Abstract
The disintegrin-metalloproteinases ADAM10 and ADAM17 mediate the release of several cell signaling molecules and cell adhesion molecules such as vascular endothelial cadherin or L-selectin affecting endothelial permeability and leukocyte transmigration. Dysregulation of ADAM activity may contribute to the pathogenesis of vascular diseases, but the mechanisms underlying the control of ADAM functions are still incompletely understood. Atherosclerosis is characterized by lipid plaque formation and local accumulation of unsaturated free fatty acids (FFA). Here, we show that unsaturated FFA increase ADAM-mediated substrate cleavage. We demonstrate that these alterations are not due to genuine changes in enzyme activity, but correlate with changes in membrane fluidity as revealed by measurement of 1,6-diphenyl-1,3,5-hexatriene fluorescence anisotropy and fluorescence recovery after photobleaching analyses. ELISA and immunoblot experiments conducted with granulocytes, endothelial cells, and keratinocytes revealed rapid increase of ectodomain shedding of ADAM10 and ADAM17 substrates upon membrane fluidization. Large amounts of unsaturated FFA may be liberated from cholesteryl esters in LDL that is entrapped in atherosclerotic lesions. Incubation of cells with thus modified LDL resulted in rapid cleavage of ADAM substrates with corresponding functional consequences on cell proliferation, cell migration, and endothelial permeability, events of high significance in atherogenesis. We propose that FFA represent critical regulators of ADAM function that may assume relevance in many biological settings through their influence on mobility of enzyme and substrate in lipid bilayers.
Keywords: ADAM, ADAMTS, Atherosclerosis, Cell Adhesion, Fatty Acid, Metalloprotease, Migration, Proliferation, Shedding
Introduction
During the past decade, ADAMs3 (A Disintegrin And Metalloproteinase) have emerged as the major proteinase family that mediates ectodomain shedding (1). This process is essential for normal cellular functions, but may also have undesirable consequences by promoting diseases such as cancer, arthritis, and atherosclerosis (2, 3). ADAM10 and ADAM17 are the best characterized ADAMs. They modulate a multitude of cellular functions through the release of transmembrane molecules. Both act as α-secretases and promote the non-amyloidogenic processing of the amyloid precursor protein (4). Surface expression of L-selectin (CD62L), which directs leukocyte accumulation, is primarily regulated by ADAM17 (5, 6). ADAM10 is, among other things, critically involved in the constitutive and regulated shedding of epithelial (E)-cadherin in primary keratinocytes (7, 8).
ADAM10 was recently identified as an important regulator of endothelial cell functions. ADAM10-mediated cleavage of vascular endothelial (VE)-cadherin regulates endothelial permeability and T cell transmigration (9), important processes in the context of cardiovascular diseases. The protease is expressed in human atherosclerotic lesions, associated with plaque progression and neovascularization (10). It is tempting to speculate that dysregulation of ADAM10 activity may contribute to defects associated with atherosclerosis or chronic edema.
Atherosclerosis is characterized by accumulation of low density lipoprotein (LDL) in the arterial walls and local release of free fatty acids (FFA) through modification of the lipoprotein. The question whether such lipid deposits might modulate ADAM activity has not been addressed. Depletion of membrane cholesterol increases ectodomain shedding of several ADAM10 and ADAM17 substrates including amyloid precursor protein (11), L1 (12) and human interleukin-6 receptor (13), but the depletion of cholesterol does not occur in atherosclerotic lesions.
FFA act both as counterparts and partners of cholesterol in creating the physical microenvironment of biological membranes. Alteration of function by changes in membrane cholesterol might thus be expected to be partly copied by FFA. Furthermore, due to their amphiphilic nature, FFA would appear far more likely candidates to modulate membrane properties than the relatively immobile cholesterol.
Therefore, we sought to explore whether FFA would influence ADAM activity or ADAM-dependent cellular function. For these analyses, a novel erythrocyte-based model was first introduced based on the unique property of the bacterial exotoxin Vibrio cholerae cytolysin (VCC) to be cleaved by ADAM10 that is present on rabbit erythrocytes (14,15). Generation of mature VCC resulted in hemolysis, which thus provided a simple and rapid readout. We found that pro-VCC (pVCC) cleavage was markedly accelerated by application of unsaturated FFA both in free form and in association with LDL, as is probably widely present in atherosclerotic lesions. The effect could be ascribed to increased fluidity of the plasma membrane but not to changes in bona fide ADAM enzymatic activity. This regulatory principle was found to be transferable to nucleated cells, and cleavage of ADAM substrates was followed by corresponding functional consequences on cell proliferation, cell migration, and endothelial permeability. Thus, our work identifies FFA as potentially important control elements of the interaction of membrane-anchored enzymes and their substrates that may assume high relevance in many biological settings.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
VCC and polyclonal rabbit antisera against VCC were produced as described (15). Monoclonal antibodies against the cytoplasmic domain of E-cadherin were purchased from BD Bioscience. Monoclonal antibody against VE-cadherin and peroxidase-conjugated immunglobulins to mouse or rabbit IgG were obtained from Santa Cruz Biotechnology. ADAM10 was detected using a polyclonal antiserum from Chemicon. sCD62L was quantified with an ELISA from Diaclone (Besancon, France). TAPI-0, GM6001, cholesterol, β-methyl cyclodextrin (MCD), FFA, and phenethyl alcohol (PhA) were obtained from Sigma. MCD/cholesterol was prepared as described (16). LDL was isolated from human plasma and modified as described (17). Hydroxamate-based inhibitors GW280264 ((2R,3S)-3-(formyl-hydroxyamino)-2-(2-methyl-1-propyl) hexanoic acid [(1S)-5-benzyloxycarbamoylamino-1-(1,3-thiazol-2-ylcarbamoyl)-1-pentyl] amide) and GI254023 ((2R,3S)-3-(formyl-hydroxyamino)-2-(3-phenyl-1-propyl) butanoic acid [(1S)-2,2-dimethyl-1-methylcarbamoyl-1-propyl] amide) were synthesized as described in United States patents US 6 172 064, US 6 191 150, and US 6 329 400. They were kindly provided by Dr. A. Ludwig, UK Aachen, Germany) and are described in detail elsewhere (18).
Cell Culture
Human umbilical vein endothelial cells (HUVECs) were obtained from Provitro and cultured in Endothelial Cell Growth Medium (PromoCell). All experiments were performed with HUVECs from passages 3–5. HaCaT cells were provided by Dr. N. E. Fusenig (DKFZ, Heidelberg, Germany) (19). HaCaT and HEK293 cells were grown in DMEM (PAA, Linz, Austria) supplemented with 10% FCS, 1% Hepes, and 1% penicillin/streptomycin.
Hemolysis Experiments
Washed rabbit erythrocytes suspended in PBS (109 cells/ml) were incubated on ice with pVCC (4–6 μg/ml final concentration) for 30 min. Cells were washed three times with ice-cold PBS and resuspended to 5 × 108 cells/ml, and aliquots received the individual ADAM-modulating agents at the given final concentrations. The Eppendorf tubes were transferred to a rotating prewarmed thermostat block, and 0.2-ml samples were removed at the given times. Hemolysis was read at 412 nm in the supernatants, and pellets were processed to white membranes by repeated washings with ice-cold 5 mm phosphate buffer, pH 8.0, in a cooled Eppendorf centrifuge. Total membrane protein was determined using the Bradford assay to guarantee equal loading of the SDS-gels for the Western blots. Every one of the depicted experiments was reproduced with no single deviant pattern of results three to eight times, and each representative example depicts matching hemolysis curves and Western blots from the same experiment.
