Abstract
The multiple transferable resistance (mTR) pump from Neisseria gonorrhoeae MtrCDE multidrug pump is assembled from the inner and outer membrane proteins MtrD and MtrE and the periplasmic membrane fusion protein MtrC. Previously we established that while there is a weak interaction of MtrD and MtrE, MtrC binds with relatively high affinity to both MtrD and MtrE. MtrD conferred antibiotic resistance only when it was expressed with MtrE and MtrC, suggesting that these proteins form a functional tripartite complex in which MtrC bridges MtrD and MtrE. Furthermore, we demonstrated that MtrC interacts with an intraprotomer groove on the surface of MtrE, inducing channel opening. However, a second groove is apparent at the interface of the MtrE subunits, which might also be capable of engaging MtrC. We have now established that MtrC can be cross-linked to cysteines placed in this interprotomer groove and that mutation of residues in the groove impair the ability of the pump to confer antibiotic resistance by locking MtrE in the closed channel conformation. Moreover, MtrE K390C forms an intermolecular disulfide bond with MtrC E149C locking MtrE in the open channel conformation, suggesting that a functional salt bridge forms between these residues during the transition from closed to open channel conformations. MtrC forms dimers that assemble into hexamers, and electron microscopy studies of single particles revealed that these hexamers are arranged into ring-like structures with an internal aperture sufficiently large to accommodate the MtrE trimer. Cross-linking of single cysteine mutants of MtrC to stabilize the dimer interface in the presence of MtrE, trapped an MtrC-MtrE complex with a molecular mass consistent with a stoichiometry of 3:6 (MtrE3MtrC6), suggesting that dimers of MtrC interact with MtrE, presumably by binding to the two grooves. As both MtrE and MtrD are trimeric, our studies suggest that the functional pump is assembled with a stoichiometry of 3:6:3.
Keywords: Antibiotics, Bacteria, Membrane Proteins, Multidrug Transporters, Protein Assembly, Neisseria gonorrhoeae
Introduction
Gram-negative bacteria utilize homologous tripartite transport systems to pump cytotoxic compounds, including antibiotics, metals, and protein toxins, from the cell (1–6). These assemblies are composed of an inner membrane protein (IMP),6 which transduces electrochemical energy, and a conduit-forming outer membrane protein (OMP), which are connected by a periplasmic membrane fusion protein (MFP), anchored to the inner membrane (1–6). There are structures for all of the individual components but not for a fully assembled tripartite complex. For example, in the case of the AcrABTolC multidrug pump and CusABC copper pump from Escherichia coli, the structures of the RND transporters AcrB (7, 8) and CusA (9), of their cognate MFPs, AcrA (10) and CusB (11), and of OMPs, TolC (12), and CusC (13), have been determined. IMPs, represented by AcrB and CusA, as well as the OMPs, such as TolC and CusC, function as trimeric assemblies. There is evidence that the periplasmic domains of AcrB and TolC contact one another (14, 15), whereas AcrA stabilizes this interaction by binding across these proteins (14, 16). Cross-linking studies indicate that the α-helical coiled-coil hairpin7 of AcrA fits into intraprotomer grooves of the open state of TolC, whereas its β-lipoyl domains interact with the periplasmic domain of AcrB at the interface between adjacent subunits (17, 18). The latter interaction contrasts with that proposed for the CusA, which has been modeled as interacting with the surface of the CusB subunits, rather than at their interface (9). In such models, the stoichiometry is 3:3:3, yet a fully assembled tripartite complex has not been cross-linked. It is also notable that the hairpin of CusB is relatively short and could not make the same extensive contacts with CusC as AcrA can with TolC, suggesting that the interaction would primarily be at the periplasmic tip of CusC. Recent studies of the MacABTolC macrolide pump from E. coli indicate that the MFP MacA forms a hexameric channel that connects the periplasmic tips of MacB and TolC (19, 20), implying that the tripartite assembly occurs with a stoichiometry of 3:6:3. Most recently, the structure of a CusA-CusB binary complex was reported, revealing a 3:6 assembly, further suggesting that the MFP forms a channel between the IMP and OMP (21).
Structural analysis of the OMPs reveals that each protomer forms a structural repeat, which has arisen from an early gene duplication event. Thus, each trimer of OMP in fact possesses a pseudo-6-fold symmetry that translates into the creation of three pairs of grooves, two pairs per protomer, one on the intraprotomer, and one on the junction between the protomers, each of which could act as an MFP interaction site. Several studies, involving ITC (14), surface plasmon resonance (22), and fluorescence recovery after photobleaching (23) measurements of MFP binding to the OMP, indicate that there are multiple binding sites, and some cross-linking studies suggest that the MFP binds to a pair of grooves on the OMP (24). Also supporting this proposal, a number of operons have been identified that encode the components of tripartite pumps that include the genes for a pair of MFPs (25, 26). Because in some cases both of these MFPs are required for function of the pump (25), this suggests that it is necessary for the OMP to bind both MFPs, leading to the hypothesis that two MFPs bind to the intra- and interprotomer grooves, respectively (26). Two recent studies on the assembly of the AcrABTolC pump, reported while our work was in progress, indicate that AcrA forms dimers, but in one study, these were shown to bind to AcrB and TolC, which interacted with one another (27), whereas in the other study, these dimers were reported to form an hexameric channel, interconnecting AcrB to TolC (28).
The determination of the stoichiometry of the pump assembly and the dynamics of the interactions between the pump components will be crucial in providing an understanding of the molecular mechanism of their action that would aid in developing novel pharmaceutical agents against drug resistance, which is an emerging global problem. We have investigated the assembly of the MtrCDE tripartite multidrug efflux pump from Neisseria gonorrhoeae, which has the advantage that all the components of the pump are encoded by the same operon and, consequently, the interactions between the components are likely to be specific, facilitating analyses of pump assembly (29, 30). We present evidence for assembly of the MtrD-MtrC-MtrE complex with a stoichiometry of 3:6:3, in which MtrC binds to both intraprotomer and interprotomer grooves on the surface of MtrE.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
The E. coli strains and plasmids used are described in supplemental Table 1, and the primers used for construction of plasmid vectors are described in supplemental Table 2.
Site-directed Mutagenesis of mtrE
A QuikChange XL kit (Stratagene) was used for site-directed mutagenesis of mtrC and mtrE. For in vivo functional analyses of the pump, the pACYCDuet-mtrC/mtrD/mtrE plasmid was used as the template, whereas for the purification of MtrC and MtrE derivatives, the pET-mtrE plasmid was used as the template, with the primers given in supplemental Table 3 used to introduce the required mutations into mtrC and mtrE. PCR reactions were performed using the following cycling parameters: initial activation, 95 °C for 1 min; denaturation, 95 °C for 50 s; annealing, 55 °C for 50 s; and extension, 68 °C for 10 min for 17 cycles, with a final extension at 68 °C for 10 min. After PCR, the template was digested with DpnI for 3 h, 5 μl of the PCR mix was transformed into NovaBlue E. coli, and colonies were selected for chloramphenicol resistance. Plasmids were extracted from the colonies, and their DNA was sequenced.
Protein Overexpression and Purification
All the proteins used in this study were purified as fusion proteins with a six-histidine tag from E. coli overexpressing strains according to previously published protocols (30).
Growth Curve Analyses
E. coli cells, of strain (ΔtolC) TG1(DE3) (30, 31), harboring plasmids were grown at 37 °C (200 rpm) until the cell density reached an A600 of 0.5, and then induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside. The cells were grown for a further 3 h and diluted with 2×YT medium containing antibiotic, and the growth curve was recorded; cells for minimum inhibitory concentration (MIC) measurements were monitored for growth 24 h later.
In Vitro Cross-linking
MtrE and MtrC (8 μm in 20 mm Na2HPO4, pH 7.5, 100 mm NaCl, 10% glycerol, 0.05% DDM) were mixed with 1 mm bis(maleimido)hexane (BMH), 1 mm bis(maleimido)ethane, or 0.75 mm dibromobimane (DBBR) (all freshly prepared in N,N-dimethylformamide) and incubated at 25 °C for 1 h. The samples were reduced with 50 mm DTT, mixed with 4× LDS sample buffer, boiled for 5 min, and separated on 4–12% gradient SDS NuPAGE precast gel with MOPS buffer. Similarly, 1 mm ethylene glycol bis-succinimidyl succinate (EGS) cross-linker was used to test for dimers and higher order oligomers of MtrC. Unless stated otherwise, the cysteine insertion mutants were prepared using the genes encoding the Cys-less MtrE C21S and NT-MtrC derivatives as the target for further mutagenesis.
