nucleus. Coactivation occurs in the nucleus, which raised the question how the mainly cytoplasmic AIB1-Δ4 had such potent effects on transcription. The first question to address was if AIB1-Δ4 enters the nucleus. We observed that AIB1-Δ4 accumulated in the nucleus after blockade of nuclear export suggesting that it was indeed being imported into the nucleus. The next question to address was whether AIB1-Δ4 had a functional role in the nucleus. We saw that AIB1-Δ4 was recruited as efficiently as AIB1 to ERE in estrogen-regulated genes in the nucleus despite significantly lower steady state nuclear levels of AIB1-Δ4. The mainly cytoplasmic location of AIB1-Δ4 is potentially due to either an inefficient nuclear import mechanism or through a rapid nuclear export mechanism. We argue for the former scenario given that AIB1-Δ4 lacks the N-terminal NLS and there is no alteration in the nuclear export sequence of AIB1-Δ4. It is known that molecules larger than 40 kDa have to be actively transported through the nuclear pore complex through interaction of the NLS with nuclear importins (46). Another potential mechanism for nuclear import termed “piggybacking” was demonstrated for various proteins such as eIF4E, IκBα, and CDK2, and BRCA1 (47–52). BRCA1 has a naturally occurring splice variant that lacks an NLS, and it is able to localize to the nucleus through interaction with another protein BARD1, which contains a canonical NLS. We propose that AIB1-Δ4 is able to similarly piggyback onto p300/CBP and/or other NLS-containing proteins and thus enter the nucleus. Alternatively, NLS-containing coactivators could up-regulate genes involved in Ran-independent nuclear import mechanisms such as via calmodulin (53). Whatever the mechanism, it appears to be an inefficient process likely accounting for the largely cytoplasmic distribution of AIB1-Δ4 in the cell. Also consistent with this hypothesis is the difference in the kinetics of recruitment of AIB1 and AIB1-Δ4. We saw a delay in the recruitment of AIB1-Δ4 to the ERE relative to AIB1 suggesting that the mechanism of nuclear import of AIB1-Δ4 is less efficient than that of AIB1.
The fact that we saw more effective coactivation by AIB1-Δ4 despite having much more AIB1 in the nucleus than AIB1-Δ4 suggests that there is a regulation of AIB1 coactivator activity that does not exist for AIB1-Δ4. Because we observed that the levels of AIB1-Δ4 interacting with p300 were higher than the levels of AIB1 associated with p300 (Fig. 4a) and p300 is generally found in active transcriptional complexes, we believe that AIB1-Δ4 is highly recruited to sites of active transcription. The increased coactivator activity of AIB1-Δ4 was confirmed by a higher increase in endogenous estrogen-regulated gene expression in cells transfected with AIB1-Δ4. These data suggest that the large amount of AIB1 that resides in the nucleus is not in active transcriptional complexes, and the reason why AIB1-Δ4 is a more potent coactivator than AIB1 can best be explained by the presence of an inhibitory domain in the N-terminal 223 amino acids of AIB1. Alternatively, loss of the N terminus could cause steric changes that increase the affinity for other coactivators. We conjectured that this N-terminal fragment containing the bHLH and PAS A domains would be able to bind N-terminal repressors of AIB1. We found that the N-terminal fragment containing the bHLH and PAS A domains of AIB1 when cotransfected with AIB1 is able to relieve repression of the coactivator function of the AIB1 protein. This effect was dose-dependent, and the more AIB1 N-terminal fragment added to the cells the less repression there was on the AIB1 protein. Interestingly, transfection of the fragment alone showed an increase in luciferase activity, which is probably not due to an inherent coactivator function of this fragment because most of the transcriptional activity of the AIB1 and AIB1-Δ4 proteins resides in their recruitment of p300/CBP in the C terminus (36). This increase in transcription is most likely due to a relief of repression of the endogenous AIB1 in the COS-7 cells because we are able to detect endogenous AIB1 protein by Western blot (17). Previous studies from our group and others have suggested that the N-terminal region containing the bHLH and PAS A domains contains an inhibitory domain that represses activity in both the nucleus and cytoplasm (17, 18, 21, 26, 36). Our studies and those from Chen et al. (36) have shown that the loss of the N-terminal region leads to potent coactivation of nuclear hormone receptor-mediated transcription. Expression of AIB1-Δ4 (loss of amino acids 1–223 of AIB1) or ACTR38 (loss of amino acids 1–447 of AIB1) leads to potent coactivation of nuclear receptor-dependent transcription from estrogen, progesterone, retinoic acid, thyroid, glucocorticoid, vitamin D, and retinoid X receptors. Interestingly, data from Li et al. (26) showed expression of AIB1 constructs containing mutations in either or both NLS (NLS amino acids 16–19 and 35–38 of AIB1) or with the bHLH domain deleted (amino acids 16–88 of AIB1) had no coactivator activity presumably because of lack of import into the nucleus. The other possibility is that they still retained amino acids 88–224, which would support the evidence that there is an inhibitory domain in this region. Loss of these amino acids in AIB1-Δ4 and ACTR38 allows these proteins to be potent coactivators. A number of proteins have been shown to bind to the bHLH PAS domain of p160 SRC family members. These have been described as having coactivator, corepressor, and phosphatase activity (54–59) depending on the context in which they are examined. It remains to be determined if these proteins play a role in repression or loss of activation of AIB1 on endogenous genes or conversely whether they have lost affinity or changed interaction with AIB1-Δ4.
