Abstract
As a consequence of their bacterial origin, mitochondria contain β-barrel proteins in their outer membrane (OMM). These proteins require the translocase of the outer membrane (TOM) complex and the conserved sorting and assembly machinery (SAM) complex for transport and integration into the OMM. The SAM complex and the β-barrel assembly machinery (BAM) required for biogenesis of β-barrel proteins in bacteria are evolutionarily related. Despite this homology, we show that bacterial β-barrel proteins are not universally recognized and integrated into the OMM of human mitochondria. Selectivity exists both at the level of the TOM and the SAM complex. Of all of the proteins we tested, human mitochondria imported only β-barrel proteins originating from Neisseria sp., and only Omp85, the central component of the neisserial BAM complex, integrated into the OMM. PorB proteins from different Neisseria, although imported by the TOM, were not recognized by the SAM complex and formed membrane complexes only when functional Omp85 was present at the same time in mitochondria. Omp85 alone was capable of integrating other bacterial β-barrel proteins in human mitochondria, but could not substitute for the function of its mitochondrial homolog Sam50. Thus, signals and machineries for transport and assembly of β-barrel proteins in bacteria and human mitochondria differ enough to allow only a certain type of β-barrel proteins to be targeted and integrated in mitochondrial membranes in human cells.
Keywords: Bacteria, Membrane Proteins, Mitochondria, Mitochondrial Transport, Protein Assembly, β-Barrel, Omp85, SAM Complex, Outer Membrane
Introduction
Mitochondria are organelles of bacterial origin, surrounded by an outer (OMM)4 and an inner membrane (IMM). The OMM contains β-barrel proteins, a class of pore-forming proteins additionally found only in chloroplasts and Gram-negative bacteria (1, 2). Similar to the majority of other mitochondrial proteins, β-barrel proteins are synthesized in the cytosol and have to be imported into mitochondria with the help of the translocase of the outer mitochondrial membrane (TOM) complex. For the membrane integration and assembly into complexes, β-barrel proteins require additional proteinaceous machinery in the OMM. This is so called sorting and assembly machinery (SAM), also known as topogenesis of outer membrane β-barrel proteins (TOB complex) (3, 4).
The central component of the SAM complex is Sam50/Tob55, a protein with a function that has been conserved from bacteria to human (4–6). Other components of the SAM complex include the Sam35/Tob38/Tom38 (7–9) and Sam37/Mas37/Tom37 (3, 10) proteins in yeast, and Metaxin 1 and Metaxin 2 (11) in mammalian mitochondria.
The signals in β-barrel proteins that are recognized by the TOM complex belong to internal targeting signals and are not yet fully understood. However, a specific signal has been identified in the C-terminal part of mitochondrial β-barrel proteins that directs them to the SAM complex to be properly sorted and integrated into the OMM (12). Unlike in yeast, this signal in mammalian β-barrel proteins is always present at the extreme C terminus of the protein, and the addition of even a short stretch of amino acids interferes with its recognition (13).
Many parallels exist between integration and assembly of β-barrel proteins in bacterial and mitochondrial outer membranes (1). In bacteria, proteins are first transported into the periplasmic space, where the signal peptide is cleaved off. A specific sequence in the C terminus of the protein is then recognized by the β-barrel assembly machinery (BAM), which integrates these proteins into the bacterial outer membrane (14). The major component of the BAM complex is YaeT/BamA in Escherichia coli (15), or Omp85/BamA in Neisseria meningitidis (16). These proteins belong to the Omp85 family, a member of which is also mitochondrial Sam50 (6). Additional components of the BAM complex differ to some extent between different bacteria. In N. meningitidis these include accessory lipoproteins RmpM, BamC, ComL/BamD, and BamE (17).
A recent report shows that the mitochondrial β-barrel protein, voltage-dependent anion-selective channel (VDAC) of Neurospora crassa can be assembled into the bacterial outer membrane (18). Similarly, it has been reported that bacterial β-barrel proteins have retained the ability to be imported and assembled into the OMM of yeast mitochondria (19). It would seem, therefore, that the basic mechanisms and signal recognition during the import and assembly of β-barrel proteins have been conserved between bacteria and mitochondria.
