Abstract
Leishmaniasis is a major health problem that affects populations of ∼90 countries worldwide, with no vaccine and only a few moderately effective drugs. Here we report the structure/function characterization of sterol 14α-demethylase (CYP51) from Leishmania infantum. The enzyme catalyzes removal of the 14α-methyl group from sterol precursors. The reaction is essential for membrane biogenesis and therefore has great potential to become a target for antileishmanial chemotherapy. Although L. infantum CYP51 prefers C4-monomethylated sterol substrates such as C4-norlanosterol and obtusifoliol (Vmax of ∼10 and 8 min−1, respectively), it is also found to 14α-demethylate C4-dimethylated lanosterol (Vmax = 0.9 min−1) and C4-desmethylated 14α-methylzymosterol (Vmax = 1.9 min−1). Binding parameters with six sterols were tested, with Kd values ranging from 0.25 to 1.4 μm. Thus, L. infantum CYP51 is the first example of a plant-like sterol 14α-demethylase, where requirements toward the composition of the C4 atom substituents are not strict, indicative of possible branching in the postsqualene portion of sterol biosynthesis in the parasite. Comparative analysis of three CYP51 substrate binding cavities (Trypanosoma brucei, Trypanosoma cruzi, and L. infantum) suggests that substrate preferences of plant- and fungal-like protozoan CYP51s largely depend on the differences in the enzyme active site topology. These minor structural differences are also likely to underlie CYP51 catalytic rates and drug susceptibility and can be used to design potent and specific inhibitors.
Keywords: Cytochrome P450, Enzyme Kinetics, Enzyme Turnover, Protein Folding, Protein Structure, CYP51, Leishmania, sterol 14alpha-Demethylase (CYP51), Sterol Biosynthesis
Introduction
Leishmaniasis is widespread on all populated continents: 12 million people in 88 countries are reported to be infected, with 1–2 million new cases and 60,000 deaths occurring each year. The infection is caused by unicellular eukaryotic organisms that form the genus Leishmania (family Trypanosomatidae, order Kinetoplastida). Similar to many other protozoan parasites, Leishmania has a complex life cycle, including two morphologically different stages and using insects (phlebotomine sand flies) as vector and a variety of mammals as hosts. The insect stage of Leishmania (promastigote) is extracellular: growing in the sand fly intestine. The infection is transmitted to mammals by the sand fly bite (1). One of the most remarkable features of Leishmania is that their mammalian stage (amastigotes) lives inside macrophages, the phagocytic mammalian cells that are responsible for killing invaders (2).
There are ∼25 species of Leishmania known to be pathogenic for humans (3, 4). According to the World Health Organization taxonomic scheme, Leishmania species are currently grouped into several complexes, based on their genetic relations and disease manifestation (5). In humans, the disease occurs in at least four major forms: 1) cutaneous, in which parasites remain at the site of infection and cause localized long term ulceration (e.g. Leishmania major); 2) diffuse cutaneous, in which disseminated lesions are subject to relapse after treatment (Leishmania panamensis); 3) mucocutaneous, a chronic destruction of mucosal tissue (Leishmania braziliensis); and 4) visceral (systemic), also known as kala-azar, or black fever (Leishmania donovani complex). Visceral leishmaniasis is the most severe form of the disease, nearly always fatal if untreated. The symptoms include splenomegaly, hepatomegaly, profound cachexia, anemia, bone marrow damage, and a sharp decrease of resistance to secondary infections (6). Post-kala-azar dermal leishmaniasis develops as a sequel to successful kala-azar treatment in 20–60% of patients depending on the geographic area (7). The severity of all types of leishmaniases strongly depends on the patient's immune response (5), HIV coinfection being an emerging problem of particular concern.
There are currently no effective vaccines for leishmaniasis (7). The major clinical drugs used worldwide are two pentavalent antimonials (meglumine antimoniate and stibogluconate): pentamidine and amphotericin B (4). Except for amphotericin B, which depletes ergosterol from the parasite membranes, the mechanisms of their action remain unclear. It has been shown that inhibitors of fungal sterol biosynthesis, azoles, are often helpful against leishmaniasis (4, 6, 8–10), but none of them have yet been included into the clinically available regimens.
Sequencing the genomes of three Leishmania species (L. major, L. braziliensis, and Leishmania infantum) has been completed, whereas sequencing of L. donovani, Leishmania mexicana, and Leishmania tarentolae is still in progress. Although the species within the Leishmania genus are considered to have diverged 20–100 million years ago, their genomes preserve rather high average conservation (∼92%) and synteny. The parasites have a 32–33-Mb haploid genome organized into 36 chromosomes that contain ∼8,200 coding genes (3). Blast searches in the Leishmania genome database reveal the presence of all the genes encoding the enzymes required for sterol biosynthesis, including sterol 14α-demethylase (CYP51; EC 1.14.13.70 or ERG11),2 the ortholog of which serves as the major target for antifungal chemotherapy (11–13). The amino acid sequence identity of CYP51s from Leishmania species to sterol 14α-demethylases from fungi is very low (below 25%), and no data on direct characterization of a CYP51 from Leishmania have been reported.
Here we describe cloning, expression in Escherichia coli, purification, spectral properties, substrate binding parameters, catalytic analysis, and structure/function characterization of CYP51 from L. infantum (synonym L. chagasi) (5). The parasite belongs to the L. donovani complex and causes visceral leishmaniasis, predominantly in children in the Mediterranean region and in South America. As has been predicted from the CYP51 sequence analysis (14, 15), the enzyme from Leishmania demonstrates catalytic preferences toward the C4-monomethylated sterol substrates obtusifoliol (Obt) and C4-norlanosterol (Nls), which is in good agreement with the presence of the plant-specific phenylalanine (Phe-104) in the B′ helix (15). With lower efficiency, it binds and metabolizes C4-dimethylated sterols (such as eburicol (Ebr) and lanosterol (Lns)) and even C4-desmethylated 14α-methylzymosterol (Mzs). Therefore, in its substrate preferences, L. infantum CYP51 is more similar to the plant-like I105F mutant of Trypanosoma cruzi CYP51 (15) than the sterol 14α-demethylases from Trypanosoma brucei or from plants: these enzymes, at least under the standard reaction conditions in vitro, strictly require a single methyl group at the C4 position and do not show any sign of Lns or Ebr 14α-demethylation (16–20).
Thus, L. infantum CYP51 is the first example of a natural plant-like sterol 14α-demethylase with much less strict structural requirements toward substrates. Comparative analysis of three Trypanosomatidae CYP51 x-ray structures suggests a possible connection between the relatively broader substrate specificity of the L. infantum CYP51 ortholog and the volume/topology of its substrate binding cavity. The data support the idea that in Leishmania species, sterol biosynthesis downstream of squalene formation is likely to bifurcate into more than one branch (9, 21) so that the subsequent 14α-demethylation, 4-demethylation, and C24 methylation reactions might vary in their order. This can extend the sterol flow as far as possible down the pathway if one of the enzyme activities is not sufficient at any moment and implies that the other post-squalene sterol biosynthetic enzymes in Leishmania, such as sterol 24C-methyltransferase (EC 2.1.1.41 or ERG6) and sterol C4-demethylase (EC 1.14.13.72 or ERG25) are also likely to have less strict substrate requirements.
