Abstract
We evaluated the metabolic impact of farnesoid X receptor (FXR) activation by administering a synthetic FXR agonist (GW4064) to mice in which obesity was induced by a high fat diet. Administration of GW4064 accentuated body weight gain and glucose intolerance induced by the high fat diet and led to a pronounced worsening of the changes in liver and adipose tissue. Mechanistically, treatment with GW4064 decreased bile acid (BA) biosynthesis, BA pool size, and energy expenditure, whereas reconstitution of the BA pool in these GW4064-treated animals by BA administration dose-dependently reverted the metabolic abnormalities. Our data therefore suggest that activation of FXR with synthetic agonists is not useful for long term management of the metabolic syndrome, as it reduces the BA pool size and subsequently decreases energy expenditure, translating as weight gain and insulin resistance. In contrast, expansion of the BA pool size, which can be achieved by BA administration, could be an interesting strategy to manage the metabolic syndrome.
Keywords: Bile Acid, Diabetes, Energy Metabolism, Nuclear Receptors, Obesity, FXR
Introduction
Bile acids (BAs)3 are essential constituents of bile that facilitate dietary lipid absorption and cholesterol catabolism. Besides their well established roles in dietary lipid absorption and cholesterol homeostasis, BAs also function as biological signaling molecules, endowing them with an endocrine function (1). For instance, BAs activate MAPK signaling pathways (2, 3) and are natural ligands that activate the nuclear hormone receptor farnesoid X receptor (Fxr, NR1H4) (4–6), a transcription factor that controls both the biosynthesis and enterohepatic recycling of BAs (7). FXR regulates the expression of the short heterodimer partner (Shp, NR0B2) that inhibits the activity of several other nuclear receptors and contributes to the negative feedback regulation of BA biosynthesis. SHP inhibits the activity of liver X receptor α and β (liver X receptor α, NR1H3; liver X receptor β, NR1H2) and liver receptor homolog-1 (LRH-1, NR5A2), both necessary for transcriptional induction of the rate-limiting enzyme in BA biosynthesis, cholesterol 7α-hydroxylase (CYP7A1) (8–13). The FXR-mediated induction of FGF19 (FGF15 in mouse) in intestinal epithelial cells also contributes to the feedback repression of BA biosynthesis. FGF15/19 passes via the portal vein to the hepatocytes and acts on FGFR4 (14–16). Likewise, the hepatic FXR-mediated SHP induction after BA administration inhibits fatty acid and triglyceride (TG) biosynthesis and VLDL production (17).
A final pathway through which BAs signal recently emerged and involves the binding and activation of G-protein-coupled receptors such as TGR5 (also named GPR131 or M-BAR) (18, 19). Through the activation of this G-protein-coupled receptor, BAs induce intracellular cAMP levels in brown adipose tissue (BAT) and skeletal muscle. This leads to activation of type 2 iodothyronine deiodinase (D2), the enzyme that converts inactive thyroxine into active 3,5,3′-triiodothyronine (20), which determines thyroid hormone receptor saturation, as well as the activation of the master regulator of mitochondrial biogenesis, the peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α). Via this pathway, energy expenditure in BAT and skeletal muscle is stimulated, translating into weight loss and insulin sensitization (21). Furthermore, TGR5 was recently shown also to enhance insulin secretion through its stimulating effects on GLP-1 release in entero-endocrine L-cells (22). Taken together, these observations build a strong case that BAs have effects beyond the strict control of BA homeostasis and function as general metabolic integrators.
In view of the complex signaling cascades induced by BAs, it is not all that surprising that the effects of BAs are not totally mimicked by those of synthetic FXR agonists. In fact, we previously reported that high fat fed mice treated with the synthetic FXR agonist GW4064 have a tendency to increase body weight (21). In this study, we provide compelling evidence that FXR activation with a synthetic agonist, GW4064, induces obesity and diabetes in high fat fed C57BL/6J mice, an effect that is diametrically opposite to the beneficial impact of the natural BA, cholic acid (CA), in this mouse model. We trace the metabolic differences between natural and synthetic FXR agonists to their opposing actions on BA pool size.
EXPERIMENTAL PROCEDURES
Materials
CA was obtained from Sigma. GW4064 was a generous gift of Mitsubishi Pharmaceuticals.
Animal Studies
Animal experiments were approved by the institutional review board at Keio University. Male C57BL/6J mice, 6–7 weeks of age, were obtained from Charles River Laboratories (l'Arbresle, France) and CLEA Japan, Inc. (Tokyo, Japan). All mice were maintained in a temperature-controlled (23 °C) facility with a 12-h light/dark cycle and were given free access to food and water. The control diet and high fat diet were obtained from Research Diets, Inc. The control diet (D12450D) contained 26.2% protein, 67.3% carbohydrate, and 4.3% fat, and the high fat diet (D12492) contained 26.2% protein, 26.3% carbohydrate, and 34.9% fat. For treatment with CA, mice were fed diets with 0.5% (w/w) CA. GW4064 (180 mg/kg w/w) was mixed with diets. Based on a daily food intake of 5 g, this resulted in a daily dose of 15 mg/kg GW4064. The mice were fasted 6 h before harvesting blood for subsequent blood measurements and tissues for RNA isolation and histology. Oxygen consumption was measured using the Oxymax apparatus (Columbus Instruments, Columbus, OH) (23).
Morphological Studies
Pieces of mouse tissues were fixed in Bouin's solution, dehydrated in ethanol, embedded in paraffin, and cut at a thickness of 5 μm. Sections were deparaffinized, rehydrated, and stained with hematoxylin and eosin.
mRNA Expression Analysis by Quantitative RT-PCR
Expression levels were analyzed in cDNA synthesized from total mRNA using real time PCR as described previously (21). The sequences of the primer sets used are displayed in Table 1.