In contrast to these reports, we show here that, unlike yeast mitochondria, mitochondria of human cells possess surprising selectivity toward foreign β-barrel proteins. Of all the β-barrel proteins tested, only those from Neisseria sp. translocated into mitochondria. Neisserial PorB proteins, however, were not recognized by the SAM complex, but would accumulate in the intermembrane space of mitochondria, causing fragmentation and loss of mitochondrial membrane potential (Δψ), as shown before (20). Neisserial Omp85, on the other hand, was the only bacterial β-barrel protein tested that was recognized by both the TOM and the SAM complex of human mitochondria and assembled into the complexes in the OMM. We show for the first time that a bacterial Omp85 is capable of functioning in a mitochondrial membrane. It could integrate into the OMM PorB proteins from Neisseria sp., but could not substitute for the function of its mitochondrial homolog Sam50. Our results indicate that the human and yeast TOM and SAM complexes have diverged, as well as that neisserial Omp85 can function alone in the OMM, a possible important prerequisite for the evolution of mitochondrial OMM transport and assembly machineries.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa cells and human embryonic kidney (HEK) 293T cells were cultivated in RPMI 1640 medium (Invitrogen) and DMEM (Invitrogen), respectively, supplemented with 10% FCS (Biochrom) and penicillin/streptomycin (Invitrogen). HeLa cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. 293T cells were transfected using calcium phosphate precipitation. In short, CaCl2 (0.25 m) and HBS buffer (50 mm HEPES, pH 7.05, 140 mm NaCl, 1.5 mm Na2HPO4) were mixed with plasmid DNA and added to HEK 293T cells. Medium was exchanged the next morning, and cells were harvested 24–36 h after transfection. Cell lines inducibly overexpressing Omp85 protein were produced using Lenti-X Tet-On Advanced Inducible Expression System (Clontech) according to the manufacturer's instructions.
Microscopy
Immunofluorescence microscopy was performed essentially as described before (21). For transmission electron microscopy, a standard procedure as described in the supplemental Methods was used.
Sequence alignment of PorB proteins from different Neisseria species was performed using the ClustalW2 program.
Biochemical Methods
Genes for proteins used in this study were obtained by PCR from the total DNA prepared from the corresponding bacterial strain. Proteins were cloned into pcDNA3 vector (Invitrogen) with an N-terminal FLAG or Myc tag.
Mitochondrial isolation and carbonate extraction using 100 mm Na2CO3, pH 11.5, were performed as described previously (5, 11, 22). For opening of the OMM, freshly prepared mitochondria were incubated in isotonic (250 mm sucrose, 1 mm EDTA, 10 mm Tris, pH 7.6) or hypotonic (1 mm EDTA, 10 mm Tris, pH 7.6) buffer. Mitochondria were then treated with 50 μg/ml protease K, inhibited later by addition of 2 mm PMSF.
Mitochondrial and cytosolic fractions were obtained by the following procedure. Harvested transfected cells were opened by homogenization, and crude mitochondrial fraction was pelleted by centrifugation for 10 min at 14,000 rpm. The remaining fraction was centrifuged for 1 h at 100,000 × g, and the supernatant was precipitated with trichloroacetic acid to obtain pure cytosolic fraction. Blue native (BN)-PAGE and Western blotting were performed as described previously (11). Samples for BN-PAGE analysis were solubilized in 1% digitonin buffer (1% digitonin (Sigma) in 20 mm Tris-HCl, 0.1 mm EDTA, 1 mm PMSF, 50 mm NaCl, 10% (v/v) glycerol, pH 7.4).
For immunoprecipitation experiments, protein A-Sepharose CL-4B beads (GE Healthcare) were washed twice with lysis buffer (1% digitonin, 50 mm NaCl, 2 mm EDTA, 1 mm PMSF, 10 mm Tris-HCl, pH 7.4) and incubated with antibodies, preimmune serum, or PBS (137 mm NaCl, 2.7 mm KCl, 8.1 mm Na2HPO4, 1.76 mm KH2PO4, pH 7.4) for 30 min at room temperature, then washed three times with lysis buffer. 200 μg of mitochondrial protein was resuspended in 1 ml of lysis buffer for 10 min on ice, centrifuged for 10 min at 14,000 g, and 250 μl of supernatant was incubated with beads for 1 h at 4 °C. Beads were then washed twice with lysis buffer, once with 10 mm Tris-HCl, pH 7.4, and analyzed by SDS-PAGE and Western blotting.
Antibodies
Tom20 and Tim23 antibodies were purchased from BD Biosciences. VDAC antibody was from Abcam, Hsp60 antibody from Stressgen Bioreagents, Tom40 antibody from Santa Cruz Biotechnology, isocitrate dehydrogenase antibody from Biogenesis, succinate dehydrogenase complex subunit A (Complex II) antibody from Invitrogen, c-myc antibody from Gramsch Laboratories, and FLAG antibody from Sigma. Sam50 and Metaxin 1 antibodies were raised in rabbits against the His-tagged, full-length proteins. Fluorochrome-coupled secondary antibodies used for immunofluorescence microscopy were purchased from Jackson ImmunoResearch.
RESULTS
Human Mitochondria Selectively Import Neisserial β-Barrel Proteins
We tested the import of several bacterial β-barrel proteins into mitochondria of human cells, considering a recent report that all bacterial β-barrel proteins expressed in yeast universally targeted mitochondria and assembled into the OMM (19). We cloned OmpA, OmpC, PhoE, BamA from E. coli, YaeT/BamA from Salmonella enterica, and Omp85/BamA from Neisseria gonorrhoeae (Omp85Ngo) into a mammalian expression vector with an N-terminal FLAG tag. Previously, an N-terminally FLAG-tagged N. gonorrhoeae PorB (PorBNgo) construct in the same expression vector was created (20). HEK 293T cells were transfected with the plasmids for the purpose of cell fractionation and HeLa cells for the purpose of microscopy studies. OmpA protein was not expressed in either cell type (data not shown). After separating transfected HEK 293T cells into a cytosolic and crude mitochondrial fraction, we could find the β-barrel proteins from E. coli and S. enterica in both fractions, whereas PorBNgo and Omp85Ngo could be detected only in the mitochondrial fraction (Fig. 1). Microscopy studies showed that only the last two proteins co-localized with mitochondria. The β-barrel proteins from E. coli and S. enterica could be seen distributed in the cytosol, sometimes in the form of large aggregates, but never exclusively co-localizing with mitochondria (supplemental Fig. S1).