EXPERIMENTAL PROCEDURES
CYP51 Gene Identification, Cloning, and Expression
Sequence data for the L. infantum genome was from the GeneDB website. A tblastn homology search was carried out using T. cruzi CYP51 (NCBI accession number AY856083) as a template. Alignment was performed by Clustal W1.81 and analyzed in GeneDoc. The putative CYP51 gene (chromosome 11, gene number LinJ11.1100) was PCR-amplified from L. infantum genomic DNA using a FailSafe PCR premix selection kit (Epicenter). The upstream primer 5′-CGCCATATGGCTGGCGAGCTACTCC-3′ contained a unique NdeI cloning site (underlined) and modification of the second codon to alanine (bold) to optimize expression in E. coli as done previously for T. brucei CYP51 (16). The rest of the sequence corresponded to the L. infantum CYP51 cDNA. The downstream primer 5′-CGCAAGCTTCAGTGATGGTGATGAGCAGCCGCCTTCTTCTTC-3′ incorporated a unique HindIII cloning site (underlined) followed by a stop codon (bold) and the C-terminal four-histidine tag (italic), the remainder being complementary to the L. infantum CYP51 sequence. The PCR conditions are described in the supplemental materials. The products were purified from an agarose gel and subcloned into pGEM-T Easy vector (Promega). The correctness of the insert was confirmed by DNA sequencing. The L. infantum cDNA and protein sequences were deposited into the NCBI database (accession number EF192938). To obtain the expression construct, the L. infantum CYP51 gene insert was excised from pGEM-T by digestion with NdeI and HindIII (New England Biolabs) and subcloned into pCW (22) as described for T. cruzi CYP51 (15). The expression plasmid was sequenced and transformed into E. coli strain HMS174 (DE3) (Novagen). For the purpose of crystallization, the full-length L. infantum CYP51 sequence was modified by replacing its N-terminal membrane anchor sequence upstream of P32 with MAKKTSSKGKL- (23). This N-terminal truncation increased the L. infantum CYP51 expression level from ∼70 to >1000 nmol/liter E. coli culture. Expression and purification procedures were similar to those described in Ref. 15; the details are provided in the supplemental materials. The molecular weight and purity of the protein were confirmed by SDS-PAGE. The average yields after the nickel column and CM-Sepharose were 93 and 75% of the P450 detected following membrane solubilization, respectively.
Quantification of P450 Concentration, Spin State Transition, and Response to Ligand Binding
UV-visible absorption spectra of L. infantum CYP51 (1.5–10 μm) were recorded at 25 °C on a dual-beam Shimadzu UV-240IPC spectrophotometer. Cytochrome P450 concentration was determined from the absolute absorbance of the Soret band, using ϵ417 = 117 mm−1 cm−1 for the low spin ferric form of the protein (16) and then confirmed by reduced CO difference spectra taken in the presence of a ∼5-molar excess of sterol substrates (see “Results”), Δϵ450–490 = 91 mm−1 cm−1 (24). The heme content was calculated as the of ratio P450/total protein (15). The spin state of the P450 samples was estimated from the absolute absorbance spectra as the ratio (ΔA393–470/ΔA417.5–470) (25); the values 0.4 and 2.0 correspond to 100% low and 100% high spin iron, respectively. In the difference spectra, the percentage of low to high spin transition was calculated using the extinction coefficient Δϵ390–420 = 110 mm−1 cm−1. Titration with sterols was carried out at ∼1.5 μm P450 concentration in a 50 mm potassium phosphate buffer, pH 7.4, containing 200 mm NaCl and 0.1 mm EDTA. Substrate binding was monitored as a difference type I spectral response reflecting low to high spin transition of the P450 heme iron (blue shift in the Soret band maximum from 417.5 to 393 nm). Sterol substrates synthesized as described in Ref. 15 were added to the sample cuvette in the concentration range 0.25–5.0 μm from 1 mm stock solutions in 45% hydroxypropyl-β-cyclodextrin (Cyclodex) (16). The apparent dissociation constants (Kd) of the enzyme-substrate complex were calculated by plotting the absorbance changes in the difference spectra (ΔA390–420) upon titration against free ligand concentration and fitting the data to a rectangular hyperbola in Sigma plot statistics. The concentrations of free substrate were calculated using the following equation,
where [S] and [E] are the concentrations of the sterol substrates and the enzyme used for the titration, respectively; ΔA is the difference in the absorption observed at given sterol concentrations; and ΔAmax is the difference in the absorption at 100% low to high spin transition in the heme iron (26). Titration with fluconazole was carried out and quantified in a similar way. Fluconazole was added from a 1 mm stock solution in Me2SO, the inhibitor concentration range being 0.5–30 μm. Type II spectral response reflects absorbance difference between ligand-free and azole coordinated CYP51 (red shift in the Soret band maximum) producing a peak at ∼428 nm and a trough at 409 nm.
Reconstitution of CYP51 Activity
The standard reaction mixture (16) contained 1 μm L. infantum CYP51 and 3 μm T. brucei cytochrome P450 reductase. After the addition of the radiolabeled sterol substrates (15), the mixture was preincubated for 5 min at 37 °C in a shaking water bath (GCA Precision Scientific). The reaction was initiated by the addition of 100 μm NADPH and stopped by extraction of the sterols with ethyl acetate. The reaction products were dried, dissolved in methanol, and analyzed by a reverse phase HPLC system (Waters) equipped with a β-RAM detector (INUS Systems, Inc.) using a Nova Pak C18 column (particle size 4 μm, 3.9 × 150 mm) and linear gradient water:acetonitrile:methanol (1.0:4.5:4.5) (solvent A) methanol (solvent B) from 0 to 100% B for 30 min at a flow rate 1 ml/min. Retention times for Mzs, Nls, Obt, and Lns were 21, 23, 25, and 26 min, respectively. Retention time for the reaction intermediate (14α-aldehyde derivative (16)) was ∼9–12 min, depending on the sterol substrate. Time course experiments were carried out at 50 μm concentration of sterol substrates, with 250-μl aliquots being taken over time. No loss of P450 CO spectra was observed under these conditions. For steady-state kinetic analysis, reactions were run for 5 min (except for Lns, for which a 10-min reaction was used), sterol concentration range being 6–75 μm. Michaelis-Menten parameters were calculated using Sigma Plot Statistics, with the reaction rates (nmol/nmol/min) being plotted against total substrate concentration.
