Abstract
Botulinum neurotoxin (BoNT) belongs to a large class of toxic proteins that act by enzymatically modifying cytosolic substrates within eukaryotic cells. The process by which a catalytic moiety is transferred across a membrane to enter the cytosol is not understood for any such toxin. BoNT is known to form pH-dependent pores important for the translocation of the catalytic domain into the cytosol. As a first step toward understanding this process, we investigated the mechanism by which the translocation domain of BoNT associates with a model liposome membrane. We report conditions that allow pH-dependent proteoliposome formation and identify a sequence at the translocation domain C terminus that is protected from proteolytic degradation in the context of the proteoliposome. Fluorescence quenching experiments suggest that residues within this sequence move to a hydrophobic environment upon association with liposomes. EPR analyses of spin-labeled mutants reveal major conformational changes in a distinct region of the structure upon association and indicate the formation of an oligomeric membrane-associated intermediate. Together, these data support a model of how BoNT orients with membranes in response to low pH.
Keywords: Bacterial Toxins, Botulinum Toxin, Electron Paramagnetic Resonance (EPR), Fluorescence, Neurotoxin
Introduction
Botulinum neurotoxin (BoNT)3 inhibits the release of acetylcholine at peripheral cholinergic nerve terminals and causes the potentially lethal, flaccid paralytic condition known as botulism (1). It is produced by Clostridium botulinum as a single chain 150-kDa protein in one of seven antigenically distinct forms (serotypes A–G) (2). It is then cleaved to form a dichain molecule in which a 50-kDa light chain (LC) and a 100-kDa heavy chain (HC) remain linked by a disulfide bond. The LC is a zinc metalloprotease that cleaves components of the synaptic membrane fusion complex and blocks neuronal exocytosis (3). The C-terminal half of the HC (HC receptor-binding domain (HCR)) binds neuronal receptors (4), whereas the N-terminal half of the HC (HC translocation domain (HCT)) mediates the translocation of the LC into the cytosol (5).
After binding to its receptors, BoNT undergoes receptor-mediated endocytosis and is transported to the endosomal compartment. It is thought that the acidic pH of the endosome triggers HCT pore formation and LC translocation (6). In vitro studies have shown that BoNT (and the isolated BoNT HCT) undergoes pH-dependent membrane insertion and pore formation (7–13), and single molecule translocation events have been observed in excised patches of neuronal cells (14, 15). These studies support a model in which the HCT acts as both a conduit and a chaperone for the transit of the LC protease across the membrane (5, 11, 16).
The HCT structure, visualized in x-ray crystal structures of BoNT/A, BoNT/B, and BoNT/E (17–21), is unique and bears no resemblance to structures observed in other toxins known to mediate pH-dependent translocation events (e.g. anthrax toxin and diphtheria toxin). The HCT contains a pair of kinked α-helices (>100 Å in length) surrounded by several loops and shorter helical regions (see Fig. 1A). The HCT also contains a >50-amino acid “belt” that wraps around the LC. A sequence with amphipathic character (BoNT/A residues 659–681) has been proposed as a putative transmembrane helix in the pore structure of BoNT (22) and has been observed as an extended structure on the surface of the BoNT HCT (see Fig. 1A). The details of how the HCT changes structure in response to low pH and membranes and which elements are involved in membrane association and insertion are unclear.
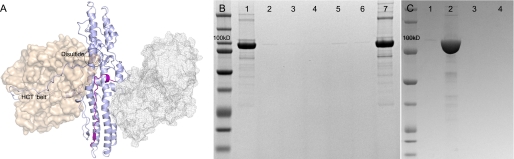
FIGURE 1.
BoNT/A LC-HCT associates irreversibly with liposomes at low pH. A, structure of BoNT/A. Coordinates are from Protein Data Bank code 3BTA with the HCR (not present in the sample) in mesh. The LC domain is shown in wheat, and the HCT is shown in light blue. The disulfide and belt that tether the LC and HCT structures together are highlighted to provide a context of how the overall structure is arranged. The hydrophobic sequence 659–681 is colored purple. B, 10 μg of LC-HCT (10 mg/ml) was added to 100 μl of POPC/POPG large unilamellar vesicles in 1 ml of 20 mm sodium acetate (pH 4.4) and 100 mm NaCl. Proteoliposomes were isolated by spinning at 10,000 × g for 10 min at room temperature. Half of the resuspended pellet (25 μl) was reserved for analysis (lane 1), whereas the remaining half was washed four times (four cycles of resuspension and centrifugation) and then analyzed (lane 7). Supernatants from the initial proteoliposome isolation (concentrated to 50 μl) and the four subsequent washes (concentrated to 25 μl) were analyzed (lane 2 and lanes 3–6, respectively). C, 10 μg of LC-HCT (10 mg/ml) was added to 100 μl of POPC/POPG large unilamellar vesicles in 1 ml of 20 mm Tris-HCl (pH 8.0) and 100 mm NaCl. Liposomes were isolated at 100,000 × g for 45 min. Half of the resuspended pellet (25 μl) was reserved for analysis (lane 1), and the other was resuspended in 1 ml of 20 mm Tris-HCl (pH 8.0) and 100 mm NaCl and reisolated (lane 4). The supernatants from the initial proteoliposome isolation and subsequent wash were concentrated to 25 μl and analyzed (lanes 2 and 3, respectively). All proteins were separated by SDS-PAGE and visualized by Coomassie Blue staining.