Measurements of ADAM Enzymatic Activity
0.1 ml of washed erythrocyte membranes suspended in 25 mm Tris, 100 mm NaCl, pH 8.0, at 109 cells/ml were incubated with a fluorogenic ADAM-peptide substrate Mca-Pro-Leu-Ala-Gln-Ala-Val-Dpa-Arg-Ser-Ser-Ser-Arg-NH2 (10 μm final concentration; R&D Systems) for 20 min at 37 °C. The reaction was stopped by the addition of 0.6 ml of an aqueous 0.5% SDS solution, and fluorescence emission spectra were recorded in a SPEX Fluoromax-3 spectrofluorometer (Jobin Yvon Inc., Munich, Germany) at 320-nm excitation. Temperature dependence was assessed by incubating samples at the given temperatures in thermostat blocks in a cold room (4–20 °C) or in the laboratory (26–50 °C). To assess ADAM activity on neutrophil granulocytes, 3 × 106 cells/ml were incubated in Hanks' buffered salt solution with 10 μm substrate for 15 min, 37 °C. Samples were centrifuged at 3000 rpm for 5 min in a cooled table top centrifuge, and supernatants were analyzed in the spectrofluorometer.
Stimulation and Inhibition Experiments
Stock solutions of fatty acids (70 mm in ethanol) were stored at −20 °C. They were applied at final concentrations of 70 μm to erythrocytes (up to 60 min) and neutrophil granulocytes (15 min), and 70–100 μm to other nucleated cells (30 min). Exposure times were longer (up to 48 h) in functional experiments (see Fig. 4). These concentrations were ascertained to be noncytotoxic in all cases. If not indicated the broad spectrum metalloprotease inhibitors GM6001 and TAPI-0 were co-incubated at a concentration of 10 μm.
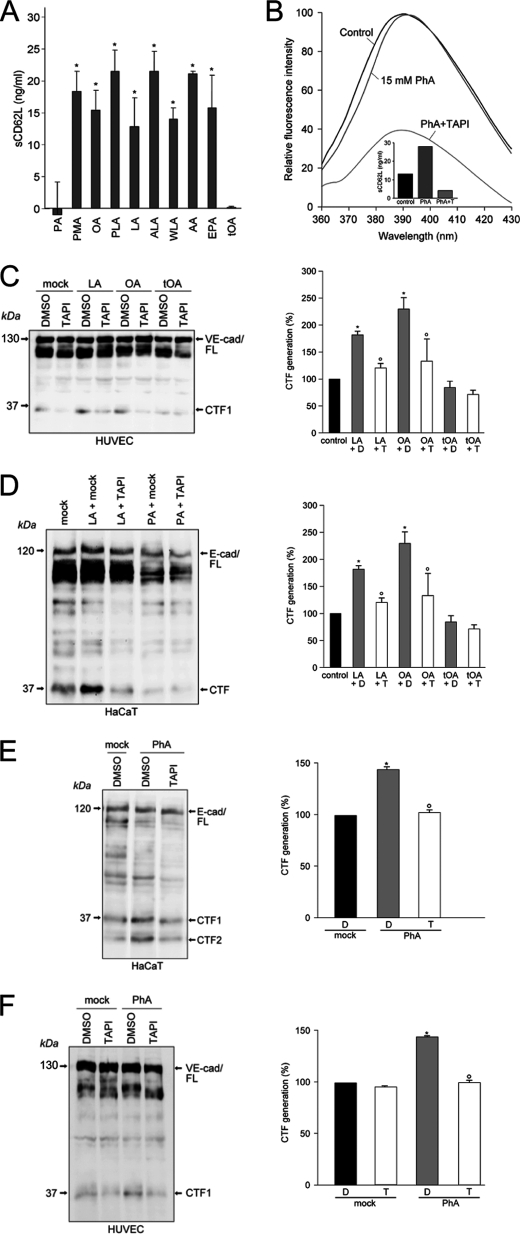
FIGURE 4.
Unsaturated fatty acids induce ADAM-dependent substrate cleavage in nucleated cells. A, neutrophil granulocytes were stimulated with 100 nm phorbol ester (PMA), or with 70 μm OA, PA, palmitoleic acid (PLA), linoleic acid (LA), α-linolenic acid (ALA), γ-linolenic acid (WLA), arachidonic acid (AA), eicosapentaenoic acid (EPA), or trans-OA (tOA) for 15 min, 37 °C, and release of soluble CD62L (sCD62L) relative to unstimulated controls was determined. *, significant increase compared with control (n = 3 ± S.D. (error bars)), p < 0.05, t test. B, PhA (15 mm) induced shedding of CD62L without altering enzyme activity. C, treatment of HUVECs with 70 μm linoleic acid or OA significantly (*) augmented generation of VE-cadherin CTF products compared with the vehicle control. Compared with DMSO (D) treated cells, TAPI (T) significantly (*) abolished the stimulation effect. Right panel shows quantification of three independent experiments (n = 3 ± S.E.; p < 0.05). D, linolenic acid (70 μm) significantly increased ADAM-dependent shedding of E-cadherin (n = 3 ± S.E.; *, p < 0.05). E, PhA (30 mm) accelerated E-cadherin CTF1 and CTF2 generation significantly. This effect was abrogated in the presence of TAPI (T) F, TAPI significantly inhibits PhA-induced VE-cadherin proteolysis in HUVECs (n = 3 ± S.E.; *, ○, p < 0.05).
Membrane Fluidity
Erythrocyte membranes (1 ml, 5 × 108 cells) were incubated for 30 min at 20 °C or at 32 °C with diphenyl hexatriene (DPH, 4 μm). Unbound washed membranes were then diluted 1:10 with PBS and transferred to a thermostated cuvette under continuous stirring. Anisotropy was measured in a Quantamaster (PTI) fluorometer using Glan-Thompson polarizers at excitation and emission wavelengths of 360 ± 4 nm and 430 ± 4 nm, respectively, including a GG390 cutoff filter at the emission site. Steady-state fluorescence anisotropy (r) was determined according to r = (IVV − IVHG)/(IVV + 2 IVHG), where IVV and IVH are the fluorescence intensities observed with the excitation polarizer in the vertical position, and the analyzing emission polarizer in both the vertical (IVV) and horizontal (IVH) configurations. Factor G corrects for the unequal transmission of differently polarized light.