Both AIB1 and AIB1-Δ4 have been shown to have effects in the epidermal growth factor (EGF) signaling. Data from our laboratory has shown that loss of both AIB1 and AIB1-Δ4 proteins together can lead to a decrease in EGF receptor (EGFR) phosphorylation (6), and AIB1-Δ4 is able to potently coactivate EGF-stimulated transcription (17). Long et al. (21) have shown that AIB1-Δ4 acts as a bridging molecule between EGFR and FAK, and this interaction facilitates the motility and metastatic capability of MDA-MB-231 breast cancer cells. They also show that AIB1-Δ4 is phosphorylated by p21-activated kinase (PAK1), which increases the association of AIB1-Δ4 with EGFR and FAK. Intriguingly, in this latter study the N-terminal region of AIB1 inhibited the interaction of AIB1 with FAK and therefore was unable to stimulate the EGF-induced migration of cancer cells, suggesting that even in the cytoplasm the N-terminal region of AIB1 was repressive. Expression of an NLS mutant of AIB1, which resides predominantly in the cytoplasm, showed much weaker interaction with FAK in cells. Another recent publication by Cai et al. (60) shows in non-small cell lung cancer that AIB1 expression is correlated with poor prognosis, and knockdown of AIB1 in non-small cell lung cancer cell line resistant to gefitinib treatment restored sensitivity to EGFR inhibition by gefitinib. Taken together, these data indicate that there is a signaling loop existing between AIB1/AIB1-Δ4 and EGFR where EGFR can affect AIB1-Δ4 through PAK1 activation and AIB1/AIB1-Δ4 can affect EGFR signaling and transcription as well. It is unclear if the siRNA used to target AIB1 in the gefitinib study also targeted AIB1-Δ4 as well, so the effects of AIB1-Δ4 on EGFR signaling are not currently known. Overall, the contribution of nuclear versus cytoplasmic function of AIB1-Δ4 to steroid and growth factor signaling needs to be further explored.
Given that we have found that the expression of AIB1-Δ4 at the protein level is higher in breast and pancreatic cancer cells (17, 61), we were interested to investigate if AIB1-Δ4 is regulated at the protein level. We have previously shown that AIB1 protein levels are greatly reduced in response to growth of cells at high confluence and the removal of growth factors (62). Interestingly, we found that the AIB1-Δ4 isoform is not regulated in the same fashion as AIB1 protein (supplemental Fig. 5). AIB1-Δ4 is resistant to proteasomal degradation induced by high confluence. This is probably due to loss of a site of regulation in the N-terminal 223 amino acids. The proteasomal regulation of AIB1 has been well characterized (57, 62, 63). Interestingly, regulation of a phospho-degron at Ser102 by protein phosphatase 1 (PP1) was shown to be important for regulating the activity of AIB1. PP1 overexpression was able to inhibit the reporter activity as well as the cell proliferative ability of AIB1. The Ser102 site is also a site of regulation by the ubiquitin ligase SPOP (64). This site is lost in AIB1-Δ4 and could explain the high coactivator activity and stability of the AIB1-Δ4 protein.
A major question that arises from these studies on AIB1-Δ4 is whether there are distinct biological functions for this isoform. To begin to investigate this, we took advantage of the unique splice junction sequence that exists in the AIB1-Δ4 transcript to develop a new technique to specifically measure the amounts of AIB1-Δ4 mRNA independent of AIB1 transcript. By utilizing Scorpion primer technology, we see higher expression of AIB1-Δ4 in cancer cell lines relative to normal cell lines and higher expression in more metastatic cancer cell lines. Observations on the role of AIB1 in disease do not make the distinction between the relative contribution of AIB1 or AIB1-Δ4 to the phenotype. Studies that analyze mRNA expression of AIB1 utilize primers that detect both AIB1 and AIB1-Δ4. We believe that it will be important to dissect out the contribution to phenotypes attributed to AIB1-Δ4 independent of AIB1 and vice versa in future clinical studies. We have also developed affinity-purified antibodies based on the knowledge of the unique N-acetylated N-terminal sequence of AIB1-Δ4, which will serve as another resource to determine the expression of AIB1-Δ4 at the protein level at various stages of tumorigenesis.