EPR spectroscopy has proven to be an effective method in probing the structures of other pH-dependent pore-forming toxins (23). In brief, the proteins are engineered to contain single cysteine residues at defined locations within the structure. The proteins are labeled with a sulfhydryl-specific nitroxide reagent and inserted into model membrane liposomes. The paramagnetic resonance of the nitroxide side chain provides information on the mobility (24) and solvent accessibility (25) of the side chain. Studies of this nature have identified the α-helical transmembrane elements in colicin E1 and diphtheria toxin (26, 27).
In this study, we explored the structure of a BoNT/A LC-HCT as it interacts with the membrane. A reconstituted liposome system showed that the protein binds liposomes in a pH-dependent manner. Using EPR, we show that the predicted transmembrane region (residues 659–681) does not form a transmembrane helix but does undergo large structural changes in the formation of the proteoliposome. We identify a region at the HCT C terminus (residues 805–837) that is protected from pepsin upon interaction with the liposome. Further characterization of this region by site-specific nitrobenzoxadiazole (NBD) fluorescence spectroscopy suggests that part of this protected C-terminal region (residues 826–835) shifts from a polar to a non-polar environment as a result of membrane association at low pH. Finally, we present evidence to suggest that BoNT/A LC-HCT assembles in the membrane as oligomers. This work provides a framework for understanding how the structural rearrangements of this unique HCT aid in the process of membrane association, pore formation, and, ultimately, LC translocation into the cytosol.
EXPERIMENTAL PROCEDURES
Recombinant LC-HCT Expression and Purification
A plasmid encoding the BoNT/A LC-HCT (residues 1–870 with a C-terminal deca-His tag (14)) was transformed into Escherichia coli BL21(DE3)-RIL cells. A 100-ml overnight culture was added to 1 liter of Terrific Broth and grown for 75 min. The culture was diluted 2-fold into fresh medium and induced with 1 mm isopropyl β-d-thiogalactopyranoside. Cells were harvested after overnight growth at 18 °C and stored at −80 °C. Pellets were resuspended in 20 mm Tris-HCl (pH 8.0) and 150 mm NaCl and lysed using a French press. Supernatants were clarified by centrifugation at 20,000 × g for 25 min and filtered with a 0.45-μm syringe disc filter. The supernatant was added to a 10-ml bed volume of TALON metal affinity resin (Clontech) charged with Co2+. The column was washed with 50 ml of 20 mm Tris-HCl (pH 8.0) and 150 mm NaCl, followed by 50 ml of 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 10 mm imidazole. The LC-HCT protein was eluted in 20 ml of 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 200 mm imidazole; concentrated to 5 ml; and purified on a Q-Sepharose Fast Flow column (GE Healthcare) using an elution buffer gradient of 20 mm Tris-HCl (pH 8.0) from 0 to 500 mm NaCl. The peak from the Q-Sepharose column was concentrated to 1 ml and run over a Superdex S-200 preparative grade size exclusion column (GE Healthcare) in 20 mm Tris-HCl (pH 8.0) and 150 mm NaCl for final purification. The retention volume was consistent with the size of a monomer based on the retention profiles of gel filtration standards (Bio-Rad).
Construction of Single Cysteine Mutants
We mutated the three free cysteines (Cys-134, Cys-166, and Cys-790) from the wild-type LC-HCT construct to alanines to create a LC-HCT 3CA template (plasmid pBL242). pBL242 was used as the background for all single cysteine mutations. All mutations were made using a standard QuikChange site-directed mutagenesis protocol (Stratagene) and verified by sequencing. Mutant proteins were expressed and purified as described above.
Liposome Preparation
A 2:1 mol ratio mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPC/POPG; Avanti) in chloroform was dried under N2 and placed under vacuum for 6 h. The dried lipid mixture was rehydrated overnight with constant stirring to a final lipid concentration of 13 mm in 10 mm HEPES and 100 mm KCl (pH 7.5). Rehydrated lipids were subjected to three rounds of freeze/thaw, followed by extrusion through a 100-nm filter to obtain large unilamellar vesicles.
Association and Proteoliposome Isolation
10 μg of LC-HCT (10 mg/ml) was added to 100 μl of POPC/POPG in 1 ml of 20 mm sodium acetate (pH 4.4) and 100 mm NaCl. Proteoliposomes were isolated by spinning at 10,000 × g for 10 min at room temperature. The supernatant was removed, and the proteoliposomes were resuspended in 1 ml of 20 mm sodium acetate (pH 4.4) and 100 mm NaCl and then reisolated by centrifugation. This step was repeated four times to wash away unbound or loosely bound protein. The high pH association was performed as described above with two changes: the buffer was 20 mm Tris-HCl (pH 8.0) and 100 mm NaCl, and liposomes were isolated at 100,000 × g for 45 min.
Labeling Single Cysteine Mutants with S-(2,2,5,5-Tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl Methanesulfonothioate (MTSL)
Purification for labeling of single cysteine mutants was the same as described above except that 1 mm DTT was added during lysis and removed on the TALON resin. Immediately after elution from the TALON resin, a 10-fold molar excess of MTSL dissolved in dimethylformamide was added and incubated for 4 h at room temperature. After incubation, a 5-fold molar excess of MTSL was added and incubated overnight at 4 °C. Unbound MTSL was removed during further purification.