Fluorescence Recovery after Photobleaching (FRAP) Measurements
FRAP was performed with HEK293 cells expressing the EGFP-tagged human oxytocin receptor at the cell surface. Cells grown on poly-l-lysine-coated coverslips were transferred into a home-made thermostated chamber system on the stage of a confocal microscope (Leica TSCSP). The cells were preincubated for 10 min at 20 °C with the indicated compounds in imaging buffer (10 mm Hepes, pH 7.4, 145 mm NaCl, 4.5 mm KCl, 1.2 mm MgCl2, 1.2 mm CaCl2, 10 mm glucose). Imaging was performed at 30 °C using a 63×, 1.3 NA oil immersion objective (2 Airy-disk pinhole). A region of interest (ROI) was defined for scanning where a spot of the plasma membrane (5 μm) was photobleached using full power of the argon laser (488 nm). Before and after photobleaching, images were acquired (scanning interval ∼1 s) using low intensity illumination. Average pixel values of the bleached ROI were background-subtracted using a cell-free ROI and were analyzed to calculate the mobile fraction M and the diffusion coefficient (D). The mobile fraction M was calculated according to the equation M = c [(Fasym − F0)/(Fi − F0)] where Fasym is the ROI fluorescence recovery asymptote, Fi is the ROI prebleaching intensity, F0 is the ROI immediate postbleaching fluorescence intensity, and c is the correction factor for whole cell photobleaching, calculated by the ratio of the whole cell prebleaching intensity to the whole cell fluorescence-recovery asymptote. Normalized FRAP curves were fitted with the one-phase exponential equation F = Fmax(1 − e−τt). The parameters represent F, fluorescence; Fmax, the asymptote of the ROI fluorescence recovery curve; τ, rate constant for fluorescence recovery; t, time. D was determined from the equation D = ω2/4τ, where ω is the actual radius of the bleached ROI.
Release of Soluble L-Selectin (sL-Selectin) from Neutrophil Granulocytes
PMN were isolated from heparinized peripheral blood of healthy donors using PolymorphprepTM (Axis-Shield, Oslo, Norway) according to the manufacturer's recommendations. Residual erythrocytes were removed using lysis buffer containing NH4Cl. PMN purity was >90% as determined by flow cytometric analysis and by counting in a hematocytometer. Stock suspensions of the cells (20–30 × 106/ml) were kept in PBS on ice and used within 2 h. Experiments were conducted in Hanks' buffered salt solution using 3 × 106 cells/ml. Background sCD62L concentrations were subtracted.
Immunoblotting
Erythrocyte membranes were solubilized in 2% SDS, equal amounts of protein were electrophoresed in each experiment, and immunoblot analyses of pVCC and VCC were undertaken as described (15). Immunoblots for the analysis of VE-cadherin and E-cadherin were performed as described elsewhere (7, 9). Signals were recorded by a luminescent image analyzer (Fuji image reader LAS1000) and analyzed with image analyzer software (GEL-PROANALYZER; Media Cybernetics, Silver Spring, MD).
In Vitro Wound Healing
HaCaT cells were seeded in 12-well plates (Sarstedt) and grown until they reached confluence (48 h). A cell-free area was introduced by scraping the monolayer with a pipette tip (10 μl; Sarstedt). To avoid a proliferative effect, cells were treated with 4 mm hydroxyurea (Sigma-Aldrich) for 24 h. For stimulation experiments, FCS-free medium containing TAPI or DMSO was added. After a 30-min preincubation, the cells were treated with enzymatically modified LDL (eLDL) (20 μg/ml) or oleic acid (OA; 20 μg/ml). Cells were photographed before stimulation and 24 h after stimulation by using an inverted phase-contrast microscope (Zeiss).
Cell Proliferation Assays
HaCaT cells were cultured overnight in medium containing FCS before starvation for 48 h. Subsequently, medium containing various agents was added, and cells were incubated for 24 h. Propidium iodide cell cycle analysis was performed as previously described (20).
Endothelial Permeability Assay
Endothelial permeability was measured as described elsewhere (9). In brief, HUVECs (3 × 105) were grown on collagen-coated Transwell filters (0.4-μm pore size; Costar) until they reached confluence. Cells were preincubated for 15 min in the presence of metalloproteinase inhibitor GM6001 (10 μmol/liter) or DMSO in RPMI 1640 medium. Afterward, FITC-dextran (Mr 40,000; Sigma) at a final concentration of 1 mg/ml was added to the upper chamber. After 10 min, 50-μl samples were taken from the lower compartment and measured at an excitation wavelength of 485 nm and an emission wavelength of 530 nm using a fluorescence plate reader (Lambda Fluoro 320; MWG Biotech).
Statistical Analysis of C-terminal Fragment (CTF) Generation
All values are expressed as means ± S.E. or as otherwise indicated. The S.E. values indicate the variation between mean values obtained from at least three independent experiments. The assumptions for normality (Kolmogorov-Smirnov test) and equal variance (Levene median test) were verified with the SigmaStat 3.1 software (Erkrath, SYSSTAT, Germany). Variance analysis was performed with one-way ANOVA. Pairwise comparison procedures were performed with the Tukey test. p values < 0.05 were classified as statistically significant.
RESULTS
ADAM10-mediated Cleavage of pVCC Leads to Hemolysis of Rabbit Erythrocytes
To distinguish whether changes in ADAM activity might be due to enhanced cell surface expression of the enzymes or to a bona fide increase in enzyme activity or to accelerated substrate cleavage occurring through biophysical alterations of the membranes, we developed a simple erythrocyte test system. In this model, cleavage of the cell-bound pVCC and a soluble fluorogenic ADAM substrate were studied in parallel. The 79-kDa pVCC binds to rabbit erythrocytes and is cleaved by a cell-associated metalloprotease to yield mature 65-kDa VCC that causes hemolysis (Fig. 1A). As previously reported (15), proteolysis was blocked by the broad spectrum metalloprotease inhibitor TAPI-0 and was here further found to be effectively suppressed by GI254023X, a preferential inhibitor of ADAM10, and GW280264, an inhibitor of ADAM10 and ADAM17 (18) (Fig. 1B).
FIGURE 1.
ADAM10 activity on rabbit erythrocytes. A, pVCC cleavage leads to pore formation and hemolysis. B, hemolysis is inhibited by the broad spectrum metalloprotease inhibitor TAPI-0, GI254023X (GI, preferential ADAM10 inhibitor), and GW280623X (GW, ADAM10 and ADAM17 inhibitor). C, cellular ADAM activity can be determined on the erythrocyte surface using a fluorogenic peptide substrate assay. D, all metalloprotease inhibitors decreased ADAM activity. E, immunological detection of ADAM10 in HaCaT keratinocytes, H4 neuroglioma cells, in rabbit brain homogenisate (rBrain) and rabbit (raE) but not human erythrocytes (huE) is shown; p, pro-ADAM10; m, mature ADAM10. F, temperature dependence of ADAM10 activity. Erythrocyte membranes were incubated with fluorogenic ADAM-peptide substrate at the given temperatures, and relative enzyme activity was determined. The temperature dependence of ADAM activity correlated with processing of membrane-bound pVCC and could be completely inhibited with GI254023X (inset).
A novel protocol was established for assessing enzyme activity using a fluorogenic ADAM peptide substrate. Erythrocyte membranes were incubated with the peptide, the reaction was terminated by solubilization of membranes with SDS, and substrate cleavage was assessed by recording emission spectra in a spectrofluorometer. This assay enabled otherwise undetectable ADAM activity to be quantified (Fig. 1C). ADAM activity could be suppressed by GI254023X and GW280264. TAPI inhibited the cleavage of pVCC as well as the proteolysis of the fluorogenic substrate in a range between 3 and 30 μm (here only shown for 30 μm, Fig. 1D).
Human erythrocytes bind but do not efficiently process pVCC, which suggested that ADAM10 may be differentially expressed on red cells of different species. Immunoblot analysis indeed indicated the presence of ADAM10 on rabbit but not on human erythrocytes (Fig. 1E).