The precise role of AIB1-Δ4 in tumorigenesis is not known. It is intriguing that AIB1-Δ4 is not degraded under conditions of low growth (e.g. high confluence), although full-length AIB1 is rapidly lost. AIB1-Δ4 protein expression may provide a selective advantage for a cancer cell to continue to grow in conditions unfavorable for proliferation for both its metastatic and transcriptional function. Therefore, further studies into the regulation and possible targets of AIB1-Δ4 in tumorigenesis need to be pursued.
Supplementary Material
Acknowledgments
Mass spectrometric analysis was performed in the Proteomics and Metabolomics Shared Resource, and microscopy was performed in the Microscopy and Imaging Shared Resource, which are supported in part by National Institutes of Health NCI Grant P30-CA051008. We also thank Geoffrey Storchan for help with chromatin immunoprecipitation experimental design.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA113477 from NCI (to A. T. R.). This work was also supported by Department of Defense Breast Cancer Research Program Grant BC083320 (to C. D. C.) and Center of Excellence Grant W81XWH-06-10590 (to V. C. Jordan, A. W., and A. T. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- ERα
- estrogen receptor-α
- EGFR
- epidermal growth factor receptor
- NLS
- nuclear localization sequence
- bHLH
- basic helix loop helix
- FAK
- focal adhesion kinase
- ANOVA
- analysis of variance
- IP
- immunoprecipitation
- IMEM
- Iscove's modified Eagle's medium
- CCS
- charcoal-stripped serum
- ERE
- estrogen-responsive element
- S
- sense
- AS
- antisense
- HMEC
- human mammary epithelial cell
- MMTV
- mouse mammary tumor virus
- PAS
- Per Arnt Sim domain.
REFERENCES
- 1. Xu J., Wu R. C., O'Malley B. W. (2009) Nat. Rev. Cancer 9, 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. (1997) Science 277, 965–968 [DOI] [PubMed] [Google Scholar]
- 3. Lahusen T., Henke R. T., Kagan B. L., Wellstein A., Riegel A. T. (2009) Breast Cancer Res. Treat. 116, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. List H. J., Reiter R., Singh B., Wellstein A., Riegel A. T. (2001) Breast Cancer Res. Treat. 68, 21–28 [DOI] [PubMed] [Google Scholar]
- 5. Oh A., List H. J., Reiter R., Mani A., Zhang Y., Gehan E., Wellstein A., Riegel A. T. (2004) Cancer Res. 64, 8299–8308 [DOI] [PubMed] [Google Scholar]
- 6. Lahusen T., Fereshteh M., Oh A., Wellstein A., Riegel A. T. (2007) Cancer Res. 67, 7256–7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. York B., O'Malley B. W. (2010) J. Biol. Chem. 285, 38743–38750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fereshteh M. P., Tilli M. T., Kim S. E., Xu J., O'Malley B. W., Wellstein A., Furth P. A., Riegel A. T. (2008) Cancer Res. 68, 3697–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuang S. Q., Liao L., Wang S., Medina D., O'Malley B. W., Xu J. (2005) Cancer Res. 65, 7993–8002 [DOI] [PubMed] [Google Scholar]
- 10. Kuang S. Q., Liao L., Zhang H., Lee A. V., O'Malley B. W., Xu J. (2004) Cancer Res. 64, 1875–1885 [DOI] [PubMed] [Google Scholar]
- 11. Qin L., Liao L., Redmond A., Young L., Yuan Y., Chen H., O'Malley B. W., Xu J. (2008) Mol. Cell. Biol. 28, 5937–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Torres-Arzayus M. I., Font de Mora J., Yuan J., Vazquez F., Bronson R., Rue M., Sellers W. R., Brown M. (2004) Cancer Cell 6, 263–274 [DOI] [PubMed] [Google Scholar]
- 13. Bautista S., Vallès H., Walker R. L., Anzick S., Zeillinger R., Meltzer P., Theillet C. (1998) Clin. Cancer Res. 4, 2925–2929 [PubMed] [Google Scholar]