EPR of Single Cysteine Mutants
EPR spectra were collected at 23 °C on a Bruker EMX spectrometer (X-band) at an incident power of 10 milliwatts and 1.6-G modulation amplitude. Spectra of soluble LC-HCT single cysteine mutants were taken at 10 mg/ml protein. Proteoliposome samples were made by combining the pellets from four individual insertions as described above. The combined pellets were washed four times with 20 mm sodium acetate (pH 4.4) and 100 mm NaCl and resuspended in 40 μl of the same buffer. Power saturation experiments were carried out on a Bruker ELEXSYS spectrometer equipped with a dielectric resonator (Bruker BioSpin). Samples were loaded in gas-permeable methylpentene polymer TPX® capillaries, and the measurements were carried out under nitrogen gas alone, in the presence of 20% oxygen, or under nitrogen gas with 10 mm nickel-diaminediacetic acid (NiEDDA). The data were analyzed to obtain the parameter P½ from a nonlinear least-squares fit of the power saturation curves in the program Origin (OriginLab Inc.). The EPR accessibility parameter (∏) was calculated as described (28).
Protection of LC-HCT by Liposome
50 μl of 5 mg/ml pepsin in 10 mm HCl (pH 2.2) was added to 10 μl of soluble LC-HCT (10 mg/ml) or LC-HCT proteoliposomes and incubated at 37 °C for 12 min. Pepsin was quenched by the addition of loading dye (soluble LC-HCT) or 1 ml of 20 mm sodium acetate (pH 4.4) and 100 mm NaCl (LC-HCT proteoliposomes). The proteoliposome sample was washed three times with 20 mm sodium acetate (pH 4.4) and 100 mm NaCl to remove pepsin.
Peptide Identification by Mass Spectrometry
Isolated proteoliposomes after pepsin treatment were resuspended in 20 μl of SDS-PAGE loading buffer and 2 μl of 10% SDS. Resuspended proteoliposomes were run on a NuPAGE 10% BisTris-HCl precast gel. Peptide bands were excised, trypsin-digested, and extracted as described (29). Digested protein samples were desalted using Michron CapTrap desalting cartridges (Michron Bioresources) using 2% acetonitrile and 0.1% TFA and eluted in a volume of 50 μl using 95% acetonitrile and 0.1% TFA. The eluate was frozen on dry ice and lyophilized. Dried samples were then resuspended in 15 μl of 0.5% acetic acid in mass spectrometry-grade water. Samples were loaded onto 100-μm (internal diameter) fused silica columns packed with 12 cm of reverse-phase resin (Synergi 4u Hydro-RP 80a, Phenomenex) and equilibrated using 5% acetonitrile and 0.1% TFA. Peptides were eluted using a 120-min linear gradient from 0 to 80% buffer containing 0.1% formic acid and 80% acetonitrile at a flow rate of 500 nl/min. Eluted ions were analyzed by nano-electrospray ionization LC-MS/MS using a Thermo LTQ linear ion trap tandem mass spectrometer as described (30). All acquired MS/MS data were searched against the C. botulinum proteome using the SEQUEST algorithm. Search results were processed and analyzed using BIGCAT (31).
Labeling Single Cysteine Mutants with N-((2-(Iodoacetoxy)ethyl)-N-methyl)amino-7-nitrobenz-2-oxa-1,3-diazole (IANBD)
Purification for labeling of single cysteine mutants was the same as described above except that 1 mm tris(2-carboxyethyl)phosphine was added at all steps and removed during the size exclusion purification. The LC-HCT from the size exclusion column was concentrated to 1 ml (∼1–5 μm) and labeled overnight at 4 °C with a 10-fold molar excess of IANBD (Molecular Probes). Excess label was removed by passing the sample over a PD10 desalting column (GE Healthcare). Labeled samples were concentrated to 30 μm, and the labeling efficiency was determined using ϵ478 nm = 25,000 m−1 cm−1 for IANBD and ϵ280 nm = 103,280 m−1 cm−1 for the LC-HCT protein. Samples not used immediately were flash-frozen in liquid nitrogen and stored at −80 °C.
Fluorescence Spectroscopy
All fluorescence measurements were taken using a HORIBA Jobin Yvon FluoroMax-3 (HORIBA Scientific). The excitation wavelength for all measurements was 470 nm with a 4-nm slit width. Emission scans for both soluble and proteoliposome samples were taken from 500 to 600 nm in 1-nm intervals using a 4-nm slit width. A buffer background emission scan was taken by adding 246 μl of 20 mm sodium acetate (pH 4.4) and 100 mm NaCl to a round quartz cuvette (Model 1924 micro cell, HORIBA Scientific). 4 μl of 30 μm NBD-labeled LC-HCT was added and mixed by inversion five times and scanned again. The maximum fluorescence intensity at 530 nm was defined as Fsol. LC-HCT proteoliposome spectra were taken by adding 204 μl of 20 mm sodium acetate (pH 4.4) and 100 mm NaCl and 42 μl of liposomes to a round quartz cuvette and taking an emission scan for the background. 4 μl of 30 μm NBD-labeled LC-HCT was added and mixed by inversion five times. The maximum fluorescence intensity at 530 nm for this sample was defined as Fmem.
RESULTS
Isolation of BoNT/A LC-HCT Proteoliposomes
The BoNT/A LC-HCT was purified as a soluble recombinant protein at pH 8.0. Previous studies have indicated that this protein is functional in in vitro substrate cleavage assays, cell-based intoxication assays, and single molecule pore formation and LC translocation assays (14). The protein was incubated at pH 4.4 or 8.0 in the presence or absence of POPC/POPG liposomes. Although pH had no visible effect on the solubility of either protein or liposomes, the low pH incubation of the mixture resulted in aggregation, similar to what has been reported in asolectin liposomes (32). The aggregated liposomes were isolated by centrifugation and shown to contain the BoNT/A LC-HCT by SDS-PAGE (Fig. 1B, lane 1). The minimum loss of LC-HCT following multiple wash and reisolation steps showed that the interaction was irreversible at low pH (Fig. 1B, lane 7). The lack of LC-HCT isolated with liposomes at high pH showed that low pH was required for the LC-HCT/liposome interaction (Fig. 1C, lane 1).