Crystallization, Data Collection, Structure Determination, and Analysis
The initial screening of crystallization conditions was performed using Hampton Research crystallization kits. The crystals were obtained in hanging drops by vapor diffusion at 25 °C, from equal mixtures of a 400 μm P450 solution in 20 mm potassium phosphate buffer, pH 7.2, containing 500 mm NaCl, 10% glycerol, 0.1 mm EDTA, and 5.8 mm TCEP (Hampton Research) preincubated with 1.68 mm fluconazole and 9.2 mm 3,6,9,12,15-pentaoxatricosan-1-ol (C8E5) (Anatrace) against a well solution containing 0.1 m Tris-HCl, pH 8.0, and 9% PEG 6,000. Crystals were soaked briefly in a 30% glycerol cryo-buffer and flash-cooled in liquid nitrogen. The data were collected at the Advanced Photon Source of Argonne National Laboratory (LS-CAT, Beamline 21ID-G) and processed with the HKL2000 software package. The structure was solved by molecular replacement using ligand-free T. brucei CYP51 (3g1q) as a search ensemble. Model building and refinement were performed with COOT (27) and REFMAC5 (28), respectively. Supplemental Table S1 summarizes the diffraction and refinement data statistics. The electron density map of the L. infantum CYP51 active site cavity with bound fluconazole is shown in supplemental Fig. S1. The coordinates and structure factors have been deposited at the RCSB Protein Data Bank under code 3L4D. The Protein Data Bank codes of other CYP51 structures discussed in this work are 3GW9 (T. brucei), 3K1O (T. cruzi), and 3LD6 (human). Structure superpositions were done in LSQkab of the CCP4 suite. The substrate was modeled in the active sites of the superimposed CYP51s in the orientation corresponding to that of a substrate analog inhibitor 14α-methylene-cyclopropyl-24.25-dihydrolanosterol in complex with T. brucei CYP513 using Coot. No operations on energy minimization were conducted to preserve experimentally derived atomic coordinates in the CYP51 active site cavities. Molecular volumes and surface areas were calculated in an Accelrys Discovery Studio Visualizer 2.5 (probe radius 1.4 Å). The figures were prepared with Accelrys and Chimera.
RESULTS
L. infantum CYP51 Sequence/Structure Analysis
CYP51 from L. infantum consists of 480 amino acid residues, has a molecular mass of ∼54 kDa, and has an isoelectric point of 7.7. Currently CYP51 sequences from five Leishmania species are available, and amino acid identities to that from L. infantum are: 95% in L. braziliensis, 96% in L. major, and 97% in L. mexicana and Leishmania amazonensis. The identity of L. infantum CYP51 to its orthologs in trypanosomes, T. brucei and T. cruzi, is also rather high, 76 and 75%, respectively. However, it drops to only 26% when compared with the human CYP51 and to 24–25% and 21–23% in comparison with the sequences of plant and fungal sterol 14α-demethylases, respectively. On the other hand, all 36 conserved residues, which we have predicted to play important roles in CYP51 structure/function, particularly the CYP51 B′ and I helix signatures (29, 30), are present in the leishmanial sequences. Strict structural conservation across phylogeny is seen for the length and location of the secondary structural elements (supplemental Fig. S2) as well as for the overall three-dimensional fold, so that the root mean square deviation for the Cα atoms between the L. infantum and human CYP51s is only ∼1.3 Å (supplemental Fig. S3).
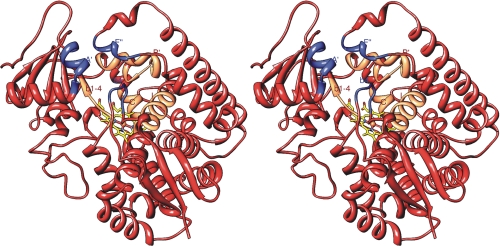
The heme iron in L. infantum CYP51 is ligated to Cys-422, and the porphyrin ring propionates are supported by six hydrogen bonds with five residues (Tyr-102 and Arg-360 with ring A and Tyr-115, Arg-123, and His-420 with ring D). As in all other (eukaryotic) CYP51s, the entrance into the substrate access channel is formed by helices A′ (which is a substrate recognition site (SRS) (31) to date unique for CYP51), F″ (SRS2), and β4 hairpin (SRS6). The substrate binding cavity is located between the distal surface of the heme, helix B′, B′/C loop (SRS1), helix C (SRS unique for CYP51), helix I (upstream of SRS4), helix K/β1–4 loop, and β strand 1–4 (SRS5) (Fig. 1 and Table 1). Quite interestingly, one C8E5 detergent molecule has been found in the asymmetric unit (supplemental Fig. S4). Surrounded by the structural elements that form the access channels entrances in all four L. infantum CYP51 molecules (helix A′ at both ends and helices F″/G′ in the middle part), the detergent appears to mimic the hydrophobic lipid environment around the substrate channel entrance.
FIGURE 1.
Substrate binding area in the structure of L. infantum CYP51. Shown is a stereo view of the three secondary structural elements forming the entrance into the substrate access channel (blue). The four elements forming the binding cavity (orange) are labeled. Fluconazole was deleted for clarity.
TABLE 1.
SRS in eukaryotic CYP51s
| Residues (L. infantum numbering) | Secondary structural elementsa |
SRS | |
|---|---|---|---|
| Substrate channel entrance | Substrate binding cavity | ||
| 45–52 | αA′ | Unique for CYP51 | |
| 101–115 | αB′-B′/C loop | SRS1 | |
| 123–129 | αC | Unique for CYP51 | |
| 207–213 | αF″ | SRS2 | |
| 283–294 | αI | SRS4 | |
| 354–360 | β1–4 | SRS5 | |
| 456–461 | β4 hairpin | SRS6 | |
a Residues from these elements can be seen on the L. infantum electron density map in supplemental Fig. S1.
Spectral Characteristics
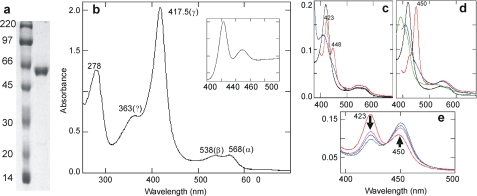
Purified L. infantum CYP51 (Fig. 2a) had a Soret (γ) maximum at 417.5 nm with a spectrophotometric index of A417.5/278 = 1.63 (specific heme content 17.7 nmol/mg of protein, which corresponds to >95% content of the P450 holoenzyme) (Fig. 2b). Based on the ratio of ΔA393–470/ΔA417.5–470 (0.42), the absence of any absorption peak in the 650-nm region, and equal intensities for the distinguishable α-(568 nm) and β-(538 nm) bands (25), the P450 is in the oxidized enzymatically active form with more than 98% of the heme iron in the low spin state.
FIGURE 2.
Purified L. infantum CYP51. a, 12% PAGE, left lane, rainbow marker (Amersham Biosciences); right lane, L. infantum CYP51 after CM-Sepharose. b–e, absolute absorbance spectra of L. infantum CYP51. b, oxidized ligand-free (17.4 μm) (inset, its reduced CO difference spectrum); c, reduction (blue) and CO complex formation (red) in a ligand-free state (black), 1.7 μm; d, reduction (blue) and CO complex formation (red) after substrate binding (green) to the ligand-free enzyme (black), 1.7 μm; e, P450 recovery when the substrate is added to the reduced CO complex (red curve in c). The arrows show the directions of changes over time.
Alterations in the heme absorbance upon addition of sodium dithionite indicate that the iron in L. infantum CYP51 can be reduced in the absence of substrate (Fig. 2c). However, the L. infantum CYP51 CO complexes revealed a predominant maximum at 423 nm (Fig. 2b, inset). Because some CYP51s display decreased stability upon reduction in the substrate-free state (32–34), the experiments were repeated in the presence of Obt (predicted as the preferred L. infantum CYP51 substrate (14)), producing highly stable absorbance spectra with the normal P450 maximum at 450 nm (Fig. 2d). Moreover, we observed that the substrate not only prevents the L. infantum CYP51 from forming P420 CO complexes when it is added before the reduction, but it also can “recover” the 450 nm peak even if added after the CO complex formation (Fig. 2e). This is the first observation of P450 recovery for a CYP51, although a reversible conversion of P420 into P450 has been seen before in other CYPs (e.g. EpoK (35) and CYP142 (36)), suggesting substrate-dependent structural rearrangements that lead to deprotonation of the heme-coordinating cysteine so that the thiolate (P450) instead of the neutral thiol (P420) (37) remains the proximal ligand upon reduction of the heme iron. More detailed studies are required to clarify the mechanism of this phenomenon in L. infantum CYP51. Here we used this substrate stabilizing effect on the L. infantum CYP51 CO complexes to confirm substrate binding (as shown below).
