Abstract
Protection against infection with Mycobacterium tuberculosis demands IFN-γ. SOCS1 has been shown to inhibit responses to IFN-γ and might thereby play a central role in the outcome of infection. We found that M. tuberculosis is a highly efficient stimulator of SOCS1 expression in murine and human macrophages and in tissues from infected mice. Surprisingly, SOCS1 reduced responses to IL-12, resulting in an impaired IFN-γ secretion by macrophages that in turn accounted for a deteriorated intracellular mycobacterial control. Despite SOCS1 expression, mycobacteria-infected macrophages responded to exogenously added IFN-γ. SOCS1 attenuated the expression of the majority of genes modulated by M. tuberculosis infection of macrophages. Using a conditional knockdown strategy in mice, we found that SOCS1 expression by macrophages hampered M. tuberculosis clearance early after infection in vivo in an IFN-γ-dependent manner. On the other hand, at later time points, SOCS1 expression by non-macrophage cells protected the host from infection-induced detrimental inflammation.
Keywords: Bacteria, Immunology, Inflammation, Interferon, Macrophages, Interleukins, SOCS, Tuberculosis
Introduction
Tuberculosis, an infectious disease caused by Mycobacterium tuberculosis, remains a leading public health problem worldwide. The global incidence of tuberculosis is rising, with 8.8 million new cases and 2 million deaths each year (1).
In most cases, the human immune system is able to control bacterial replication and prevent development of active disease. The rate of progression from colonization to disease is low, and ∼90% of infected individuals never develop clinical disease. However, M. tuberculosis is able to prevent the host immune response from totally eliminating the microorganism. Thus, the host becomes chronically infected.
A host counters mycobacterial infections primarily via Th1 immune responses involving cellular effector mechanisms, such as macrophage activation. IFN-γ is known to be an important mediator of macrophage activation and intracellular control of pathogens, including mycobacteria (2). Disseminated M. tuberculosis is observed in IFN-γ-deficient mice (3, 4). Individuals with defects in genes involved in the secretion or signaling of IFN-γ are even susceptible to weakly virulent mycobacteria (5). However, the expression of selected IFN-γ-inducible genes is paradoxically decreased in macrophages infected with mycobacteria (6–8). It remains unclear how mycobacteria-induced signaling contributes to macrophage dysfunction.
Immune and inflammatory systems are controlled by multiple cytokines, including interleukins and interferons. Many cytokines exert their biological function through Janus kinases (JAK) and signal transducers and activators of transcription (STAT). SOCS (suppressor of cytokine signaling) is a family of eight intracellular proteins. SOCS proteins function in a negative feedback loop to inhibit cytokine signaling by binding to either JAK or the cytokine receptor, either inhibiting JAK activity directly or targeting the receptor complex for ubiquitination and subsequent proteasome-mediated degradation (9).
SOCS1, one of the better described of the family members, has been reported to inhibit STAT1-mediated responses (10, 11). The vital importance of SOCS1 is stressed by the fact that SOCS1−/− mice die within 3 weeks after birth with severe lymphopenia, necrosis of the liver, and mononuclear infiltration of several organs (12, 13). The neonatal defects exhibited by SOCS1−/− mice appear to be due to increased production of IFN-γ by T and NKT cells and uncontrolled IFN-γ signaling in myeloid cells (12–14). Accordingly, T cell activation and differentiation is also regulated by SOCS1 (15, 16). Besides regulating IFN-γ signaling, SOCS1 is crucial in attenuating STAT1-mediated IFN-α/β signaling (17) and has also been shown to attenuate IL-12 (18), IL-4 (19), and other γc-dependent cytokine (20) signaling in myeloid and lymphoid cells.
SOCS1 is induced during infection of human macrophages with Mycobacterium avium (21) and in J774 cells infected with BCG3 (22). Knockdown of SOCS1 with shRNA has been recently shown to improve mycobacterial clearance in peripheral blood mononuclear cells (23). Given the indicated role of IFN-γ in the control of mycobacterial infections, we hypothesized that SOCS1 could play a central role by fine tuning the balance between control of infection and generation of pathology during M. tuberculosis infection.
We studied the regulation and role of SOCS1 during mycobacterial infection of human and mouse macrophages as well as during infection in vivo. Infection efficiently stimulated SOCS1 expression in macrophages. Surprisingly, SOCS1 expression by macrophages regulated IL-12 rather than IFN-γ responses, resulting in diminished IFN-γ secretion and a less efficient bacterial control in vitro and early after infection of mice in vivo. Later after infection in vivo, SOCS1 expression by non-macrophage cells hampered a severe infection-induced inflammation.
EXPERIMENTAL PROCEDURES
Ethics Statement
All animal experiments were conducted in accordance with the guidelines of Karolinska Institutet and were approved by Stockholm's District Ethical Committee of Animal Research, permit numbers 302/10 and 415/08.
Mice
Mutant mouse strains with genomic deficiency in SOCS1 (24), MyD88 (25), IRF3 (26), TLR2 (27), TLR4 (28), NOD2 (29), IFN-γ (4), IFN-γR (30), IFN-α/βR (31), and RAG1 (32) were generated by homologous recombination in embryonic stem cells. Animals were bred and kept under specific pathogen-free conditions. All mice were backcrossed to C57Bl/6 genetic background that was used as a control.
RAG1−/−/SOCS1−/− mice were obtained by crossing RAG1−/− and SOCS1−/+ mice (33). Similarly, IFN-γ−/−/SOCS1−/− mice were generated by crossing of IFN-γ−/− and SOCS1+/− mice.
Tissue-specific SOCS1-deficient mice were generated using the cre/loxP system by breeding SOCS1fl/fl mice, which carry a SOCS1 allele flanked by loxP sites (34), with mice expressing cre under the endogenous lysozyme M promoter (LysM-cre) (35), resulting in mice in which floxed SOCS1 was deleted in myeloid cells. SOCS1fl/fl mice were generated on a C57BL/6 genetic background, whereas all other mice were 5th to 10th generation backcrosses to C57BL/6. SOCS1fl/fl littermates were used as controls in our experiments. The deletion of the SOCS1 gene in macrophages but not in T cells from SOCS1fl/fl LysM cre mice was confirmed by PCR analysis (data not shown).
Generation of Mouse Bone Marrow-derived Macrophages and Dendritic Cells
Mouse bone marrow-derived macrophages (BMM) and dendritic cells (BMDC) were differentiated as described previously (36).
Generation of Human Monocyte-derived Macrophages
CD14+ cells were isolated from peripheral blood from healthy donors by Ficoll-Hypaque, selected with anti-CD14 MACS beads (Miltenyi Biotech, Auburn, CA), and cultured in presence of GM-CSF as described (37).
Infection and Infectivity Assay
BCG Montreal and M. tuberculosis Harlingen and H37Rv were grown in Middlebrook 7H9 (Difco) supplemented with albumin, dextrose, catalase, and, for BCG cultures, 50 μg/ml hygromycin (Sigma).
BMM and BMDC were infected at the indicated MOI, and after 2 h, cells were washed twice with PBS to remove extracellular bacteria. Mice were infected intravenously with 1 × 106 BCG bacteria.
M. tuberculosis Harlingen strain was inoculated by the aerosol route using a nose-only exposure unit (Intox Products, Albuquerque, NM) as described previously (38). A 15-ml suspension of 0.5 × 106 M. tuberculosis was loaded into a nebulizer, and animals were exposed to the bacterial aerosol for 20 min. Bacteria were quantified on Middlebrook 7H11 agar containing a 10% enrichment of oleic acid, albumin, dextrose, catalase, 5 μg/ml amphotericin B, and 8 μg/ml polymyxin B grown for 3 weeks at 37 °C.
Real-time PCR
Transcripts were quantified by real-time PCR as described previously (39). The primer sequences used are shown in the supplemental Experimental Procedures.
Hprt was used as a control gene to calculate the ΔCt values for individual samples. The relative amount of cytokine/Hprt transcripts was calculated using the 2−(ΔΔCt) method as described. These values were then used to calculate the relative expression of cytokine mRNA in uninfected and infected cells and tissues.
Histopathological Analysis
Formalin-fixed samples of left lungs of mice experimentally inoculated with M. tuberculosis were blocked on paraffin. From each lung sample, four sections were obtained, one longitudinal along the long axis of the lobe and three across/transversal of the remaining piece of lung.
The blocks were processed, and sections were stained with hematoxylin-eosin. All sections were interpreted by the same pathologist and scored semiquantitatively, blinded to the variables of the experiment. The following features were scored: 1) lung area occupied with granulomas (percentage of the total area of the section); 2) lung area free of lesions or area of healthy lung (percentage of the total area of the section); 3) abundance of lymphocytes within the tuberculous lesions, ranging from 0 (not observed) to 4 (very large aggregates, often forming nodules); 4) abundance of lymphocytes in perivascular cuffs, ranging from 0 (not observed) to 4 (very thick lymphocytic cuffs); and 5) extension of necrosis, raging from 0 (no necrosis observed) to 4 (extensive necrosis and necrotic centers with mineralization).