FIGURE 1.
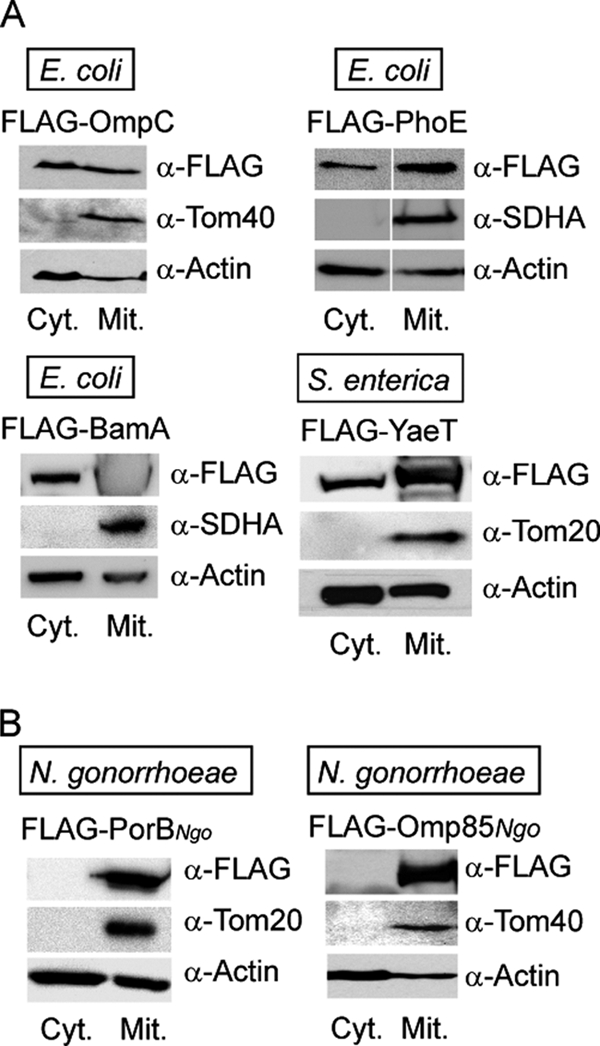
Neisserial, but not enterobacterial β-barrel proteins are located exclusively to mitochondria. HEK 293T cells were transfected with plasmids containing genes for β-barrel proteins originating from enterobacteria E. coli and S. enterica (A) or from N. gonorrhoeae (B), with an N-terminal FLAG tag. After 24–36 h of overexpression, cells were harvested and separated into the crude mitochondrial (Mit.) and cytosolic (Cyt.) fraction. 50 μg of mitochondrial protein and a corresponding amount of precipitated cytosolic fraction were analyzed by SDS-PAGE and immunodetection using antibodies against FLAG tag, actin, or an indicated mitochondrial protein. Tom40 and Tom20, components of the mitochondrial TOM complex; SDHA, succinate dehydrogenase complex subunit A, flavoprotein.
Neisserial β-barrel proteins behaved differently in respect to mitochondria. Overexpression of PorBNgo led to fragmentation and loss of Δψ (supplemental Fig. S1), a consequence of this protein not being recognized by the SAM complex and integrated into the OMM (20). Overexpression of Omp85Ngo, however, had no effect on mitochondria; their morphology was unchanged as well as their Δψ, as assessed by confocal and electron microscopy (supplemental Figs. S1 and S2). We conclude that the enterobacterial β-barrel proteins we tested are not imported by human mitochondria and that the signal detected in the mitochondrial fraction after cell fractionation probably comes from cytosolic protein aggregates. On the other hand, neisserial β-barrel proteins PorBNgo and Omp85Ngo are readily transported into human mitochondria upon overexpression but exhibit different effects on mitochondrial morphology and Δψ.
Omp85Ngo Forms Complexes in the OMM
We then analyzed mitochondria isolated from cells overexpressing the mentioned bacterial β-barrel proteins by BN-PAGE and Western blotting to detect possible mitochondrial complexes of these proteins. We observed such complexes only in the case of mitochondria containing overexpressed FLAG-Omp85Ngo (data not shown and Fig. 2A). The protein was found in three complexes of molecular masses ranging from >700 kDa to ∼300 kDa (Fig. 2A).
FIGURE 2.