- 14. Harigopal M., Heymann J., Ghosh S., Anagnostou V., Camp R. L., Rimm D. L. (2009) Breast Cancer Res. Treat. 115, 77–85 [DOI] [PubMed] [Google Scholar]
- 15. Thorat M. A., Turbin D., Morimiya A., Leung S., Zhang Q., Jeng M. H., Huntsman D. G., Nakshatri H., Badve S. (2008) Histopathology 53, 634–641 [DOI] [PubMed] [Google Scholar]
- 16. Osborne C. K., Bardou V., Hopp T. A., Chamness G. C., Hilsenbeck S. G., Fuqua S. A., Wong J., Allred D. C., Clark G. M., Schiff R. (2003) J. Natl. Cancer Inst. 95, 353–361 [DOI] [PubMed] [Google Scholar]
- 17. Reiter R., Wellstein A., Riegel A. T. (2001) J. Biol. Chem. 276, 39736–39741 [DOI] [PubMed] [Google Scholar]
- 18. Reiter R., Oh A. S., Wellstein A., Riegel A. T. (2004) Oncogene 23, 403–409 [DOI] [PubMed] [Google Scholar]
- 19. Tilli M. T., Reiter R., Oh A. S., Henke R. T., McDonnell K., Gallicano G. I., Furth P. A., Riegel A. T. (2005) Mol. Endocrinol. 19, 644–656 [DOI] [PubMed] [Google Scholar]
- 20. Nakles R. E., Shiffert M. T., Díaz-Cruz E. S., Cabrera M. C., Alotaiby M., Miermont A. M., Riegel A. T., Furth P. A. (2011) Mol. Endocrinol. 25, 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Long W., Yi P., Amazit L., LaMarca H. L., Ashcroft F., Kumar R., Mancini M. A., Tsai S. Y., Tsai M. J., O'Malley B. W. (2010) Mol. Cell 37, 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oh A. S., Lahusen J. T., Chien C. D., Fereshteh M. P., Zhang X., Dakshanamurthy S., Xu J., Kagan B. L., Wellstein A., Riegel A. T. (2008) Mol. Cell. Biol. 28, 6580–6593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hurtado A., Holmes K. A., Geistlinger T. R., Hutcheson I. R., Nicholson R. I., Brown M., Jiang J., Howat W. J., Ali S., Carroll J. S. (2008) Nature 456, 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Driessen H. P., de Jong W. W., Tesser G. I., Bloemendal H. (1985) CRC Crit. Rev. Biochem. 18, 281–325 [DOI] [PubMed] [Google Scholar]
- 25. Polevoda B., Sherman F. (2002) Genome Biol. 3, reviews0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li C., Wu R. C., Amazit L., Tsai S. Y., Tsai M. J., O'Malley B. W. (2007) Mol. Cell. Biol. 27, 1296–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qutob M. S., Bhattacharjee R. N., Pollari E., Yee S. P., Torchia J. (2002) Mol. Cell. Biol. 22, 6611–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yeung P. L., Zhang A., Chen J. D. (2006) Biochem. Biophys. Res. Commun. 348, 13–24 [DOI] [PubMed] [Google Scholar]
- 29. Fagotto F., Glück U., Gumbiner B. M. (1998) Curr. Biol. 8, 181–190 [DOI] [PubMed] [Google Scholar]
- 30. Matsubayashi Y., Fukuda M., Nishida E. (2001) J. Biol. Chem. 276, 41755–41760 [DOI] [PubMed] [Google Scholar]
- 31. Whitehurst A. W., Wilsbacher J. L., You Y., Luby-Phelps K., Moore M. S., Cobb M. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7496–7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu L., Chen Y. G., Massagué J. (2000) Nat. Cell Biol. 2, 559–562 [DOI] [PubMed] [Google Scholar]
- 33. Xu L., Massagué J. (2004) Nat. Rev. Mol. Cell Biol. 5, 209–219 [DOI] [PubMed] [Google Scholar]
- 34. Yokoya F., Imamoto N., Tachibana T., Yoneda Y. (1999) Mol. Biol. Cell 10, 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lodrini M., Münz T., Coudevylle N., Griesinger C., Becker S., Pfitzner E. (2008) Nucleic Acids Res. 36, 1847–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen H., Lin R. J., Schiltz R. L., Chakravarti D., Nash A., Nagy L., Privalsky M. L., Nakatani Y., Evans R. M. (1997) Cell 90, 569–580 [DOI] [PubMed] [Google Scholar]
- 37. Masiakowski P., Breathnach R., Bloch J., Gannon F., Krust A., Chambon P. (1982) Nucleic Acids. Res. 10, 7895–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sundstrom S. A., Komm B. S., Ponce-de-Leon H., Yi Z., Teuscher C., Lyttle C. R. (1989) J. Biol. Chem. 264, 16941–16947 [PubMed] [Google Scholar]