Western Blotting
Uninfected and BCG or M. tuberculosis-infected BMM were lysed and separated on 10% separating, 5% stacking SDS-polyacrylamide gels as described (15). Samples were then transferred onto nitrocellulose membranes (Bio-Rad) by electroblotting at 100 V, 250 mA for 80 min. Immunostaining was performed using polyclonal rabbit anti-phosphorylated (Tyr701) STAT1, total STAT1, or anti-actin (1:500 dilution; Sigma). Membranes were then washed and incubated with horseradish peroxidase (HRP)-conjugated polyclonal goat anti-rabbit immunoglobulin (1:2000 dilution; DAKO, Glostrup, Denmark) and developed using ECL-Plus (Amersham Biosciences) and photographed using a Fuji intelligent dark box II digital camera.
Microarray Analysis
The RNA was labeled using Quick-Amp labeling kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. The Cy3-labeled cRNA was hybridized to Whole Mouse Genome Microarrays (Agilent), and the Agilent “Feature Extraction” software was used to extract data. The analysis included four arrays on a chip per sample. The data array consisted of infected or uninfected WT and SOCS1−/− BMM. The data were normalized in GeneSpring GX by setting threshold raw signal to 1 for the median of control samples. Base line was set to the median of all the samples. Of 41,252 probes, 37,469 probes passed filtered on expression 20–100th percentile in the raw data, of which 30,127 were flagged by software as present or marginally present in at least one condition. The microarray data were deposited in the GEO data base, accession number GSE23508.
Enzyme-linked Immunosorbent Spot Assay
For the IFN-γ ELISPOT assay, 105 BMDC were infected with BCG and plated in triplicates in 96-well nitrocellulose-bottomed plates previously coated with 5 μg/ml anti-mouse IFN-γ mAb AN18 (MabTech AB, Sweden). Plates were incubated for 20 h at 37 °C. Biotinylated detector antibody (R4-6A2, MabTech) was added for 2 h and incubated at room temperature. Afterward, plates were washed and incubated for 1 h in the dark with 100 μl of avidin-peroxidase complexes (ABC-elite kit, Vector Laboratories, Burlingame, CA). The spots were developed by adding 20 mg of 3-amino-9-ethyl-carbozole (Sigma) dissolved in 2.5 ml of dimethylformamide in 47.5 ml of acetate buffer containing 0.015% H2O2.
Intracellular Cytokine Staining
Lungs were perfused with PBS through the heart before removal from mice. Following digestion with collagenase D and DNase I, erythrocytes were lysed, and single-cell suspensions were prepared by filtering lung tissue through 40-μm nylon cell strainers. Single spleen cell suspensions were obtained, and red blood cells were lysed. 106 cells were stimulated with 20 μg/ml PPD (Statens Serum Institute, Copenhagen, Denmark) overnight, followed by a 6-h incubation with brefeldin A (5 μg/ml). To determine the percentages of IFN-γ-secreting T-cells, we first stained with anti-mouse CD4 eFluor®450 (eBioscience, San Diego, CA) and CD3ϵ PerCP (BD Pharmingen, San Diego, CA) and then permeabilized the cells using leukocytic permeabilization reagent IntraPrepTM (Beckman Coulter) and stained them with anti-IFN-γ APC (eBioscience). Data were acquired in a CyAnTM ADP flow cytometer (Beckman Coulter) and analyzed using FlowJo software (Tree Star Inc., Ashland, OR).
RESULTS
Expression of SOCS1 mRNA in BCG-infected Macrophages Requires Phagocytosis and the Presence of Innate TLR, NOD2, and IFN-α/β Receptors
In a first set of experiments, regulation of SOCS1 gene expression in macrophages infected with mycobacteria was studied. SOCS1 mRNA accumulation increased in BMM infected with BCG (Fig. 1A). In order to determine whether phagocytosis was required for SOCS1 expression, macrophages were treated with cytochalasin D, an inhibitor of actin polymerization. Cytochalasin D-treated BMM showed lower SOCS1 mRNA levels than untreated controls (Fig. 1A). As a control, similar levels of SOCS1 mRNA after incubation with a TLR agonist binding to cell surface receptors were measured in cells treated or not with cytochalasin D (supplemental Fig. S1A).
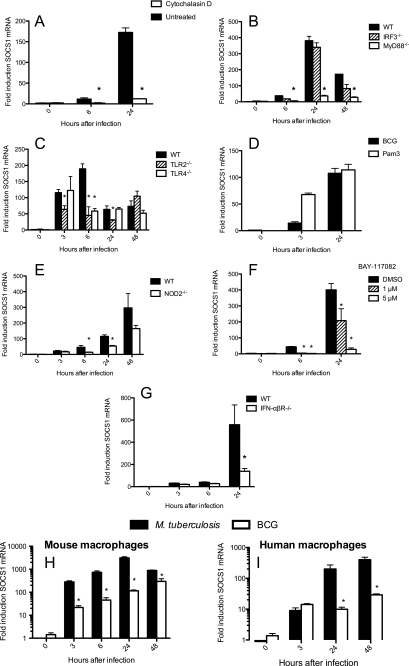
FIGURE 1.
The expression of SOCS1 mRNA in BCG-infected macrophages requires phagocytosis and the presence of innate TLR, NOD2, and IFN-α/β receptors. Mouse BMM (A–H) or monocyte-derived human macrophages (I) were infected with BCG (A–I) or M. tuberculosis (H and I). BMM were treated or not with 5 μm cytochalasin D (A) or with the indicated concentrations of BAY-117082 (F) 1 h before BCG (A–I) or M. tuberculosis (H and I) infection. Total RNA was isolated from IRF3−/− (B), MyD88−/− (B), TLR2−/− (C), TLR4−/− (C), NOD2−/− (E), IFN-α/βR−/− (G), and WT BMM (A–H) as well as from human macrophages (I) at the indicated times after infection with BCG. A MOI of 5:1 was used all over. Total RNA was also isolated after incubation of uninfected WT BMM with 1 μg/ml Pam3 (D). The accumulation of SOCS1 (A–I) and Hprt mRNA was measured by real time PCR. Duplicate determinations SOCS1 and Hprt mRNA were measured in triplicate samples for each group and time point. The mean -fold induction ± S.E. (error bars) is depicted. *, differences with WT (B, C, E, and G) or untreated (A and F) BMM are significant (p < 0.05, Student's t test). Differences with M. tuberculosis-infected mouse (H) or human (I) macrophages are significant (p < 0.05, Student's t test).
We then asked whether MyD88- and IRF3-mediated intracellular signaling pathways are involved in the augmented SOCS1 mRNA levels after infection with BCG. Similar SOCS1 mRNA levels were found in BCG-infected IRF3−/− and wild type (WT) BMM, whereas infected MyD88−/− BMM displayed significantly lower SOCS1 mRNA levels than controls (Fig. 1B). Because TLR2 and -4 have been shown to recognize mycobacterial components and mediate MyD88-dependent macrophage responses to infection (40–42), the role of these TLR in SOCS1 regulation during mycobacterial infection was studied. SOCS1 mRNA levels were diminished in TLR2−/− BMM, albeit to a lower degree than in MyD88−/− BMM (Fig. 1C). In line with this, stimulation of BMM with the TLR2 agonist Pam3 was sufficient to induce SOCS1 mRNA (Fig. 1D). In contrast, TLR4 seemed to play a minor role if any in regulation of SOCS1 mRNA levels in BCG-infected macrophages (Fig. 1C).
The NOD2 pathway is also involved in mycobacterial recognition (43). Diminished levels of SOCS1 mRNA were observed in NOD2−/− BCG-infected BMM, compared with WT controls (Fig. 1E).
Both TLR and NOD pathways will trigger activation of NF-κB. Co-incubation of BCG-infected BMM with BAY 11-7082, a pharmacological inhibitor of IκB-α phosphorylation, reduced the relative levels of SOCS1 mRNA (Fig. 1F). As expected, the NF-κB-dependent MCP-1 but not IFN-α mRNA levels were reduced in BAY 11-7082-treated BCG-infected BMM (supplemental Fig. S1B) (data not shown). IFN-α/β are also involved in SOCS1 mRNA expression during infection because IFN-α/βR−/− BMM showed lower levels of SOCS1 compared with WT controls (Fig. 1G).
Induction of SOCS1 mRNA after infection with BCG and M. tuberculosis was then compared. Murine BMM and human monocyte-derived macrophages expressed higher titers of SOCS1 mRNA after infection with M. tuberculosis than BCG (Fig. 1, H and I).
SOCS1 Hampers Efficient Growth Control of Intracellular BCG and M. tuberculosis by Macrophages
Next, we investigated whether SOCS1 plays a role in intracellular growth control of BCG. SOCS1−/− and WT BMM were infected with BCG. SOCS1−/− BMM infected at different MOI showed reduced bacterial load (Fig. 2, A–C) compared with WT controls. Accordingly, SOCS1−/− BMM showed lower levels of M. tuberculosis H37Rv or Harlingen than WT cells (Fig. 2, D and E). Similar levels of SOCS1−/− and WT BMM death were recorded during M. tuberculosis infection as measured by release of lactate dehydrogenase into the medium (Fig. 2F).