TABLE 1.
Primer sequences of genes used for quantification of mRNAs by real-time PCR
| Gene | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) |
|---|---|---|
| 18 S | GATGGGAAGTACAGCCAGGT | TTTCTTCAGCCTCTCCAGGT |
| FXR | CAAAATGACTCAGGAGGAGTACG | GCCTCTCTGTCCTTGATGTATTG |
| LXRα | GGGAGGAGTGTGTGCTGTCAG | GAGCGCCTGTTACACTGTTGC |
| SHP | CAAGGAGTATGCGTACCTGAAG | GGCTCCAAGACTTCACACAGT |
| PGC1α | AAGGGCCAAACAGAGAGAGA | GCGTTGTGTCAGGTCTGATT |
| Cyp7a1 | TACAGAGTGCTGGCCAAGAG | TTCAAGGATGCACTGGAGAG |
| IBABP | ACCATTGGCAAAGAATGTGA | GACCTCCGAAGTCTGGTGAT |
| FGF15 | GGCAAGATATACGGGCTGAT | GATGGTGCTTCATGGATCTG |
| D2 | TTCTGAGCCGCTCCAAGT | GGAGCATCTTCACCCAGTTT |
| UCP1 | GGCCCTTGTAAACAACAAAATAC | GGCAACAAGAGCTGACAGTAAAT |
| mCPT-1 | GCACTGCAGCTCGCACATTACAA | CTCAGACAGTACCTCCTTCAGGAAA |
| PPARγ | AGGCCGAGAAGGAGAAGCTGTTG | TGGCCACCTCTTTGCTCTGCTC |
| aP2 | TGCCACAAGGAAAGTGGCAG | CTTCACCTTCCTGTCGTCTG |
| Leptin | CAAGCAGTGCCTATCCAGAA | GTGAAGCCCAGGAATGAAGT |
| Adiponectin | AAGAAGGACAAGGCCGTTCTCTT | GCTATGGGTAGTTGCAGTCAGTT |
| FAS | CTGCAGAGAAGCGAGCATAC | TCTGCCAGTGAGTTGAGGAC |
| ACC1 | ACCCACTCCACTGTTTGTGA | CCTTGGAATTCAGGAGAGGA |
Biochemistry and Determination of Glucose, Lipids, and BAs
An oral glucose tolerance test (OGTT) was performed in animals that were fasted 6 h. Glucose was administered by gavage at a dose of 2 g/kg. An intraperitoneal insulin tolerance test (IPITT) was done in 6-h fasted animals. Insulin was injected at a dose of 0.75 units/kg. Glucose quantification was done with the Maxi kit glucometer 4 (Bayer Diagnostic, Puteaux, France) or glucose RTU (bioMérieux Inc., Marcy l'Etoile, France). Plasma insulin and leptin concentrations were measured using ELISA (Crystal Chem Inc., Downers Grove, IL). Serum total cholesterol, TG, and free fatty acids were measured as described previously (17). TG, free fatty acids, and total cholesterol in the 3T3-L1 cells and liver were extracted by the classical Folch method (24) and measured as described previously (17). BAs in enterohepatic organs of fed mice were determined as described previously (25, 26).
Cell Culture
3T3-L1 cell were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin in 5% CO2-humidified atmosphere. Differentiation of preadipocytes 2 days after confluence was induced by exposing them to a differentiation medium containing 5 μg/ml insulin, 1 μm dexamethasone, and 0.5 mm 3-isobutyl-1-methylxanthine in 10% FBS-supplemented DMEM for 48 h (27). Then the culture medium was replaced with DMEM supplemented with 10% FBS, 5 μg/ml insulin, and with or without 10 μm GW4064 or 20 μm chenodeoxycholic acid (CDCA). Culture medium was changed every day. Four days after the initiation of adipocyte differentiation, cells were stimulated with insulin (5 μg/ml), and the decrease of glucose in the medium was measured after 12 h.
Western Blot Analysis and Antibodies
3T3-L1 cells and frozen tissues were homogenized in lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, 2.5 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1% Triton X-100, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml phenylmethylsulfonyl fluoride). After centrifugation at 15,000 × g for 15 min, the supernatants were collected, and protein concentration was determined. Antibodies to total Akt and phosphorylated-Akt (Ser-473) were purchased from Cell Signaling Technology (Beverly, MA). Antibodies to IRS-1 and phosphorylated-IRS-1 (Ser-307) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Statistical Analysis
Values were reported as means ± S.E. Statistical differences were determined using analysis of variance (StatView software, Abacus Concepts, Inc., Berkeley, CA). Statistical significance is displayed as follows: *, p < 0.05; **, p < 0.01 versus high fat diet and as #, p < 0.05, or ##, p < 0.01, versus high fat diet supplemented with GW4064.
RESULTS
GW4064 Induces Weight Gain in C57BL/6J Mice
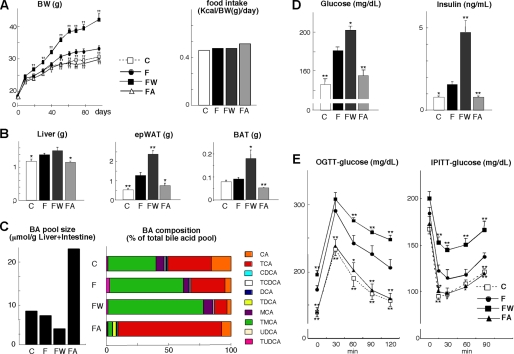
We first evaluated the metabolic effects of FXR activation with the synthetic agonist GW4064 in male C57BL/6J mice. These mice received either normal chow, a high fat diet, or a high fat diet supplemented with GW4064 or CA for 3 months. Food intake was not affected by any of these treatments (Fig. 1A). When compared with animals fed a normal chow, high fat fed animals gained more weight. The animals fed with the same high fat diet supplemented with CA gained weight at a rate comparable with mice fed the standard chow (Fig. 1A). GW4064 induced a rather spectacular increase in weight gain (Fig. 1A). At the end of the study, the weight of the liver, epididymal white adipose tissue (epWAT), and BAT of the high fat fed animals were all significantly increased (Fig. 1B). GW4064 administration induced a more pronounced increase in fat-induced changes in liver and adipose tissue mass. CA completely prevented these changes (Fig. 1B).