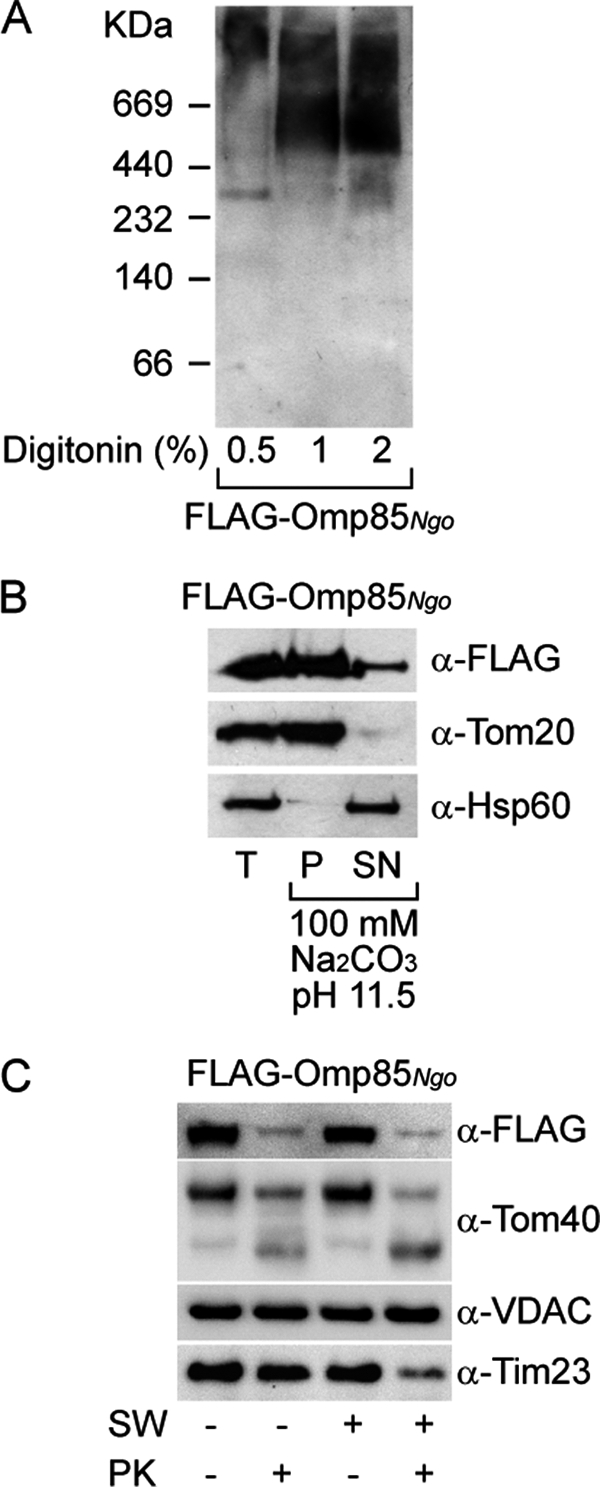
Omp85 integrates and forms complexes in the OMM. A, Omp85 from N. gonorrhoeae with an N-terminal FLAG tag (FLAG-Omp85Ngo) was overexpressed in HEK 293T cells, and crude mitochondria were isolated. Mitochondria were solubilized in 0.5, 1, and 2% digitonin and analyzed by BN-PAGE and immunodetection using antibodies against the FLAG tag. B, mitochondria from A were solubilized in 100 mm Na2CO3, pH 11.5. After centrifugation, total mitochondria (T), membrane pellet (P), and supernatant (SN) after carbonate extraction were analyzed by SDS-PAGE and immunodetection using antibodies against the FLAG tag, mitochondrial membrane protein Tom20, and soluble mitochondrial protein Hsp60. C, mitochondria from A were incubated in isotonic buffer (SW −) or hypotonic buffer (SW +) to rupture the OMM. Subsequently, mitochondria were either treated with protease K (PK +) or left untreated (PK −). Samples were analyzed by SDS-PAGE and immunodetection using antibodies against the FLAG tag, OMM β-barrel proteins Tom40 and VDAC, and IMM protein Tim23.
Membrane integration of FLAG-Omp85Ngo was assessed by carbonate extraction at pH 11.5. Most of the protein behaved as the integral OMM protein Tom20 and not as the soluble protein Hsp60 (Fig. 2B). Protease treatment of mitochondria before and after opening of the OMM by swelling in hypotonic buffer revealed that FLAG-Omp85Ngo behaved similarly to the OMM β-barrel protein Tom40, and not as Tim23, an IMM protein exposed to the intermembrane space (Fig. 2C). Omp85Ngo and Tom40 probably expose protease-sensitive loops to cytosol and are being degraded into smaller fragments, as seen for Tom40 (Fig. 2C). Of note, another OMM β-barrel protein VDAC is not affected by the protease treatment, probably due to different conformation and protection by the surrounding membrane (Fig. 2C). We conclude that Omp85Ngo, after entering mitochondria, integrates and assembles into complexes in the OMM.