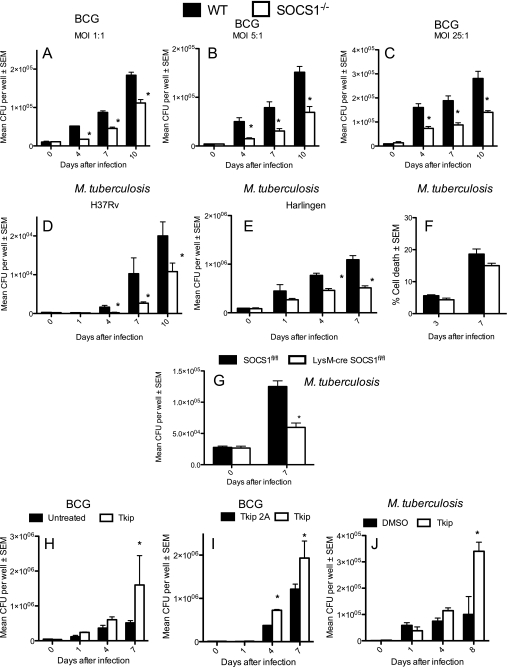
FIGURE 2.
SOCS1 hinders growth control of BCG and M. tuberculosis by macrophages. RAG1−/−/SOCS1+/+ and RAG1−/−/SOCS1−/− BMM, (indicated hereafter as WT and SOCS1−/−) were infected with different MOI of BCG (A–C) or M. tuberculosis H37Rv (D) or Harlingen (E) at an MOI of 5:1, washed after 2 h, and lysed with PBS-Triton buffer at the indicated time points after infection. The cfu were determined in triplicate cell cultures. At least two independent experiments for each panel were performed. The content of lactate dehydrogenase in supernatants from M. tuberculosis Harlingen-infected SOCS1−/− and WT BMM at an MOI of 5:1 was measured as assessment of cell damage. The mean percentage lysis ± S.E. (error bars) in triplicate cultures with respect to cells incubated with 1% Triton X-100 is depicted (F). LysM-cre SOCS1fl/fl and SOCS1fl/fl BMM were infected with M. tuberculosis in triplicate cell cultures at an MOI of 1:1. The cfu in lysates were determined. At least two independent experiments for each panel were performed (G). BMM were treated with 30 μm lipophilic JAK2 tyrosine kinase inhibitor peptide (Tkip) (H–J) or with a Tkip2A (I), an alanine substitution-containing mutant form of Tkip that does not show any biological activity. Peptides were added 30 min before infection with BCG (H and I) or M. tuberculosis H37Rv (J) and replenished after cells were extensively washed, 2 h after bacterial co-incubation. Control infected cells were incubated with DMSO (H and J). The mean cfu ± S.E. were determined in triplicate cell cultures from at least two independent experiments. *, differences with control BMM are significant (p < 0.05, Student's t test).
The SOCS1 gene was deleted in macrophages and neutrophils by crossing SOCS1fl/fl mice with LysM-cre SOCS1fl/fl mice. Infected LysM-cre SOCS1fl/fl BMM also showed reduced M. tuberculosis levels compared with SOCS1fl/fl controls (Fig. 2G). SOCS1 mRNA levels in LysM-cre SOCS1fl/fl were 40–300-fold lower than the controls. The remnant expression is probably due to non-macrophage cells in the BMM cultures or to incomplete deletion of SOCS1 in LysM-cre SOCS1fl/fl BMM (supplemental Fig. S2A). Altogether, we confirm that SOCS1-silenced BMM show decreased M. tuberculosis levels.
In agreement, increased bacterial growth was observed upon incubation of BCG or M. tuberculosis-infected BMM with Tkip, a 12-mer peptide that, similar to SOCS1, binds and inhibits autophosphorylation of the JAK2 kinase and phosphorylation of the intracellular IFN-γ receptor (44) (Fig. 2, H–J). BMM treated with a Tkip-related peptide (alanine substituted by phenylalanine at positions 8 and 11, abrogating thereby binding to JAK2 (45)) showed lower bacterial levels than those treated with the native peptide (Fig. 2I). As shown previously (44), incubation of BMM with Tkip during stimulation with recombinant IFN-γ diminished pSTAT1 levels (data not shown).
SOCS1 Inhibits IFN-γ Secretion and Thereby Precludes Efficient Clearance of BCG and M. tuberculosis in Macrophages
BCG-infected SOCS1−/− BMM (supplemental Fig. S2, B–D) and BMDC (supplemental Fig. S2, E–G) showed increased levels of IFN-β (supplemental Fig. S2, B and E), IFN-α (supplemental Fig. S2, C and F) and IFN-γ (supplemental Fig. S2, D and G) mRNA as compared with WT BMM. Also, the frequency of IFN-γ-secreting BCG-infected SOCS1−/− BMDC was higher than WT controls (supplemental Fig. S2H).
Because IFN-γ controls macrophage activation and mycobacterial growth, we studied whether the improved mycobacterial growth control in SOCS1−/− BMM is IFN-γ-dependent. For this purpose, titers of M. tuberculosis and BCG in IFN-γ−/−/SOCS1−/− and IFN-γ−/− BMM were compared. IFN-γ−/−/SOCS1−/− and IFN-γ−/− BMM showed similar BCG loads (Fig. 3A and supplemental Fig. S2I). Moreover, similar bacterial levels were measured in WT, IFN-γ−/−, and IFN-γ−/−/SOCS1−/− M. tuberculosis-infected BMM, whereas the bacterial load in SOCS1−/− BMM was lower compared with all other groups (Fig. 3B). Altogether, these results indicate that SOCS1 inhibits secretion of IFN-γ that otherwise would mediate a more effective clearance of mycobacteria in macrophages.
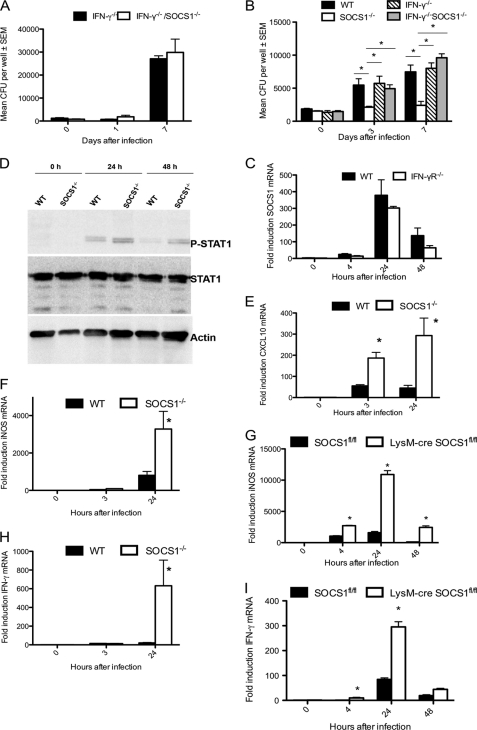
FIGURE 3.
SOCS1 hinders clearance of M. tuberculosis by macrophages in an IFN-γ-mediated manner. IFN-γ−/− and IFN-γ−/−/SOCS1−/− BMM were infected with BCG at MOI 1:1 (A). WT, SOCS1−/−, IFN-γ−/− and IFN-γ−/−/SOCS1−/− BMM were infected with M. tuberculosis H37Rv at an MOI of 1:1 (B). The cfu were determined in lysates from triplicate cultures (A and B). The mean cfu/well ± S.E. (error bars) from one of two independent experiments is depicted. Total RNA was isolated from IFN-γR−/− and WT BMM at different time points after infection with M. tuberculosis H37Rv at an MOI of 5:1 (C). The accumulation of SOCS1 and Hprt mRNA was measured by real-time PCR. Protein extracts from WT and SOCS1−/− BMM were prepared at the indicated time points after infection with M. tuberculosis and separated by SDS-PAGE, and phosphorylated STAT1, total STAT1, and actin were detected by Western blot (D). Total RNA was extracted from SOCS1−/− and WT (E, F, and H) or LysM-cre SOCS1fl/fl and SOCS1fl/fl (G and I) BMM at the indicated time points after infection with M. tuberculosis H37Rv. The relative concentrations of CXCL10 (E), iNOS (F and G), IFN-γ (H and I), and Hprt mRNA were determined by real-time PCR. The mean -fold induction of these transcripts ± S.E. is depicted. *, differences with control BMM are significant (p < 0.05 Student's t test).
Whether infection-stimulated SOCS1 expression is IFN-γ-dependent was next investigated. We found that IFN-γ was not required for SOCS1 expression because similar levels of SOCS1 mRNA were detected in BCG- or M. tuberculosis-infected WT and IFN-γR−/−-deficient BMM (Fig. 3C and supplemental Fig. S2J).