FIGURE 1.
FXR ligands affect metabolic control in mice fed a high fat diet. A, body weight (BW) change and cumulative food intake of C57BL/6J mice during 3 months on different diets/treatments. Food intake was measured daily for 1 week at day 14. C denotes control diet; F denotes high fat diet; FW denotes high fat diet + GW4064, and FA denotes high fat diet + CA. B, comparison of liver, epWAT, and BAT tissue weight after 3 months of treatment with different diets. C, comparison of the BA pool size and composition in enterohepatic organs of mice treated with different diets for 2 months. Abbreviations used are as follows: T, Tauro; U, Urso; and M, Muri. D, serum glucose and insulin levels after 6 h of fasting and 70 days on the indicated treatments. E, glucose levels during an OGTT and IPITT in mice on the different diets for 72 and 80 days, respectively. Data are expressed as the mean ± S.E. (n = 5). *, p < 0.05; **, p < 0.01 versus high fat diet (F).
GW4064 Decreases BA Pool Size and Induces Insulin Resistance
FXR regulates BA homeostasis. Therefore, we analyzed BA composition in the enterohepatic organs of the mice subjected to the different treatments. In mice fed the high fat diet mixed with GW4064, BA pool size was reduced, and taurocholic acid and CA content were markedly decreased. The relative contribution of tauromuricholic acid increased upon GW4064 treatment (Fig. 1C). Conversely, BA pool size was increased in mice fed a high fat diet mixed with CA due to elevated taurocholic acid and CA content. Tauromuricholic acid was decreased, consistent with our previous report (Fig. 1C) (21).
Fasting glucose and insulin levels were both increased by feeding a high fat diet and reflected the presence of insulin resistance. Although these parameters were improved by CA, GW4064 increased fasting glucose and insulin levels (Fig. 1D). During an OGTT, administration of CA to high fat fed animals normalized their glucose tolerance, whereas glucose tolerance was significantly poorer after GW4064 treatment (Fig. 1E). Moreover, the slope of the decrease in serum glucose during an IPITT was steeper in mice supplemented with CA than in control high fat fed mice. GW4064 treatment flattened this slope (Fig. 1E).
In addition, serum-free fatty acid, TG, and leptin levels were all increased by the addition of GW40464 to the high fat diet, and they were normalized by CA supplementation (Table 2). These data indicate that in a model for diet-induced obesity, GW4064 aggravates and CA improves glucose tolerance and insulin sensitivity.
TABLE 2.
Serum and liver lipid levels and serum leptin level in the four treatment groups
Data are expressed as mean ± S.E. (n = 5).
| High fat diet | High fat diet + GW4064 | High fat diet + CA | Control diet | |
|---|---|---|---|---|
| Serum | ||||
| TG (mg/dl) | 62.89 ± 5.75 | 83.04 ± 5.88a | 40.16 ± 4.07a | 45.77 ± 6.38 |
| Total cholesterol (mg/dl) | 177.01 ± 14.57 | 200.91 ± 13.92 | 125.88 ± 9.85a | 113.87 ± 7.93b |
| Free fatty acids (μEq/liter) | 628.09 ± 22.19 | 714.44 ± 82.58 | 585.78 ± 31.47 | 597.30 ± 22.27 |
| Leptin (pg/ml) | 6.79 ± 1.63 | 13.04 ± 2.34b | 2.79 ± 0.20a | 3.22 ± 0.46a |
| Liver | ||||
| TG (mg/g liver) | 88.97 ± 7.22 | 159.66 ± 16.36b | 55.50 ± 5.90a | 54.77 ± 5.87a |
| TC (mg/g liver) | 7.01 ± 0.55 | 12.78 ± 1.62b | 15.21 ± 1.73b | 5.32 ± 0.76 |
| Free fatty acids (μEq/g liver) | 41.94 ± 6.89 | 56.10 ± 6.67 | 45.99 ± 6.75 | 31.07 ± 1.47 |
a p < 0.05 versus high fat diet.
b p < 0.01 versus high fat diet.
GW4064 Increases Lipid Accumulation in Liver and Adipose Tissue
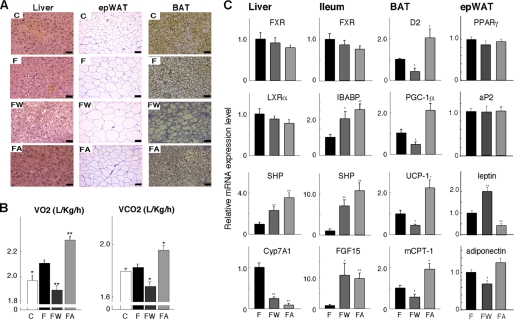
To identify the tissue(s) contributing to the changed metabolic profile after the various treatments of the C57BL/6J mice, we performed a detailed histological survey of the key metabolic tissues (Fig. 2A). This enabled us to compare the effects of GW4064 with those of CA, which we previously characterized in this model (17, 21). Liver sections of high fat fed animals showed more unstained inclusions, indicating steatosis, which was even more pronounced in livers of GW4064-treated mice. Such inclusions were absent when the high fat diet was supplemented with CA (Fig. 2A) (17). Indeed, analysis of hepatic lipid levels revealed that GW4064 increased the high fat diet-induced TG accumulation, whereas this was completely prevented by the supplementation of CA (Table 2). Both treatments increased hepatic cholesterol levels, consistent with the inhibition of BA biosynthesis. The high fat diet caused significant adipocyte hypertrophy in both WAT, characterized by a larger adipocyte volume, and BAT, typified by the presence of larger lipid vacuoles within the cells (Fig. 2A). Significantly higher amounts of lipids accumulated in the animals that received GW4064 (Fig. 2A). This adipocyte hypertrophy was decreased when the high fat diet was supplemented with CA (Fig. 2A).