The hitherto undescribed effect of temperature on ADAM activity was determined and optimal cleavage of the fluorogenic peptide substrate was found to occur at 32 °C-38 °C (Fig. 1F). When processing of membrane-bound pVCC was examined at 26, 32, and 38 °C, a satisfactory correlation was noted (Fig. 1F, inset).
Unsaturated Fatty Acids and Cholesterol Depletion Enhance ADAM-mediated pVCC Cleavage but Not ADAM Activity
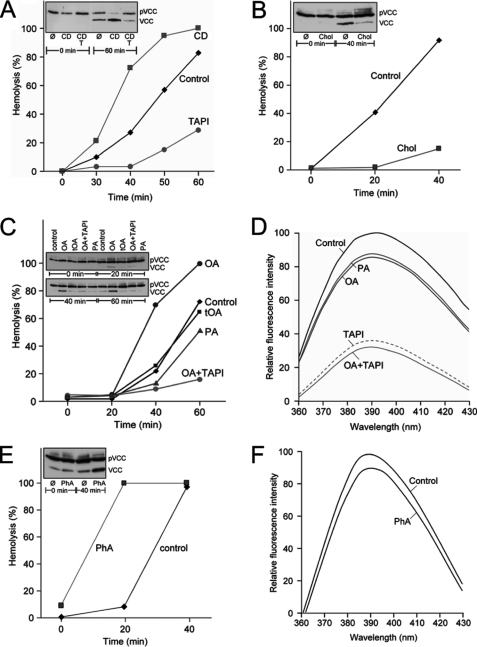
Cholesterol depletion with MCD fluidizes membranes and augments ectodomain shedding of ADAM substrates from nucleated cells (11–13). Analogously, incubation of pVCC-laden erythrocytes with MCD was found to accelerate toxin processing and hemolysis (Fig. 2A). Vice versa, sterol enrichment of plasma membranes with MCD/cholesterol retarded pVCC cleavage and hemolysis (Fig. 2B). Externally applied unsaturated fatty acids spontaneously intercalate into bilayers and increase membrane fluidity (21). When 70 μm OA was introduced to the medium, acceleration of the hemolytic process ensued (Fig. 2C). In marked contrast, trans-OA was without effect, a key finding that established the critical role of the fluidizing cis-double bond in enhancing ADAM function. In accord with this contention, pVCC cleavage and hemolysis were slightly retarded upon addition of palmitic acid (PA) to the cells.
FIGURE 2.
Unsaturated fatty acids enhance pVCC proteolysis and hemolysis without changing ADAM activity. A and B, cholesterol depletion with 30 mm β-methyl cyclodextrin (CD) accelerated toxin processing and hemolysis (A) whereas sterol enrichment with MCD/cholesterol (Chol) retarded the process (B). C, 70 μm OA accelerated pVCC processing whereas trans-OA (tOA) and PA were without effect. D, neither OA nor PA significantly altered ADAM enzymatic activity. E and F, PhA (30 mm) augmented toxin cleavage and hemolysis (E) but not ADAM activity (F).
A central finding surfaced when the influence of fatty acids on ADAM activity was assessed with the soluble peptide substrate. Strikingly, it was then found that neither OA nor PA altered ADAM activity to any substantial extent that would have explained the large effects on pVCC processing (Fig. 2D). It followed that the observed alterations in biological function did not derive from any genuine increase in surface expression or activity of the enzyme. The simplest alternative explanation was that enzyme function was governed by membrane fluidity. PhA spontaneously partitions into membranes and fluidizes lipid bilayers. Indeed, incubation of pVCC-laden erythrocytes with PhA again resulted in acceleration of toxin cleavage and hemolysis (Fig. 2E), but not in increased ADAM activity (Fig. 2F).
FFA Modulate Membrane Fluidity
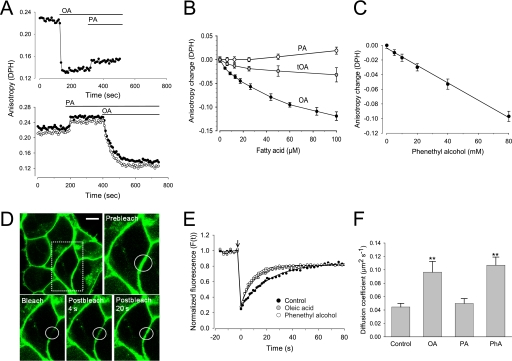
Obviously, no biochemical specificity could be assigned to the observed modulation of ADAM function. In contrast, the link to membrane fluidity became readily apparent through measurements of the rotational diffusion of fatty acyl chains in the membrane interior using steady-state fluorescence anisotropy with DPH as a probe. In erythrocyte ghosts, a rapid and striking decrease of the fluorescence anisotropy was observed in response to OA (Fig. 3A). In contrast, administration of PA slightly increased fluorescence anisotropy of DPH. Anisotropy experiments were also performed at 32 °C (Fig. 3A, open circles), the optimum temperature of ADAM10 activity. Compared with the data obtained at 20 °C (filled circles), the higher temperature led to a small decrease (approximately 0.01–0.02 unit) of all anisotropy values but did not change the effects of the different compounds. Trans-OA induced only a very modest decrease of anisotropy, markedly less than OA (Fig. 3B).
FIGURE 3.
FFA increase membrane fluidity. A, anisotropy measurements were performed with erythrocyte ghosts loaded with 4 μm DPH. 70 μm OA or PA was added at the indicated time. Experiments were performed at 20 °C (filled circles) and at 32 °C (open circles, lower panel). B and C, membranes were incubated with the indicated concentrations of fatty acids (B) or PhA (C) for 5 min at 32 °C, and the corresponding anisotropy data were acquired. Mean changes of anisotropy (n = 3) were displayed as a function of the concentration of the substances. D–F, FRAP was performed using confocal microscopy with HEK293 cells expressing the EGFP-tagged human oxytocin receptor at the cell surface. Cells were incubated for 10 min at 20 °C with ethanol (100 mm) as control, OA (50 μm), PA (50 μm), or PhA (40 mm) in imaging buffer. The agents were removed and the photobleaching experiment started. Representative images show bleaching spot (D) (scale bar, 5 μm) and typical recovery curves (E). F, diffusion coefficients (D) were significantly increased in response to the fluidizers OA (>70% of the cells, n>50) and PhA (>90% of the cells, n>40) (mean values ± S.D. (error bars)). **, significant difference compared with control, t test, p < 0.01.
Membrane fluidization already occurred with OA concentrations in the low micromolar range. Importantly, OA caused its fluidizing effect within a minute or less, which perfectly matched the fast time scale observed for OA-induced modulation of ADAM function. Application of PhA also led to a strong and dose-dependent decrease of DPH anisotropy (Fig. 3C). The concentrations required to induce membrane fluidization were roughly 1000-fold higher for PhA compared with OA. Ethanol at 100 mm induced no change of membrane fluidity. OA and PhA increased fluidity in membranes of nucleated cells to an extent similar to that in erythrocyte ghosts (data not shown) (21, 22).
The influence of the fluidizers OA and PhA on lateral mobility of a fluorescent transmembrane protein, the EGFP-tagged G protein-coupled oxytocin receptor expressed in HEK293 cells (16), was then analyzed using FRAP. Circular spots in the plasma membrane were selected for photobleaching in cells exposed to OA, PA, PhA, or ethanol (as control) (Fig. 3D). The recovery of EGFP fluorescence is displayed in typical FRAP curves (Fig. 3E), from which the parameters diffusion coefficient (D) and mobile fraction were derived by fitting the data to exponential kinetics. Recoveries characteristic of lateral diffusion, with high mobile fractions, were observed. The D values in controls were comparable with those reported for G protein-coupled receptors and other transmembrane proteins (23, 24). Pretreatment of the cells with OA and PhA led to a >2-fold increase of the diffusion coefficient, whereas PA and ethanol had no significant effect on D (Fig. 3F).