For pulldown of cross-linked complexes of MtrE and MtrC, ∼10 μl of 8 μm MtrE was mixed with 0.5 ml of crude cell lysate, which contained NT-MtrC, and the proteins were incubated with 1 mm N-succinimidyl-3-(2-pyridyldithio)-propionate (SPDP) or succinimidyl 6-(3-[2-pyridyldithio]-propionamido)-hexanoate (LC SPDP) at 25 °C for 1 h, immobilized on Ni2+-Sepharose beads (50 μl), and washed with increasing concentrations of imidazole (10–40 mm) before elution with 50 μl of 500 mm imidazole. Protein complexes were identified by Western blotting using both anti-His tag and anti-S tag antibodies.
In Vivo Cross-linking
E. coli cells (strain ΔtolC TG1) were grown in 2×YT broth to an A600 nm of 0.5, induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside, and incubated at 25 °C for a further 2 h. Cells were harvested, washed twice in cross-linking buffer (20 mm Na2HP04 (pH 7.5), 300 mm NaCl, 1 mm EDTA, and 10% glycerol), resuspended in the same buffer with 1 mm BMH, and incubated at 25 °C for 1 h. Excess cross-linker was then removed by washing the cells twice in the same buffer. The cells were disrupted by sonication in the presence of 2 mg/ml N-ethylmaleimide, cell debris was removed by super-speed centrifugation (using a TLA 100.4 rotor at 15k rpm), and the membranes were isolated by ultracentrifugation (using a TLA 100.4 rotor at 50,000 rpm). The membranes were solubilized in cross-linking buffer, with 8 m urea and 2% (w/v) βDDM, for 2 h at room temperature; 50 μl of Ni2+-Sepharose beads was mixed with the membrane proteins and incubated at room temperature for 1 h; the beads were washed with cross-linking buffer containing 10–40 mm imidazole to remove nonspecific contaminants, and target protein was eluted with 500 mm imidazole buffer containing 50 mm DTT followed by boiling the beads at 100 °C for 10 min. The protein mixture was separated on 4–12% NuPAGE (Invitrogen) and immunoblotted onto PVDF membranes, and cross-linked complexes were identified with anti-His antibodies. Unless stated otherwise, the cysteine insertion mutants were prepared using the genes encoding the Cys-less MtrE C21S and MtrC C25S derivatives as the target for further mutagenesis.
Microcalorimetry
ITC measurements were carried out at 25 °C using a VP-ITC microcalorimeter (MicroCal). MtrC was prepared in 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 10% v/v glycerol, whereas MtrE was prepared in the same buffer with 0.1% w/v βDDM (buffer 1). After adding βDDM to NT-MtrC to a final concentration of 0.1% (w/v), it was dialyzed, in different dialysis cassettes, at the same time with MtrE against buffer 1. An ITC experiment involved making a single 2-μl injection and a series of 8-μl injections of NT-MtrC from the syringe into the cell, which contained the MtrE. All data were corrected for the heat changes arising from injection of proteins into buffer before data analysis with ORIGIN software (MicroCal). Experiments were carried out in triplicate, and a statistical average was taken.
Disc Diffusion Assays
E. coli cells were grown to an A600 of about 0.5 and induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside, the cells were diluted to 0.1, and then 200 μl of cell culture was spread on an Müller-Hinton agar plate previously incorporated with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside; antibiotic discs were placed on the plate medium and incubated at 37 °C overnight. The zone of culture growth inhibition was determined. The discs used were impregnated with 1 μg/ml nafcillin, 30 μg/ml tetracycline, 10 μg/ml erythromycin, 5 μg/ml novobiocin, and 30 μg/ml vancomycin.
Mass Spectrometry
Analyses were performed in a nanoflow electrospray mass spectrometer QSTAR and Q-ToF-2 (Waters Corp., Milford, MA). The following experimental parameters were used on QSTAR to record mass spectra of the proteins (generally at 10–20 mg/ml): capillary voltage up to 1.2 kV, declustering potential 150 V, focusing potential 150 V, declustering potential-2 15 V, and collision energy up to 70 V. In MS mode, the following QToF-2 parameters were used for analysis of the Mtr proteins: capillary voltage 1668 V, sample cone 150 V, extractor cone 0 V, accelerating voltage into the collision cell (termed collision energy as per the manufacturer's terminology) 150 V, ion transfer stage pressure 6.1 × 10−3 mbar, and TOF analyzer pressure 1.0 × 10−4 mbar.
To identify MtrC and MtrE in samples run on SDS-PAGE gels, MALDI-TOF Peptide Mass Fingerprinting was performed using a Voyager-DETM STR BioSpectrometryTM workstation (Applied Biosystems, Warrington, UK). De-isotoped and calibrated spectra were then used to generate peak lists, which were searched using the MASCOT mass spectrometry database search software to identify the proteins.
Atomic Force Microscopy
Proteins for atomic force microscopy were diluted to a final concentration of 1 μg/ml, and 45 μl of the sample was allowed to adsorb to freshly cleaved mica. Imaging in air was performed with a multimode atomic force microscope (Digital Instruments, Santa Barbara, CA) in tapping mode. The silicon cantilevers containing a diamond-like extratip had a drive frequency of ∼300 kHz and a specified spring constant of 40 newtons/m (MikroMasch, Portland, OR), and the applied imaging force was kept as low as possible (target amplitude ∼1.6–1.8 V and amplitude set point ∼1.3–1.5 V). The molecular volumes of the protein particles were determined from particle dimensions based on atomic force microscopy images using the SPIP software (Image Metrology).
Electron Microscopy
Protein samples (0.05 mg/ml, mixed with glutaraldehyde (2.5% (v/v) final)) were applied to electron microscope hydrophilic grids coated with carbon film and stained with 2% uranyl acetate. The preparations were examined using a Philips CM120 electron microscope equipped with a LaB6 filament, with an acceleration voltage of 120 kV. Electron micrographs were taken at a magnification of ×45,000. Selected images were digitized with a step size of 12.5 μm on a Super COOLSCAN 9000 Nikon. The WEB and SPIDER software package (Wadsworth Center, New York) was used for all image processing.
Molecular Modeling
A molecular model for MtrE has previously been reported (30). A homology model for MtrC was designed using the MexA (1T5E.pdb) and AcrA (2F1M.pdb) structures as templates and the methodology previously described for the MtrE model (30). Essentially, this consisted of creating combined sequence and structural alignments using T-Coffee (32) and Expresso3D (33), respectively, which were manually analyzed, followed by homology model building with Modeler (34), using the MexA (1T5E.pdb) and AcrA (2F1M.pdb) structures as templates.
RESULTS
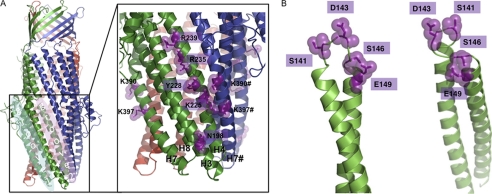
Previous cross-linking studies indicate that the MFP binds within an intraprotomer groove, bounded by helices 3, 7, and 8 of the OMP (17, 18). However, a second groove is apparent at the interface of the subunits within the OMP trimer, bounded by helices 4 and 7 of adjacent subunits (12, 35), which could act as a second site for binding the MFP. We sought to investigate the role of the interprotomer groove of MtrE, using a structural model (Fig. 1A) that we had previously generated (30), to guide mutagenesis of the groove residues. Using such an approach, we previously established that MtrC binds to the intraprotomer groove of MtrE (30). Furthermore, our studies indicate that MtrC communicates the binding of drugs to MtrD to MtrE, presumably via a conformational change that induces opening of the MtrE channel.
FIGURE 1.
Homology models of the structure of MtrE and MtrC. To generate molecular models of MtrE and MtrC, combined sequence and structural alignments were made using T-Coffee and Expresso3D, respectively, which were manually analyzed, followed by homology model building with Modeler, using the MexA (1T5E.pdb) and AcrA (2F1M.pdb) structures as templates. A, the molecular model shows the position of the intra- and interprotomer grooves (groove 1 and groove 2, respectively) and of the mutations introduced into the intersubunit groove (in the expanded panel) of the OMP MtrE. B, the molecular model shows the position of the mutations introduced into the α-helical coiled-coil hairpin domain of the MFP MtrC.
Residues within the Intersubunit Groove of the MtrE Trimer Affect the Pump Function
A number of point mutations in the intersubunit groove of MtrE were constructed based upon a comparative analysis of the modeled structure of MtrE, identifying conserved, solvent-accessible residues likely to be positioned on the surface of the groove (Fig. 1A). The antibiotic sensitivity of these mutants was tested; this initial analysis indicated that there was a modest increase in the sensitivity of these mutants to a range of antibiotics (Table 1). This is perhaps not surprising because single point mutations would not be expected to block binding to the groove surface, which presumably involves a number of interactions. Consequently, we tested whether this effect was cumulative by constructing double mutations; these double mutants were highly susceptible to antibiotics, confirming that residues in the interprotomer groove are important for pump function (Table 1).