FIGURE 2.
Changes in liver, epWAT, and BAT histology and energy expenditure induced by FXR ligands. A, hematoxylin and eosin (HE)-stained liver, epWAT, and BAT sections of mice treated with control or high fat diet and exposed to the same treatments as specified in Fig. 1B. Abbreviations are as in Fig. 1. Scale bar, 50 μm. B, O2 consumption (VO2) and CO2 production (VCO2) by indirect calorimetry in mice on the different diets for 3 months. The acclimation time in the chambers was 2 h. The VO2 was normalized to body weight to the 0.75 power. The right panel represents the averaged VO2 and VCO2. C, mRNA expression levels of indicated genes in liver, ileum, BAT, and epWAT of mice after 2 months of treatment with different diets. Data are expressed as the mean ± S.E. (n = 5). *, p < 0.05, or **, p < 0.01 versus high fat diet (F).
Indirect calorimetry was then performed to establish whether the facilitating effect of GW4064 on the development of obesity and lipid accumulation in the tissues was mediated by decreased energy expenditure. A higher CO2 production and O2 consumption was evident in animals fed a high fat diet supplemented with CA compared with animals on a high fat diet (Fig. 2B) (21). GW4064, however, remarkably decreased both O2 consumption and CO2 production, indicating that GW4064 reduced energy expenditure.
GW4064 Decreases the Expression of Genes Involved in BA Biosynthesis and Energy Expenditure
To better understand the molecular mechanisms underlying the reduced energy expenditure induced by GW4064, we analyzed gene expression in different tissues using quantitative RT-PCR. We focused our attention on the liver, the main metabolic hub and tissue controlling BA biosynthesis, on the ileum, as the site of BA absorption, on the BAT, the tissue coordinating energy expenditure, and on the epWAT, which is a major site for fat storage (Fig. 2C). Hepatic gene expression reflected the changes in BA pool size and its consequences on BA production and cholesterol homeostasis. The mRNA level of Shp was increased, and consequently Cyp7A1 expression was decreased after treatment with either GW4064 or CA, consistent with previous reports (9, 11). In contrast, mRNA levels of Fxr and Lxrα, which regulate Shp and Cyp7A1 expression, remained unchanged (Fig. 2C). In the ileum, mRNA levels of Ibabp, Shp, and Fgf15 were increased in mice treated with either GW4064 or CA. These three genes have established FXR-response elements in their promoters.
In contrast to the parallel changes in liver and ileum, GW4064 and CA had opposing effects in BAT. The mRNA levels of D2 and Pgc-1α were decreased by GW4064, whereas expression of these genes was increased by CA treatment (Fig. 2C). D2 and PGC-1α are two principal players, which facilitate the induction of genes involved in BAT thermogenesis (28, 29). Consequently, the expression of the mRNAs for uncoupling protein (Ucp) 1 and muscle-type carnitine palmitoyltransferase I (mCPT-I) were significantly decreased after GW4064 treatment, whereas mRNA levels of these genes were increased by CA (Fig. 2C). In addition, expression of the adipogenesis markers, Pparγ and aP2, in epWAT was not changed in response to GW4064 or CA treatment. The expression of leptin and adiponectin reflected the degree of adipocyte hypertrophy in response to the different diets (Fig. 2C). In line with our previous observations (21), these results suggest that BAT is the primary target organ that underpins the inhibitory effects of GW4064-induced decreased BA pool size on energy expenditure.
GW4064 Does Not Affect Adipocyte Differentiation and Insulin Signaling
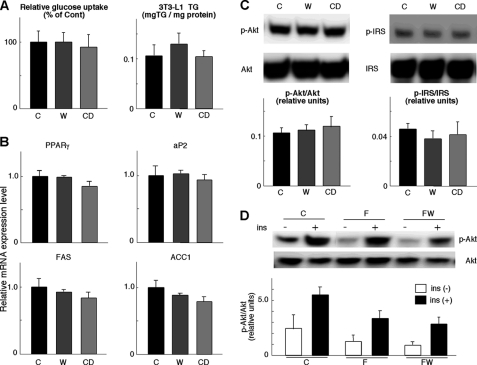
Several papers suggest that FXR activation enhances insulin signaling and adipogenesis in WAT and 3T3-L1 cells (30–32), whereas some evidence of decreased insulin signaling was found in WAT (30) and liver (32) from fxr−/− mice. It remains, however, contested whether synthetic FXR agonists, such as GW4064, increase insulin signaling in WAT, given that the expression of Fxr is low in WAT (±2% of liver) (30). We therefore activated FXR by the addition of GW4064 or CDCA (a natural ligand of FXR) to the medium of differentiated 3T3-L1 cells. Glucose uptake into 3T3-L1 cells during 12 h, 5 days after the induction of adipocyte differentiation, is unchanged by either GW4064 or CDCA treatment (Fig. 3A). Both compounds did not increase TG accumulation in 3T3-L1 adipocytes (Fig. 3A) nor did they alter the mRNA expression of adipocyte marker genes, aP2, Pparγ, fatty-acid synthase (Fas), and acetyl-CoA carboxylase 1 (Acc1) (Fig. 3B). Furthermore, we investigated the phosphorylation of Akt/PKB on serine 473 and IRS-1 on serine 307 in 3T3-L1 cells as markers of insulin sensitivity and resistance, respectively (33, 34). Phosphorylation levels of Akt/PKB and IRS-1 were similar between the 3T3-L1 cells differentiated for 5 days in the presence of GW4064 or CDCA (Fig. 3C). We also investigated Akt/PKB phosphorylation in vivo. Consistent with our data in 3T3-L1 cells, the levels of serine 473 Akt/PKB phosphorylation were not induced in epWAT after 76 days of GW4064 administration (Fig. 3D). Taken together, our data suggest that FXR agonists had no effect on insulin signaling and adipogenesis in 3T3-L1 cells and WAT (Fig. 2C).