Omp85Ngo Complexes Function in the Assembly of PorBNgo into the OMM
Overexpressed FLAG-Omp85Ngo seems to be capable of integration into the OMM, where it forms complexes (Figs. 2A and 3A, lane 1). If the Omp85Ngo follows the same import route as mitochondrial β-barrel proteins, then its assembly into the OMM would depend on the recognition of the β-sorting signal at its C terminus by the SAM complex (12). We therefore overexpressed FLAG-Omp85Ngo-12, a construct lacking the last 12 amino acids where the β-sorting signal is located, in HEK 293T cells. Microscopy analysis showed that when overexpressed FLAG-Omp85Ngo-12 was partially found in mitochondria, which mostly fragmented and lost Δψ, in agreement with the protein not being properly sorted to the OMM (supplemental Fig. S3). No protein complexes could be detected in mitochondria (Fig. 3A, lane 2), indicating that the intact β-sorting signal is required for Omp85 membrane assembly. However, a large portion of FLAG-Omp85Ngo-12 did not co-localize with mitochondria (supplemental Fig. S3), pointing to the importance of the β-signal in the initial targeting to mitochondria.
FIGURE 3.
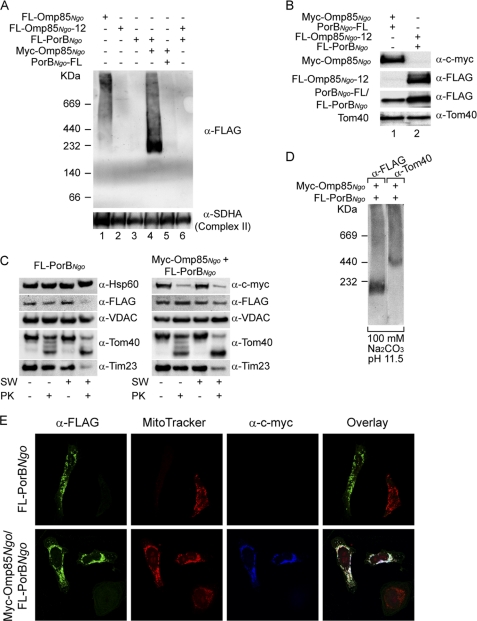
Mitochondrial Omp85 assists the assembly of PorB into complexes and prevents its toxicity to mitochondria. A, HEK 293T cells were transfected with plasmids carrying genes for FLAG-Omp85Ngo, FLAG-Omp85Ngo-12 (lacking the last 12 amino acids), FLAG-PorBNgo, combinations of Myc-Omp85Ngo and N- or C-terminally FLAG-tagged PorBNgo (FLAG-PorBNgo and PorBNgo-FLAG), or a combination of FLAG-Omp85Ngo-12 and FLAG-PorBNgo. Mitochondria were isolated and analyzed by BN-PAGE and immunodetection using antibodies against FLAG tag and succinate dehydrogenase complex subunit A (SDHA; Complex II). B, mitochondria from A (Myc-Omp85Ngo + PorBNgo-FLAG and FLAG-Omp85Ngo-12 + FLAG-PorBNgo) were analyzed by SDS-PAGE and immunodetection using antibodies against Myc tag, FLAG tag, and mitochondrial protein Tom40. C, HEK 293T cells were transfected with plasmid carrying gene for FLAG-PorBNgo or with the combination of Myc-Omp85Ngo- and FLAG-PorBNgo-encoding plasmids. Mitochondria were isolated and incubated in isotonic buffer (SW −) or hypotonic buffer (SW +) to rupture the OMM. Subsequently, mitochondria were either treated with protease K (PK +) or left untreated (PK −). Samples were analyzed by SDS-PAGE and immunodetection using antibodies against the FLAG tag, Myc tag, OMM β-barrel proteins Tom40 and VDAC, IMM protein Tim23, and matrix protein Hsp60. D, mitochondria isolated from HEK 293T cells simultaneously overexpressing Myc-Omp85Ngo and FLAG-PorBNgo were subjected to carbonate extraction in 100 mm Na2CO3, pH 11.5. After centrifugation, pellet was resuspended in 1% digitonin buffer and analyzed by BN-PAGE and Western blotting, using antibodies against the FLAG tag and Tom40, a component of the TOM complex in the OMM. E, HeLa cells were grown on coverslips and transfected with the plasmid containing the gene for N-terminally FLAG-tagged PorB (FLAG-PorBNgo) or with a combination of plasmids carrying genes for FLAG-PorBNgo and Myc-Omp85Ngo. Cells were decorated with Δψ-sensitive dye MitoTracker (red), fixed and analyzed by immunofluorescence using antibodies against FLAG tag (green) and Myc tag (blue) and confocal microscopy.