TABLE 1.
The inhibitory zone determined by disc diffusion method for mtrE mutants expressed with or without mtrC or mtrCD in E. coli (ΔtolC) TG1(DE3) cells
The indicated Mtr proteins were expressed from pACYC and/or pET21a in (ΔtolC) TG1(DE3) E. coli cells. The inhibitory zone diameter is given in mm. The abbreviations are: Naf, nafcillin; Tet, tetracycline; Ery, erythromycin; Nov, novobiocin; Van, Vancomycin; NI, no inhibition of growth; NG, no growth due to complete growth inhibition; CL, cysteine-less; WT, wild type. * was used to signify when proteins were expressed from pET21a, which would confer resistance to nafcillin. All disc diffusion assays were undertaken in triplicate, and the average result is given.
| MtrE derivative | Naf (1 μg/ml) | Tet (30 μg/ml) | Ery (10 μg/ml) | Nov (5 μg/ml) | Van (30 μg/ml) |
|---|---|---|---|---|---|
| Vector control: pACYC | 20 | 35 | 31 | 27 | NI |
| MtrCD | 12 | 26 | 22 | 19 | NI |
| MtrCDE | 12 | 25 | 20 | 19 | NI |
| MtrCDE-K225E | 14 | 30 | 31 | 24 | NI |
| MtrCDE-Y228F | 16 | 32 | 30 | 20 | NI |
| MtrCDE-R235E | 13 | 30 | 28 | 21 | NI |
| MtrCDE-R239E | 14 | 29 | 27 | 25 | NI |
| MtrCDE-K390E | 15 | 34 | 20 | 23 | NI |
| MtrCDE-K397E | 14 | 29 | 29 | 21 | NI |
| MtrCDE-K225E/R239E | 20 | 39 | 36 | 30 | NI |
| MtrCDE-K225E/K390E | 19 | 37 | 33 | 27 | NI |
| MtrCE | 17 | 31 | 25 | 20 | NI |
| MtrCE-K225E/R239E | 22 | 38 | 30 | 25 | NI |
| MtrE-K225E/R239E | NI* | 26 | 26 | 17 | NI |
| Vector control: pACYC + pET21a | NI* | 40 | 35 | 25 | NI |
| MtrC(CL)/MtrE(CL) + MtrD(WT) | NI* | 26 | 21 | 17 | NI |
| MtrC(CL)/MtrE(K390C) + MtrD(WT) | NI* | 26 | 21 | 17 | NI |
| MtrC(E149C)/MtrE(CL) + MtrD(WT) | NI* | 26 | 21 | 17 | NI |
| MtrC(E149C)/MtrE(K390C) + MtrD(WT) | NI* | 40 | 35 | 25 | 11 |
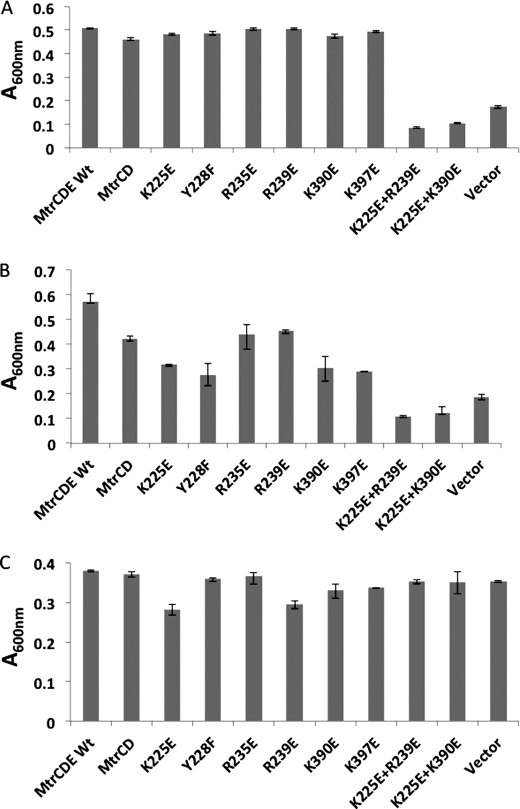
We sought to further substantiate these findings by performing a growth curve analysis for each mutant in the presence of tetracycline and nafcillin. These antibiotics were specifically chosen because they might identify how these mutations impair pump function; tetracycline targets the ribosome, and its removal from the cytoplasm would be sufficient to confer resistance, whereas nafcillin targets penicillin-binding proteins in the periplasm, so that its removal from the cell would be required to confer resistance. Our expectation was that mutations that affected the interaction of MtrC with MtrE would have a more pronounced effect on the growth of cells treated with nafcillin than tetracycline. In the case of the single mutations, the cells were clearly more susceptible to growth inhibition by nafcillin (Fig. 2B) than tetracycline (Fig. 2A). This suggests that these MtrE derivatives were impaired in their ability to interact with MtrCD. Considering that changing one residue in the intersubunit groove might not be enough to severely perturb the interaction of MtrE with MtrCD, we tested whether introducing double mutations would compound the effect on the sensitivity of the cells to antibiotics. None of the double mutants afforded any resistance to nafcillin (Fig. 2B). A Western blot analysis indicated that this cannot be attributed to a failure of the MtrE double mutants to express; the MtrE K225E/K390E and K225E/R239E derivatives were expressed at comparable levels to wild-type MtrE, as were MtrC and MtrD with which the MtrE derivatives were co-expressed (data not shown). The nafcillin sensitivity of the MtrE double mutants might hence be attributable to their inability to interact with MtrCD. However, an ITC analysis revealed that MtrC not only was capable of binding to the MtrE K225E/R239E derivative but did so with a 3-fold higher affinity than for the wild-type MtrE (supplemental Fig. 1). Furthermore, we found that the double mutants were also susceptible to tetracycline (Fig. 2A), which is surprising because one might have expected that MtrCD could confer resistance to tetracycline even in the absence of MtrE. These data imply that the MtrE derivative interacts with MtrCD but that the assembled MtrCDE pump is non-functional. The nature of this assembly is, however, debatable. The increase in affinity of the MtrE K225E/R239E derivatives for MtrC might simply be attributable to MtrC being bound more tightly at the interprotomer groove; however, it is possible that with the interprotomer groove having been disrupted, MtrC only binds to the intraprotomer groove, which has a higher affinity than the interprotomer groove for the MFP. The non-functionality of the pump might be attributable to MtrE adopting a constitutively open or closed state. In an attempt to distinguish between these possibilities, we tested the susceptibility of the cells to vancomycin, which is too big to enter cells by passive diffusion but can gain entry through an open OMP (30, 35). None of the mutants were susceptible to vancomycin (Table 1 and Fig. 2C), indicating that the channel of MtrE was closed. One possibility is that the MtrE derivatives are impaired in their ability to open the channel and, as a consequence, the mutants are susceptible to tetracycline and nafcillin, which can diffuse into the cell, but are insensitive to vancomycin, which cannot enter the cells. Indeed, we note that cells expressing MtrCD alone confer greater resistance to nafcillin and tetracycline than cells expressing MtrCD with MtrE K225E/K390E and K225E/R239E (Fig. 2), indicating that these MtrE derivatives interact with MtrCD to impair pump function. Considering that MtrCD can confer elevated resistance suggests that they interact with an E. coli OMP other than TolC; such promiscuous behavior has been noted for other tripartite pumps (36). Presumably then, MtrE K225E/K390E and K225E/R239E are bound more tightly by MtrCD than this E. coli OMP, impairing pump function. Although cells expressing MtrCE K225E/R239E were insensitive to vancomycin, they had increased sensitivity to a range of transported antibiotics (Table 1), suggesting that the pump is leaky but not fully open (to vancomycin).
FIGURE 2.
Mutations in the interprotomer grooves of MtrE impair the pump. A–C, bar charts showing the relative growth (measured as the A600 nm) over 4 h for ΔtolC TG1(DE3) cells expressing the mtrCD and mtrE genes, including wild-type mtrE and a series of mtrE mutants in the interprotomer groove as indicated, in the presence of 16 μg/ml nafcillin (A), 3 μg/ml tetracycline (B), and 160 μg/ml vancomycin (C). The same strain transformed with the empty vector was used as a control. Each bar is the average of at least three measurements, with error bars representing the S.D. of these measurements from the average.