Infection of BMM with M. tuberculosis led to STAT1 activation as measured by phosphorylation of the transcription factor. Higher levels of pSTAT1 were observed in SOCS1−/− compared with WT BMM when measured 24 and 48 h after infection (Fig. 3D). In agreement with higher levels of STAT1 activation, the titer of the STAT1-regulated CXCL10 and iNOS transcripts were increased in M. tuberculosis-infected SOCS1−/− as compared with respective controls (Fig. 3, E and F). Moreover and similar to observations in BCG-infected BMM, the level of IFN-γ mRNA was augmented in M. tuberculosis-infected SOCS1−/− BMM (Fig. 3H). Both iNOS and IFN-γ mRNA levels were higher in LysM-cre SOCS1fl/fl BMM compared with controls (Fig. 3, G and I), confirming that SOCS1 expression in macrophages hampers IFN-γ expression.
Because IL-12 is known to be a main stimulus for IFN-γ secretion, we next investigated whether an M. tuberculosis-induced SOCS1-diminished expression of IL-12 could account for the increased IFN-γ secretion in SOCS1−/− BMM. However, similar mRNA levels of IL-12 p35 and IL-12 p40, constituents of biologically active IL-12 dimers, were found in M. tuberculosis-infected SOCS1−/− and WT BMM (Fig. 4, A and B), suggesting that the response to but not the secretion of IL-12 could account for the increased IFN-γ release in SOCS1−/− BMM. In order to test this hypothesis, the concentration of IFN-γ mRNA and protein was measured in WT and SOCS1−/− BMM stimulated with recombinant IL-12. SOCS1−/− BMM stimulated with IL-12 contained enhanced IFN-γ mRNA and protein levels compared with WT controls (Fig. 4, C and D).
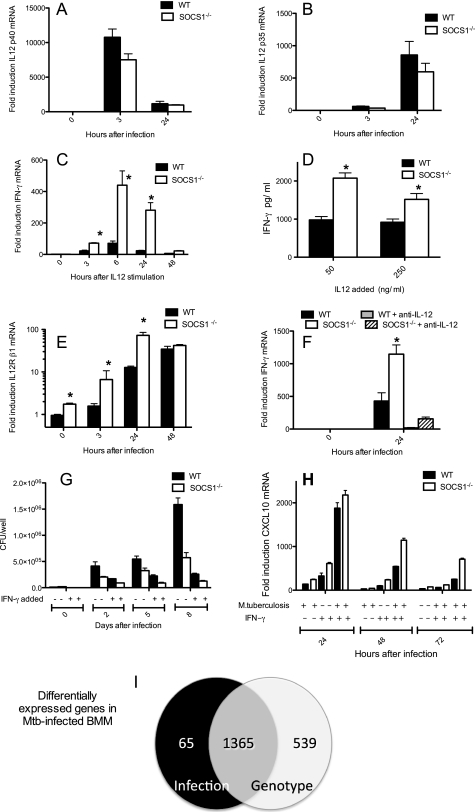
FIGURE 4.
SOCS1 inhibits response to but not the secretion of IL-12, does not hamper responses to IFN-γ, and regulates the global transcriptional responses to infection of macrophages with M. tuberculosis. Total RNA was extracted from SOCS1−/− and WT BMM at the indicated time points after infection with M. tuberculosis H37Rv (A, B, E, and F) or after stimulation with 20 ng/ml recombinant IL-12 p70 (C). The accumulation of IL-12 p40 (A), IL-12 p35 (B), IFN-γ (C and F), IL-12Rβ1 (E), and Hprt mRNA was measured by real time PCR. The mean -fold accumulation of the transcripts of triplicate cultures per time point in relation to Hprt ± S.E. (error bars) is depicted. The level of IFN-γ in the supernatant of triplicate cultures of IL-12-stimulated SOCS1−/− or WT BMM was measured by ELISA (D). *, differences with control BMM are significant (p < 0.05, Student's t test). The levels of IFN-γ mRNA in SOCS1−/− and WT BMM incubated or not with 10 μg/ml anti-p40/p70 IL-12-neutralizing antibodies (BD Biosciences) 1 h before infection with M. tuberculosis (F) were determined as described above. Twenty-four h after infection with M. tuberculosis, SOCS1−/− or WT BMM were co-incubated with 100 units of recombinant IFN-γ or left untreated. The number of cfu ± S.E. in lysates from triplicate cultures at each time point after infection is depicted (G). SOCS1−/− and WT BMM were treated with IFN-γ 24 h after infection with M. tuberculosis. Infected and mock controls were harvested at the indicated time points after infection. The accumulation of CXCL10 (G) and Hprt mRNA was measured by real-time PCR. The mean -fold accumulation of CXCL10 mRNA of triplicate cultures per time point in relation to Hprt ± S.E. is depicted (H). Differences between IFN-γ-treated and untreated cells are significant (p < 0.05, Student's t test). To obtain the microarray data, RNA was isolated from M. tuberculosis-infected or uninfected, WT or SOCS1−/− BMM cultures, with four independent samples in each group. The Venn diagram illustrates the number of genes altered by pathogen exposure (independently of the genotype) or differentially expressed by SOCS1−/− and WT BMM (independently of infection) (I). The statistical significance of differentially expressed probes was identified by two-way analysis of variance (p < 0.05), including the Benjamini and Hochberg false discovery rate correction (5%).
The increased response to IL-12 of SOCS1−/− BMM was associated with higher levels of the IL-12Rβ1 (IL-12 receptor β1) but not IL-12Rβ2 mRNA in both M. tuberculosis-infected and uninfected SOCS1−/− compared with WT BMM (Fig. 4E) (data not shown).
The effect of IL-12 neutralization in IFN-γ expression by M. tuberculosis-infected SOCS1−/− BMM was then studied. The addition of anti-IL-12 antibodies reduced IFN-γ mRNA levels in M. tuberculosis-infected SOCS1−/− and WT BMM (Fig. 4F), indicating that IL-12 is required for the enhanced IFN-γ secretion by the infected SOCS1−/− BMM.
We then studied if the increased responses to IL-12 of SOCS1−/− BMM are due to uncontrolled IL-12 signaling and/or to increased IL-12R expression. We found that IL-12Rβ1 mRNA accumulation in M. tuberculosis-infected IFN-γ−/− and IFN-γ−/−/SOCS1−/− BMM was similar (supplemental Fig. S3A). This result suggests that hyperresponses to IL-12 during infection of SOCS1−/− BMM are primarily due to an intrinsically increased IL-12 signaling because IL-12Rβ1 expression is IFN-γ-dependent.
Whether SOCS1 further impedes the responses of M. tuberculosis-infected BMM to IFN-γ was then studied. Incubation of either SOCS1−/− or WT BMM with IFN-γ 24 h after infection with M. tuberculosis resulted in diminished bacterial load as compared with respective IFN-γ-untreated infected controls (Fig. 4G). Endorsing the previous result, CXCL10 or iNOS mRNA levels were increased in IFN-γ-treated, infected WT or SOCS1−/− BMM cells as compared with uninfected controls or with infected cells in the absence of IFN-γ stimulation. Uninfected SOCS1−/− BMM showed higher levels of CXCL10 mRNA after stimulation with IFN-γ as compared with WT controls (Fig. 5H) (data not shown). NO is generated by iNOS, which requires both NF-κB and STAT1 for its activation, explaining thereby the lack of iNOS mRNA expression in uninfected, IFN-γ-treated WT or SOCS1−/− BMM (supplemental Fig. S3C). Of importance, levels of CXCL10 and iNOS mRNA were higher in SOCS1−/− BMM stimulated or not with IFN-γ 3 or 24 h after infection with M. tuberculosis, as compared with the respective WT control (Fig. 4H and supplemental Fig. S3, B and C).
FIGURE 5.
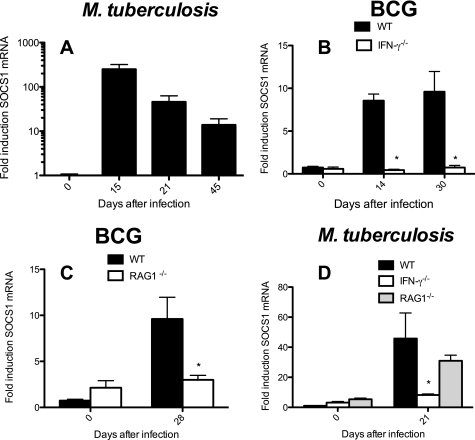
IFN-γ-dependent control of SOCS1 mRNA levels in lungs from mice infected with M. tuberculosis or BCG. Total RNA was extracted from lungs of individual WT (A–D), IFN-γ−/− (B and D), RAG1−/− (C and D) mice after infection intravenously with 106 BCG (B and C) or via the aerosol route with 250 M. tuberculosis Harlingen strain (D). The mean -fold accumulation of SOCS1 transcripts ± S.E. (error bars) in lungs from infected mice (n ≥ 5 mice/infected group) is depicted. *, differences with WT-infected mice in B–D are significant (p < 0.05, Student's t test).