We next wondered whether Omp85Ngo complexes in the OMM retain their function, which is to integrate other bacterial β-barrel proteins into the bacterial outer membrane (16). When FLAG-PorBNgo was expressed alone, it formed no visible complexes in mitochondria (Fig. 3A, lane 3). However, when Myc-Omp85Ngo and FLAG-PorBNgo were overexpressed simultaneously in HEK 293T cells (Fig. 3B, lane 1), we could detect a complex of ∼200 kDa with a FLAG tag antibody (Fig. 3A, lane 4). This complex was specific because it did not form when the C terminus of PorBNgo was masked with a FLAG tag (Fig. 3A, lane 5), and it is known that the signal in the C terminus and especially a phenylalanine at the last position are essential for assembly of bacterial β-barrel proteins into complexes (23). This complex also did not form when FLAG-Omp85Ngo-12 construct was overexpressed simultaneously with FLAG-PorBNgo (Fig. 3A, lane 6, and Fig. 3B, lane 2), indicating that functional Omp85Ngo complexes in mitochondria are a prerequisite for the formation of PorBNgo complexes. As a loading control, immunodetection with an antibody against a subunit of mitochondrial Complex II (Fig. 3A) and Tom40 (Fig. 3B) was performed.
We then analyzed the behavior of FLAG-PorBNgo after swelling and protease treatment in the presence or in the absence of Myc-Omp85Ngo. When Myc-Omp85Ngo was not present, FLAG-PorBNgo in mitochondria behaved similarly to the intermembrane space-exposed IMM protein Tim23 and was degraded by a protease when the OMM was opened (Fig. 3C, left). However, when Myc-Omp85Ngo was overexpressed together with FLAG-PorBNgo, the latter behaved as the OMM β-barrel protein VDAC and was resistant to protease treatment under every condition (Fig. 3C, right). The protein complex formed by FLAG-PorBNgo when co-expressed with Myc-Omp85Ngo was also resistant to extraction with 100 mm Na2CO3 at pH 11.5, showing that it is membrane-integrated, similar to the TOM complex (Fig. 3D). As mentioned before, overexpression of PorBNgo leads to fragmentation of mitochondria and loss of Δψ due to protein mislocalization (Fig. 3C) (20). However, in cells overexpressing both Myc-Omp85Ngo and FLAG-PorBNgo we could detect only a limited mitochondrial fragmentation and no loss of Δψ (Fig. 3C). Taken together, these results suggest that Omp85Ngo follows the import route of mitochondrial β-barrel proteins, which involves the recognition of the C-terminal β-sorting signal by the SAM complex. OMM complexes of Omp85Ngo can function in membrane complex assembly of PorBNgo which is its natural substrate, and this assembly alleviates the effects that overexpression of PorBNgo have on mitochondrial morphology and Δψ.
Omp85Ngo Can Assemble Other Neisserial PorB Proteins into the OMM
Can Omp85Ngo assist the assembly of other similar β-barrel proteins into the OMM? We cloned PorB proteins from several commensal neisserial strains, Neisseria lactamica (PorBNla), N. sicca (PorBNsi), and N. cinerea (PorBNci), into a mammalian expression vector with an N-terminal FLAG tag. All proteins could be found in mitochondrial fraction upon overexpression (Fig. 4). Microscopy studies showed that all proteins co-localized with mitochondria, where they caused fragmentation and loss of Δψ, similar to PorBNgo (data not shown). This indicated that they were not recognized by the SAM complex and integrated into the OMM, which was supported by the fact that no complexes could be detected after BN-PAGE and Western blotting of mitochondria isolated from HEK 293T cells where these proteins were overexpressed (Fig. 4, lanes 1–3). However, when FLAG-PorBNla, FLAG-PorBNsi, and FLAG-PorBNci were overexpressed together with FLAG-Omp85Ngo, we could detect several complexes (Fig. 4). Higher ones (marked with an asterisk) correspond to Omp85Ngo complexes (Fig. 2A). The lower molecular mass complexes resemble the one detected after simultaneous overexpression of Omp85Ngo and PorBNgo (Fig. 3A). The intensity of the signal corresponds to the level of similarity between PorBNgo and PorB from other Neisseria sp. (supplemental Fig. S4). Therefore, Omp85Ngo most efficiently assembles the PorBNla, which is the most similar to its original substrate PorBNgo, whereas other two PorB proteins are assembled less efficiently, or almost not at all.
FIGURE 4.