FIGURE 3.
FXR agonists do not affect adipocyte differentiation and insulin sensitivity in vitro and in vivo. A, insulin-stimulated glucose uptake after adipocyte differentiation of 3T3-L1 cells with or without GW4064 or CDCA. Cells were stimulated with insulin (5 μg/ml) for 12 h, and the decrease of glucose in the medium was measured (left panel). TG content in these differentiated 3T3-L1 cells is shown in the right panel. C denotes control medium; W indicates medium with 10 μm GW4064; CD indicates medium with 20 μm CDCA. B, mRNA expression levels of Pparγ, aP2, Fas, and ACC1 in 3T3-L1 cells, as specified in A. C, determination of Akt/PKB or IRS-1 phosphorylation levels by Western blotting in differentiated 3T3-L1 cells. Cells were cultured with or without GW4064 or CDCA and stimulated with 5 μg/ml insulin for 5 min. Western blotting was performed with anti-Ser-473-phosphorylated Akt, total Akt, anti-Ser-307-phosphorylated IRS-1, or total IRS-1 antibodies. D, determination of Akt/PKB phosphorylation levels by Western blotting with anti-Ser-473-phosphorylated Akt or total Akt antibody in epWAT. Protein extracts were prepared from epWAT of C57BL/6J mice, which were fed a high fat diet with or without GW4064 for 76 days.
CA Can Overcome the Negative Impact of GW4064 on Metabolic Homeostasis
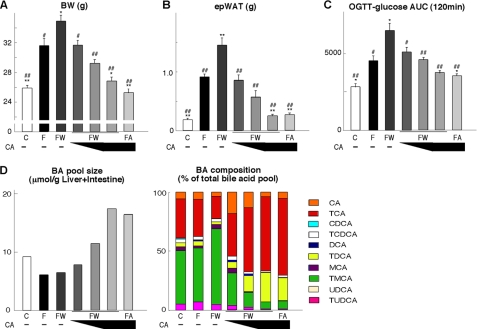
The apparent discrepancy between the metabolic effects of the synthetic (GW4064) and natural (CA) FXR agonists on metabolic function could be due to their differential effects on BA pool size (Fig. 1C). Therefore, we tested whether we could overcome the negative metabolic impact of GW4064 administration to the high fat diet by supplementing this diet with or without different doses of CA (0.125, 0.25, or 0.5% w/w) for 76 days. An additional control group of high fat fed mice that only received CA was included as well. Again, GW4064 administration accentuated the body weight gain induced by high fat feeding, whereas CA completely eliminated weight gain (Fig. 4A). When different amounts of CA were given to mice fed a high fat diet supplemented with GW4064, a dose-dependent normalization of body weight gain was observed. Strikingly, the highest dose of CA was able to revert body weight gain to similar levels as seen upon chow feeding. These differences in body weight gain were paralleled by similar changes in epWAT mass (Fig. 4B). Not surprisingly, also glucose tolerance followed the same trend, with a striking reduction of the abnormalities induced by GW4064 in a CA dose-dependent manner (Fig. 4C). The fact that CA could revert the metabolic abnormalities induced by GW4064 was clearly linked to the fact that it increased BA pool size and changed its composition (Fig. 4D).
FIGURE 4.
CA coadministration overcomes the negative effects of GW4064 by restoring the BA pool. A, body weight of C57BL/6J mice after 76 days on different diets/treatments. Abbreviations are as in Fig. 1. Different doses of CA (0.125, 0.25, or 0.5% w/w) were supplemented to a high fat diet + GW4064. B, comparison of epWAT tissue weight after 76 days on the different diets. C, AUC of glucose levels during an OGTT after 60 days on the different diets. D, comparison of BA pool size and composition in enterohepatic organs in C57BL/6J mice treated with different diets for 10 days. Data are expressed as the mean ± S.E. (n = 5). *, p < 0.05, or **, p < 0.01, versus F and #, p < 0.05, or ##, p < 0.01, versus high fat diet + GW4064.
DISCUSSION
This study demonstrates that prolonged FXR activation (for 3 months) with the synthetic agonist, GW4064, induced obesity and diabetes in high fat fed C57BL/6J mice. Long term FXR activation by GW4064 reduced basal metabolism, increased body weight gain, and enhanced TG accumulation in WAT, BAT, and liver. This resulted in glucose intolerance and insulin resistance. We furthermore show that GW4064 induces such a detrimental metabolic profile via a reduction in hepatic BA synthesis and a subsequent decrease of the BA pool size. Administration of CA, a natural FXR agonists, improves metabolism. Most importantly, CA prevents the negative metabolic effects of GW4064 during coadministration, as it replenishes the BA pool size. This proves that the mechanism behind the effects of GW4064 is mediated via FXR activation and the subsequent lowering of the BA pool size (Fig. 5).