Evidence for the Binding of MtrC to the Interprotomer Groove of MtrE
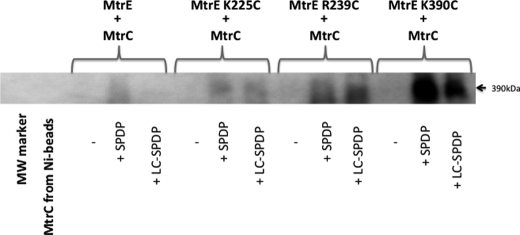
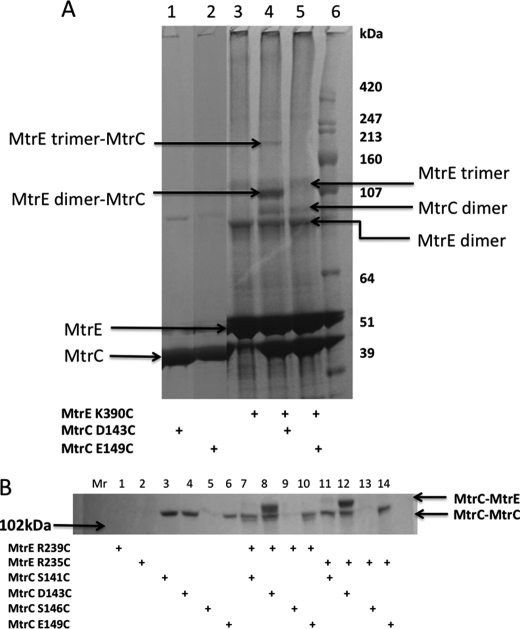
To test whether MtrC interacts directly with the intersubunit groove of MtrE, we replaced residues within the groove with cysteines for cross-linking studies (Fig. 3). Hetero-bifunctional cross-linkers SPDP and LC-SPDP, which react at one end with a sulfhydryl group and at the other with a primary amine group, were used to cross-link the His-tagged MtrE cysteine derivatives to S-tagged NT-MtrC, which was present in the crude extract from disrupted cells. Cross-linked MtrE-MtrC complexes were purified using Ni2+-Sepharose beads and identified by Western blotting using anti-S tag antibodies. The K225C, R239C, and K390C derivatives of MtrE were all found to form cross-linked dimers of MtrE-MtrC (Fig. 3), indicating that MtrC binds along the length of the intersubunit groove. Interestingly, the most abundant band had a molecular mass of about 390 kDa, which would be too large for a 3:3 assembly but consistent with a 6:3 assembly of MtrC and MtrE.
FIGURE 3.
A Western blot analysis of the cross-linking of MtrE derivatives to NT-MtrC. His-tagged, cysteine-free (C21S) MtrE and its derivatives were used as bait for S-tagged NT-MtrC; interactions were stabilized by SPDP or LC SPDP, and bound NT-MtrC was detected using anti-S tag antibodies. Approximately 10 μl of 8 μm MtrE derivative was mixed with 0.5 ml of crude cell lysate containing NT-MtrC and incubated with 1 mm SPDP or LC SPDP at 25 °C for 1 h; 50 μl of Ni2+-Sepharose beads was added, washed with 10–40 mm imidazole, before proteins were eluted with 50 μl of 500 mm imidazole, from which 25 μl was applied to a 4–12% polyacrylamide gel. The blot shows the high molecular mass (390 kDa) MtrE-MtrC complex. The samples in the gel lanes are as follows: 1) marker (MW marker); 2) MtrC incubated with nickel-Sepharose (Ni-beads); 3) MtrE-MtrC; 4) MtrE-MtrC + SPDP; 5) MtrE-MtrC + LC SPDP; 6) MtrE K225C-MtrC; 7) MtrE K225C-MtrC + SPDP; 8) MtrE K225C-MtrC + LC SPDP; 9) MtrE R239C-MtrC; 10) MtrE R239C-MtrC + SPDP; 11) MtrE R239C-MtrC + LC SPDP; 12) MtrE K390C-MtrC; 13) MtrE K390C-MtrC + SPDP; and 14) MtrE K390C-MtrC + LC SPDP. The positions of relevant molecular mass markers are shown on the gel; the absence and addition of cross-linkers is indicated by − and +, respectively.
MtrC Oligomerizes to Form a Hexameric Structure
Most models suggested that the MFPs function independently, each bridging one protomer of the IMP and OMP, to form a tripartite assembly composed of a trimeric IMP and OMP that are coupled by three MFPs (18). Such a model would not require that the MFPs interact with one another.
We used EGS to test for interactions, in vitro, between MtrC hairpins, trapping complexes up to a hexamer (Fig. 4A). Nanoflow electrospray mass spectrometry was used to determine the molecular mass of the MtrC hairpin, revealing peak series consistent with the hairpin monomer (9563 ± 0 Da; the calculated molecular mass of the MtrC hairpin is 9558.62 Da), dimer (19,170 ± 1 Da), and hexamer (57,522 ± 17 Da) (Fig. 4B). It was possible to visualize individual hairpins and ring-like structure oligomers in solution by atomic force microscopy (Fig. 4C). These particles had average molecular volumes of 41 ± 1 and 243 ± 5 nm3, respectively, which is also compatible with hexameric hairpins. Similarly, cross-linking studies (Fig. 4D) and the mass spectrum of NT-MtrC (N-terminal truncated, Δ1–34, MtrC) indicated that it exists as a monomer (42,438 ± 3 Da; the calculated molecular mass for MtrC is 43,822.75 Da) and that it exists at oligomeric states up to the hexamer (254,709 ± 10 Da) (Fig. 4E). Interestingly, for both the MtrC hairpin and NT-MtrC, following the appearance of monomers and dimers, the hexameric state is the most abundant oligomeric species.
FIGURE 4.
MtrC and its hairpin domain forms hexamers. A and D, 1 mm EGS was added to 8 μm MtrC hairpin (lane 2) (A) and 8 μm NT-MtrC (lanes 2 and 3) (D) and analyzed by SDS-PAGE. Lane 1 in each gel indicates the molecular mass standard. MtrC and its hairpin were found to form multimers up to and including a hexamer. B and E, the mass spectra of the MtrC hairpin (B) and NT-MtrC (E) indicate that they form hexameric assemblies. The charge states of the MtrC hairpin (labeled 16+ to 14+) that correspond to a measured mass of 57,522 ± 17 Da (B) and oligomerization scheme are shown. In E, the charge states corresponding to both the hexamer (labeled 35+ to 30+) with a measured mass of 254,709 ± 10 Da and a low abundant pentamer with a mass of 212,259 ± 10 Da are illustrated. C, atomic force microscopy-generated three-dimensional pictures of low magnification images of the hairpin (left) and its multimer (right),below which are shown the frequency distribution of molecular volumes of MtrC hairpin monomers (left) and hexamers (right). In each case, the curve indicates a fitted Gaussian function. The peaks correspond to volumes of 41 ± 1 nm3 (n = 100) and 243 ± 5 nm3 (n = 20), indicating that the hairpin forms hexamers. F, EM-generated picture of the average top view of 336 selected images of NT-MtrC particles (top left panel), with the protein density contour level highlighted (top right panel) and after the imposition of 6-fold symmetry. The box size is 280 Å, and the scale bar is 65 Å. The images indicate that MtrC forms a hexameric ring structure, with outer and inner diameters of ∼160 and 55 Å, correspondingly.
In addition, we were able to visualize the MtrC hexamers by electron microscopy (EM), revealing a ring structure (Fig. 4F). An inspection of the image of an averaged ring reveals that it contains six semicircular subunits, which give rise to 12 protrusions (two per subunit). Applying a self-rotation function and imposition of 6- and 12-fold averaging improved the detail of the image, but the imposition of 3- or 9-fold averaging led to an asymmetric image. These data are consistent with MtrC forming a hexameric ring structure that could bridge MtrD and MtrE. This partial dissociation of the helices of the hexameric assembly could provide a mechanism to allow the helices to overlap and engage with the helices of the OMP trimer because this diameter (of 55 Å) would in principle be able to accommodate the MtrE trimer.