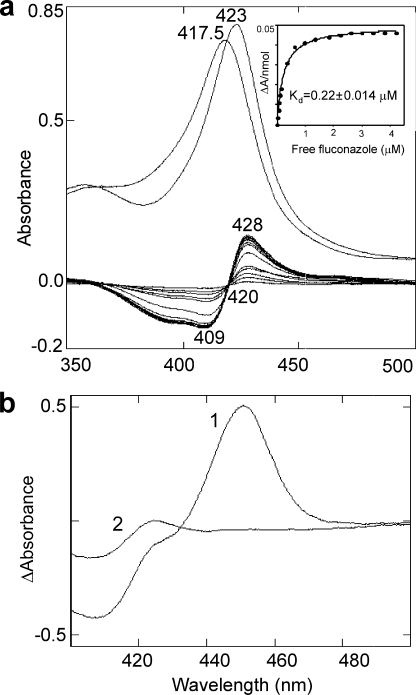
The addition of azole derivatives to oxidized L. infantum CYP51 leads to a typical sterol 14α-demethylase type II spectral response with the red shift in the Soret band maximum to 422–425 nm, depending on the inhibitor. Fig. 3a shows an example with fluconazole, an antifungal drug that has been reported to cure cutaneous leishmaniasis (10). Interestingly, when fluconazole is added to the preformed substrate-stabilized CO complexes of L. infantum CYP51, the P450 peak disappears (Fig. 3b), which indicates that the inhibitor not only replaces the substrate in the enzyme active site but most likely displaces the CO from the iron coordination sphere. On the other hand, a 10-fold molar excess of fluconazole over the enzyme, required to achieve the effect in Fig. 3b, implies that the potency of fluconazole to inhibit L. infantum CYP51 can be rather moderate. This has been proven in the reconstituted CYP51 reaction in vitro (supplemental Fig. S5). The inhibitor/enzyme ratios causing a 2-fold decrease in the CYP51 activity are 2 (initial rate) and 25 (60-min reaction), confirming our previous observation that CYP51 inhibitor potencies can be ranked by their influence on P450-CO complexes (29). Studies of the inhibitory effects of several different azole derivatives on the L. infantum enzyme activity are currently being conducted and will be published elsewhere.
FIGURE 3.
Interaction of L. infantum CYP51 (6.1 μm) with fluconazole. a, upper spectra, absolute absorbance in the Soret band region (417.5 nm, no ligand; 423 nm, plus 30 μm fluconazole). Lower spectra, difference spectra (type II response) upon titration with fluconazole. The inhibitor concentration range is 0.5–30 μm. Inset, the titration curve. b, CO difference spectra in the presence of a 2.5-fold molar excess of Nls (spectrum 1) and after addition of a 10-fold molar excess of fluconazole to spectrum 1 (spectrum 2). Absolute CO binding spectra are shown in supplemental Fig. S6.
Substrate Preferences
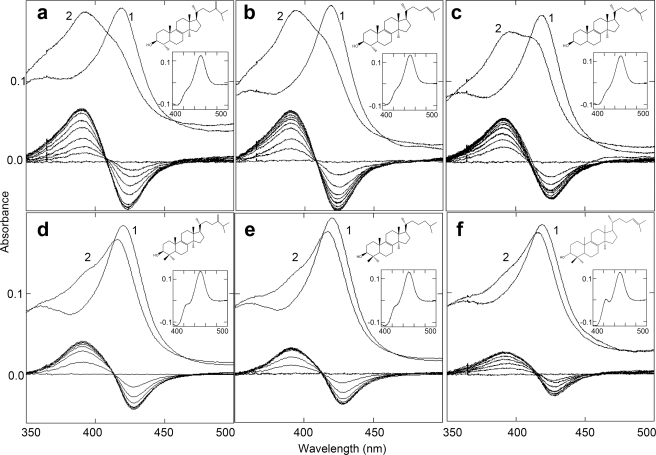
Binding of Obt to L. infantum CYP51 causes a classic type I P450 spectral response, with the peak, isosbestic point, and trough in the difference spectra at 390, 408, and 423 nm, respectively (Fig. 4 and Table 2). The sterol induces low to high spin transition in ∼95% of the ferric iron, which is the greatest high spin content ever seen in a CYP51. Previously, the highest low to high spin transition observed was 55% in the T. brucei CYP51 ortholog, also in response to Obt (16). Another protozoan CYP51 (T. cruzi) exhibits up to 35% high spin content upon binding of its preferred substrate Ebr (15), whereas spectral responses of fungal or plant CYP51s have been reported to be much weaker, usually not exceeding 10% low to high spin transition (18, 33, 34, 38, 39).
FIGURE 4.
Interaction of L. infantum CYP51 (1.5–1.6 μm) with sterol substrates. The ranges of sterol concentrations are 0.25–3.0 μm (0.25 μm increments) for Obt (a), Nls (b), and Mzs (c) and 0.5–5.0 μm (0.5 μm increments) for Ebr (d), Dhl (e), and Lns (f). In each panel, the upper spectra show the absolute absorbance in the Soret band region. Spectrum 1, ligand free; spectrum 2, after saturation with the sterol substrates. Lower spectra, difference spectra (type I response). Insets, reduced CO difference spectra; the corresponding sterol structures are shown.
TABLE 2.
L. infantum CYP51 substrate binding and catalytic parameters
| Sterol | Substrate binding (spectral response) |
Catalytic parameters |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Soret band, difference spectra (nm) |
Binding parameters |
Michaelis-Menten (substrate conversion, I+P)a |
Vmax expb |
|||||||||
| Max | Min | Isosbestic point | ΔAmax/nmol P450 | High spin content | Kd | ΔAmax/Kd | Vmax | Km | Vmax/Km | I formation | P formation | |
| o.u.c | % | μm | μm | μm | min−1 μm−1 | min−1 | min−1 | |||||
| Obt | 390 | 423 | 408 | 0.103 ± 0.005 | 94 | 0.27 ± 0.01 | 0.38 | 8.10 ± 0.32 | 11.31 ± 0.04 | 0.72 | 0.32 ± 0.01 | 7.3 ± 0.2 |
| Nls | 390 | 423 | 408 | 0.099 ± 0.005 | 90 | 0.45 ± 0.02 | 0.22 | 10.11 ± 0.41 | 18.52 ± 0.07 | 0.55 | 2.02 ± 0.04 | 6.8 ± 0.2 |
| Mzs | 391 | 425 | 411 | 0.074 ± 0.004 | 67 | 0.58 ± 0.03 | 0.13 | 1.92 ± 0.07 | 9.11 ± 0.06 | 0.21 | 1.27 ± 0.02 | 0.92 ± 0.05 |
| Lns | 391 | 428 | 413 | 0.034 ± 0.003 | 31 | 1.42 ± 0.09 | 0.02 | 0.91 ± 0.04 | 12.04 ± 0.08 | 0.07 | 0.59 ± 0.03 | 0.26 ± 0.01 |
| Ebr | 391 | 428 | 413 | 0.052 ± 0.004 | 46 | 0.91 ± 0.06 | 0.06 | NTd | NT | |||
| Dhl | 391 | 428 | 413 | 0.043 ± 0.002 | 39 | 0.82 ± 0.05 | 0.05 | NT | NT | |||
a The values were calculated from Michaelis-Menten curves.
b Experimentally reached values.
c Optical units.
d NT, not tested.