At this juncture, the results indicated that simple alterations in the physical properties of lipid bilayers might influence ADAM function due to the dependence of molecular mobility on membrane fluidity. Through their capacity to rapidly intercalate into cell membranes, FFA represented prime candidates to assume this regulatory function in biological settings. Experiments with nucleated cells were conducted to test this hypothesis.
Unsaturated Fatty Acids Induce ADAM-dependent Substrate Cleavage
Neutrophil granulocytes were stimulated with phorbol ester (phorbol 12-myristate 13-acetate) or incubated with 70 μm PA or a series of unsaturated fatty acids shown in Fig. 4A for 10 min, and the release of the preferential ADAM17 substrate L-selectin (6, 25) was quantified in the supernatants. All cis-unsaturated fatty acids induced rapid release of sL-selectin to a similar extent as phorbol 12-myristate 13-acetate. Again, and in marked contrast, trans-OA was without effect. The increased release of sL-selectin could be inhibited with GW280623X but not with GI254023X, indicating that ADAM17 is the major protease involved in this cleavage (data not shown). Nevertheless, other proteases such as ADAM8 have been described to release L-selectin (26). Thus, we cannot exclude that these proteases are also activated through membrane fluidization and contribute to the observed effects.
Because FFA trigger diverse cell signaling pathways, such downstream effects on ADAM expression or enzyme activity could not definitively be excluded. However, OA treatment did not increase cell surface expression of ADAMs. Moreover, a battery of cell signaling inhibitors including ERK inhibitor, JNK3 inhibitor, PKC inhibitor, NADPH oxidase inhibitor, Ca2+ chelator, and inhibitor of phosphoinositide 3-kinases did not reduce the OA-induced release of sL-selectin (data not shown).
Moreover, PhA similarly provoked rapid, maximal shedding of L-selectin without alterations of enzyme activity (Fig. 4B). Thus, membrane fluidity also appeared to be the decisive factor underlying rapid modulation of ADAM function in nucleated cells.
Even more important, treatment of HUVECs with unsaturated FFA such as linoleic acid or OA also increased VE-cadherin proteolysis in contrast to trans-OA as shown by immunoblot analysis (Fig. 4C, left) and densitometric quantification of three independent experiments (Fig. 4C, right).
To test the generality of our findings, processing of E-cadherin as additional preferential ADAM10 substrate (7) was investigated in a human keratinocyte cell line. Indeed, comparable results were obtained when HaCaT cells were stimulated with linoleic acid and PA, respectively (Fig. 4D). In accordance with the previous data, PhA accelerated the cleavage of E-cadherin and VE-cadherin (Fig. 4, E and F). E-cadherin is a substrate for sequential proteolysis (7). After metalloprotease-mediated cleavage the remaining fragment is immediately processed through intramembranous cleavage by γ-secretase leading to the formation of a second CTF (CTF2). In HaCaTs, induction of PhA-induced ADAM-mediated cleavage was accompanied by increased γ-secretase processing of the remaining membrane stubs and increased CTF2 generation (Fig. 4E).
Enzymatically Modified LDL Induces ADAM-mediated Shedding
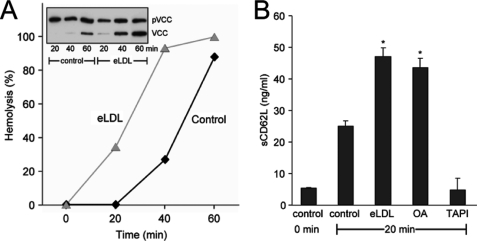
LDL entrapped in the arterial intima has to undergo biochemical alterations to become atherogenic. eLDL constitutes the major fraction of lipid extracted from early human atherosclerotic lesions (27–29). eLDL of defined composition with the same properties as lesional LDL can readily be produced (17, 28), and it was of interest to examine the bioavailability of FFA contained therein and to determine whether the modified lipoprotein might thus be able to drive ADAM-dependent events relevant to the pathogenesis of atherosclerosis. Incubation of pVCC-laden erythrocytes with 20 μg/ml eLDL markedly accelerated ADAM-dependent toxin processing and hemolysis (Fig. 5A). In neutrophil granulocytes, eLDL provoked rapid shedding of L-selectin to the same extent as OA (Fig. 5B).
FIGURE 5.
eLDL stimulates ADAM-dependent proteolysis on erythrocytes and neutrophil granulocytes. A, incubation of pVCC-laden erythrocytes with 20 μg/ml eLDL accelerated ADAM-dependent toxin processing and hemolysis. B, eLDL and OA provoked rapid shedding of sCD62L in neutrophil granulocytes (*, significant increase compared with control (n = 3 ± S.D. (error bars)), p < 0.005, t test).
eLDL-induced Shedding Is Not Cell Type-dependent
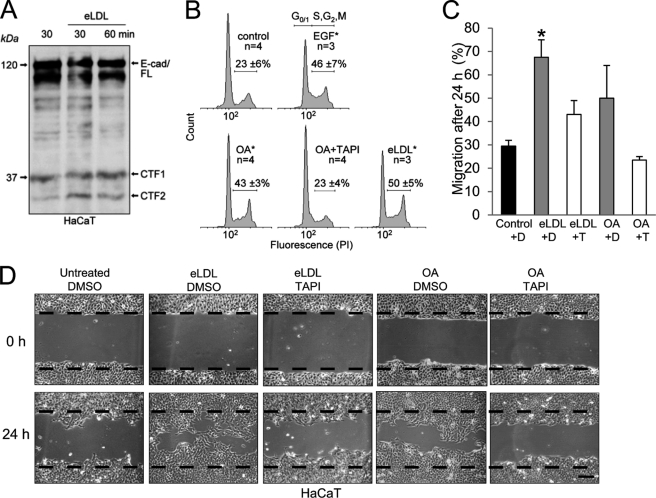
The results obtained before were transferable to all other cells analyzed in this study, and functional consequences (7–9) followed in the wake of enhanced ADAM-dependent processing. The shedding of E-cadherin has been shown to modulate β-catenin subcellular localization and downstream signaling leading to effects on cell migration and proliferation (7). Indeed, incubation of HaCaT keratinocytes with eLDL resulted in increased E-cadherin proteolysis (Fig. 6A), and this was followed by stimulation of cell proliferation (Fig. 6B), as determined by propidium iodide cell cycle analysis (Fig. 6B; S + G2/M (%): control 22.7 ± 5.8; EGF 46.3 ± 7.4; OA 43.4 ± 3.3; OA + TAPI 23.3 ± 3.6; eLDL 50 ± 4.9) and by enhanced epithelial cell migration as evidenced by in vitro wound healing assays (Fig. 6, C and D). Single FFA like OA also stimulated keratinocyte migration, but the effect was less drastic than eLDL stimulation (Fig. 6, C and D).