Accessibility of Paramagnetic Nitroxide Probe to Hydrophobic and Hydrophilic Quenchers
Three of the five native LC-HCT cysteines (Cys-133, Cys-164, and Cys-790) were mutated to alanine to create a template for site-directed cysteine substitutions (LC-HCT 3CA). The two remaining cysteines (Cys-429 and Cys-454) are involved in a very stable disulfide that tethers the LC and HCTs together (Fig. 1A). This bond was not reduced under our labeling conditions (data not shown). Individual cysteine substitutions were made in the LC-HCT 3CA background at amino acid positions throughout the HCT with an emphasis on residues in sequence 659–681. Each mutant protein was purified to homogeneity, labeled with MTSL (a thiol-reactive nitroxide spin label), and subjected to EPR spectroscopic analysis in both its soluble (pH 8.0) and proteoliposome (pH 4.4) states. EPR power saturation experiments were conducted in the presence of water-soluble NiEDDA and membrane-soluble molecular oxygen O2. The NiEDDA accessibility (reported as ∏) was reduced for proteoliposome forms of the protein at all but two of the tested sites (Table 1). (The accessibility of F613C-MTSL and D725C-MTSL did not change significantly.) However, the accessibility to O2 was also reduced at most sites; only N644C-MTSL, F657C-MTSL, and D725C-MTSL showed slight increases (Table 1). The location of each tested residue within the soluble LC-HCT structure is included in supplemental Fig. 1. In brief, the analysis did not readily point to a region involved in BoNT/A pore formation.
TABLE 1.
NiEDDA and O2 accessibility of MTSL-labeled LC-HCT single cysteine mutants
Δ∏ values are calculated from ∏ proteoliposome (PL) − ∏ soluble (sol). Percent changes represent Δ∏/∏sol × 100 and are included for those residues that showed a significant change in accessibility. Most residues decreased in both O2 and NiEDDA accessibilities when associated with liposomes. The few exceptions all had low accessibility to O2 in both the soluble and liposome-associated forms and are therefore not thought to lie within the membrane. Residues in the 659–681 region are in bold.
| Residue | NiEDDA |
O2 |
||||||
|---|---|---|---|---|---|---|---|---|
| ∏PL | ∏sol | Δ∏ | Change | ∏PL | ∏sol | Δ∏ | Change | |
| % | % | |||||||
| 581 | 2.79 | 4.29 | −1.5 | −34.9 | 0.84 | 1.14 | −0.30 | −26.3 |
| 605 | 1.04 | 1.78 | −0.74 | −41.5 | 0.37 | 0.68 | −0.31 | −45.6 |
| 613 | 0.02 | 0.01 | 0.01 | 0.04 | 0.10 | −0.06 | ||
| 615 | 1.96 | 4.28 | −2.32 | −54.2 | 0.78 | 1.47 | −0.69 | −46.9 |
| 631 | 1.04 | 2.52 | −1.48 | −58.7 | 0.59 | 0.73 | −0.14 | −19.1 |
| 644 | 0.04 | 0.13 | −0.09 | 0.26 | 0.10 | 0.16 | ||
| 647 | 1.82 | 2.70 | −0.88 | −32.6 | 0.80 | 0.83 | −0.03 | −3.6 |
| 654 | 0.43 | 0.90 | −0.53 | −58.9 | 0.30 | 0.51 | −0.21 | −41.2 |
| 657 | 0.26 | 0.36 | −0.10 | −27.8 | 0.51 | 0.19 | 0.32 | 168 |
| 662 | 1.34 | 3.17 | −1.83 | −57.7 | 0.53 | 0.83 | −0.30 | −36.1 |
| 665 | 0.09 | 0.31 | −0.22 | −71 | 0.18 | 0.24 | −0.06 | −25.0 |
| 666 | 0.14 | 0.63 | −0.49 | −77.8 | 0.17 | 0.34 | −0.17 | −50.0 |
| 667 | 0.61 | 3.15 | −2.54 | −80.6 | 0.43 | 1.20 | −0.77 | −64.1 |
| 669 | 3.05 | 4.02 | −0.97 | −24.1 | 1.02 | 1.33 | −0.31 | −23.3 |
| 670 | 0.57 | 1.59 | −1.02 | −64.2 | 0.46 | 0.49 | −0.03 | −6.1 |
| 671 | 1.32 | 3.14 | −1.82 | −58 | 0.70 | 0.96 | −0.26 | −27.1 |
| 674 | 1.90 | 5.21 | −3.31 | −63.5 | 1.04 | 1.48 | −0.44 | −29.7 |
| 679 | 3.37 | 4.12 | −0.75 | −18.2 | 1.01 | 1.21 | −0.20 | −16.7 |
| 725 | 0.02 | 0.01 | 0.01 | 0.06 | 0.04 | 0.02 | ||
| 750 | 1.55 | 2.21 | −0.66 | −29.8 | 0.67 | 0.95 | −0.28 | −29.5 |
| 770 | 0.43 | 1.09 | −0.66 | −60.6 | 0.29 | 0.43 | −0.14 | −32.6 |
| 774 | 0.81 | 1.66 | −0.85 | −51.2 | 0.47 | 0.81 | −0.34 | −42.0 |
| 775 | 0.15 | 0.37 | −0.22 | −59.5 | 0.21 | 0.27 | −0.06 | −22.2 |
| 817 | 1.50 | 1.95 | −0.45 | −23.1 | 0.73 | 1.16 | −0.43 | −37.1 |
| 819 | 0.89 | 2.93 | −2.04 | −69.6 | 0.48 | 1.04 | −0.56 | −53.8 |
| 823 | 1.35 | 2.46 | −1.11 | −45.1 | 0.78 | 1.37 | −0.59 | −43.1 |
| 825 | 1.16 | 3.18 | −2.02 | −63.5 | 0.70 | 1.26 | −0.56 | −44.4 |
| 827 | 2.34 | 4.21 | −1.87 | −44.4 | 1.04 | 1.24 | −0.20 | −16.1 |
| 829 | 0.78 | 3.13 | −2.35 | −75.1 | 0.94 | 1.03 | −0.09 | −8.7 |
| 833 | 1.14 | 2.81 | −1.67 | −59.4 | 0.82 | 1.23 | −0.41 | −33.3 |
| 835 | 1.26 | 3.50 | −2.24 | −64.0 | 0.84 | 1.46 | −0.62 | −42.5 |
Identification of Protease-protected Fragment within BoNT/A LC-HCT Proteoliposomes