The type I response of L. infantum CYP51 to another C4-monomethylated sterol, Nls, identified in Leishmania species in vivo (21), is comparable with that induced by Obt with high spin form content reaching 90%. The slightly higher Kd value (0.45 μm versus 0.27 μm) might suggest a somewhat lower binding efficiency, especially when the ratios ΔAmax/Kd are compared.
Interestingly, we found that L. infantum CYP51 can bind Mzs, a C4-desmethylasted analog of Nls, ΔAmax/nmol = 0.074 (up to 67% high spin), Kd = 0.58 μm. This sterol has not been previously reported to interact with a CYP51 or to serve as a CYP51 substrate but was identified in Leishmania cells (40). In the Mzs-induced difference spectra, the peak, isosbestic point, and trough are all slightly red-shifted.
Finally, the C4-dimethylated sterols, Ebr, Lns, and 24,25-dihydrolanosterol (Dhl), that serve as natural CYP51 substrates in fungi, yeast, and vertebrates (41) also cause obvious type I spectral responses in the L. infantum enzyme. Although the amplitudes of the spectral change are significantly lower (corresponding to 31–46% high spin content) and all the difference spectra are red-shifted even more than in the case of Mzs, with the trough at 428 nm and the isosbestic point at 413 nm, the increase in their apparent Kd values is not large, with ∼2-fold molar excess of the sterols over the enzyme being sufficient to reach saturation. Reduced CO complexes with the P450 (450 nm) maximum clearly confirm binding of all six tested sterols to the L. infantum CYP51. This is quite opposite to the plant (18, 20, 38) and T. brucei (16) CYP51s, which display either no or very weak affinity to the C4-dimethylated sterols.
Because lower values for the amplitude of the spectral response at substrate saturation with similar apparent affinities imply the possibility of an alternative orientation (42), it appears that the CYP51 from L. infantum can accommodate each of the sterol substrates regardless of the number of the methyl groups at the C4 position. Only the equilibrium in enzyme-substrate complexes is shifted, with the portion with the substrate orientation that displaces the water molecule from the iron coordination sphere decreasing from C4-mono- to C4-des- to C4-double-methylated sterols. Although not so obvious, the binding parameters suggest that the presence of the methylene group at the C24 position (Obt and Ebr) might also be slightly preferable, although the effect could be due to higher flexibility of the side chain in Ebr and Obt in comparison with the more rigid isopropylene moiety (Δ24–25 double bond) in Lns and Nls.
Catalytic Activity
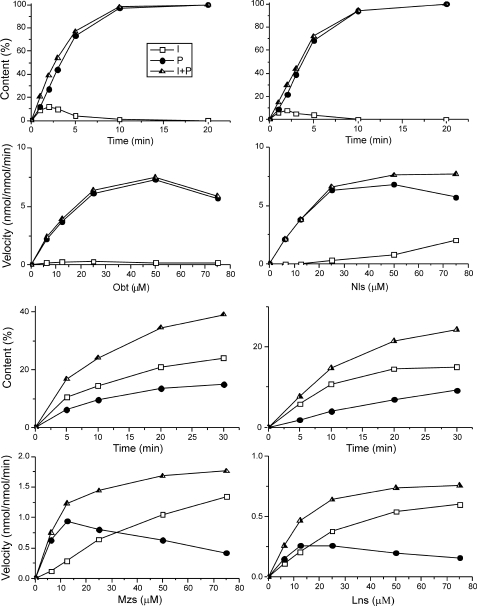
Sterol 14α-demethylation is a sequence of three stereospecific oxidations catalyzed by a single cytochrome P450 enzyme. During the three sequential catalytic turnover cycles, the 14α-methyl group of a sterol substrate is oxygenated to the 14α-alcohol and then to the 14α-aldehyde, with the third step eliminating formic acid leading to formation of the Δ14–15 double bond in the sterol core (43). In this work, four of the six sterols described in the previous section, C4-mononmethylated Obt and Nls, C4-desmethylated Mzs, and C4-dimethylated Lns, were further tested as potential substrates in the reconstituted L. infantum CYP51 reaction, and we found that the enzyme was able to metabolize all of them (Fig. 5 and Table 2). The fastest turnover number (Vmax = 10.1 min−1 for total substrate conversion) was calculated for Nls. Because of the differences in Km values, however, the highest catalytic efficiency was achieved with Obt (Vmax/Km = 0.72 versus 0.55 for Nls). For both of these substrates, only a trace of the 14α-aldehyde intermediate (16) was detected. Although the presence of this intermediate causes the slight lag of product (P) formation, its content rapidly decreases over time, implying that in the reaction with Obt and Nls, the first step of 14α-demethylation should be rate-limiting (k1 ≪ k2).
FIGURE 5.
Catalytic activity of L. infantum CYP51 with four sterol substrates. Upper graphs, time course; lower graphs, steady-state kinetics. I, the 14α-aldehyde intermediate; P, 14α-demethylated product; I+P, total content of the reaction metabolites.
Mzs was 14α-demethylated significantly more slowly (Vmax = 1.9 min−1), and the rate of 14α-demethylation for Lns was less than 10% of that of Nls/Obt (Vmax = 0.9 min−1). In both cases, the reaction equilibrium was notably shifted toward the formation of the aldehyde intermediate, with its content increasing over time. The shapes of the steady-state kinetic curves suggest that although the process of the total metabolite formation still follows the Michaelis-Menten equation (fits rectangular hyperbola), the kinetics of product release in both cases becomes quite typical for substrate inhibition. In other words, if a sterol substrate is not preferred (Mzs and Lns), the enzyme affinity to its aldehyde intermediate seems to be even lower than the affinity to the initial substrate, so that k1 ≫ k2. Therefore, higher excess of the substrate results in accumulation of the intermediate and not the reaction product.
Thus, in good agreement with the results of the titration experiments, catalytic parameters of L. infantum CYP51 confirm that the enzyme prefers C4-monomethylated sterol substrates but can metabolize C4-double and C4-desmethylated sterols and therefore displays the broadest substrate preferences among the known plant-like sterol 14α-demethylases. Catalytically preferred substrates, however, bind predominantly in a way that allows the enzyme to maintain the sterol molecule in the favorable orientation during the successive regiospecific three-step 14α-demethylation.