FIGURE 6.
eLDL augments E-cadherin proteolysis in HaCaT keratinocytes. A, keratinocytes were treated with 20 μg/ml eLDL, and E-cadherin proteolysis was determined by immunblotting. B, increased E-cadherin proteolyis in HaCaTs was accompanied by significant changes in cell cycle after treatment with 30 μm OA or 5 μg/ml eLDL; EGF (4 nm) was used as a positive control, TAPI-0 (10 μm) application abolished the OA effect. The numbers of independent measurements (n) and the average percentage of cells in S + G2/M phase ± S.D. (error bars) are shown. *, significance compared with control, p ≤ 0.005, t test. C, quantification of three independent in vitro wound healing assays. A cell-free area was introduced by scratching HaCaT cells with a pipette tip, and cells were washed and either mock-treated or stimulated with 20 μg/ml eLDL or 20 μg/ml OA in the presence or absence of TAPI (T) or DMSO (D). Migration was evaluated after 24 h. D, micrographs of one representative of three independent experiments are shown. Scale bar, 100 μm.
eLDL Enhances ADAM10-mediated VE-cadherin Release
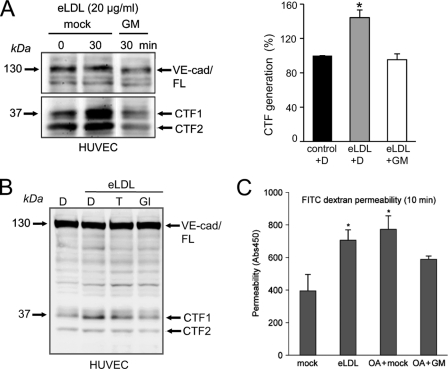
VE-cadherin represents the major adhesion molecule of endothelial adherence junctions. It plays an essential role in controlling endothelial permeability, vascular integrity, and leukocyte transmigration in health and disease. To determine whether eLDL would also activate ADAM-mediated VE-cadherin shedding, endothelial cells were treated with eLDL for 30 or 60 min. Indeed, exposure of these cells to eLDL augmented proteolysis of VE-cadherin, and this could be completely blocked by broad spectrum metalloprotease inhibitor GM6001 (Fig. 7A) but also by the ADAM10 inhibitor GI254023X (Fig. 7B), indicating that ADAM10 is responsible for this process. As predictable from a previous study (9), acceleration of VE-cadherin cleavage correlated functionally with an increased endothelial permeability for macromolecules (Fig. 7C).
FIGURE 7.
Influence of eLDL on human endothelial cells. A, HUVECs were treated with 20 μg/ml eLDL for 30 min in the presence or absence of the broad spectrum metalloprotease inhibitor GM6001 (GM, 10 μm). eLDL significantly augmented metalloprotease-dependent VE-cadherin cleavage. Densitometric quantification of three independent experiments is shown in the right panel (n = 3 ± S.E. (error bars); p < 0.05). eLDL-induced VE-cadherin proteolysis could be inhibited with TAPI (10 μm) and was completely abrogated with GI254023X (GI, 3 μm). C, HUVECs were grown to confluence on collagen-coated Transwell filters and pretreated with DMSO or broad spectrum metalloprotease inhibitor GM6001 (GM, 10 μm) for 30 min. Then, cells were stimulated with eLDL (20 μg/ml), and permeability for FITC-dextran (40 kDa) was measured after 10 min in a fluorescence plate reader (λEX 485 nm; λEM 525 nm). One representative of three experiments is shown. Data are expressed as mean ± S.D. *, p < 0.05.
DISCUSSION
Atherosclerosis is a complex process, closely associated with dysregulated lipid metabolism and endothelial cell dysfunction. ADAM10 and ADAM17 contribute to the regulation of endothelial cell homeostasis through proteolysis of several substrates such as VE-cadherin, CX3CL1, or JAM-A. Increased ADAM-mediated proteolysis is assumed to correlate with decreased vascular integrity and increased production of cytokines and chemokines and with increased transmigration of leukocytes. Thus, understanding the regulation of ADAM activity is of central interest.
ADAM-mediated shedding occurs constitutively and after activation with a broad variety of stimuli. The shedding process can be regulated at different levels including transcriptional control, posttranslational modifications, enzyme stability, and cellular localization (30, 31). Stimulation of ADAM-mediated shedding occurs very quickly. The accessibility of the catalytic site plays an important role for ADAM17 activity (32).
Due to the complexity and interdependence of biological networks in nucleated cells, precise definition of regulatory mechanisms is often more difficult than possible. Cholesterol depletion induces ADAM-dependent ectodomain shedding (11–13), probably due to changes in membrane fluidity or to sterol-dependent allocation of ADAMs to specific membrane microdomains. The latter is difficult to prove and must be reconciled with the findings indicating that ADAM10 is mostly excluded from, whereas ADAM17 appears sequestered in lipid rafts (33). Whether and how changes in membrane organization or fluidity might be responsible for the regulation of ADAM functions remains a matter of debate. In this study, we offer a more general model of membrane-enzyme regulation proposing that cleavage of membrane-anchored substrates by their respective cellular enzymes can be directly and simply influenced by changing the physical membrane properties.
The establishment of the erythrocyte model, which allowed the activity of the enzyme and cleavage of a membrane-bound substrate to be differentially assessed, was of key importance for these analyses. ADAM10 has recently been identified by mass spectrometry on rabbit erythrocyte membranes (14), and the erythrocyte model is singularly attractive for obvious reasons. First, ADAM activation downstream of classical signaling pathways can be excluded. Second, enzyme activity is strictly immobilized to the cell surface. Third, activity is independent of cell viability and can be assessed in membranes of lysed cells. Fourth, the fluorometric method for assessing enzyme activity introduced herein is far more sensitive than conventional assays for ADAM activity, and the use of pVCC as the sensor for cleavage of a membrane bound substrate is unsurpassed with regard to simplicity, sensitivity, and reproducibility. Striking findings surfaced. Unsaturated FFA dramatically accelerated cleavage of pVCC by ADAM10. Remarkably, this occurred in the absence of a bona fide increase in enzymatic activity. Measurement of DPH fluorescence anisotropy and FRAP analyses bore out the anticipation that FFA-dependent increase in bilayer fluidity accelerated lateral membrane movement and was responsible for the observed effects. This contention was corroborated by many findings. Acceleration of pVCC cleavage was nonspecifically invoked by all unsaturated fatty acids tested as well as by the membrane fluidizer PhA. A decisive finding was that trans-OA, which lacked membrane-fluidizing properties, was without effect. Further, as predictable by the model, decreasing membrane fluidity decelerated pVCC cleavage. This was demonstrated through application of PA and by cholesterol enrichment of membranes. Finally, and in perfect accord with the earlier findings in nucleated cells, cholesterol depletion with accompanying increase in fluidity of the erythrocyte membrane markedly accelerated pVCC cleavage by ADAM10.
Classical models were selected to provide proof of principle in nucleated cells, whereby FFA were offered in free form or in lipoprotein-bound state and at concentrations that conformed to physiological situations. Thus, plasma levels of nonesterified fatty acids are generally low but can become considerably elevated to reach millimolar concentrations, for example in patients with insulin-resistant diabetes mellitus (34) and sepsis (35). Preferential accumulation of unsaturated FFA must occur locally in atherosclerotic lesions due to enzymatic cleavage of cholesteryl esters in LDL (27, 28). eLDL contains ∼3 μmol of FFA/mg cholesterol, of which ∼75% are unsaturated (36). Accordingly, 40–80 μm unsaturated FFA correspond to eLDL concentrations of 20–40 μg/ml cholesterol, as used in this study.