SOCS1 Regulates the Macrophage Global Transcriptional Responses to Infection with M. tuberculosis
To investigate how SOCS1 shapes the macrophage response to M. tuberculosis at the transcriptome level, a genome-wide expression analysis was performed. For this purpose, total RNA was isolated from M. tuberculosis-infected or uninfected SOCS1−/− and WT BMM, and cRNA was transcribed, labeled, and hybridized to genome-wide high density microarrays. The expression levels of 1430 genes, or 6.1% of the mouse genome, differed in the infected compared with uninfected BMM, independent of their genotype. From these genes, 1365 tally within the 1904 genes differentially regulated in SOCS1−/− compared with WT BMM (independent of their infection status) (Fig. 4I). The majority, 987 genes, showed diminished levels of expression in SOCS1−/−-infected compared with WT-infected BMM, whereas 371 were increased in SOCS1−/−-infected BMM. Moreover, titers of 1804 genes diverged in SOCS1−/− compared with WT-infected BMM. 1306 genes from this subgroup were regulated by M. tuberculosis infection and the genotype.
Differences in levels of 40 IFN-regulated transcripts were found in SOCS1−/−- compared with WT-infected BMM (Table 1). Thirty-eight of these transcripts, involved in antigen presentation, chemotaxis, and effector mechanisms, were increased in the SOCS1−/−-infected BMM, whereas only two were found to be reduced (Table 1). IFN-γ- and IFN-β-regulated as well as IFN-α/β-regulated transcripts were increased in M. tuberculosis-infected SOCS1−/− BMM (Table 1). The differences recorded are a consequence of M. tuberculosis infection because levels of these transcripts in WT and SOCS1−/− BMM before infection were similar. Moreover, M. tuberculosis induced expression of the selected IFN-regulated genes because 34 of these transcripts were increased in infected compared with uninfected WT BMM, whereas none decreased after infection (Table 1).
TABLE 1.
IFN-regulated genes in M. tuberculosis-infected SOCS1−/− and WT BMM
Shown are genes significantly increased or decreased due to the genotype and the infection (two-way analysis of variance, p < 0.05) and further selected for a -fold difference between M. tuberculosis-infected SOCS1−/− and WT BMM of ≥2. The selection was made with regard to the GO annotation and their known function. The ratios of housekeeping gene-normalized mean levels of transcripts selected, in SOCS1−/− and WT-uninfected BMM, and of infected and uninfected WT BMM are also shown.
| Gene symbols and functions | Description | SOCS1 KO-infected/WT-infected | SOCS1 KO-uninfected/WT-uninfected | WT-infected/WT-uninfected |
|---|---|---|---|---|
| Effector mechanisms and unknown functions | ||||
| Iigp2 | Interferon-inducible GTPase 2 | 2.08 | 1.00 | 1.90 |
| Ifih1 | Interferon induced with helicase C domain 1 | 2.20 | 1.00 | 2.90 |
| Ifi202b | Interferon-activated gene 202B | 2.28 | 1.21 | 13.30 |
| Gvin1 | GTPase, very large interferon-inducible 1 | 2.38 | 1.04 | 1.82 |
| Ifi47 | Interferon γ-inducible protein 47 | 2.65 | 1.04 | 3.10 |
| Ifi203 | Interferon-activated gene 203 | 2.84 | 1.21 | 1.30 |
| Ifi205 | Interferon-activated gene 205 | 2.93 | 0.96 | 41.86 |
| Ifit1 | Interferon-induced protein with tetratricopeptide repeats 1 | 4.59 | 1.03 | 7.19 |
| Ifit2 | Interferon-induced protein with tetratricopeptide repeats 2 | 3.20 | 0.83 | 3.68 |
| Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | 3.30 | 0.95 | 3.17 |
| Iigp1 | Interferon-inducible GTPase 1 | 4.85 | 1.13 | 3.89 |
| Ifi44 | Interferon-induced protein 44 | 5.01 | 1.02 | 6.70 |
| Eg240921 | Interferon-inducible protein p204 | 5.70 | 1.11 | 5.90 |
| Mpa2l | Macrophage activation 2-like | 2.03 | 1.08 | 23.18 |
| Noxa1 | NADPH oxidase activator 1 | 2.02 | 1.50 | 3.02 |
| Defb11 | Defensin β 11 | 2.26 | 1.04 | 0.49 |
| Stat1 | Signal transducer and activator of transcription 1 | 2.25 | 1.00 | 3.38 |
| Irg1 | Immunoresponsive gene 1 | 3.31 | 1.10 | 178.28 |
| Antigen presentation | ||||
| Cd1d1 | CD1d1 antigen | 2.30 | 1.01 | 3.13 |
| Cd1d2 | CD1d2 antigen | 3.61 | 1.42 | 3.09 |
| H2-T24 | Histocompatibility 2, T region locus 24 | 2.29 | 0.92 | 6.08 |
| Cd86 | CD86 antigen | 2.29 | 1.04 | 1.56 |
| Ciita | Class II transactivator | 0.43 | 1.07 | 0.27 |
| Cytokine and chemokines, receptors, SOCS | ||||
| Socs1 | Suppressor of cytokine signaling 1 | 73.04 | ||
| Socs3 | Suppressor of cytokine signaling 3 | 2.07 | 1.05 | 131.15 |
| Cish | Cytokine-inducible Src homology 2-containing protein | 2.45 | 0.89 | 51.10 |
| Ifng | Interferon γ | 5.95 | 1.23 | 10.66 |
| Il12rb1 | Interleukin 12 receptor, β 1 | 3.19 | 1.36 | 14.00 |
| Il1b | Interleukin 1 β (Il1b), mRNA | 4.08 | 1.00 | 97.05 |
| Cxcr3 | Chemokine (CXC motif) receptor 3 | 0.36 | 1.02 | 0.46 |
| Cxcl9 | Chemokine (CXC motif) ligand 9 | 4.81 | 0.86 | 9.93 |
| Cxcl10 | Chemokine (CXC motif) ligand 10 | 4.32 | 1.08 | 16.58 |
| Type I IFN-stimulated genes | ||||
| Isg15 | ISG15 ubiquitin-like modifier | 3.54 | 1.03 | 15.11 |
| Ifnb1 | Interferon β 1, fibroblast | 5.33 | 1.18 | 25.49 |
| Oas1g | 2′-5′ oligoadenylate synthetase 1 | 2.27 | 0.93 | 23.25 |
| Oasl2 | 2′-5′ oligoadenylate synthetase-like 2 | 2.45 | 0.95 | 2.90 |
| Oas2 | 2′-5′ oligoadenylate synthetase 2 | 3.00 | 1.06 | 1.44 |
| Gbp5 | Guanylate nucleotide binding protein 5 | 2.48 | 0.92 | 8.20 |
| Mx1 | Myxovirus (influenza virus) resistance 1 | 3.39 | 1.06 | 4.60 |
| Mx2 | Myxovirus (influenza virus) resistance 2 | 3.40 | 1.04 | 4.86 |
| Ddx58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (Ddx58) | 2.27 | 1.03 | 2.92 |
Higher Levels of SOCS1 mRNA after Infection in Vivo with M. tuberculosis
Levels of SOCS1 mRNA in lung cells from WT mice infected with M. tuberculosis or BCG were augmented as compared with uninfected controls (Fig. 5A) (data not shown).
SOCS1 mRNA levels were, on the contrary, not increased in lungs from BCG-infected RAG1−/− or IFN-γ−/− mice (Fig. 5, B and C). This suggests that IFN-γ secretion and adaptive immune cells are required for the elevated SOCS1 mRNA levels registered after BCG infection in WT mice. SOCS1 transcript titers in lungs from M. tuberculosis-infected IFN-γ−/− and IFN-γR−/− but not RAG-1−/− mice were lower than in WT-infected controls (Fig. 5D). On the other hand, SOCS1 mRNA was increased in RAG1−/− and IFN-γ−/− mice compared with uninfected controls (supplemental Fig. S4, D and E).
SOCS1 Expression in Macrophages Hampers M. tuberculosis Control Early after Infection
Whether SOCS1 could affect the outcome of mycobacterial infection in vivo was next explored.
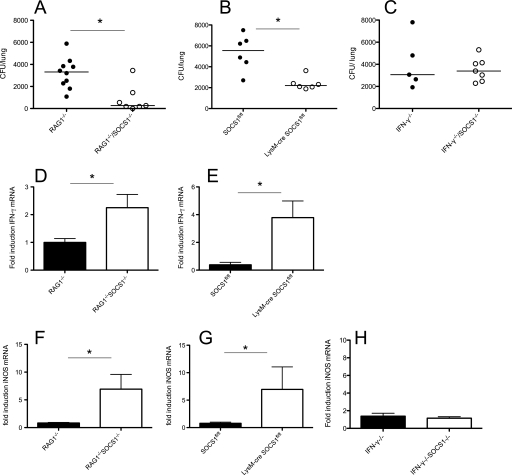
We found lower bacterial levels in lungs of both LysM-cre SOCS1fl/fl and RAG1−/−/SOCS1−/− mice 7 days after aerosol infection with 250 M. tuberculosis bacteria in comparison with respective controls (Fig. 6, A and B). On the contrary, similar bacterial titers were measured in lungs from IFN-γ−/−/SOCS1−/− and IFN-γ−/− mice (Fig. 6C). Dissemination of bacteria to the spleen remained undetectable at this time point.