DISCUSSION
There are a few other P450s, all involved in steroidogenesis, that, similar to the CYP51, also catalyze three-step reactions, e.g. CYP11, CYP17, and CYP19. Whether the multistep mechanism is processive or distributive (with or without dissociation of intermediates, respectively) remains under discussion (44). Accumulation of the CYP51 reaction intermediates (predominantly the 14α-aldehyde derivative) in vitro is mostly known as a result of nonoptimal reaction conditions (pH, reducing equivalents, inhibitors) that were used in the experiments to elucidate the reaction mechanism (45, 46). In vivo, the aldehyde intermediate of the CYP51 reaction was isolated from human tissues and hypothesized to serve as a natural regulator (inhibitor) of sterol biosynthesis (47). We found that the 14α-aldehyde intermediate is the only compound that can be formed from Lns by the CYP51 ortholog from T. brucei, which we believe might also play a regulatory role in vivo by allowing the bloodstream forms of the parasite to switch the major sterol production from the endogenous carbon source to the usage of the host cholesterol (16). Leishmania cannot utilize host cholesterol for membrane biogenesis (21); therefore, it is not clear whether the 14α-aldehyde formation might take place in Leishmania species in vivo. Formation of the intermediate in vitro, inversely correlating with substrate preferences, suggests that the portion of enzyme-substrate complexes where the substrate does not induce the iron spin-state transition might be catalytically unfavorable, especially for the reaction to proceed to the final stage. It can therefore be proposed that in vivo, CYP51 most likely acts using the processive mechanism, so the 14α-demethylated product of the reaction can be included into the further conversions on the pathway of sterol biosynthesis, unless regulatory needs for the aldehyde derivative appear (16). In vitro, at optimized reaction conditions, the preferred (physiological) substrates are converted successively into the 14α-demethylated product, but for “weaker,” nonpreferred substrates, such as Mzs and Lns in the case of L. infantum CYP51, the reaction might proceed as distributive.
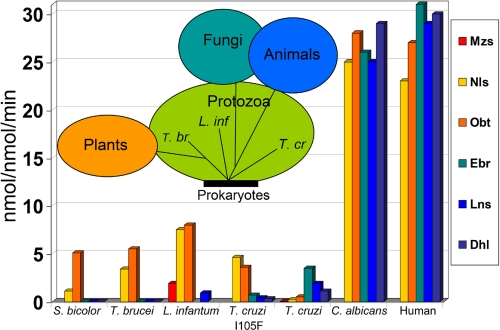
The CYP51 catalysis occurs in the endoplasmic reticulum membrane and requires a balanced ratio between the enzyme, its electron donor partner (cytochrome P450 reductase), initial substrate, lipid environment, availability of NADPH, and molecular oxygen. In vitro, reconstitution of the CYP51 activity often remains problematic; the Km and Vmax values reported from different research groups strongly depend on the reaction conditions, sometimes varying more than 3 orders of magnitude (e.g. Vmax for Candida albicans CYP51, 26 min−1 (16) versus 0.04 min−1 (33)). When catalytic parameters are determined using identical conditions (Fig. 6), their comparison becomes more meaningful. Thus, the fungal and human CYP51s are certainly the fastest and can metabolize all of the tested sterols. Plant CYP51s seem to have evolved in a direction to avoid metabolism of any C4-dimethylated sterols, which is quite reasonable because in vivo in plants (or more broadly, in photosynthetic organisms (48)), squalene cyclization into cycloartenol produces only C4-monomethylated sterol precursors. Although T. brucei as well as other Trypanosomatidae cyclize squalene into lanosterol, turnover numbers and substrate preferences of Sorghum bicolor and T. brucei CYP51s are very similar; both of them display strict requirements toward the single α-methyl group as a substituent at the C4-atom. Interestingly, the three CYP51s from Trypanosomatidae (the organisms diverged ∼2.5 billion years ago (3)), regardless of their relatively high, >75%, amino acid identity, display the greatest variety in substrate preferences within any phylum.
FIGURE 6.
Catalytic rates of CYP51 orthologs from different phyla with different sterol substrates. Rates are shown for Trypanosomatidae (L. infantum, T. brucei, and T. cruzi), plant (S. bicolor), fungi (C. albicans), and humans. Inset, CYP51 substrate preference-based phylogenetic tree.
Comparison of the CYP51 catalytic parameters across phyla predicts a phylogenetic tree with four branches (Fig. 6, inset). As suggested by Yoshida et al. (41), the root of the tree must be of prokaryotic origin, although, most likely, the closest prokaryotic ancestor of eukaryotic CYP51s has yet to be identified. To date the only biologically relevant sterol 14α-demethylase has been found in Methylococcus capsulatus (49), with 26% sequence identity to protozoan CYP51, and containing the T. cruzi-like isoleucine in the B′ helix (Ile-81 versus T. cruzi Ile-105). Among the other 11 sequenced bacterial CYP51 family members, Mycobacteria (nine sequences), Nocardia (one sequence), and Rhodococcus (one sequence) did not have sterol biosynthetic pathways. Profound structural differences of Mycobacteria tuberculosis CYP51, including a more than 4-fold larger volume of its substrate binding cavity, suggest the possibility that this P450 can accommodate alternative structures and could have acquired another function (23).
Among eukaryotic CYP51s, the T. brucei/plant branch is directed toward developing the strictest substrate requirements. The animal/fungi branch can provide the fastest sterol flow. The T. cruzi branch is currently the only example of a CYP51 expressing clear preferences toward C4-dimethylated sterols, whereas L. infantum must be placed somewhere between the T. brucei/plants and animal/fungi CYP51s. Actually, the substrate preferences and catalytic parameters of L. infantum CYP51 are closest to the I105F mutant of T. cruzi CYP51; substitution of the isoleucine in this enzyme to the plant-specific phenylalanine (I105F) sharply increases the ability of the mutant to 14α-demethylate Nls (150-fold) and Obt (60-fold) and yet does not abolish demethylation of Lns, Ebr, and Dhl (15).
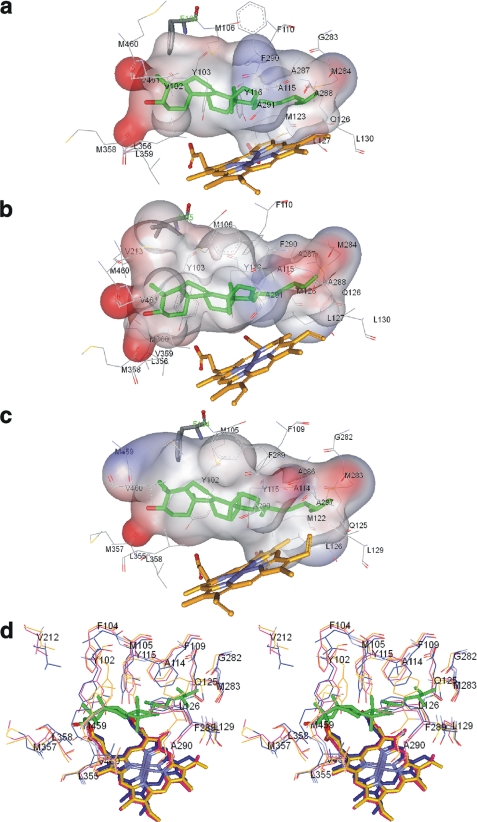
Eukaryotic CYP51s, with their striking structural similarity across phylogeny and rigidity upon ligand binding (30), proved to be very different from most other P450s, particularly xenobiotic metabolizing forms, where the structural fold is usually accepted to be highly flexible (50–53). Therefore, searching for the explanation to catalytic differences observed for the three protozoan CYP51s, we focused on the comparison of the composition of their substrate binding cavities (Fig. 7). The most obvious differences observable include volume, shape, surface area, and distribution of electrostatic potential. Active site volume and surface area, smallest in the T. brucei and largest in the T. cruzi CYP51, could well be the basis for the strictest substrate requirements exhibited by the T. brucei enzyme ortholog. Comparable cavity volumes in the T. cruzi and L. infantum CYP51 structures (both larger than that in T. brucei) can explain catalytic similarity between the L. infantum ortholog and the plant-like I105F mutant of T. cruzi CYP51: bulky phenylalanine decreases the mutant cavity, allowing it to properly position the C4-monomethylated sterol substrates. One more example in support of this assumption: L. infantum CYP51 can bind C4-dimethylated and C4-desmethylated sterols with its substrate binding cavity best “carved” for the C4-monomethylated structures. Therefore, for Nls and Obt, the arrangement around the C24 atom in the sterol arm does not matter much (Table 2). With the less favorable substrates, however, the enzyme seems to prefer a more flexible arm (Ebr versus Lns), perhaps because of its easier fit into the shape of the cavity. In the same way it explains domination of the processive mechanism, which is clearly used by L. infantum CYP51 with the preferred (C4-monomethylated) substrates: if a substrate molecule fits perfectly, it remains in a catalytic orientation during all three P450 catalytic cycles.