Local accumulation of FFA might affect endothelial cell function but also the recruitment of leukocytes. ADAM10 and ADAM17 have been implicated in the release of several cell adhesion molecules that mediate the interaction of leukocytes with the endothelial cell wall such as L-selectin, CX3CL1, or CXCL16. Moreover, they are critically involved in the regulation of interendothelial cell adhesion through proteolysis of adhesion molecules such as VE-cadherin or JAM-A. To analyze the effect of FFA on nucleated cells we chose two models: the ADAM17-dependent shedding of L-selectin in PMN and the ADAM10-dependent release of soluble VE-cadherin on HUVECs. To verify the generality of the activation mechanisms, we additionally investigated the ADAM10-mediated cleavage of E-cadherin in keratinocytes. Uniform results concordant with the proposed regulatory principle were obtained in all cases. In PMN, membrane fluidizers promoted rapid shedding of L-selectin. Again, trans-OA was without effect. A battery of signaling inhibitors did not suppress the effect, which was shown to occur in the absence of any increase in cell surface expression of the enzyme. Analogous findings with regard to cleavage of E- and VE-cadherin were made in keratinocytes and endothelial cells. All substrates analyzed in this study are preferential substrates for ADAM10 or ADAM17, and our pharmacological approaches indeed suggest that these are the major enzymes responsible for the observed effects. However, most transmembrane proteins can be cleaved by several proteases, and we cannot rule out that additionally other proteases might contribute to the increased proteolysis after membrane fluidization.
Very satisfactorily, downstream events that have been linked to cadherin cleavage also followed. The proliferation assays unveiled the mitogenic properties of unsaturated FFA in eLDL, and this raises the possibility that eLDL may be at least partially responsible for local cell proliferation in atherosclerotic lesions. Single fatty acids also stimulated cell migration, but they could not reach the drastic eLDL effect, probably a combination of several FFA would have been more effective. Cleavage of VE-cadherin led to enhanced endothelial permeability, and this finding may be linked to the events of early atherogenesis including the insudation of plasma components and the immigration of white blood cells to areas of LDL accumulation (Fig. 8).
FIGURE 8.
Schematic model of the influence of FFA on ADAM activity in the context of atherosclerosis. Atherosclerosis is triggered by accumulation of LDL in the vessel wall, which is, among other things, modified by hydrolytic enzymes. FFA are liberated from this eLDL by combined action of proteases and cholesterylesterase. FFA increase membrane fluidity of surrounding cells and augment ADAM10-dependent VE-cadherin proteolysis thus contributing to increased endothelial permeability. Because ADAMs also regulate the transmigration of leukocytes through the endothelium, protease activation may favor the invasion of these cells. Moreover, it is likely that local accumulation of FFA also activates ADAM-dependent proteolytic events on invading monocytes, such as release of TNF-α and triggers proliferation of surrounding smooth muscle cells (SMC), thus additionally contributing to the pathologic process of atherosclerosis.
Implications of the present work for other important diseases are self-evident. For example, cell membrane fluidity appears to be diminished in Alzheimer disease. Higher membrane fluidity favors the nonamyloidogenic α-secretase cleavage of amyloid precursor protein, whereas lower membrane fluidity is associated with preferential β-secretase cleavage, leading to an increase in toxic Aβ fragments. In addition, oligomeric Aβ can disturb the structure and function of cell membranes and decrease membrane fluidity (37, 38). Polyunsaturated fatty acids are emerging as potential agents for prevention of cognitive decline and treatment of Alzheimer disease (39). Preclinical studies suggest that docosahexaenoic acid maintains membrane fluidity and improves synaptic functions. Epidemiological studies support the association between omega-3 polyunsaturated fatty acid consumption and a lower prevalence of dementia, and it has been suggested that low levels of unsaturated lipids in the brain contribute to Alzheimer disease risk (40). It is tempting to speculate that our present findings have a direct bearing on all these observations.
The present study uncovers a novel principle of enzyme regulation and indicates that membrane fluidity is fundamentally important for the function of ADAMs. It identifies FFA as regulators thereof and indicates that such processes could bear high relevance. Future studies will soon disclose whether extracellular and intracellular FFA and other modifiers of membrane fluidity control other enzyme-substrate and also perhaps receptor-ligand interactions in cell membranes.
Acknowledgments
We thank Björn Ahrens, Fatima Boukhallouk, Silvia Weis, and Miriam Lemmer for excellent technical assistance. We are grateful to the Blood Transfusion Center of the University of Mainz for providing blood samples from healthy donors. We thank Prof. Dr. W. Schunack for critical discussion and for pointing to the importance of using trans-OA as appropriate control for our hypothesis.
Note Added in Proof
Yang et al. (41) have recently reported that treatment of human neuroblastoma cells with secretory phospholipase A2 type III enhances α-secretase-dependent amyloid precursor protein processing through alterations of membrane fluidity. Their proposal that this is due to release of FA from membrane phospholipids accords with the conclusions reached in the present study.
This work was supported by the Deutsche Forschungsgemeinschaft Sonderforschungsbereich Grant 490 (to S. B. and M. H.), Grant CRC877 (to K. R.), and by the Cluster of Excellence “Inflammation at Interfaces” (to K. R.).
- ADAM
- a disintegrin and metalloproteinase
- CTF
- C-terminal fragment
- DMSO
- dimethyl sulfoxide
- DPH
- diphenyl hexatriene
- E-cadherin
- epithelial cadherin
- EGFP
- enhanced GFP
- eLDL
- enzymatically modified LDL
- FFA
- free fatty acids
- FRAP
- fluorescence recovery after photobleaching
- HUVEC
- human umbilical vein endothelial cell
- MCD
- β-methyl cyclodextrin
- OA
- oleic acid
- PA
- palmitic acid
- PhA
- phenethyl alcohol
- PMN
- polymorphonuclear neutrophil
- pVCC
- pro-VCC
- ROI
- region of interest
- sL-selectin
- soluble L-selectin
- VCC
- Vibrio cholerae cytolysin
- VE-cadherin
- vascular endothelial cadherin.