Soluble LC-HCT and LC-HCT proteoliposomes were treated with 5 mg/ml pepsin in 10 mm HCl (pH 2.2). Pepsin completely proteolyzed the soluble LC-HCT, but two low molecular mass bands were observed when the LC-HCT was proteolyzed in the context of proteoliposomes (Fig. 2A). Both bands were subjected to in-gel tryptic digest and mass spectrometry analysis. Recovered tryptic peptides were separated by reversed-phase chromatography and analyzed by electrospray ionization mass spectrometry (Fig. 2B). The majority of the peptides mapped to the C terminus of the HCT (residues 805–820 and 826–835) and localized within one region of the LC-HCT structure (Fig. 3D, inset, green).
FIGURE 2.
Liposomes protect LC-HCT peptides from pepsin cleavage. A, 20 μg of soluble LC-HCT without (lane 1) and with (lane 2) pepsin, 40 μg of LC-HCT in proteoliposomes (lane 3), 100 μg of LC-HCT in proteoliposomes treated with pepsin (lane 4), and 8 μg of pepsin (lane 5). Arrows indicate LC-HCT peptides protected by the liposome. B, in-gel tryptic digest of bands A and B followed by mass spectrometry suggests sequences that were protected from protease degradation in the context of the proteoliposome. The two most abundant peptides (green) overlap to one region of the LC-HCT primary sequence: residues 805–821 and 826–836.
FIGURE 3.
Fluorescence intensity of NBD attached to cysteine-substituted LC-HCT in soluble and liposome-associated forms. Emission scans are shown of the soluble (red lines) and liposome-bound (black lines) forms of I830C-NBD (A) and I684C-NBD (B) at pH 4.4. The large increase in intensity and blue shift to 530 nm when I830C-NBD bound to liposomes is consistent with movement of NBD into a non-polar environment. C, compilation of the NBD intensity changes (Fmem/Fsol) observed at 530 nm for specific LC-HCT residues when proteins in the soluble (Fsol) and liposome-associated (Fmem) forms are compared. Residues were selected throughout the HCT structure with an emphasis on the protease-protected region identified in Fig. 2 (residues 805–836; green). Residues from the neighboring loop (sequence 682–688; orange) are included to show that this change is specific to the protease-protected region. D, close-up of the location of tested residues within the context of the LC-HCT structure. Residues with Fmem/Fsol > 3 are shown in blue, and residues with Fmem/Fsol < 3 are shown in red.
Steady-state Fluorescence of NBD-labeled LC-HCT in Soluble versus Proteoliposome Forms
The emission properties of the NBD fluorophore are sensitive to the polarity of the local environment: as the dye moves from an aqueous to a non-aqueous environment, the fluorescence intensity increases, and the wavelength of the maximum emission intensity decreases (blue shift). Therefore, when introduced as a site-specific label, NBD can be used to differentiate between residues that are in hydrophobic and hydrophilic environments. We generated a panel of NBD-labeled proteins with a special emphasis on two regions; sequence 659–681 is the extended hydrophobic sequence identified by previous primary sequence analyses (22), and sequences 805–820 and 826–835 were shown to be protease-resistant in the context of proteoliposomes (Fig. 2). Each NBD-labeled LC-HCT mutant was examined spectroscopically for changes in NBD fluorescence intensity between soluble and liposome-associated forms. Typical spectra for a residue that showed an increase in NBD fluorescence upon association with liposomes (I830C-NBD) and a residue that showed no change in environment (I684C-NBD) are shown in Fig. 3 (A and B, respectively).
Changes in fluorescence intensity were analyzed by dividing the NBD fluorescence intensity of the liposome-associated LC-HCT (Fmem) by the NBD fluorescence intensity of the soluble LC-HCT (Fsol) at 530 nm (Fig. 3C). A large value meant that the side chain had shifted into a more hydrophobic environment upon association with the liposome. Residues in region 826–835 showed Fmem/Fsol values that were >3 and mapped to one tip of the HCT helical axis, specifically a loop connecting two α-helices (Fig. 3, C and D). Mutation and testing of residues from a neighboring loop (Fig. 3D, orange) did not reveal a similar increase in NBD fluorescence. The location of each tested residue within the soluble LC-HCT structure is included in supplemental Fig. 2.