FIGURE 6.
Reduced bacterial load and increased IFN-γ and iNOS mRNA in M. tuberculosis-infected SOCS1-deficient mice. RAG1−/− SOCS1−/−, RAG1−/− (A, D, and F), LysM-cre SOCS1fl/fl, SOCS1fl/fl (B, E, and G), IFN-γ−/−/SOCS1−/−, and IFN-γ−/− (C and H) mice were infected with M. tuberculosis H37Rv via the aerosol route. Animals were sacrificed 1 week after infection, and cfu per lung and spleen were assessed. The cfu/lung of individual mice and the median/group at the indicated time points after infection are depicted (A–C). *, differences in cfu are significant (p < 0.05, Mann-Whitney U test). Total RNA was extracted from lungs, and the mean -fold accumulation of IFN-γ (D and E) and iNOS (F–H) transcripts ± S.E. (error bars) in lungs from infected mice (n ≥ 5/group) was calculated. *, differences with controls are significant (p < 0.05, Student's t test).
In agreement with in vitro data, lungs from RAG1−/−/SOCS1−/−- and LysM-cre SOCS1fl/fl-infected mice showed higher accumulation of both IFN-γ and iNOS transcripts compared with controls (Fig. 6, D–G). In contrast there was no enhanced accumulation of iNOS transcripts in IFN-γ−/−/SOCS1−/−- and IFN-γ−/−-infected mice compared with uninfected controls (Fig. 6H).
Increased Severity of Pulmonary Inflammation in SOCS1−/− Mice
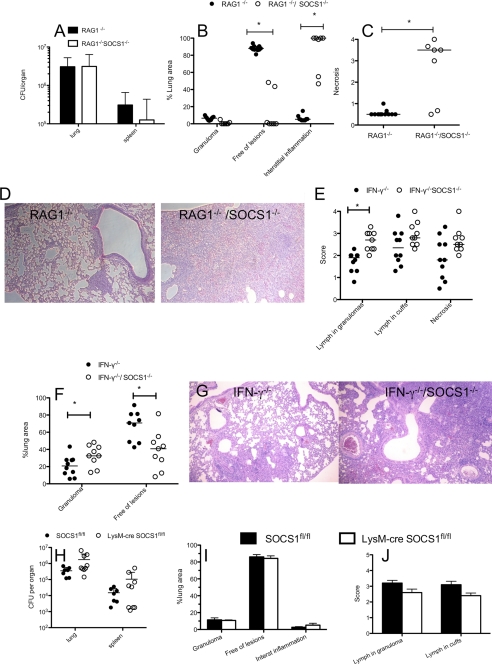
On the other hand, lungs and spleens from LysM-cre SOCS1fl/fl and RAG1−/−/SOCS1−/− mice showed no reduction of the bacterial numbers at later time points after infection (Fig. 7, A and H). Increased levels of IFN-γ and iNOS mRNA were measured in lungs from RAG1−/− and RAG1−/−/SOCS1−/− at 3 weeks after infection as compared with uninfected controls. However, titers of IFN-γ and iNOS mRNA in lungs from RAG1−/−/SOCS1−/− and RAG1−/− mice were similar (supplemental Fig. S4, A and B). We then asked whether SOCS1 expression by macrophages could affect antigen-specific T cell responses. Similar frequencies of PPD-stimulated IFN-γ-secreting CD4+ T cells in spleens and lungs from M. tuberculosis-infected LysM-cre SOCS1fl/fl and SOCS1fl/fl controls were found, whereas cells from uninfected mice showed no PPD responses (supplemental Fig. S4C).
FIGURE 7.
SOCS1 deficiency in non-macrophage cells mediates increased severity of pulmonary inflammation. Shown are bacterial load in lungs and spleens of mice (A) and histopathological scoring of hematoxylin-eosin-stained paraffin lung sections (B–D) from RAG1−/−/SOCS1−/− and RAG1−/− mice measured 24 days after aerosol infection with M. tuberculosis. E–G, pulmonary histopathology of lung sections from IFN-γ−/−/SOCS1−/− and IFN-γ−/− mice sacrificed 4 weeks after infection. *, differences with the control group are significant (p < 0.05, Mann-Whitney U test). Bacterial load in lungs and spleens (H) and histopathological scoring of hematoxylin-eosin-stained paraffin lung sections from LysM-cre SOCS1fl/fl and SOCS1fl/fl (I and J) mice measured 24 days after infection with M. tuberculosis. Error bars, S.E.
Lungs from RAG1−/− mice at 24 days after M. tuberculosis infection showed small amounts of granulomatous lesions that, altogether, made up a minor percentage of the lung area. The lungs from these mice remained otherwise free of inflammatory lesions. In contrast, RAG1−/−/SOCS1−/− mice showed a spectacular pathology, where most of the lung parenchyma was consolidated, showing an atypical proliferative interstitial pneumonitis formed by hyperplastic type II pneumocytes, macrophages, and neutrophils. Large numbers of neutrophils were observed within large areas of necrosis as well as around the blood vessels (Fig. 7, B–D).
Similarly, lungs from IFN-γ−/−/SOCS1−/− mice 3 weeks after M. tuberculosis infection showed interstitial pneumonia with thickening of the alveolar septa, leaving less area of lung parenchyma free of lesions than in IFN-γ−/− controls (Fig. 7, E–G). Lungs from IFN-γ−/− and IFN-γ−/−/SOCS1−/− had, in contrast to RAG1−/− or RAG1−/−/SOCS1−/− mice, well limited granulomas formed by aggregates of macrophages or epithelioid cells with marked infiltration of neutrophils and variable numbers of lymphocytes. The granulomas were generally rounder, smaller, and better defined in IFN-γ−/− than in IFN-γ−/−/SOCS1−/− mice. Thus, the total area of lung parenchyma occupied by granulomas was larger and lymphocytes within the granulomas were more abundant in IFN-γ−/−/SOCS1−/− than in IFN-γ−/− mice (Fig. 7, E–G). As expected, M. tuberculosis-infected IFN-γ−/− and IFN-γ−/−/SOCS1−/− lungs showed no increased iNOS, CXCL9, or CXCL10 mRNA levels compared with uninfected controls, whereas transcript levels were increased in lungs from WT-infected mice (supplemental Fig. S4, F–H).
Importantly, lungs from LysM-cre SOCS1fl/fl showed similar histopathological features as SOCS1fl/fl mice 3 and 6 weeks after M. tuberculosis infection, with only the exception of a slightly increased interstitial inflammation 6 weeks after infection (Fig. 7, I and J, and supplemental Fig. S4, I and J). Altogether, SOCS1 expression by macrophages or neutrophils thus seems to hamper control of M. tuberculosis in vivo early after infection, whereas SOCS1 expression in non-macrophage cells can protect mice from infection-induced damaging inflammation.
DISCUSSION
We here report that infection with virulent or avirulent mycobacteria induces SOCS1 expression in mouse and human macrophages and dendritic cells in vitro, and in vivo in a murine model. SOCS1 expression in macrophages required phagocytosis of mycobacteria and was largely mediated by MyD88/TLR2 and NOD2 receptors. The ensuing NF-κB pathway downstream of both MyD88 and NOD2 receptor signaling was also required. Thus, TLR and non-TLR signals cooperate in SOCS1 mRNA expression by mycobacteria-infected BMM. IFN-α/β signaling was also required for mycobacterial induced SOCS1 expression by BMM. SOCS1 has been shown to hamper growth control of intracellular infections by Chlamydia and Leishmania (12, 33), explained by the essential role of SOCS1 in the control of macrophage activation by regulating TLR signaling (24, 46).
We here showed by using knock-out, conditional knockdown, and SOCS1 mimetic peptides that SOCS1 also inhibited growth control of M. tuberculosis and BCG by macrophages. Surprisingly, SOCS1 inhibition of BCG and M. tuberculosis growth control in BMM was mediated by its ability to obstruct IFN-γ secretion from these cells rather than by an inhibition of responses to IFN-γ. This SOCS1-mediated inhibition of bacterial control was not mediated by a diminished release of IL-12. Instead, SOCS1-deficient BMM proved to be hyperresponsive to IL-12. Neutralization of IL-12 hindered IFN-γ expression in infected SOCS1−/− BMM, suggesting a novel mechanism of mycobacterial inhibition of phagocyte activation. The increased response to IL-12 of SOCS1−/− BMM was associated with increased levels of IL-12Rβ1 mRNA before as well as after infection with M. tuberculosis. IL-12Rβ1 is expressed on a variety of immune cells, including T, NK, macrophages, and dendritic cells (47–49); is up-regulated by IL-12 and IFN-γ signaling (49); and serves in humans to enhance immunity to mycobacterial pathogens (50, 51). However, increased responses to IL-12 by BMM were not due to an SOCS1-mediated defect in IL-12Rβ1 expression because increased expression of IL-12 is IFN-γ-dependent. In concurrence with our results, SOCS1-deficient T cells and dendritic cells showed higher IFN-γ secretion in response to IL-12 (16, 52, 53). JAK2 and TYK2 kinases that are associated with IL-12R will activate and phosphorylate STAT4 upon binding of IL-12. SOCS1 has been shown to bind to JAK2 and inhibit both the kinase activity and the subsequent IL-12 signaling (54).