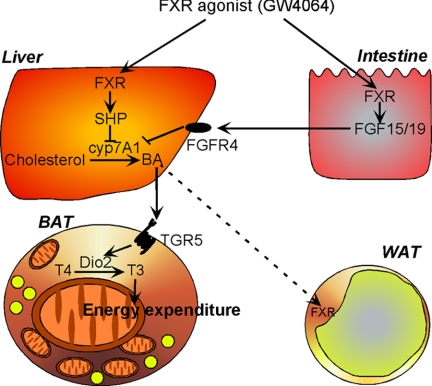
FIGURE 5.
Changes in energy metabolism after FXR activation using GW4064. Administration of a synthetic FXR agonist to high fat fed mice leads to a reduction in bile acid synthesis and as a consequence to a reduced bile acid pool size. This translates into reduced energy expenditure in brown adipose tissue, TG accumulation in WAT, BAT, and liver, and insulin resistance.
Our results are in apparent contrast to several earlier studies, which have shown that treatment with synthetic FXR agonists either improved insulin sensitivity in db/db (32), KK-Ay (32), and ob/ob mice (30) or had no effect on glucose homeostasis in basal conditions or during OGTT in C57BL/6J mice (30, 32). In this study, we used high fat fed C57BL/6J mice as a model for diet-induced obesity and diabetes, which is opposed to previous studies that all relied on mice with genetic predisposition to metabolic disease, such as the ob/ob, db/db, and KK-Ay mice. Other differences between our study and the previous studies are the long duration of our study and the administration of a relatively low dose via admixing to the diet. Despite these differences, we show increased expression of well characterized FXR target genes in liver and intestine, proving consistent FXR activation in our study.
Several previous publications suggested that FXR activation improves insulin signaling via Akt/PKB phosphorylation and adipogenesis in WAT by PPARγ-dependent and -independent mechanisms (30, 31). Likewise, a decrease in insulin signaling was observed in WAT (30) and liver (32) from fxr−/− mice, whereas insulin signaling was increased in hepatocytes from mice that express FXR after adenoviral infection (32). To clarify whether FXR, which is weakly expressed in WAT and BAT, has functions in adipose tissue that could contribute to the GW4064-induced increase in body weight, we evaluated the effect of FXR agonists in WAT and 3T3-L1 cells. The activation of FXR with either synthetic or natural FXR ligands in the 3T3-L1 adipocyte cell line had no effect on glucose uptake, TG accumulation, IRS1, and Akt/PKB phosphorylation, and expression of adipocyte differentiation marker genes, indicating that FXR activation did not increase 3T3-L1 adipocyte differentiation and/or insulin signaling. GW4064 had also no effect on Akt/PKB phosphorylation and adipogenesis markers in WAT in vivo. Our results hence are in line with a previous report, showing no difference in Pparγ and aP2 mRNA expression between FXR−/− and wild type mice (30), and suggest that the obesity and diabetes seen after long term GW4064 treatment in vivo is independent of alterations in insulin signaling in adipose tissue.
Other potential candidates to explain the metabolic effects of FXR activation are FGF15/19 and phosphoenolpyruvate carboxykinase. FXR activation increases Ppeck expression via a mechanism that involves PPARα and as such could have a negative impact on glucose homeostasis (35). As discussed before, short term treatment with GW4064, however, improves or does not change glucose homeostasis again indicating that after long term GW4064 administration most effects are mediated by the increased body weight gain we describe. FGF15/19 was reported to improve metabolism and decrease adiposity by the direct inhibition of hepatic Acc2 and stearoryl-coenzyme A desaturase-1 expression and an increase in fatty acid oxidation (36, 37). Furthermore, FGF19 was shown to increase glucose uptake in 3T3-L1 adipocytes and decrease WAT mass in vivo (38). Thus, FGF15 induction in the primary FXR signaling tissue, the ileum (16, 39), could mediate in an endocrine manner part of the effects of BAs on energy homeostasis. However, despite the fact that both GW4064 and CA induce the expression of FGF15 in ileum, their divergent effect on energy expenditure argues against an important contribution of FGF15 to the metabolic effects of GW4064.
The differences between the metabolic, morphological, and molecular changes induced by the synthetic FXR agonist, GW4064, and the natural FXR agonist, CA (21), suggested that FXR-independent mechanisms, perhaps related to alterations in the BA pool size or circulating BA levels, could underpin the observed metabolic alterations. The molecular feedback loops that control BA biosynthesis depend on FXR and involve the induction of Shp expression in the liver and of Fgf15/19 expression in the ileum (9, 11, 14, 15), effects that are similar after administration of both GW4064 or CA. As expected, BA pool size was increased by CA administration, but interestingly GW4064 decreased BA pool size. The reversal of the metabolic abnormalities induced by GW4064 in high fat diet fed mice by coadministration of CA provided unequivocal proof of concept that the absence of detrimental metabolic effects were due to maintenance of BA pool size. This is also in line with our previous mouse studies in which we established that administration of CA, which increases the BA pool size, raises energy expenditure in BAT via induction of the cAMP-dependent thyroid hormone activating enzyme D2 (21). Several recent studies support that increased BA levels and/or BA pool size have beneficial metabolic effects. In mice that lack FXR specifically in the intestine, a high BA pool is associated with weight loss (16). In humans, BA levels are also tightly correlated to increased insulin sensitivity. Patients after bariatric surgery to correct for obesity have higher circulating levels of BAs, which are positively correlated with metabolic improvements (40, 41). This latter observation was recently confirmed in obese patients, who have a decreased postprandial BA response in comparison with normal weight subjects (42). In contrast to this body of evidence that suggests that increasing BA levels benefit metabolic homeostasis, GW4064 decreased the expression levels of genes involved in energy expenditure in BAT, underscoring that these effects are not directly caused by FXR activation but rather are the consequence of reduced TGR5 activation subsequent to the reduction in the BA pool.