- 39. Nordeen S. K., Kühnel B., Lawler-Heavner J., Barber D. A., Edwards D. P. (1989) Mol. Endocrinol. 3, 1270–1278 [DOI] [PubMed] [Google Scholar]
- 40. Taveau M., Stockholm D., Spencer M., Richard I. (2002) Anal. Biochem. 305, 227–235 [DOI] [PubMed] [Google Scholar]
- 41. Thelwell N., Millington S., Solinas A., Booth J., Brown T. (2000) Nucleic Acids Res. 28, 3752–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bos P. D., Zhang X. H., Nadal C., Shu W., Gomis R. R., Nguyen D. X., Minn A. J., van de Vijver M. J., Gerald W. L., Foekens J. A., Massagué J. (2009) Nature 459, 1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordón-Cardo C., Guise T. A., Massagué J. (2003) Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 44. Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bruns C. J., Harbison M. T., Kuniyasu H., Eue I., Fidler I. J. (1999) Neoplasia 1, 50–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stewart M. (2007) Nat. Rev. Mol. Cell Biol. 8, 195–208 [DOI] [PubMed] [Google Scholar]
- 47. Dostie J., Ferraiuolo M., Pause A., Adam S. A., Sonenberg N. (2000) EMBO J. 19, 3142–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fabbro M., Rodriguez J. A., Baer R., Henderson B. R. (2002) J. Biol. Chem. 277, 21315–21324 [DOI] [PubMed] [Google Scholar]
- 49. Jans D. A., Hübner S. (1996) Physiol. Rev. 76, 651–685 [DOI] [PubMed] [Google Scholar]
- 50. Moore J. D., Yang J., Truant R., Kornbluth S. (1999) J. Cell Biol. 144, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thompson M. E. (2010) FEBS J. 277, 3072–3078 [DOI] [PubMed] [Google Scholar]
- 52. Turpin P., Hay R. T., Dargemont C. (1999) J. Biol. Chem. 274, 6804–6812 [DOI] [PubMed] [Google Scholar]
- 53. Hanover J. A., Love D. C., Prinz W. A. (2009) J. Biol. Chem. 284, 12593–12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Belandia B., Parker M. G. (2000) J. Biol. Chem. 275, 30801–30805 [DOI] [PubMed] [Google Scholar]
- 55. Goel A., Janknecht R. (2004) J. Biol. Chem. 279, 14909–14916 [DOI] [PubMed] [Google Scholar]
- 56. Kim J. H., Li H., Stallcup M. R. (2003) Mol. Cell 12, 1537–1549 [DOI] [PubMed] [Google Scholar]
- 57. Li C., Liang Y. Y., Feng X. H., Tsai S. Y., Tsai M. J., O'Malley B. W. (2008) Mol. Cell 31, 835–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu X., Li H., Chen J. D. (2001) J. Biol. Chem. 276, 23962–23968 [DOI] [PubMed] [Google Scholar]
- 59. Zhang A., Yeung P. L., Li C. W., Tsai S. C., Dinh G. K., Wu X., Li H., Chen J. D. (2004) J. Biol. Chem. 279, 33799–33805 [DOI] [PubMed] [Google Scholar]
- 60. Cai D., Shames D. S., Raso M. G., Xie Y., Kim Y. H., Pollack J. R., Girard L., Sullivan J. P., Gao B., Peyton M., Nanjundan M., Byers L., Heymach J., Mills G., Gazdar A. F., Wistuba I., Kodadek T., Minna J. D. (2010) Cancer Res. 70, 6477–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Henke R. T., Haddad B. R., Kim S. E., Rone J. D., Mani A., Jessup J. M., Wellstein A., Maitra A., Riegel A. T. (2004) Clin. Cancer Res. 10, 6134–6142 [DOI] [PubMed] [Google Scholar]
- 62. Mani A., Oh A. S., Bowden E. T., Lahusen T., Lorick K. L., Weissman A. M., Schlegel R., Wellstein A., Riegel A. T. (2006) Cancer Res. 66, 8680–8686 [DOI] [PubMed] [Google Scholar]
- 63. Li X., Lonard D. M., Jung S. Y., Malovannaya A., Feng Q., Qin J., Tsai S. Y., Tsai M. J., O'Malley B. W. (2006) Cell 124, 381–392 [DOI] [PubMed] [Google Scholar]
- 64. Li C., Ao J., Fu J., Lee D. F., Xu J., Lonard D., O'Malley B. W. (2011) Oncogene, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.