The expression microarray analysis showed, in agreement with previous reports, that a significant segment of the macrophage transcriptome is altered after infection with M. tuberculosis (55, 56). Unexpectedly, the majority of infection-regulated genes overlapped with those differentially regulated by SOCS1, strongly suggesting a major role of SOCS1 in the control of metabolic activity of macrophages during the infection with M. tuberculosis. Equally surprising was the finding that the majority of these differentially expressed genes were down-regulated in infected compared with uninfected BMM. The expression of most of these genes was further down-regulated in M. tuberculosis-infected SOCS1−/− BMM compared with WT controls. On the contrary, the majority of genes involved in defense or immune responses (data not shown) and almost all IFN-regulated immune genes as well as IFN-β and IFN-γ themselves showed higher levels in the SOCS1−/−-infected cells (supplemental Table SI). Altogether, our data suggest that SOCS1 attenuates both the negative and the positive regulation of the majority of the genes changing their expression after infection of macrophages with M. tuberculosis.
Of importance, we demonstrated that SOCS1 expression by macrophages impaired M. tuberculosis clearance and IFN-γ and iNOS expression in mice, when measured 7 days after infection. At this early stage of infection, when increased secretion of IFN-γ is not detected in lungs from infected WT mice, a higher level of IFN-γ secreted by SOCS1-deficient macrophages is sufficient to impair bacterial growth in vivo.
Instead, the inability of SOCS1 to hamper bacterial control at later time points, when IFN-γ secretion by T cells is prominent, may thus be explained by the above mentioned capacity of infected, SOCS1-expressing macrophages to respond to IFN-γ. In line with this, similar levels of IFN-γ and IFN-γ-regulated gene transcripts were present in lungs from SOCS1−/− mice 3 weeks after infection. Relevant to our observation, SOCS1 expression in blood is increased in pulmonary tuberculosis patients that in parallel express an IFN-regulated gene signature (57).
We also observed an overwhelming infection-induced pulmonary inflammation in RAG1−/−/SOCS1−/− and IFN-γ−/−/SOCS1−/− mice. Because LysM-cre SOCS1fl/fl mice, in contrast, showed no increased pathology, we conclude that SOCS1-expressing non-macrophage cells are the main controllers of detrimental inflammation. SOCS1 silencing in macrophages or dendritic cells has been shown to potentiate anti-tumor immune responses (34), whereas SOCS1 deficiency in whole organs except for T and B cells enhances inflammation-mediated colon tumor development (58).
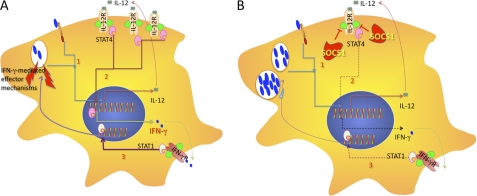
In summary, we propose a model in which M. tuberculosis actively promotes SOCS1 expression by macrophages to counteract effective mycobacterial control at early time points after infection in vivo, before initiation of IFN-γ secretion by NK or T cells. SOCS1 facilitates mycobacterial growth by hampering macrophage IFN-γ secretion in response to infection-induced IL-12 (Fig. 8). At later time points after infection, despite SOCS1 expression, macrophages respond to IFN-γ secreted by T cells or NK cells and will not counteract bacterial control to any further extent. Instead, SOCS1 in non-macrophage cells protects mice from severe inflammation.
FIGURE 8.
Macrophage responses during M. tuberculosis infection in the absence (A) or presence (B) of SOCS1. Infection of macrophages with M. tuberculosis induces SOCS1 and secretion of IL-12 in an TLR2/MyD88- and NOD2-mediated manner. SOCS1 probably hampers STAT4 activation and reduces IFN-γ secretion in response to M. tuberculosis-stimulated IL-12. Decreased IFN-γ levels account for diminished levels of activated STAT1 and IFN-regulated effector molecules in the presence of SOCS1. IFN-γ-dependent IL-12Rβ1 expression is also decreased in the presence of SOCS1. As a consequence, higher intracellular mycobacterial levels are observed. Of importance, SOCS1 does not hinder responses to IFN-γ. The secretion of IFN-α/β is also inhibited.
The mechanisms behind the anti-inflammatory properties of SOCS1 during M. tuberculosis remain to be studied. We suggest that the efficient stimulation of SOCS1 expression reflects an evolutionary adaptation that may be associated with the ability of M. tuberculosis to establish a chronic infection.
Supplementary Material
Acknowledgments
We thank Dr. Fredric Carlsson for comments, Dr. Andzej Pawlowski for help and advice with M. tuberculosis infections, and Berit Olsson and Helene Braxenholm for excellent technical assistance (all from Karolinska Institutet); Dr. Robert Geffers (HZI, Braunschweig) for the analysis of the microarray data; Ewa Westergren (Swedish University of Agricultural Sciences) for the preparation of histological slides; Dr. H. Johnson (University of Florida) for the gift of SOCS1 mimetic peptides; and Dr. T. Naka and T. Kishimoto (Osaka University, Japan) for kindly providing SOCS1+/− mice.
This work was supported by European Community Grant 200732 HOMITB, the Karolinska Institutet, and the Swedish Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Table S1 and Figs. S1–S4.
- BCG
- bacille Calmette-Guerin
- BMM
- bone marrow-derived macrophage(s)
- BMDC
- bone marrow-derived dendritic cell(s)
- MOI
- multiplicity of infection.
REFERENCES
- 1. World Health Organization (2006) Global Tuberculosis Control: Surveillance, Planning, Financing, p. 1, World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Nathan C. F., Murray H. W., Wiebe M. E., Rubin B. Y. (1983) J. Exp. Med. 158, 670–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cooper A. M., Dalton D. K., Stewart T. A., Griffin J. P., Russell D. G., Orme I. M. (1993) J. Exp. Med. 178, 2243–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flynn J. L., Chan J., Triebold K. J., Dalton D. K., Stewart T. A., Bloom B. R. (1993) J. Exp. Med. 178, 2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casanova J. L., Abel L. (2002) Annu. Rev. Immunol. 20, 581–620 [DOI] [PubMed] [Google Scholar]
- 6. Hmama Z., Gabathuler R., Jefferies W. A., de Jong G., Reiner N. E. (1998) J. Immunol. 161, 4882–4893 [PubMed] [Google Scholar]
- 7. Hussain S., Zwilling B. S., Lafuse W. P. (1999) J. Immunol. 163, 2041–2048 [PubMed] [Google Scholar]
- 8. Wojciechowski W., DeSanctis J., Skamene E., Radzioch D. (1999) J. Immunol. 163, 2688–2696 [PubMed] [Google Scholar]
- 9. Yoshimura A., Naka T., Kubo M. (2007) Nat. Rev. Immunol. 7, 454–465 [DOI] [PubMed] [Google Scholar]
- 10. Kile B. T., Alexander W. S. (2001) Cell Mol. Life Sci. 58, 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sakamoto H., Yasukawa H., Masuhara M., Tanimura S., Sasaki A., Yuge K., Ohtsubo M., Ohtsuka A., Fujita T., Ohta T., Furukawa Y., Iwase S., Yamada H., Yoshimura A. (1998) Blood 92, 1668–1676 [PubMed] [Google Scholar]
- 12. Alexander W. S., Starr R., Fenner J. E., Scott C. L., Handman E., Sprigg N. S., Corbin J. E., Cornish A. L., Darwiche R., Owczarek C. M., Kay T. W., Nicola N. A., Hertzog P. J., Metcalf D., Hilton D. J. (1999) Cell 98, 597–608 [DOI] [PubMed] [Google Scholar]
- 13. Marine J. C., Topham D. J., McKay C., Wang D., Parganas E., Stravopodis D., Yoshimura A., Ihle J. N. (1999) Cell 98, 609–616 [DOI] [PubMed] [Google Scholar]