Evidence for the Formation of an MtrE(Trimer)-MtrC(Hexamer) Complex
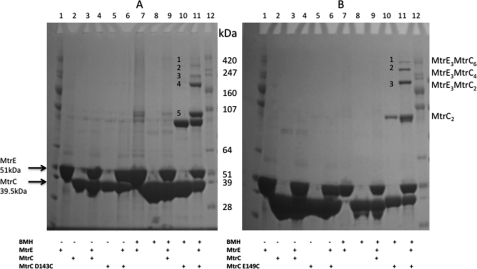
Considering that there is a tendency for the hairpin domains of MtrC to interact, we sought to test the effects of stabilizing this interaction on the ability of MtrC (dimers) to interact with MtrE. The D143C and E149C derivatives of MtrC, in which a single cysteine was inserted into the hairpin domain, underwent intermolecular cross-linking with BMH to form dimers (Fig. 5, A and B, lane 10). In the presence of MtrE, BMH cross-linking of MtrC trapped a number of high molecular mass complexes (Fig. 5, A and B, lane 11), which mass spectrometry analyses confirmed to contain both MtrC and MtrE (MASCOT data not shown). Once formed, these complexes appeared to be stable (supplemental Fig. 2). In the case of E149C, these complexes were calculated to have masses of ∼250, 330, and 400 kDa (Fig. 5B, lane 11), whereas D143C had two bands, with apparent masses of 220 and 270 kDa, instead of a band at 250 kDa. Considering that the MtrC dimer and MtrE timer have molecular masses of ∼88 and 149 kDa, respectively, a reasonable explanation for the occurrence of these complexes is that they correspond to the MtrE trimer binding one, two, and three dimers of MtrC (i.e. with predicted molecular masses of 236, 324, and 412 kDa, respectively). Furthermore, considering that the intra- and interprotomer grooves of MtrE are about 16 Å apart, they are sufficiently close to allow BMH to bridge MtrC bound to these two grooves. In contrast, these complexes could not be trapped with the shorter bifunctional cross-linker bis(maleimido)ethane (data not shown).
FIGURE 5.
Evidence for the formation of an MtrC-MtrE complex with a stoichiometry of 6:3. A and B, an SDS-PAGE gel analysis of the cross-linking of 8 μm MtrC D143C (A) and 8 μm MtrC E149C (B) with 1 mm BMH in the absence and presence of 8 μm Cys-less (C21S) MtrE was undertaken with the following samples: MtrE (lane 2), MtrC (lane 3), MtrE-MtrC (lane 4), MtrC D143C or E149C (lane 5), and MtrE-MtrC D143C or E149C (lane 6) in the absence of BMH and MtrE (lane 7), MtrC (lane 8), MtrE-MtrC (lane 9), MtrC D143C or E149C (lane 10), and MtrE-MtrC D143C or E149C (lane 11) in the presence of BMH. Lanes 1 and 12, and molecular mass standards. Complexes of MtrC and MtrE, identified by MS, are denoted as 1–5 in panel A and 1–3 in panel B. For each gel lane, 20 μl of 8 μm of each protein/1 mm BMH solution was loaded onto 4–12% polyacrylamide gel. The positions of relevant molecular mass markers are shown on the gel.
Moreover, when the MtrC D143C and E149C derivatives were mixed with MtrE C21S K225C, these high molecular mass complexes were further stabilized by BMH cross-linking; six bands with a molecular mass greater than the 247-kDa marker were apparent, which were shown by mass spectrometry to consist of both MtrC and MtrE (supplemental Fig. 3). We found similar cross-linking patterns for MtrC D143C and MtrC E149C in combination with MtrE C21S K390C (data not shown). These data are consistent with formation of cross-linked complexes of the MtrE trimer with up to six MtrC molecules. This behavior is expected because the MtrC derivatives can cross-link both as dimers, stabilizing the 2:3, 4:3, and 6:3 complexes, and with the MtrE derivatives, to additionally stabilize the 1:3, 3:3, and 5:3 complexes of MtrC-MtrE. If the MtrC:MtrE stoichiometry was 3:3, then the molecular mass of these complexes would be about 270 kDa, much less than the molecular mass of the largest complexes trapped by cysteine cross-linking. Our findings are consistent with an MtrC:MtrE stoichiometry of 6:3.
In Vivo Cross-linking of MtrC and MtrE Impairs Pump Function
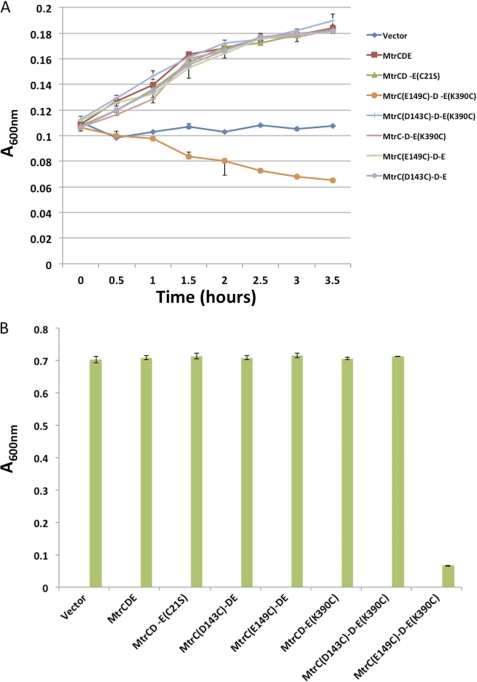
Considering that MtrC D143C and MtrC E149C could be cross-linked to the interprotomer groove of MtrE, via K225C and K390C, with BMH in vitro, these residues should come sufficiently close to allow direct disulfide bond formation. We expressed each of the mtrC D143C, mtrC E149C, mtrE K225C, and mtrE K390C mutants with their cognate wild-type mtrC, mtrD, and mtrE genes in ΔtolC TG1 cells and tested these for tetracycline resistance. The mtrC D143C, mtrC E149C, and mtrE K390C mutants had resistance to tetracycline comparable with cells expressing the wild-type proteins, but the mtrE K225C mutant had reduced resistance. Consequently, we focused further analyses on mtrE K390C, which was expressed with mtrC D143C and mtrC E149C and wild-type mtrD (Fig. 6A). Although the mtrE K390C/mtrC D143C mutant retained resistance, the mtrE K390C/mtrC E149C mutant was sensitive to tetracycline. This behavior indicates that there is a detrimental interaction, presumably via the formation of a disulfide bond, between MtrC E149C and MtrE K390C. We tested all of the mutants for their susceptibility to vancomycin, revealing that, in contrast to the other mutants, the mtrE K390C/mtrC E149C mutant was sensitive to vancomycin (Fig. 6B). These data indicate that the interaction of the MtrC E149C and MtrE K390C derivatives locks MtrE in the open channel conformation. Indeed, if MtrC E149C failed to interact with MtrE K390C, then the cells would be insensitive to vancomycin.
FIGURE 6.
An interaction between MtrC E149C and MtrE K390C induces opening of the MtrE channel. A and B, A series of growth curves (A) and bar charts (B), showing the relative growth measured as the A600 nm over 3.5 h, for E. coli cells (strain ΔtolC TG1(DE3)) expressing the mtrCDE genes, including wild-type mtrC and mtrE as well as the mtrC D143C, mtrC E149C, and mtrE K390C mutants alone and in combination, in the presence of 3 μg/ml tetracycline (A) and 160 μg/ml vancomycin (B). The same strain transformed with the empty vector was used as a control. The cysteine insertion mutants of mtrC and mtrE were constructed in the Cys-less mtrC C25S and mtrE C21S mutants so that disulfide bonds can only occur between the inserted cysteines. Each bar is the average of at least three measurements, with error bars representing the S.D. of these measurements from the average.
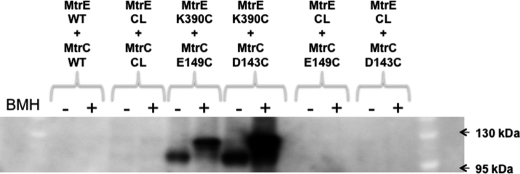
Considering that MtrC E149C interacts with MtrE K390C in vivo, we sought to confirm this interaction by cross-linking these residues with BMH in vivo (Fig. 7). The His tag on MtrC was used to purify and identify, by Western blotting, the cross-linked protein complexes. Two unique bands due to protein cross-linking were detected in samples containing MtrC E149C and MtrE K390C; no corresponding band was apparent when this MtrC derivative was expressed with Cys-less MtrE, indicating that these bands arise from cross-linking of MtrC to MtrE. The lower molecular mass band was present whether or not the samples had been treated with BMH, whereas the other slightly higher molecular mass band was only present in those samples treated with BMH. Probably the complex with cross-linker moved at a slightly higher molecular weight on the gel, as seen for other proteins (37). MtrC D143C was also cross-linked to MtrE K390C in a similar manner. Interestingly, both MtrC D143C and E149C residues cross-linked to MtrE K390C in the absence of BMH, indicative of the formation of intermolecular disulfide bonds, which were resistant to reduction by 50 mm DTT. Other periplasmic proteins have been reported that form disulfide bonds that are resilient to reduction by reducing agents (37). One possibility is that the residues forming the disulfide bond become buried, in this case within the interprotomer groove, so that they are shielded from the DTT.
FIGURE 7.