FIGURE 7.
CYP51 substrate binding cavities. The ligand binding surface includes protein atoms located within 4 Å from Ebr modeled into the CYP51 active site. a, T. brucei. Surface volume is 1,433 Å3, and the surface area is 866 Å2. b, T. cruzi. The surface volume is 1,560 Å3, and the surface area is 1016 Å2. c, L. infantum. The surface volume is 1,524 Å3, and the surface area is 934 Å2. Ebr (green), heme, and the substrate preference-defining residues (Phe/Ile) are presented as sticks, and the other residues located within van der Waals contacts are shown as lines. The surface is colored by electrostatic potential. d, stereo view of the active sites residues in the superimposed protozoan CYP51 structures. Magenta, L. infantum; blue, T. cruzi; orange, T. brucei. L. infantum CYP51 numbering is used. In T. cruzi CYP51, in addition to Ile-105 (Phe-104 in L. infantum), there is also a Val instead of Leu in the β1–4 strand (Leu-358 in L. infantum). All other residues in the three CYP51 orthologs are identical.
In summary, Trypanosomatidae represent the phylum where different CYP51s display the greatest differences in substrate preferences. This might result from the fact that protozoa have diverged much earlier than other eukaryotes. L. infantum CYP51 is the first example of a plant-like sterol 14α-demethylase that can also metabolize C4-double and C4-desmethyated sterol substrates (although it is not excluded that CYP51s with broader substrate preferences will be found among plant orthologs as well). Because the preferred L. infantum CYP51 substrates are C4-monomethylated, in vivo elimination of the 14α-methyl group in Leishmania most likely occurs after the first C4 demethylation. This supports the idea (9, 21) that C24 methylation in these parasites can take place both before and after the CYP51 reaction (as shown in supplemental Scheme S1). Bifurcation of sterol biosynthesis into several branches can be important in the parasites to allow the pathway to proceed as far as possible toward production of the sterols suitable as membrane components. This suggests that other enzymes in the postsqualene portion of the pathway in Leishmania, such as sterol 24C-methyltransferase and sterol C4-demethylase, may have relatively broader substrate specificity as well, and therefore combinatory therapy aimed at simultaneous inhibition of several sterol biosynthetic steps might be especially beneficial for treatment of leishmaniasis. High similarity in the overall CYP51 structure with local differences in organization of the substrate binding cavity assume that CYP51 family evolution could have been directed at preserving overall structure/function relations with concomitant introduction of minor differences in the topology of their substrate binding cavity. These minor differences are also likely to underlie the enzyme substrate preferences, catalytic turnover rate, and drug susceptibility and can be used in designing potent and specific inhibitors to cure human diseases. Relatively weak inhibition of L. infantum CYP51 by fluconazole implies that more potent inhibitors of the enzyme can be found, their curative effects for leishmaniasis significantly exceeding the effect reported (10) for fluconazole.
Supplementary Material
Acknowledgment
We thank F. Buckner (University of Washington, Seattle) for the L. infantum genomic DNA.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 067871 (to M. R. W. and G. I. L.). This work was also supported by Vanderbilt Institute of Chemical Biology Pilot Project Grant 2011 (to G. I. L.) and National Science Foundation Grant MCB-0929212 (to W. D. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Table S1, Scheme S1, and Figs. S1–S6.
The atomic coordinates and structure factors (code 3L4D) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
G. I. Lepesheva, T. Y. Hargrove, M. R. Waterman, and Z. Wawrzak, unpublished data.
- CYP
- cytochrome P450
- CYP51
- sterol 14α-demethylase
- Obt
- obtusifoliol (4α,14α-dimethyl-5α-ergosta-8,24(24′)-dien-3β-ol)
- Nls
- C4-norlanosterol (4α,14α-dimethylcholesta-8,24-dien-3β-ol)
- Lns
- lanosterol (lanosta-8,24-dien-3β-ol)
- Dhl
- 24,25-dihydrolanosterol (lanosta-8-en-3β-ol)
- Ebr
- eburicol (24-methylenelanosta-8-en-3β-ol)
- Mzs
- 14α-methylzymosterol (14α-methylcholesta-8,24-dien-3β-ol)
- SRS
- substrate recognition site.