Conformational Changes in HCT Structure Revealed by EPR Spectroscopy
The observation that protected residues are located at one tip of the BoNT/A HCT (Fig. 3D) suggests the possibility that this region merely “dips” into the membrane without undergoing a significant structural change. To assess the extent of HCT structural change for residues throughout the structure, we collected EPR line-shape spectra for soluble and liposome-associated LC-HCTs using proteins labeled with MTSL at site-specific locations (Fig. 4 and supplemental Figs. 3 and 4). Qualitative assessments of relative changes in side chain mobility were based on 1) the overall breadth of the spectrum along the horizontal magnetic field axis and 2) the line width of the central resonance. An increase in breadth is interpreted as a decrease in molecular ordering and/or motional frequency (24).
FIGURE 4.
Line-shape measurements of MTSL attached to cysteine-substituted LC-HCT in soluble and proteoliposome forms. A, line-shape changes in the previously described hydrophobic sequence (residues 659–681; purple) and protease-protected region (residues 805–836; green). Side chains are colored according to large (red) or small (blue) changes when comparing line shapes from soluble and proteoliposome samples. Region 659–680 shows greater line-shape changes compared with region 805–836. B–E, representative line-shape overlays of soluble (sol; red lines), pH 4.4 proteoliposome (PL; blue lines), and pH 5.0 proteoliposome (black lines) samples for mutants F666C (B), I667C (C), K805C (D), and L829C (E). A 200-G scan width was used for F666C, I667C, and L829C. A 250-G scan width was used for K805C. All spectra were normalized to the same number of spins.
An example of a mobile surface-exposed residue that did not change its mobility upon conversion to the proteoliposome state is L829C-MTSL (Fig. 4E). The lack of line-shape change for this residue (and the neighboring residues G827C-MTSL and R835C-MTSL) (supplemental Fig. 4) suggests that, despite the change to a hydrophobic environment, residues within loop 826–835 are not conformationally restricted within the proteoliposome. An example of a residue that undergoes a large change in mobility upon formation of the proteoliposome is F666C-MTSL (Fig. 4B). In the soluble structure, this residue is buried at a turn within the hydrophobic sequence 659–681 and makes significant contact with the LC (Fig. 4A). The EPR line-shape analysis confirmed that F666C-MTSL is immobile in the protein's soluble form (Fig. 4B, red) but is highly mobile after liposome incorporation at pH 4.4 (Fig. 4B, blue). An experiment conducted at pH 5.0 revealed a mixture of both mobile and immobile conformations (Fig. 4B, black). Of interest, the neighboring residue I667C-MTSL is mobile in the soluble form of the protein and becomes restricted in the context of the proteoliposome (Fig. 4C). The I667C-MTSL proteoliposome spectrum at pH 5.0 suggests the presence of spin-spin coupling, a physical event requiring two spin labels in close proximity. Spin-spin coupling was also observed for K805C-MTSL at pH 4.4 (Fig. 4D) and suggests the formation of oligomeric structures within the liposome.
DISCUSSION
In this study, the recombinant BoNT/A LC-HCT was expressed as a soluble protein. When added to Neuro2A cells, this protein forms channels and supports the cleavage of endogenous SNAP25 (14). We have shown here that the BoNT/A LC-HCT associates with liposomes in a pH-dependent manner. At high pH, there was no proteoliposome formation; all of the LC-HCT remained in the supernatant and was not pelleted with the liposomes. In contrast, a low pH incubation resulted in liposome aggregation and the formation of stable proteoliposomes. Although liposome aggregation has been reported in similar studies of both BoNT (32) and diphtheria toxin (26), the physical basis for the effect is not clear. We demonstrated that the BoNT/A LC-HCT did not precipitate when incubated with liposomes at pH 8.0 or when treated at low pH in the absence of liposomes (even at pH 2, the condition used for pepsin cleavage) and that the liposomes did not aggregate when treated at low pH in the absence of protein.
The expectation for the EPR spin-label accessibility experiments was that they would reveal HCT residues that insert into the membrane. For membrane proteins, there are effectively two solvents, the aqueous phase and the fluid lipid bilayer. The environment of nitroxide-labeled residues within a membrane protein can therefore be defined through the use of paramagnetic quenchers that are either lipid-soluble (O2) or water-soluble (NiEDDA). The collision frequency between the spin label and these quenchers reflects the local environment of the residue tested. Because the HCT is large (residues 432–872 in BoNT/A), we decided to focus our initial experiments on the hydrophobic sequence 659–681. In the course of probing this region, we observed numerous pH-dependent MTSL line-shape changes, suggesting that structure 659–681 undergoes significant conformational change as BoNT associates with the membrane. We did not observe any residues where conversion from the soluble to proteoliposome form revealed a significant increase in O2 accessibility (Table 1). Expanding the analysis to include additional sites revealed only one residue that underwent an increase in O2 accessibility (F657C-MTSL) Furthermore, although many of the spin labels showed a decrease in NiEDDA accessibility upon conversion to the proteoliposome, the ∏ values were often higher than what one might expect for a membrane-inserted residue (Table 1). In short, the EPR accessibility measurements did not provide a clear model for what region of the structure is inserting into the liposome.
As an alternative approach for assessing the regions of the protein that interact with the membrane, we conducted a protease protection experiment. Pepsin-treated proteoliposomes were isolated, and the associated peptides were separated by SDS-PAGE. Although the equivalent concentration of pepsin resulted in complete digestion of the soluble LC-HCT, we consistently observed ∼14- and ∼5-kDa fragments that were resistant to protease digestion in the context of the liposome. These fragments were subjected to trypsin digest and mass spectrometry analysis. The predominant peptides emerging from this analysis were located in the C terminus of the protein (amino acids 805–820 and 826–835). Fluorescence quenching experiments using NBD-labeled proteins indicated that many of these residues (positions 826–830, 832, and 834–835) move into a hydrophobic environment upon conversion of the LC-HCT from the soluble to proteoliposome-associated form. These data are consistent with a model in which these residues are protected at the membrane upon conversion to proteoliposome.