REFERENCES
- 1. Reiss K., Saftig P. (2009) Semin. Cell Dev. Biol. 20, 126–137 [DOI] [PubMed] [Google Scholar]
- 2. Murphy G. (2008) Nat. Rev. Cancer 8, 929–941 [DOI] [PubMed] [Google Scholar]
- 3. Pruessmeyer J., Ludwig A. (2009) Semin. Cell Dev. Biol. 20, 164–174 [DOI] [PubMed] [Google Scholar]
- 4. Deuss M., Reiss K., Hartmann D. (2008) Curr. Alzheimer Res. 5, 187–201 [DOI] [PubMed] [Google Scholar]
- 5. Li Y., Brazzell J., Herrera A., Walcheck B. (2006) Blood 108, 2275–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peschon J. J., Slack J. L., Reddy P., Stocking K. L., Sunnarborg S. W., Lee D. C., Russell W. E., Castner B. J., Johnson R. S., Fitzner J. N., Boyce R. W., Nelson N., Kozlosky C. J., Wolfson M. F., Rauch C. T., Cerretti D. P., Paxton R. J., March C. J., Black R. A. (1998) Science 282, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 7. Maretzky T., Reiss K., Ludwig A., Buchholz J., Scholz F., Proksch E., de Strooper B., Hartmann D., Saftig P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9182–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maretzky T., Scholz F., Köten B., Proksch E., Saftig P., Reiss K. (2008) J. Invest Dermatol. 128, 1737–1746 [DOI] [PubMed] [Google Scholar]
- 9. Schulz B., Pruessmeyer J., Maretzky T., Ludwig A., Blobel C. P., Saftig P., Reiss K. (2008) Circ. Res. 102, 1192–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Donners M. M., Wolfs I. M., Olieslagers S., Mohammadi-Motahhari Z., Tchaikovski V., Heeneman S., van Buul J. D., Caolo V., Molin D. G., Post M. J., Waltenberger J. (2010) Arterioscler. Thromb. Vasc. Biol. 30, 2188–2195 [DOI] [PubMed] [Google Scholar]
- 11. Kojro E., Gimpl G., Lammich S., Marz W., Fahrenholz F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5815–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maretzky T., Schulte M., Ludwig A., Rose-John S., Blobel C., Hartmann D., Altevogt P., Saftig P., Reiss K. (2005) Mol. Cell. Biol. 25, 9040–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matthews V., Schuster B., Schütze S., Bussmeyer I., Ludwig A., Hundhausen C., Sadowski T., Saftig P., Hartmann D., Kallen K. J., Rose-John S. (2003) J. Biol. Chem. 278, 38829–38839 [DOI] [PubMed] [Google Scholar]
- 14. Wilke G. A., Bubeck Wardenburg J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13473–13478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valeva A., Walev I., Weis S., Boukhallouk F., Wassenaar T. M., Endres K., Fahrenholz F., Bhakdi S., Zitzer A. (2004) J. Biol. Chem. 279, 25143–25148 [DOI] [PubMed] [Google Scholar]
- 16. Gimpl G., Fahrenholz F. (2000) Eur. J. Biochem. 267, 2483–2497 [DOI] [PubMed] [Google Scholar]
- 17. Bhakdi S., Torzewski M., Paprotka K., Schmitt S., Barsoom H., Suriyaphol P., Han S. R., Lackner K. J., Husmann M. (2004) Circulation 109, 1870–1876 [DOI] [PubMed] [Google Scholar]
- 18. Ludwig A., Hundhausen C., Lambert M. H., Broadway N., Andrews R. C., Bickett D. M., Leesnitzer M. A., Becherer J. D. (2005) Comb. Chem. High Throughput Screen. 8, 161–171 [DOI] [PubMed] [Google Scholar]
- 19. Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. (1988) J. Cell Biol. 106, 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haugwitz U., Bobkiewicz W., Han S. R., Beckmann E., Veerachato G., Shaid S., Biehl S., Dersch K., Bhakdi S., Husmann M. (2006) Cell. Microbiol. 8, 1591–1600 [DOI] [PubMed] [Google Scholar]
- 21. Merrill A. R., Aubry H., Proulx P., Szabo A. G. (1987) Biochim. Biophys. Acta 896, 89–95 [DOI] [PubMed] [Google Scholar]
- 22. Nagy E., Balogi Z., Gombos I., Akerfelt M., Björkbom A., Balogh G., Török Z., Maslyanko A., Fiszer-Kierzkowska A., Lisowska K., Slotte P. J., Sistonen L., Horváth I., Vígh L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kenworthy A. K., Nichols B. J., Remmert C. L., Hendrix G. M., Kumar M., Zimmerberg J., Lippincott-Schwartz J. (2004) J. Cell Biol. 165, 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pucadyil T. J., Kalipatnapu S., Harikumar K. G., Rangaraj N., Karnik S. S., Chattopadhyay A. (2004) Biochemistry 43, 15852–15862 [DOI] [PubMed] [Google Scholar]
- 25. Le Gall S. M., Bobé P., Reiss K., Horiuchi K., Niu X. D., Lundell D., Gibb D. R., Conrad D., Saftig P., Blobel C. P. (2009) Mol. Biol. Cell 20, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gómez-Gaviro M., Domínguez-Luis M., Canchado J., Calafat J., Janssen H., Lara-Pezzi E., Fourie A., Tugores A., Valenzuela-Fernández A., Mollinedo F., Sánchez-Madrid F., Díaz-González F. (2007) J. Immunol. 178, 8053–8063 [DOI] [PubMed] [Google Scholar]
- 27. Seifert P. S., Hugo F., Tranum-Jensen J., Zâhringer U., Muhly M., Bhakdi S. (1990) J. Exp. Med. 172, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhakdi S., Dorweiler B., Kirchmann R., Torzewski J., Weise E., Tranum-Jensen J., Walev I., Wieland E. (1995) J. Exp. Med. 182, 1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Torzewski M., Klouche M., Hock J., Messner M., Dorweiler B., Torzewski J., Gabbert H. E., Bhakdi S. (1998) Arterioscler. Thromb. Vasc. Biol. 18, 369–378 [DOI] [PubMed] [Google Scholar]
- 30. Edwards D. R., Handsley M. M., Pennington C. J. (2008) Mol. Aspects Med. 29, 258–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murphy G. (2009) Semin. Cell Dev. Biol. 20, 138–145 [DOI] [PubMed] [Google Scholar]
- 32. Le Gall S. M., Maretzky T., Issuree P. D., Niu X. D., Reiss K., Saftig P., Khokha R., Lundell D., Blobel C. P. (2010) J. Cell Sci. 123, 3913–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tellier E., Canault M., Rebsomen L., Bonardo B., Juhan-Vague I., Nalbone G., Peiretti F. (2006) Exp. Cell Res. 312, 3969–3980 [DOI] [PubMed] [Google Scholar]
- 34. Boden G. (1997) Diabetes 46, 3–10 [PubMed] [Google Scholar]
- 35. Nogueira A. C., Kawabata V., Biselli P., Lins M. H., Valeri C., Seckler M., Hoshino W., Júnior L. G., Bernik M. M., de Andrade Machado J. B., Martinez M. B., Lotufo P. A., Caldini E. G., Martins E., Curi R., Soriano F. G. (2008) Shock 29, 342–348 [DOI] [PubMed] [Google Scholar]
- 36. Suriyaphol P., Fenske D., Zähringer U., Han S. R., Bhakdi S., Husmann M. (2002) Circulation 106, 2581–2587 [DOI] [PubMed] [Google Scholar]
- 37. Müller W. E., Koch S., Eckert A., Hartmann H., Scheuer K. (1995) Brain Res. 674, 133–136 [DOI] [PubMed] [Google Scholar]
- 38. Müller W. E., Eckert G. P., Scheuer K., Cairns N. J., Maras A., Gattaz W. F. (1998) Amyloid. 5, 10–15 [DOI] [PubMed] [Google Scholar]
- 39. Carrié I., Abellan Van Kan G., Rolland Y., Gillette-Guyonnet S., Vellas B. (2009) Curr. Pharm. Des. 15, 4173–4185 [DOI] [PubMed] [Google Scholar]
- 40. Leskovjan A. C., Kretlow A., Miller L. M. (2010) Anal. Chem. 82, 2711–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang X., Shang W., He Y., Cui J., Haidekker M. A., Sun G. Y., Lee J. C. (2010) J. Lipid Res. 51, 957–966 [DOI] [PMC free article] [PubMed] [Google Scholar]