REFERENCES
- 1. Bates P. A. (2007) Int. J. Parasitol. 37, 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodríguez N. E., Gaur U., Wilson M. E. (2006) Cell. Microbiol. 8, 1106–1120 [DOI] [PubMed] [Google Scholar]
- 3. Peacock C. S., Seeger K., Harris D., Murphy L., Ruiz J. C., Quail M. A., Peters N., Adlem E., Tivey A., Aslett M., Kerhornou A., Ivens A., Fraser A., Rajandream M. A., Carver T., Norbertczak H., Chillingworth T., Hance Z., Jagels K., Moule S., Ormond D., Rutter S., Squares R., Whitehead S., Rabbinowitsch E., Arrowsmith C., White B., Thurston S., Bringaud F., Baldauf S. L., Faulconbridge A., Jeffares D., Depledge D. P., Oyola S. O., Hilley J. D., Brito L. O., Tosi L. R., Barrell B., Cruz A. K., Mottram J. C., Smith D. F., Berriman M. (2007) Nat. Genet. 39, 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh S., Sivakumar R. (2004) J. Infect. Chemother. 10, 307–315 [DOI] [PubMed] [Google Scholar]
- 5. Bañuls A. L., Hide M., Prugnolle F. (2007) Adv. Parasitol. 64, 1–109 [DOI] [PubMed] [Google Scholar]
- 6. Murray H. W., Berman J. D., Davies C. R., Saravia N. G. (2005) Lancet 366, 1561–1577 [DOI] [PubMed] [Google Scholar]
- 7. Kedzierski L. (2010) J. Glob. Infect. Dis. 2, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balaña-Fouce R., Reguera R. M., Cubría J. C., Ordóñez D. (1998) Gen. Pharmacol. 30, 435–443 [DOI] [PubMed] [Google Scholar]
- 9. Beach D. H., Goad L. J., Holz G. G., Jr. (1988) Mol. Biochem. Parasitol. 31, 149–162 [DOI] [PubMed] [Google Scholar]
- 10. Alrajhi A. A., Ibrahim E. A., De Vol E. B., Khairat M., Faris R. M., Maguire J. H. (2002) N. Engl. J. Med. 346, 891–895 [DOI] [PubMed] [Google Scholar]
- 11. Zonios D. I., Bennett J. E. (2008) Semin. Respir. Crit. Care Med. 29, 198–210 [DOI] [PubMed] [Google Scholar]
- 12. Petrikkos G., Skiada A. (2007) Int. J. Antimicrob. Agents 30, 108–117 [DOI] [PubMed] [Google Scholar]
- 13. Lepesheva G. I., Waterman M. R. (2004) Mol. Cell. Endocrinol. 215, 165–170 [DOI] [PubMed] [Google Scholar]
- 14. Lepesheva G. I., Hargrove T. Y., Ott R. D., Nes W. D., Waterman M. R. (2006) Biochem. Soc. Trans. 34, 1161–1164 [DOI] [PubMed] [Google Scholar]
- 15. Lepesheva G. I., Zaitseva N. G., Nes W. D., Zhou W., Arase M., Liu J., Hill G. C., Waterman M. R. (2006) J. Biol. Chem. 281, 3577–3585 [DOI] [PubMed] [Google Scholar]
- 16. Lepesheva G. I., Nes W. D., Zhou W., Hill G. C., Waterman M. R. (2004) Biochemistry 43, 10789–10799 [DOI] [PubMed] [Google Scholar]
- 17. Lepesheva G. I., Waterman M. R. (2007) Biochim. Biophys. Acta 1770, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bak S., Kahn R. A., Olsen C. E., Halkier B. A. (1997) Plant J. 11, 191–201 [DOI] [PubMed] [Google Scholar]
- 19. Cabello-Hurtado F., Taton M., Forthoffer N., Kahn R., Bak S., Rahier A., Werck-Reichhart D. (1999) Eur. J. Biochem. 262, 435–446 [DOI] [PubMed] [Google Scholar]
- 20. Taton M., Rahier A. (1991) Biochem. J. 277, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haughan P. A., Goad L. J. (1991) in Biochemical Protozoology (Coombs G., North M. eds) pp. 312–328, Taylor & Francis, London [Google Scholar]
- 22. Barnes H. J., Arlotto M. P., Waterman M. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5597–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lepesheva G. I., Park H. W., Hargrove T. Y., Vanhollebeke B., Wawrzak Z., Harp J. M., Sundaramoorthy M., Nes W. D., Pays E., Chaudhuri M., Villalta F., Waterman M. R. (2010) J. Biol. Chem. 285, 1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Omura T., Sato R. (1964) J. Biol. Chem. 239, 2370–2378 [PubMed] [Google Scholar]
- 25. Lepesheva G. I., Strushkevich N. V., Usanov S. A. (1999) Biochim. Biophys. Acta 1434, 31–43 [DOI] [PubMed] [Google Scholar]
- 26. Lepesheva G. I., Virus C., Waterman M. R. (2003) Biochemistry 42, 9091–9101 [DOI] [PubMed] [Google Scholar]
- 27. Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Lepesheva G. I., Hargrove T. Y., Anderson S., Kleshchenko Y., Furtak V., Wawrzak Z., Villalta F., Waterman M. R. (2010) J. Biol. Chem. 285, 25582–25590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lepesheva G. I., Waterman M. R. (2011) Biochim. Biophys. Acta 1814, 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gotoh O. (1992) J. Biol. Chem. 267, 83–90 [PubMed] [Google Scholar]
- 32. Lepesheva G. I., Podust L. M., Bellamine A., Waterman M. R. (2001) J. Biol. Chem. 276, 28413–28420 [DOI] [PubMed] [Google Scholar]
- 33. Warrilow A. G., Martel C. M., Parker J. E., Melo N., Lamb D. C., Nes W. D., Kelly D. E., Kelly S. L. (2010) Antimicrob. Agents Chemother. 54, 4235–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Warrilow A. G., Melo N., Martel C. M., Parker J. E., Nes W. D., Kelly S. L., Kelly D. E. (2010) Antimicrob. Agents Chemother. 54, 4225–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogura H., Nishida C. R., Hoch U. R., Perera R., Dawson J. H., Ortiz de Montellano P. R. (2004) Biochemistry 43, 14712–14721 [DOI] [PubMed] [Google Scholar]
- 36. Driscoll M. D., McLean K. J., Levy C., Mast N., Pikuleva I. A., Lafite P., Rigby S. E., Leys D., Munro A. W. (2010) J. Biol. Chem. 285, 38270–38282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perera R., Sono M., Sigman J. A., Pfister T. D., Lu Y., Dawson J. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3641–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kahn R. A., Bak S., Olsen C. E., Svendsen I., Moller B. L. (1996) J. Biol. Chem. 271, 32944–32950 [DOI] [PubMed] [Google Scholar]
- 39. Yoshida Y., Aoyama Y. (1984) J. Biol. Chem. 259, 1655–1660 [PubMed] [Google Scholar]
- 40. Rodrigues J. C., Attias M., Rodriguez C., Urbina J. A., Souza W. D. (2002) Antimicrob. Agents Chemother. 46, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoshida Y., Aoyama Y., Noshiro M., Gotoh O. (2000) Biochem. Biophys. Res. Commun. 273, 799–804 [DOI] [PubMed] [Google Scholar]
- 42. Nakayama K., Puchkaev A., Pikuleva I. A. (2001) J. Biol. Chem. 276, 31459–31465 [DOI] [PubMed] [Google Scholar]
- 43. Trzaskos J. M., Bowen W. D., Shafiee A., Fischer R. T., Gaylor J. L. (1984) J. Biol. Chem. 259, 13402–13412 [PubMed] [Google Scholar]
- 44. Sohl C. D., Guengerich F. P. (2010) J. Biol. Chem. 285, 17734–17743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shafiee A., Trzaskos J. M., Paik Y. K., Gaylor J. L. (1986) J. Lipid Res. 27, 1–10 [PubMed] [Google Scholar]
- 46. Trzaskos J. M., Fischer R. T., Favata M. F. (1986) J. Biol. Chem. 261, 16937–16942 [PubMed] [Google Scholar]
- 47. Tabacik C., Aliau S., Serrou B., Crastes de Paulet A. (1981) Biochem. Biophys. Res. Commun. 101, 1087–1095 [DOI] [PubMed] [Google Scholar]
- 48. Nes W. R., McKean M. R. (eds) (1977) Biochemistry of Steroids and Other Isopentenoids, University Park Press, Baltimore [Google Scholar]
- 49. Jackson C. J., Lamb D. C., Marczylo T. H., Warrilow A. G., Manning N. J., Lowe D. J., Kelly D. E., Kelly S. L. (2002) J. Biol. Chem. 277, 46959–46965 [DOI] [PubMed] [Google Scholar]
- 50. Ekroos M., Sjögren T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Poulos T. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13121–13122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scott E. E., He Y. A., Wester M. R., White M. A., Chin C. C., Halpert J. R., Johnson E. F., Stout C. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13196–13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Williams P. A., Cosme J., Vinkovic D. M., Ward A., Angove H. C., Day P. J., Vonrhein C., Tickle I. J., Jhoti H. (2004) Science 305, 683–686 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.