The residues identified by protease protection and fluorescence quenching are located at one end of the HCT helical axis in a loop connecting two α-helices. Most of the residues are surface-exposed in the crystal structure of the soluble protein, and spin labels introduced at these residues are unrestricted in both the soluble and proteoliposome forms. The sequence of the loop is 826RGTLIGQVDR835, which is notable in that it contains charged and polar amino acids. Arginines are of particular interest, as these residues are often observed at protein-membrane interfaces and have been implicated in voltage gating (33, 34). Accessibility measurements for MTSL-labeled residues in this region showed higher than average decreases in NiEDDA accessibility upon conversion to the proteoliposome form, although, like other accessibility measurements, a concomitant increase in O2 accessibility was not observed (Table 1).
There are several possible explanations for why the accessibility measurements do not agree with the results obtained by protease protection and fluorescence quenching. The first is that any heterogeneity resulting from inefficient incorporation into the membrane will result in a mixed population of probe environments. The assays differ significantly in how a mixed population of states would be sampled; the NBD signal is likely to be dominated by residues shifting to a non-polar environment, whereas the EPR signal is likely to be dominated by residues that are not inserted in the membrane. Second, if BoNT forms an oligomeric structure as it inserts, the interpretation of our accessibility values could become significantly more complicated. The possibility of oligomerization in this system is supported by the observation of spin-spin coupling in the I667C-MTSL and K805C-MTSL samples (Fig. 4, C and D). Third, it is possible that defined transmembrane structural elements do not exist, either in nature and/or under the conditions of our proteoliposomes. In electrophysiological measurements, the channel conductivity of BoNT requires a potential gradient, a difficult thing to implement and maintain in a liposome-based system. Despite the success in using EPR to analyze the transmembrane states of diphtheria toxin, many biophysical studies point to the role of multiple insertion states, shallow and deep, as diphtheria toxin enters the membrane (35, 36). With this in mind, region 825–837 identified here by protease protection and NBD fluorescence studies could represent a shallow insertion of the toxin.
In considering a model in which region 825–837 mediates an initial “docking” interaction and/or shallow insertion into the membrane, we need to consider the position of the BoNT HCR and what is known about the interactions of this domain with the membrane. Multiple biochemical studies have highlighted the importance of ganglioside binding to the interaction of BoNT with the neuronal membrane (4), and a crystal structure of the GT1b ganglioside headgroup bound to the BoNT/A HCR has been solved (37). Using the crystal structure of BoNT/A holotoxin in its soluble state, it is difficult to envision a membrane plane that would accommodate the ganglioside-binding site at Trp-1265 and loop 825–837 (Fig. 5A). Furthermore, the HCR position could occlude the formation of an oligomeric structure wherein I667C-MTSL and K805C-MTSL molecules would be capable of spin-spin coupling (Fig. 5B). In contrast, the position of the HCR in the crystal structure of BoNT/E is in a notably different position (19). The ganglioside-binding pocket in this structure would align easily into a membrane plane that accommodated loop 825–837. Furthermore, the structure can easily accommodate an oligomeric structure in which residues along the central helical axis can interact (Fig. 5B).
FIGURE 5.
Model of membrane orientation based on BoNT/A and BoNT/E HCR positions. A, in this view, the HCR of BoNT/A (red) is positioned to the right of the HCT (light blue), with the ganglioside-binding Trp-1265 residue positioned in the “back” of the molecule. The HCR in BoNT/E (orange) is positioned such that Trp-1265 and loop 825–830 (green) could be located in a single plane. B, this top view of the HCT highlights the position of the HCR from the BoNT/A (red) and BoNT/E (orange) structures. The HCR position in the BoNT/E structure would accommodate an oligomerization model that includes the spin-spin coupling observed with I667C-MTSL and K805C-MTSL.
The task of creating an atomic model for the BoNT pore structure remains a significant hurdle in our understanding of how BoNT directs the translocation of the LC across the membrane. The data accumulated in this study provide clear evidence that one tip of the HCT helical axis is protected from harsh protease conditions and moves into a more hydrophobic environment upon exposure to low pH and liposomes. The data also indicate other regions of the structure, notably the hydrophobic sequence 659–681, that undergo significant structural changes to form the proteoliposome structure. Directed studies to test models of oligomerization represent a significant priority for on-going study.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AI075259.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- BoNT
- botulinum neurotoxin
- LC
- light chain
- HC
- heavy chain
- HCR
- HC receptor-binding domain
- HCT
- HC translocation domain
- NBD
- nitrobenzoxadiazole
- POPC/POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- MTSL
- S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate
- NiEDDA
- nickel-diaminediacetic acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IANBD
- N-((2-(iodoacetoxy)ethyl)-N-methyl)amino-7-nitrobenz-2-oxa-1,3-diazole.