In vivo cross-linking of MtrC E149C to MtrE K390C. An SDS-PAGE gel analysis of the in vivo cross-linking of MtrC D143C and E149C to MtrE K390C is shown. The wild-type and Cys-less proteins are abbreviated WT and CL, respectively. The cells, expressing the MtrC and MtrE derivatives, were harvested from a 200-ml culture, resuspended to 1 ml, and incubated with 1 mm BMH for 1 h at 25 °C. The cells were washed to remove excess BMH and sonicated to release the membranes, which were collected by ultracentrifugation and resuspended in 1 ml of cross-linking buffer containing 8 m urea and 2% βDDM. Solubilized membrane proteins were separated from the membrane debris by ultracentrifugation. To the supernatant, 100 μl of Ni2+-Sepharose beads was added; after washing with 10–40 mm imidazole, proteins were eluted from the beads with 50 μl of 500 mm imidazole, and 40 μl was loaded onto 4–12% polyacrylamide gel. Protein samples were treated with 50 mm DTT in an attempt to reduce any disulfide bonds. The gel was blotted with anti-His tag antibodies to identify complexes of His-tagged MtrC. As a control, cells co-expressing MtrC E149C or MtrC D143C and Cys-less MtrE were treated with BMH so that any MtrC-MtrE complexes could be distinguished from MtrC dimers. Note that cross-linked bands shown were only present for cells expressing MtrC D143C or E149C and MtrE K390C. The positions of relevant molecular mass markers are shown on the gel.
Evidence That the MtrC Hairpin Tip Scans along the Intersubunit Groove of MtrE
We hypothesized that if Cys-149 of MtrC comes into close contact with Cys-390 of MtrE then we should be able to cross-link the cysteine insertion derivatives of these proteins using a bifunctional cross-linker with a much shorter spacer arm than BMH, and that such a shorter cross-linker would hinder the formation of MtrC dimers that could be positioned in the intra- and interprotomer grooves of MtrE. We tested whether DBBR, with a 5 Å spacer length, would cross-link MtrE K390C to MtrC E149C and/or D143C in vitro. Interestingly, although an SDS-PAGE analysis did not reveal a unique band that would confirm cross-linking of MtrE K390C to MtrC E149C, two unique bands were apparent for the cross-linking of MtrE K390C to MtrC D143C, with DBBR (Fig. 8A). The higher molecular mass band was subjected to a MALDI-MS analysis, revealing that it contained both MtrC and MtrE. The molecular mass of this band is suggestive of a complex of MtrC monomer bound to MtrE trimer. We extended this analysis by testing whether MtrC S141C, D143C, S146C, and E149C derivatives could cross-link with the MtrE R235C and R239C derivatives, which lie further up the intersubunit groove. MtrC D143C, but none of the other derivatives, interacted with MtrE R235C and R239C (Fig. 8B). Presumably, Asp-143, positioned at the tip of the hairpin, is in a more flexible position so that it can interact with several residues along the interprotomer groove as MtrC docks on to MtrE, whereas other residues along the hairpin may only come into close contact with groove residues as MtrC and MtrE become fully engaged in the functional pump.
FIGURE 8.
The tip of the MtrC hairpin interacts with multiple residues along the intersubunit groove of MtrE. A and B, an SDS-PAGE gel analysis of the cross-linking of MtrC D143C and E149C to MtrE K390C (A) and MtrC S141C, D143C, S146C, and E149C to MtrE R239C and R235C (B) with dibromobimane. The presence of each protein is indicated by a + below each gel lane. Note that only MtrC D143C interacted with MtrE and was cross-linked to residues at positions 235, 239, and 390. For each gel lane, 20 μl of an 8 μm of each protein/0.75 mm DBBR solution was loaded onto a 4–12% polyacrylamide gel. The positions of relevant molecular mass markers are shown on the gel.
DISCUSSION
In the absence of a structure for a fully assembled tripartite pump, the debate is still open on how the individual components interact with one another. Although some previous cross-linking studies supported modeling of the assembly with a stoichiometry of 3:3:3 and have since gained popularity (17, 18), some others, which have indicated an engagement in both grooves (24), have been somewhat overlooked. However, recent structural studies have handed further support to that hypothesis. For example, the MFP MacA was shown to form a hexamer, the assembly of which was required for the interaction with the cognate IMP MacB (19). This is consistent with the finding that the MacB transporter functions as a homodimer that interacts with trimeric TolC (38) because, although it is very difficult to construct a centrally symmetric assembly with only three MFPs in this system, if we consider a hexameric IMP assembly, then there is a trimer of MacB dimers within the tripartite assembly, with each MacB monomer binding a MacA molecule. Furthermore, it was proposed that the β-domains of MacA interact with the periplasmic domain of MacB, whereas the α-helical hairpins form a connecting channel to TolC (19, 20). Recent structural studies of a CusAB complex have revealed a hexamer of the MFP CusA bound to a trimer of the IMP CusB, consistent with this view of the MacBA assembly (9). The β-domains of CusB form a cap over the periplasmic domain of CusA, whereas the lipoyl and hairpin domains form a channel. At its periplasmic end, defined by the hairpins, the channel has an internal diameter of 56 Å but is constricted in the region defined by the lipoyl domains to about 18 Å, suggesting that it forms a connecting channel to CusC. Considering that CusC forms trimers, these data suggest that the CusABC tripartite complex is assembled with a stoichiometry of 3:6:3. Although these findings relate to the metal ion transporters, they also bring into question whether similar stoichiometry may be found in the classical multidrug pumps.
We have addressed this stoichiometry question by investigating the assembly of the MtrCDE multidrug pump. Both the IMP MtrD and the OMP MtrE form trimers (supplemental Fig. 4) (30), whereas MtrC can form hexamers (Fig. 4). Our previous studies revealed that MtrD interacts weakly with MtrE, whereas MtrC has a relatively strong interaction with MtrE and MtrD, suggesting that these proteins form a tripartite complex in which the interaction of MtrE and MtrD is stabilized by the binding of MtrC across these proteins (30). Considering that MtrC is anchored to the inner membrane by an α-helix and that the NT-MtrC has an affinity for MtrD that is an order of magnitude greater than that for MtrE, this suggests that the MtrCD complex is preformed ready for interaction with MtrE (30). Here, we have shown that MtrC readily forms dimers, which upon cross-linking are trapped in complex with MtrE, with a stoichiometry of 3:6 (Fig. 5). Consistent with this behavior, we provide evidence that MtrC binds to both intraprotomer and interprotomer grooves on the surface of MtrE (30) (Figs. 3 and 7). Accordingly, our studies are consistent with the MtrCDE tripartite pump being assembled with a stoichiometry of 3:6:3 and with a topology in which MtrD and MtrE contact one another and MtrC is bound across these proteins (supplemental Fig. 5). This positions MtrC to detect drug binding to MtrD and to communicate this to MtrE, inducing opening of its channel (30).
The emerging view of the MFPs, such as MacA and CusA, forming hexameric channels that connect the IMP to the OMP contrasts with the established view of the AcrABTolC assembly, in which the tips of the IMP AcrB and OMP TolC directly interact and the monomers of the MFP AcrA, which contact the periplasmic domains of both AcrB and TolC, do not interact with each other, rather than forming a continuous connecting channel. In support of the hypothesis that more than one MFP binds per OMP-IMP protomer, we have identified a hexameric assembly of MtrC (Fig. 4, D and E), with our EM studies revealing that the six protomers are arranged into a cylindrical structure (Fig. 4F). However, the internal aperture of the channel formed by MtrC, at ∼55 Å, is apparently larger than expected for 12 helices arranged into a cylinder. For example, the MacA hexameric assembly has an internal diameter of about 30 Å (19), whereas TolC has an internal diameter of about 35 Å (8). To confirm this difference in the apertures of the MtrC and the MacA hexamers, we also visualized the MacA hexamer, confirming that this also formed a ring structure but with a smaller internal diameter of about 34 Å (supplemental Fig. 6). As a consequence, although the diameter of the MacA hexamer is insufficient to fit TolC, the MtrC hexamer could accommodate the MtrE trimer. One possibility is that within the hexamer formed by MtrC, the interactions between the helices are stretched, possibly to the point of dissociation, but the assembly is kept together by interactions between adjacent β-domains, which could be strengthened by dimerization of adjacent hairpin domains. Consistent with such a proposal, MtrC dimers, stabilized by cross-linking, promote assembly of the MtrC-MtrE complex (Fig. 5A). It is notable that in the structure of the MacA hexamer, the β-domains are arranged into a ring with an internal diameter of 53 Å (19). If the interaction between adjacent hairpin dimers is weaker than between the β-domains, this could allow for a partial dissociation of the complex in which the hairpin dimer ”fingers“ move apart, increasing the apparent diameter of the hexameric assembly. Indeed, considering that in the MacA and AcrA structures the helical hairpins are displaced by about 20° from one another (10, 19) and that previous studies have revealed that there is conformational flexibility in AcrA that allows movement of the hairpin relative to the β-domain (39), it is feasible that the helices can move apart in the assembled hexamer. This appears to be the case for the CusB hexamer, in which the hairpins are tethered together at their base by the lipoyl domains but the hairpins are splayed out at their periplasmic ends, creating a channel with an aperture of 56 Å (9). This is sufficiently large to allow the insertion of the tip of CusC. It was proposed that this state might represent the initial interaction of CusC trimer with the CusB hexamer and that subsequently CusC slides a further 40 Å inside the CusB hexameric channel so that the CusB hairpins are totally engaged with CusC. Recent structural studies of ZneB, the MFP of the tripartite zinc efflux pump ZneABC, established that it has a compact crescent shape with bound Zn2+ but adopts an extended linear shape upon releasing the Zn2+ (40). This might provide a mechanism for ZneB to ”slide“ across the surface of the OMP ZneC. Our studies provide some support for such a two-step interaction in that we have established that a D143C derivative of MtrC can be cross-linked to the K225C, K390C, R235C, and R239C derivatives of MtrE, which are located along the intersubunit groove in this order from the periplasmic end to the equatorial region (Fig. 1, supplemental Fig. 3, and Fig. 8). Considering that Asp-143 is predicted to lie at the tip of the hairpin, this might suggest that the hairpin domain scans along the groove during engagement.