- 14. Naka T., Tsutsui H., Fujimoto M., Kawazoe Y., Kohzaki H., Morita Y., Nakagawa R., Narazaki M., Adachi K., Yoshimoto T., Nakanishi K., Kishimoto T. (2001) Immunity 14, 535–545 [DOI] [PubMed] [Google Scholar]
- 15. Chong M. M., Cornish A. L., Darwiche R., Stanley E. G., Purton J. F., Godfrey D. I., Hilton D. J., Starr R., Alexander W. S., Kay T. W. (2003) Immunity 18, 475–487 [DOI] [PubMed] [Google Scholar]
- 16. Chong M. M., Metcalf D., Jamieson E., Alexander W. S., Kay T. W. (2005) Blood 106, 1668–1675 [DOI] [PubMed] [Google Scholar]
- 17. Fenner J. E., Starr R., Cornish A. L., Zhang J. G., Metcalf D., Schreiber R. D., Sheehan K., Hilton D. J., Alexander W. S., Hertzog P. J. (2006) Nat. Immunol. 7, 33–39 [DOI] [PubMed] [Google Scholar]
- 18. Eyles J. L., Metcalf D., Grusby M. J., Hilton D. J., Starr R. (2002) J. Biol. Chem. 277, 43735–43740 [DOI] [PubMed] [Google Scholar]
- 19. Dickensheets H., Vazquez N., Sheikh F., Gingras S., Murray P. J., Ryan J. J., Donnelly R. P. (2007) Genes Immun. 8, 21–27 [DOI] [PubMed] [Google Scholar]
- 20. Cornish A. L., Chong M. M., Davey G. M., Darwiche R., Nicola N. A., Hilton D. J., Kay T. W., Starr R., Alexander W. S. (2003) J. Biol. Chem. 278, 22755–22761 [DOI] [PubMed] [Google Scholar]
- 21. Vázquez N., Greenwell-Wild T., Rekka S., Orenstein J. M., Wahl S. M. (2006) J. Leukoc. Biol. 80, 1136–1144 [DOI] [PubMed] [Google Scholar]
- 22. Imai K., Kurita-Ochiai T., Ochiai K. (2003) FEMS. Immunol. Med. Microbiol. 39, 173–180 [DOI] [PubMed] [Google Scholar]
- 23. Srivastava V., Manchanda M., Gupta S., Singla R., Behera D., Das G., Natarajan K. (2009) J. Biol. Chem. 284, 25532–25541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakagawa R., Naka T., Tsutsui H., Fujimoto M., Kimura A., Abe T., Seki E., Sato S., Takeuchi O., Takeda K., Akira S., Yamanishi K., Kawase I., Nakanishi K., Kishimoto T. (2002) Immunity 17, 677–687 [DOI] [PubMed] [Google Scholar]
- 25. Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 115–122 [DOI] [PubMed] [Google Scholar]
- 26. Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. (2000) Immunity 13, 539–548 [DOI] [PubMed] [Google Scholar]
- 27. Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 28. Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. (1999) J. Immunol. 162, 3749–3752 [PubMed] [Google Scholar]
- 29. Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 30. Huang S., Hendriks W., Althage A., Hemmi S., Bluethmann H., Kamijo R., Vilcek J., Zinkernagel R. M., Aguet M. (1993) Science 259, 1742–1745 [DOI] [PubMed] [Google Scholar]
- 31. Müller U., Steinhoff U., Reis L. F., Hemmi S., Pavlovic J., Zinkernagel R. M., Aguet M. (1994) Science 264, 1918–1921 [DOI] [PubMed] [Google Scholar]
- 32. Mombaerts P., Iacomini J., Johnson R. S., Herrup K., Tonegawa S., Papaioannou V. E. (1992) Cell 68, 869–877 [DOI] [PubMed] [Google Scholar]
- 33. Yang T., Stark P., Janik K., Wigzell H., Rottenberg M. E. (2008) J. Immunol. 180, 4040–4049 [DOI] [PubMed] [Google Scholar]
- 34. Hashimoto M., Ayada T., Kinjyo I., Hiwatashi K., Yoshida H., Okada Y., Kobayashi T., Yoshimura A. (2009) Cancer Sci. 100, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Förster I. (1999) Transgenic Res. 8, 265–277 [DOI] [PubMed] [Google Scholar]
- 36. Rothfuchs A. G., Gigliotti D., Palmblad K., Andersson U., Wigzell H., Rottenberg M. E. (2001) J. Immunol. 167, 6453–6461 [DOI] [PubMed] [Google Scholar]
- 37. Davies J. Q., Gordon S. (2005) Methods Mol. Biol. 290, 105–116 [DOI] [PubMed] [Google Scholar]
- 38. Chackerian A. A., Alt J. M., Perera T. V., Dascher C. C., Behar S. M. (2002) Infect. Immun. 70, 4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rothfuchs A. G., Trumstedt C., Mattei F., Schiavoni G., Hidmark A., Wigzell H., Rottenberg M. E. (2006) J. Immunol. 176, 6982–6990 [DOI] [PubMed] [Google Scholar]
- 40. Underhill D. M., Ozinsky A., Smith K. D., Aderem A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14459–14463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thoma-Uszynski S., Stenger S., Takeuchi O., Ochoa M. T., Engele M., Sieling P. A., Barnes P. F., Rollinghoff M., Bolcskei P. L., Wagner M., Akira S., Norgard M. V., Belisle J. T., Godowski P. J., Bloom B. R., Modlin R. L. (2001) Science 291, 1544–1547 [DOI] [PubMed] [Google Scholar]
- 42. Abel B., Thieblemont N., Quesniaux V. J., Brown N., Mpagi J., Miyake K., Bihl F., Ryffel B. (2002) J. Immunol. 169, 3155–3162 [DOI] [PubMed] [Google Scholar]
- 43. Gandotra S., Jang S., Murray P. J., Salgame P., Ehrt S. (2007) Infect. Immun. 75, 5127–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Flowers L. O., Johnson H. M., Mujtaba M. G., Ellis M. R., Haider S. M., Subramaniam P. S. (2004) J. Immunol. 172, 7510–7518 [DOI] [PubMed] [Google Scholar]
- 45. Ahmed C. M., Dabelic R., Waiboci L. W., Jager L. D., Heron L. L., Johnson H. M. (2009) J. Virol. 83, 1402–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kinjyo I., Hanada T., Inagaki-Ohara K., Mori H., Aki D., Ohishi M., Yoshida H., Kubo M., Yoshimura A. (2002) Immunity 17, 583–591 [DOI] [PubMed] [Google Scholar]
- 47. Puddu P., Fantuzzi L., Borghi P., Varano B., Rainaldi G., Guillemard E., Malorni W., Nicaise P., Wolf S. F., Belardelli F., Gessani S. (1997) J. Immunol. 159, 3490–3497 [PubMed] [Google Scholar]
- 48. Munder M., Mallo M., Eichmann K., Modolell M. (1998) J. Exp. Med. 187, 2103–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Musikacharoen T., Oguma A., Yoshikai Y., Chiba N., Masuda A., Matsuguchi T. (2005) Blood 105, 711–720 [DOI] [PubMed] [Google Scholar]
- 50. Altare F., Durandy A., Lammas D., Emile J. F., Lamhamedi S., Le Deist F., Drysdale P., Jouanguy E., Döffinger R., Bernaudin F., Jeppsson O., Gollob J. A., Meinl E., Segal A. W., Fischer A., Kumararatne D., Casanova J. L. (1998) Science 280, 1432–1435 [DOI] [PubMed] [Google Scholar]
- 51. de Jong R., Altare F., Haagen I. A., Elferink D. G., Boer T., van Breda Vriesman P. J., Kabel P. J., Draaisma J. M., van Dissel J. T., Kroon F. P., Casanova J. L., Ottenhoff T. H. (1998) Science 280, 1435–1438 [DOI] [PubMed] [Google Scholar]
- 52. Evel-Kabler K., Song X. T., Aldrich M., Huang X. F., Chen S. Y. (2006) J. Clin. Invest. 116, 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hanada T., Tanaka K., Matsumura Y., Yamauchi M., Nishinakamura H., Aburatani H., Mashima R., Kubo M., Kobayashi T., Yoshimura A. (2005) J. Immunol. 174, 4325–4332 [DOI] [PubMed] [Google Scholar]
- 54. Davey G. M., Heath W. R., Starr R. (2006) Tissue Antigens 67, 1–9 [DOI] [PubMed] [Google Scholar]
- 55. Ehrt S., Schnappinger D., Bekiranov S., Drenkow J., Shi S., Gingeras T. R., Gaasterland T., Schoolnik G., Nathan C. (2001) J. Exp. Med. 194, 1123–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shi S., Nathan C., Schnappinger D., Drenkow J., Fuortes M., Block E., Ding A., Gingeras T. R., Schoolnik G., Akira S., Takeda K., Ehrt S. (2003) J. Exp. Med. 198, 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Berry M. P., Graham C. M., McNab F. W., Xu Z., Bloch S. A., Oni T., Wilkinson K. A., Banchereau R., Skinner J., Wilkinson R. J., Quinn C., Blankenship D., Dhawan R., Cush J. J., Mejias A., Ramilo O., Kon O. M., Pascual V., Banchereau J., Chaussabel D., O'Garra A. (2010) Nature 466, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hanada T., Kobayashi T., Chinen T., Saeki K., Takaki H., Koga K., Minoda Y., Sanada T., Yoshioka T., Mimata H., Kato S., Yoshimura A. (2006) J. Exp. Med. 203, 1391–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.