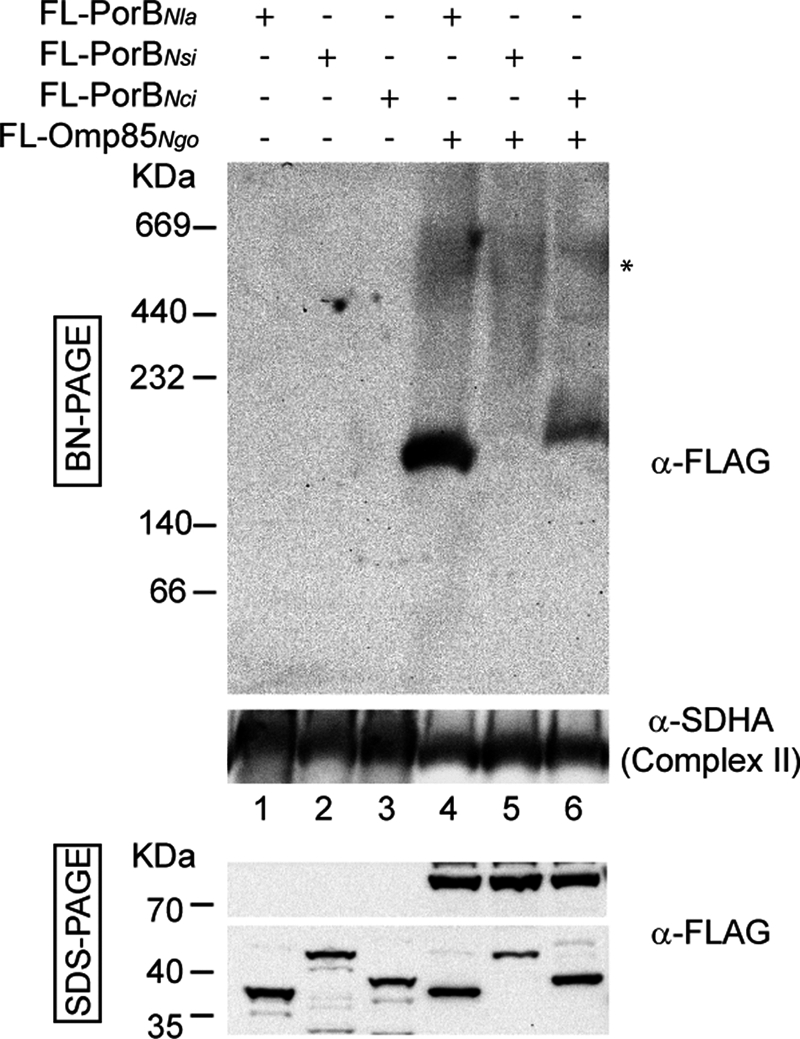
Mitochondrial Omp85 can function in the complex assembly of other related PorB proteins, although with different efficiency. HEK 293T cells were transfected with plasmids carrying genes for PorB of N. lactamica, N. sicca, or N. cinerea with an N-terminal FLAG tag (FL-PorBNla, FL-PorBNsi, and FL-PorBNci), alone or co-transfected with FLAG-Omp85Ngo. After 24–36 h of overexpression, cells were harvested, and crude mitochondria were isolated. Mitochondria were analyzed by BN-PAGE or SDS-PAGE and immunodetection using antibodies against the FLAG tag and succinate dehydrogenase complex subunit A (SDHA; Complex II).
Omp85Ngo and Sam50 Function Independently in the OMM
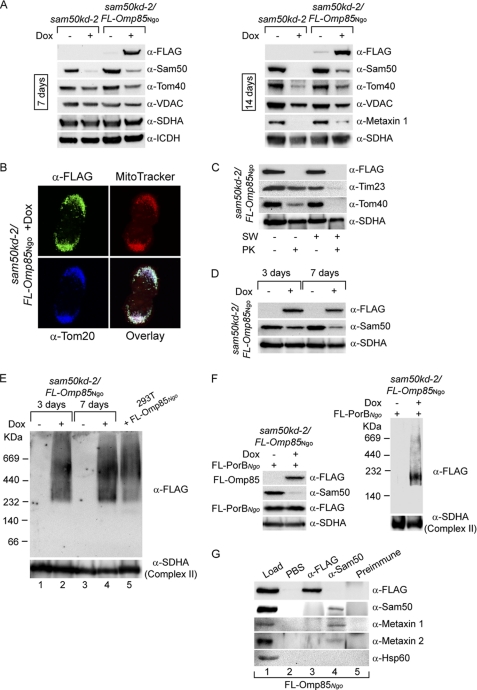
The mitochondrial SAM machinery most probably originates from the BAM-like machinery of the ancestral bacterium. Although accessory subunits of the BAM machinery of modern bacteria and the SAM complex of mitochondria show no sequence similarity (1), the central components, Omp85Ngo and Sam50/Tob55, are evolutionarily related proteins (6). We wanted to know, therefore, whether the Omp85Ngo can substitute for the function of Sam50. Based on the previously described cell line with an inducible, shRNA-mediated knockdown of Sam50 (11), we created a cell line that upon doxycycline induction overexpressed Omp85Ngo while at the same time Sam50 was depleted. We monitored levels of various mitochondrial proteins after 7 and 14 days of Sam50 depletion/Omp85Ngo overexpression. Among others, the amounts of the substrates of the SAM complex, such as Tom40 and VDAC, or the components of the SAM complex, such as Metaxin 1, were determined (Fig. 5A). The effects of Sam50 depletion, which are the reduction of both the substrates and components of the SAM complex, could not be reversed by the presence of Omp85Ngo. In addition, the growth of the cell line that overexpresses Omp85Ngo (supplemental Fig. S2) was severely impaired very soon after induction with doxycycline, indicating that larger amounts of Omp85Ngo were deleterious to mitochondrial function (data not shown). We conclude that Omp85Ngo cannot replace Sam50 in human mitochondria.
FIGURE 5.