Finally, it is interesting to point out that changes in the BA pool size may also explain some of the effects of SHP on energy homeostasis. In fact, SHP was reported to inhibit thermogenesis in BAT by decreasing Pgc-1α mRNA expression, as concluded from the resistance to diet-induced obesity and the increase of energy expenditure in fxr−/− mice (43). As Fxr, Shp mRNA is almost undetectable in mouse BAT (21, 30). A possible reason for this apparent discrepancy is that adaptive thermogenesis might be induced by the increase in BA pool size, because the expression of Cyp7A1 and BA production is facilitated in the absence of SHP (13).
In conclusion, activation of FXR with the synthetic agonist GW4064 decreases the BA pool size. Through the reduction in the BA pool size, synthetic FXR agonists will decrease the activation of TGR5 and hence facilitate the development of obesity and diabetes. This is opposite to the effects of BAs, natural FXR and TGR5 ligands, as they concurrently replenish and enlarge the BA pool and activate TGR5. These data caution against long term use of FXR agonists in the metabolic syndrome. In the absence of contraction of the BA pool, synthetic FXR agonists may have beneficial effects, which could be obtained after low dose, short term, or local treatment. Conversely, strategies to increase the BA pool size may constitute a significant therapeutic advance to combat the metabolic syndrome and suggest that such strategies are worth testing in a clinical setting.
Acknowledgments
We thank Ron Evans, Pierre Chambon, David Moore, and Satoshi Iwasaki for helpful discussions and Shinya Inoue, Nao Kaneko-Iwasaki, Tatsuya Maruyama, Nadia Messaddeq, Arno van Cruchten and Albert Bootsma for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health grants. This work was also supported by grants from the Ecole Polytechnique Fédérale de Lausanne, the Swiss National Science Foundation, the Uehara Memorial Foundation, Takeda Science Foundation, the Sumitomo Foundation, Ono Medical Foundation, Astellas Foundation for Research on Metabolic Disorders, the Novartis Foundation (Japan) for Promotion of Science, the Japan Health Foundation, Kowa Life Science Foundation and the Ministry of Education, Culture, Sports, Science and Technology of Japan.
- BA
- bile acid
- FXR
- farnesoid X receptor
- SHP
- short heterodimer partner
- TG
- triglyceride
- BAT
- brown adipose tissue
- D2
- type 2 iodothyronine deiodinase
- PPARγ
- peroxisome proliferator-activated receptor γ
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- OGTT
- oral glucose tolerance test
- IPITT
- intraperitoneal insulin tolerance test
- epWAT
- epididimal white adipose tissue.
REFERENCES
- 1. Houten S. M., Watanabe M., Auwerx J. (2006) EMBO J. 25, 1419–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gupta S., Stravitz R. T., Dent P., Hylemon P. B. (2001) J. Biol. Chem. 276, 15816–15822 [DOI] [PubMed] [Google Scholar]
- 3. Qiao L., Han S. I., Fang Y., Park J. S., Gupta S., Gilfor D., Amorino G., Valerie K., Sealy L., Engelhardt J. F., Grant S., Hylemon P. B., Dent P. (2003) Mol. Cell. Biol. 23, 3052–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. (1999) Science 284, 1362–1365 [DOI] [PubMed] [Google Scholar]
- 5. Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., Lehmann J. M. (1999) Science 284, 1365–1368 [DOI] [PubMed] [Google Scholar]
- 6. Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. (1999) Mol. Cell 3, 543–553 [DOI] [PubMed] [Google Scholar]
- 7. Houten S. M., Auwerx J. (2004) Ann. Med. 36, 482–491 [DOI] [PubMed] [Google Scholar]
- 8. Brendel C., Schoonjans K., Botrugno O. A., Treuter E., Auwerx J. (2002) Mol. Endocrinol. 16, 2065–2076 [DOI] [PubMed] [Google Scholar]
- 9. Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., Maloney P. R., Willson T. M., Kliewer S. A. (2000) Mol. Cell 6, 517–526 [DOI] [PubMed] [Google Scholar]
- 10. Kerr T. A., Saeki S., Schneider M., Schaefer K., Berdy S., Redder T., Shan B., Russell D. W., Schwarz M. (2002) Dev. Cell 2, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. (2000) Mol. Cell 6, 507–515 [DOI] [PubMed] [Google Scholar]
- 12. Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J. (2000) Cell 102, 731–744 [DOI] [PubMed] [Google Scholar]
- 13. Wang L., Lee Y. K., Bundman D., Han Y., Thevananther S., Kim C. S., Chua S. S., Wei P., Heyman R. A., Karin M., Moore D. D. (2002) Dev. Cell 2, 721–731 [DOI] [PubMed] [Google Scholar]
- 14. Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., Goodwin B., Jones S. A. (2003) Genes Dev. 17, 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A. (2005) Cell Metab. 2, 217–225 [DOI] [PubMed] [Google Scholar]