REFERENCES
- 1. Simpson L. L. (1981) Pharmacol. Rev. 33, 155–188 [PubMed] [Google Scholar]
- 2. Giménez D. F., Giménez J. A. (1993) in Botulinum and Tetanus Neurotoxins (DasGupta B. R. ed) pp. 421–431, Plenum Press, New York [Google Scholar]
- 3. Schiavo G., Rossetto O., Benfenati F., Poulain B., Montecucco C. (1994) Ann. N.Y. Acad. Sci. 710, 65–75 [DOI] [PubMed] [Google Scholar]
- 4. Binz T., Rummel A. (2009) J. Neurochem. 109, 1584–1595 [DOI] [PubMed] [Google Scholar]
- 5. Montal M. (2010) Annu. Rev. Biochem. 79, 591–617 [DOI] [PubMed] [Google Scholar]
- 6. Montecucco C., Papini E., Schiavo G. (1994) FEBS Lett. 346, 92–98 [DOI] [PubMed] [Google Scholar]
- 7. Hoch D. H., Romero-Mira M., Ehrlich B. E., Finkelstein A., DasGupta B. R., Simpson L. L. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1692–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donovan J. J., Middlebrook J. L. (1986) Biochemistry 25, 2872–2876 [DOI] [PubMed] [Google Scholar]
- 9. Shone C. C., Hambleton P., Melling J. (1987) Eur. J. Biochem. 167, 175–180 [DOI] [PubMed] [Google Scholar]
- 10. Blaustein R. O., Germann W. J., Finkelstein A., DasGupta B. R. (1987) FEBS Lett. 226, 115–120 [DOI] [PubMed] [Google Scholar]
- 11. Koriazova L. K., Montal M. (2003) Nat. Struct. Biol. 10, 13–18 [DOI] [PubMed] [Google Scholar]
- 12. Galloux M., Vitrac H., Montagner C., Raffestin S., Popoff M. R., Chenal A., Forge V., Gillet D. (2008) J. Biol. Chem. 283, 27668–27676 [DOI] [PubMed] [Google Scholar]
- 13. Lai B., Agarwal R., Nelson L. D., Swaminathan S., London E. (2010) J. Membr. Biol. 236, 191–201 [DOI] [PubMed] [Google Scholar]
- 14. Fischer A., Mushrush D. J., Lacy D. B., Montal M. (2008) PLoS Pathog. 4, e1000245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fischer A., Montal M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10447–10452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brunger A. T., Breidenbach M. A., Jin R., Fischer A., Santos J. S., Montal M. (2007) PLoS Pathog. 3, 1191–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lacy D. B., Tepp W., Cohen A. C., DasGupta B. R., Stevens R. C. (1998) Nat. Struct. Biol. 5, 898–902 [DOI] [PubMed] [Google Scholar]
- 18. Swaminathan S., Eswaramoorthy S. (2000) Nat. Struct. Biol. 7, 693–699 [DOI] [PubMed] [Google Scholar]
- 19. Kumaran D., Eswaramoorthy S., Furey W., Navaza J., Sax M., Swaminathan S. (2009) J. Mol. Biol. 386, 233–245 [DOI] [PubMed] [Google Scholar]
- 20. Masuyer G., Thiyagarajan N., James P. L., Marks P. M., Chaddock J. A., Acharya K. R. (2009) Biochem. Biophys. Res. Commun. 381, 50–53 [DOI] [PubMed] [Google Scholar]
- 21. Garcia-Rodriguez C., Levy R., Arndt J. W., Forsyth C. M., Razai A., Lou J., Geren I., Stevens R. C., Marks J. D. (2007) Nat. Biotechnol. 25, 107–116 [DOI] [PubMed] [Google Scholar]
- 22. Oblatt-Montal M., Yamazaki M., Nelson R., Montal M. (1995) Protein Sci. 4, 1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oh K. J., Altenbach C., Collier R. J., Hubbell W. L. (2000) Methods Mol. Biol. 145, 147–169 [DOI] [PubMed] [Google Scholar]
- 24. Mchaourab H. S., Lietzow M. A., Hideg K., Hubbell W. L. (1996) Biochemistry 35, 7692–7704 [DOI] [PubMed] [Google Scholar]
- 25. Hubbell W. L., Mchaourab H. S., Altenbach C., Lietzow M. A. (1996) Structure 4, 779–783 [DOI] [PubMed] [Google Scholar]
- 26. Oh K. J., Zhan H., Cui C., Hideg K., Collier R. J., Hubbell W. L. (1996) Science 273, 810–812 [DOI] [PubMed] [Google Scholar]
- 27. Shin Y. K., Levinthal C., Levinthal F., Hubbell W. L. (1993) Science 259, 960–963 [DOI] [PubMed] [Google Scholar]
- 28. Altenbach C., Froncisz W., Hemker R., Mchaourab H., Hubbell W. L. (2005) Biophys. J. 89, 2103–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 30. Link A. J., Fleischer T. C., Weaver C. M., Gerbasi V. R., Jennings J. L. (2005) Methods 35, 274–290 [DOI] [PubMed] [Google Scholar]
- 31. McAfee K. J., Duncan D. T., Assink M., Link A. J. (2006) Mol. Cell. Proteomics 5, 1497–1513 [DOI] [PubMed] [Google Scholar]
- 32. Fu F. N., Busath D. D., Singh B. R. (2002) Biophys. Chem. 99, 17–29 [DOI] [PubMed] [Google Scholar]
- 33. Aggarwal S. K., MacKinnon R. (1996) Neuron 16, 1169–1177 [DOI] [PubMed] [Google Scholar]
- 34. Börjesson S. I., Elinder F. (2008) Cell Biochem. Biophys. 52, 149–174 [DOI] [PubMed] [Google Scholar]
- 35. Wang Y., Malenbaum S. E., Kachel K., Zhan H., Collier R. J., London E. (1997) J. Biol. Chem. 272, 25091–25098 [DOI] [PubMed] [Google Scholar]
- 36. Rosconi M. P., London E. (2002) J. Biol. Chem. 277, 16517–16527 [DOI] [PubMed] [Google Scholar]
- 37. Stenmark P., Dupuy J., Imamura A., Kiso M., Stevens R. C. (2008) PLoS Pathog. 4, e1000129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.