Clearly if the coiled-coils of the hairpin domain of the MFP fit into the intra- and interprotomer grooves, then this interaction would presumably destabilize the hairpin interactions. However, this may not preclude the formation of a hexameric assembly in which three MFP dimers are tethered together by interactions between adjacent β-domains, with one subunit of the dimer interacting with the intraprotomer groove and the other with the interprotomer groove. Indeed, in the fully assembled tripartite complex, the MFPs are likely to be held together, via their β-domains, by their interaction with the IMP. Considering that the OMP and consequently the IMP bind a pair of MFPs, probably as a dimer, which is bound more tightly by the IMP than the OMP (14, 30), one can envisage a situation where the trimeric IMP tethers three MFP dimers, which can dynamically associate/dissociate. Dimerization of the hairpin domains of the MFP could be used to position the second MFP subunit for binding to the second groove, tightening the interaction of the dimer as a whole with the OMP. A recent study of the kinetics of the interaction of AcrA, AcrB, and TolC, indicated that (i) AcrA forms dimers that interact with AcrB and (ii) the binary complexes of AcrAB and AcrBTolC can form before the ternary complex of AcrABTolC, but formation of the AcrATolC complex inhibits ternary complex formation (27). These findings are suggestive of an ordered assembly of the AcrABTolC ternary complex. Our studies revealed that MtrC binds MtrD with a much higher affinity than MtrE (30), and considering that MtrC is anchored to the inner membrane by its N-terminal α-helix, this suggests that the MtrCD binary complex will form prior to its interaction with MtrE. One possibility is that MtrD contacts MtrE, allowing the MtrC dimers bound to MtrD to then bind to MtrE. These features of the MFP may enable it to serve a dual role; having a propensity to form dimers that can be assembled into hexamers, which can bridge major facilitator transporters and possibly ABC transporters to their cognate OMPs, these dimers can disassemble so that they can interact with the intra- and interprotomer grooves of the cognate OMPs of RND transporters. Indeed, although two recent studies both reported that AcrA forms dimers, they reached somewhat conflicting conclusions, with one suggesting that AcrB interacts with TolC, with AcrA dimers bridging AcrB-TolC complex (27), whereas the other indicated that the dimers assemble into a hexameric channel that intervenes between AcrB and TolC (28). Our studies of MtrCDE allow some of these conflicting findings to be reconciled and point toward a general conclusion that tripartite multidrug pumps are assembled with a stoichiometry of 3:6:3.
Previous studies have been interpreted in terms of a transport mechanism in which there are two energetically distinct steps (35). Firstly, the helical protrusions of the OMP engage with the exposed periplasmic crown of the IMP to induce a partial relaxation of helices H7/H8, resulting in partial opening of the OMP channel and exposure of the intraprotomer groove of the OMP, which forms part of the binding site for the MFP. Secondly, the engagement of the MFP with the intraprotomer groove is thought to disrupt the so-called secondary filter, which is composed of a double ring of aspartate residues in the case of TolC, allowing the full opening of the channel (35, 41, 42). Consistent with this mechanism, we established that although MtrD and MtrE can interact with one another and with MtrC, the MtrE channel remains locked in the absence of transported drug substrates (30). Similarly formation of the AcrABTolC ternary complex has been shown to be independent of drug binding (27). The mutation of residues within the intraprotomer groove had the effect of making these mutants more susceptible to nafcillin, tetracycline, and vancomycin than control cells that lack the pump (30). Furthermore, these MtrE mutants, when expressed alone, were insensitive to vancomycin, but when expressed with MtrC, they became highly susceptible to vancomycin, indicating that the intraprotomer groove has a critical role in controlling opening of the MtrE channel in response to binding MtrC, possibly by communicating its occupancy to the second selective filter, causing it to be destabilized. Here, we have found that alterations of residues within the interprotomer groove also have the effect of making the mutants more susceptible to nafcillin and tetracycline but had no effect on their vancomycin sensitivity (Table 1 and Fig. 2). Although this might suggest that these MtrE mutants simply lacked the ability to interact with MtrCD, the K225/R239E and K225E/K390E mtrE double mutants had a much greater susceptibility to nafcillin and tetracycline than cells expressing MtrCD alone (Fig. 2), indicating that the MtrCDE pump assembled but was non-functional. Our microcalorimetry studies confirmed that the K225E/R239E derivative of MtrE was capable of binding MtrC (supplemental Fig. 1). Although indicating that this derivative could be assembled into a tripartite pump, it is possible that MtrC binds exclusively to the intraprotomer groove of these derivatives because the interprotomer groove has been disrupted. In this manner, the pump could become locked in a non-functional state. Because these mutants were insensitive to vancomycin, we concluded that the pump was closed, prohibiting both cellular nafcillin/tetracycline extrusion and vancomycin entry (Fig. 2). In contrast, cells expressing MtrE K390C and MtrC E149C, with wild-type MtrD, were sensitive to both tetracycline and vancomycin, indicating that an interaction between the hairpin of MtrC and the interprotomer groove of MtrE could trap an assembly in which the MtrE channel was open (Fig. 6). Consistent with this proposal, MtrC E149C and MtrE K390C became cross-linked in vivo due to the formation of a disulfide bond (Fig. 7, lane 4). This disulfide bond appeared resistant to reduction by DTT, possibly because the cysteines involved are protected by the formed protein-protein complex at the intersubunit groove of MtrE and so inaccessible to DTT (32). However, MtrC E149C could not be cross-linked to MtrE K390C in vitro with DBBR, suggesting that this is a transient interaction, which is brought about during the closed-to-open conformational change in MtrE. Presumably, a salt bridge forms between the MtrE Lys-390 and MtrC Glu-149 during the cycling of MtrE between the closed and open channel conformations. However, this interaction cannot be essential because although the MtrE K390E derivative is impaired in function, the pump is still viable in conferring resistance, albeit reduced (Fig. 2). On the other hand, combining this mutation with another mutation, such as K225E, completely abolished the ability of the pump to confer resistance (Fig. 2). It appears that disrupting a single salt bridge impairs the ability of the pump to open and close, but disrupting two can lock the OMP in the closed conformation. In contrast, MtrC D143C, which was also capable of forming a disulfide bond with MtrE K390C (Fig. 7), did not prevent closure of the MtrE channel (Fig. 2C), and the pump was functional (Fig. 2, A and B). Moreover, D143C was the only derivative that could be cross-linked with DBBR to K390C and to R235C and R239C, which lie further along the intersubunit groove. Presumably by virtue of its position at the tip of the hairpin, Asp-143 can be accommodated at various positions within the MtrE groove without inactivating the pump. These findings highlight the highly dynamic nature of the interactions of the MFP and its cognate OMP, and it is likely that they have to reflect the dynamics of the conformational changes in the IMP, which is thought to undergo peristaltic cycling during the drug extrusion process (43, 44). Intriguingly the importance of the conformational flexibility during the pump function on the level of the IMP was also recently highlighted by the usage of conditional cysteine cross-linking (45, 46).
In summary, we demonstrate that a number of surface residues in the OMP MtrE, which are predicted to line up along the interprotomer groove, are involved in direct interactions with the hairpin of its cognate MFP MtrC. Mutations of the residues of this region appear to have a cumulative effect on the pump function, and pairwise cysteine cross-links, although not affecting the pump assembly, are able to stall the pump in an inactive state. These findings are consistent with an accumulating body of evidence that tripartite pumps may assemble in stoichiometries higher than 3:3:3 (20–24, 27, 28, 47) and indicate an active role of the MFP in controlling the conformational changes in the OMP to elicit channel opening for drug extrusion.