- 16. Stroeve J. H., Brufau G., Stellaard F., Gonzalez F. J., Staels B., Kuipers F. (2010) Lab. Invest. 90, 1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watanabe M., Houten S. M., Wang L., Moschetta A., Mangelsdorf D. J., Heyman R. A., Moore D. D., Auwerx J. (2004) J. Clin. Invest. 113, 1408–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y., Hinuma S., Fujisawa Y., Fujino M. (2003) J. Biol. Chem. 278, 9435–9440 [DOI] [PubMed] [Google Scholar]
- 19. Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Nakamura T., Itadani H., Tanaka K. (2002) Biochem. Biophys. Res. Commun. 298, 714–719 [DOI] [PubMed] [Google Scholar]
- 20. Bianco A. C., Salvatore D., Gereben B., Berry M. J., Larsen P. R. (2002) Endocr. Rev. 23, 38–89 [DOI] [PubMed] [Google Scholar]
- 21. Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., Schoonjans K., Bianco A. C., Auwerx J. (2006) Nature 439, 484–489 [DOI] [PubMed] [Google Scholar]
- 22. Thomas C., Gioiello A., Noriega L., Strehle A., Oury J., Rizzo G., Macchiarulo A., Yamamoto H., Mataki C., Pruzanski M., Pellicciari R., Auwerx J., Schoonjans K. (2009) Cell Metab. 10, 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Picard F., Géhin M., Annicotte J., Rocchi S., Champy M. F., O'Malley B. W., Chambon P., Auwerx J. (2002) Cell 111, 931–941 [DOI] [PubMed] [Google Scholar]
- 24. Folch J., Lees M., Sloane-Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 25. Mataki C., Magnier B. C., Houten S. M., Annicotte J. S., Argmann C., Thomas C., Overmars H., Kulik W., Metzger D., Auwerx J., Schoonjans K. (2007) Mol. Cell. Biol. 27, 8330–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakakura H., Suzuki M., Kimura N., Takeda H., Nagata S., Maeda M. (1993) J. Chromatogr. 621, 123–131 [DOI] [PubMed] [Google Scholar]
- 27. Student A. K., Hsu R. Y., Lane M. D. (1980) J. Biol. Chem. 255, 4745–4750 [PubMed] [Google Scholar]
- 28. de Jesus L. A., Carvalho S. D., Ribeiro M. O., Schneider M., Kim S. W., Harney J. W., Larsen P. R., Bianco A. C. (2001) J. Clin. Invest. 108, 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin J., Wu P. H., Tarr P. T., Lindenberg K. S., St-Pierre J., Zhang C. Y., Mootha V. K., Jäger S., Vianna C. R., Reznick R. M., Cui L., Manieri M., Donovan M. X., Wu Z., Cooper M. P., Fan M. C., Rohas L. M., Zavacki A. M., Cinti S., Shulman G. I., Lowell B. B., Krainc D., Spiegelman B. M. (2004) Cell 119, 121–135 [DOI] [PubMed] [Google Scholar]
- 30. Cariou B., van Harmelen K., Duran-Sandoval D., van Dijk T. H., Grefhorst A., Abdelkarim M., Caron S., Torpier G., Fruchart J. C., Gonzalez F. J., Kuipers F., Staels B. (2006) J. Biol. Chem. 281, 11039–11049 [DOI] [PubMed] [Google Scholar]
- 31. Rizzo G., Disante M., Mencarelli A., Renga B., Gioiello A., Pellicciari R., Fiorucci S. (2006) Mol. Pharmacol. 70, 1164–1173 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y., Lee F. Y., Barrera G., Lee H., Vales C., Gonzalez F. J., Willson T. M., Edwards P. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1006–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aguirre V., Werner E. D., Giraud J., Lee Y. H., Shoelson S. E., White M. F. (2002) J. Biol. Chem. 277, 1531–1537 [DOI] [PubMed] [Google Scholar]
- 34. Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. (2004) Nature 431, 200–205 [DOI] [PubMed] [Google Scholar]
- 35. Stayrook K. R., Bramlett K. S., Savkur R. S., Ficorilli J., Cook T., Christe M. E., Michael L. F., Burris T. P. (2005) Endocrinology 146, 984–991 [DOI] [PubMed] [Google Scholar]
- 36. Fu L., John L. M., Adams S. H., Yu X. X., Tomlinson E., Renz M., Williams P. M., Soriano R., Corpuz R., Moffat B., Vandlen R., Simmons L., Foster J., Stephan J. P., Tsai S. P., Stewart T. A. (2004) Endocrinology 145, 2594–2603 [DOI] [PubMed] [Google Scholar]
- 37. Tomlinson E., Fu L., John L., Hultgren B., Huang X., Renz M., Stephan J. P., Tsai S. P., Powell-Braxton L., French D., Stewart T. A. (2002) Endocrinology 143, 1741–1747 [DOI] [PubMed] [Google Scholar]
- 38. Kurosu H., Choi M., Ogawa Y., Dickson A. S., Goetz R., Eliseenkova A. V., Mohammadi M., Rosenblatt K. P., Kliewer S. A., Kuro-o M. (2007) J. Biol. Chem. 282, 26687–26695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Houten S. M., Volle D. H., Cummins C. L., Mangelsdorf D. J., Auwerx J. (2007) Mol. Endocrinol. 21, 1312–1323 [DOI] [PubMed] [Google Scholar]
- 40. Nakatani H., Kasama K., Oshiro T., Watanabe M., Hirose H., Itoh H. (2009) Metabolism 58, 1400–1407 [DOI] [PubMed] [Google Scholar]
- 41. Patti M. E., Houten S. M., Bianco A. C., Bernier R., Larsen P. R., Holst J. J., Badman M. K., Maratos-Flier E., Mun E. C., Pihlajamaki J., Auwerx J., Goldfine A. B. (2009) Obesity 17, 1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Glicksman C., Pournaras D. J., Wright M., Roberts R., Mahon D., Welbourn R., Sherwood R., Alaghband-Zadeh J., le Roux C. W. (2010) Ann. Clin. Biochem. 47, 482–484 [DOI] [PubMed] [Google Scholar]
- 43. Wang L., Liu J., Saha P., Huang J., Chan L., Spiegelman B., Moore D. D. (2005) Cell Metab. 2, 227–238 [DOI] [PubMed] [Google Scholar]