Supplementary Material
Acknowledgments
We are grateful for support in atomic force microscopy imaging from Dr. Michael Edwardson and Dr. Robert Henderson (Department of Pharmacology, University of Cambridge, UK).
This work was supported by grants from the Wellcome Trust (to M. I. B. W. and A. R. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–6.
Hairpin refers to the 83-residue MtrC α-helical coiled-coil hairpin domain, consisting of residues 103–183 of MtrC.
- IMP
- inner membrane protein
- OMP
- outer membrane protein
- MFP
- membrane fusion protein
- ABC
- ATP-binding cassette
- MF transporter
- major facilitator
- RND transporter
- resistance-nodulation-cell division transporter
- Mtr
- multiple transferable resistance
- ITC
- isothermal calorimetry
- βDDM
- β-dodecyl maltoside
- NT-MtrC
- the N-terminal truncated (Δ1–34) MtrC
- BMH
- bis(maleimido)hexane
- DBBR
- dibromobimane
- EGS
- ethylene glycol bis-succinimidyl succinate
- SPDP
- N-succinimidyl 3-(2-pyridyldithio)-propionate
- LC SPDP
- Succinimidyl 6-(3-[2-pyridyldithio]-propionamido)hexanoate.
REFERENCES
- 1. Pietras Z., Bavro V. N., Furnham N., Pellegrini-Calace M., Milner-White E. J., Luisi B. F.(2008) Curr. Drug Targets 9, 719–728 [DOI] [PubMed] [Google Scholar]
- 2. Misra R., Bavro V. N. (2009) Biochim. Biophys. Acta 1794, 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blair J. M., Piddock L. J. (2009) Curr. Opin. Microbiol. 12, 512–519 [DOI] [PubMed] [Google Scholar]
- 4. Holland I. B., Schmitt L., Young J. (2005) Mol. Membr. Biol. 22, 29–39 [DOI] [PubMed] [Google Scholar]
- 5. Rensing C., Grass G. (2003) FEMS Microbiol. Rev. 27, 197–213 [DOI] [PubMed] [Google Scholar]
- 6. Su C. C., Long F., Yu E. W. (2011) Protein Sci. 20, 6–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murakami S., Nakashima R., Yamashita E., Yamaguchi A. (2002) Nature 419, 587–593 [DOI] [PubMed] [Google Scholar]
- 8. Yu E. W., McDermott G., Zgurskaya H. I., Nikaido H., Koshland D. E., Jr. (2003) Science 300, 976–980 [DOI] [PubMed] [Google Scholar]
- 9. Long F., Su C. C., Zimmermann M. T., Boyken S. E., Rajashankar K. R., Jernigan R. L., Yu E. W. (2010) Nature 467, 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higgins M. K., Bokma E., Koronakis E., Hughes C., Koronakis V. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9994–9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Su C. C., Yang F., Long F., Reyon D., Routh M. D., Kuo D. W., Mokhtari A. K., Van Ornam J. D., Rabe K. L., Hoy J. A., Lee Y. J., Rajashankar K. R., Yu E. W. (2009) J. Mol. Biol. 393, 342–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koronakis V., Sharff A., Koronakis E., Luisi B., Hughes C. (2000) Nature 405, 914–919 [DOI] [PubMed] [Google Scholar]
- 13. Kulathila R., Kulathila R., Indic M., van den Berg B. (2011) PLoS One 6, e15610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Touzé T., Eswaran J., Bokma E., Koronakis E., Hughes C., Koronakis V. (2004) Mol. Microbiol. 53, 697–706 [DOI] [PubMed] [Google Scholar]
- 15. Tamura N., Murakami S., Oyama Y., Ishiguro M., Yamaguchi A. (2005) Biochemistry 44, 11115–11121 [DOI] [PubMed] [Google Scholar]
- 16. Tikhonova E. B., Zgurskaya H. I. (2004) J. Biol. Chem. 279, 32116–32124 [DOI] [PubMed] [Google Scholar]
- 17. Lobedanz S., Bokma E., Symmons M. F., Koronakis E., Hughes C., Koronakis V. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4612–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Symmons M. F., Bokma E., Koronakis E., Hughes C., Koronakis V. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7173–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yum S., Xu Y., Piao S., Sim S. H., Kim H. M., Jo W. S., Kim K. J., Kweon H. S., Jeong M. H., Jeon H., Lee K., Ha N. C. (2009) J. Mol. Biol. 387, 1286–1297 [DOI] [PubMed] [Google Scholar]
- 20. Xu Y., Song S., Moeller A., Kim N., Piao S., Sim S. H., Kang M., Yu W., Cho H. S., Chang I., Lee K., Ha N. C. (2011) J. Biol. Chem. 286, 13541–13549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Su C. C., Long F., Zimmermann M. T., Rajashankar K. R., Jernigan R. L., Yu E. W. (2011) Nature 470, 558–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tikhonova E. B., Dastidar V., Rybenkov V. V., Zgurskaya H. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16416–16421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reffay M., Gambin Y., Benabdelhak H., Phan G., Taulier N., Ducruix A., Hodges R. S., Urbach W. (2009) PLoS One 4, e5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stegmeier J. F., Polleichtner G., Brandes N., Hotz C., Andersen C. (2006) Biochemistry 45, 10303–10312 [DOI] [PubMed] [Google Scholar]
- 25. Mima T., Joshi S., Gomez-Escalada M., Schweizer H. P. (2007) J. Bacteriol. 189, 7600–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gristwood T., Fineran P. C., Everson L., Salmond G. P. (2008) Mol. Microbiol. 69, 418–435 [DOI] [PubMed] [Google Scholar]
- 27. Tikhonova E. B., Yamada Y., Zgurskaya H. I. (2011) Chem Biol. 18, 454–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu Y., Lee M., Moeller A., Song S., Yoon B. Y., Kim H. M., Jun S. Y., Lee K., Ha N. C. (2011) J. Biol. Chem. 286, 17910–17920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shafer W. M., Qu X., Waring A. J., Lehrer R. I. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janganan T. K., Zhang L., Bavro V. N., Matak-Vinkovic D., Barrera N. P., Burton M. F., Steel P. G., Robinson C. V., Borges-Walmsley M. I., Walmsley A. R. (2011) J. Biol. Chem. 286, 5484–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagakubo S., Nishino K., Hirata T., Yamaguchi A. (2002) J. Bacteriol. 184, 4161–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 33. Armougom F., Moretti S., Poirot O., Audic S., Dumas P., Schaeli B., Keduas V., Notredame C. (2006) Nucleic Acids Res. 34, W604–W608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 35. Bavro V. N., Pietras Z., Furnham N., Pérez-Cano L., Fernández-Recio J., Pei X. Y., Misra R., Luisi B. (2008) Mol Cell. 30, 114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Welch A., Awah C. U., Jing S., van Veen H. W., Venter H. (2010) Biochem. J. 430, 355–364 [DOI] [PubMed] [Google Scholar]
- 37. Kobayashi T., Ito K. (1999) EMBO J. 18, 1192–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin H. T., Bavro V. N., Barrera N. P., Frankish H. M., Velamakanni S., van Veen H. W., Robinson C. V., Borges-Walmsley M. I., Walmsley A. R. (2009) J. Biol. Chem. 284, 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mikolosko J., Bobyk K., Zgurskaya H. I., Ghosh P. (2006) Structure 14, 577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Angelis F., Lee J. K., O'Connell J. D., 3rd, Miercke L. J., Verschueren K. H., Srinivasan V., Bauvois C., Govaerts C., Robbins R. A., Ruysschaert J. M., Stroud R. M., Vandenbussche G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11038–11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andersen C., Koronakis E., Bokma E., Eswaran J., Humphreys D., Hughes C., Koronakis V. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11103–11108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pei X. Y., Hinchliffe P., Symmons M. F., Koronakis E., Benz R., Hughes C., Koronakis V. (2011) Proc. Natl Acad Sci U.S.A. 108, 2112–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seeger M. A., Schiefner A., Eicher T., Verrey F., Diederichs K., Pos K. M. (2006) Science 313, 1295–1298 [DOI] [PubMed] [Google Scholar]
- 44. Murakami S., Nakashima R., Yamashita E., Matsumoto T., Yamaguchi A. (2006) Nature 443, 173–179 [DOI] [PubMed] [Google Scholar]
- 45. Seeger M. A., von Ballmoos C., Eicher T., Brandstätter L., Verrey F., Diederichs K., Pos K. M. (2008) Nat. Struct. Mol. Biol. 15, 199–205 [DOI] [PubMed] [Google Scholar]
- 46. Takatsuka Y., Nikaido H. (2009) J. Bacteriol. 191, 1729–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim H. S., Nagore D., Nikaido H. (2010) J. Bacteriol. 192, 1377–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.