Omp85Ngo functions independently of Sam50 and cannot substitute for it. A, a cell line was produced that, upon induction with doxycycline exhibits simultaneous knockdown of Sam50 and overexpression of FLAG-Omp85Ngo. Cells were induced with doxycycline (+Dox) for 7 or 14 days. Mitochondria were prepared and analyzed by SDS-PAGE and immunodetection with antibodies against FLAG tag, Sam50, Tom40, Metaxin 1, VDAC, succinate dehydrogenase complex subunit A, and isocitrate dehydrogenase. Sam50 and Tom40, central components of the SAM and TOM complex; Metaxin 1, a component of the SAM complex; SDHA, succinate dehydrogenase complex subunit A, flavoprotein; ICDH, isocitrate dehydrogenase. B, cells from A were induced by doxycycline for 7 days and analyzed by immunofluorescence and confocal microscopy using a membrane potential-sensitive dye MitoTracker (red), FLAG tag antibody (green), and Tom20 antibody (blue). C, mitochondria from A were incubated in isotonic buffer (SW −) or hypotonic buffer (SW +) to rupture the OMM. Mitochondria were then treated with protease K (PK +) or left untreated (PK −). Samples were analyzed by SDS-PAGE and Western blotting using antibodies against the FLAG tag, the OMM β-barrel protein Tom40, and IMM proteins Tim23 and SDHA. D, cells from A were induced for 3 or 7 days with doxycycline, and isolated mitochondria were analyzed by SDS-PAGE and Western blotting using antibodies against the FLAG tag, Sam50, and SDHA. E, mitochondria from D were solubilized in 1% digitonin buffer and analyzed by BN-PAGE and Western blotting using antibodies against the FLAG tag and SDHA. Additionally, mitochondria isolated from HEK 293T cells overexpressing FLAG-Omp85Ngo were analyzed in the same way. F, cells from A were induced for 7 days and transfected with the plasmid carrying the gene for FLAG-PorBNgo. Mitochondria were isolated from noninduced and induced samples and analyzed by SDS-PAGE or BN-PAGE and Western blotting, using antibodies against the FLAG tag, Sam50, and SDHA. G, mitochondria from HEK 293T cells overexpressing FLAG-Omp85Ngo were solubilized in 1% digitonin-containing lysis buffer, and proteins were precipitated using antibodies against FLAG tag and Sam50. Precipitates were analyzed by SDS-PAGE and Western blotting using antibodies against the FLAG tag, Sam50, Metaxin 1, Metaxin 2, and Hsp60. Lane 1 represents total loaded mitochondria, lane 2 and 5 are control lanes where PBS or a serum from a nonimmunized rabbit corresponding to the Sam50 antibody was used.
Importantly, Omp85Ngo was effectively imported and assembled in the OMM despite the Sam50 depletion. When cells were analyzed by microscopy after 7 days of Sam50 depletion/Omp85Ngo overexpression, Omp85Ngo was found to localize to mitochondria without damaging them (Fig. 5B). Additionally, in these mitochondria Omp85Ngo was accessible to protease in a way similar to that shown for mitochondria where there was no Sam50 depletion (Figs. 2C and 5C). Omp85Ngo was sufficiently expressed even after 3 days of Sam50 depletion/Omp85Ngo overexpression, at the point where there is still enough Sam50 to import and assemble Omp85Ngo efficiently (Fig. 5D). At this time point, the protein is already present in mitochondrial complexes (Fig. 5E, lane 2), which do not change significantly after further Sam50 depletion (Fig. 5E, lane 4) and resemble complexes seen in HEK 293T cells where Omp85Ngo was overexpressed (Fig. 5E, lane 5).
We also observed that the functioning of Omp85Ngo in the OMM is independent of the presence of Sam50. Sam50 depletion/Omp85Ngo overexpression was induced in cells for 7 days, and they were transfected with a plasmid containing the gene for FLAG-PorBNgo. The expressed FLAG-PorBNgo was effectively assembled in a complex in the absence of Sam50 (Fig. 5F). In addition, immunoprecipitation analysis using mitochondria isolated from HEK 293T cells overexpressing FLAG-Omp85Ngo showed that the FLAG antibody precipitated only FLAG-Omp85Ngo, and not Sam50, Metaxins, or the matrix protein Hsp60 (Fig. 5G, lane 3). Sam50 antibody precipitated Sam50 and Metaxins, but not FLAG-Omp85Ngo or Hsp60 (Fig. 5G, lane 4). In control lanes, where PBS or a serum of a nonimmunized rabbit was used none of these proteins could be detected (Fig. 5G, lanes 2 and 5). Considering also that the depletion of Sam50 does not affect complexes of Omp85Ngo, we conclude that Sam50, Metaxin 1, and Metaxin 2 are not the part of observed Omp85Ngo complexes and that Omp85Ngo and Sam50 function independently of each other.
DISCUSSION
The full complexity of the assembly of β-barrel proteins in bacteria and mitochondria has been elucidated only recently (1, 24). New machineries necessary for membrane integration of β-barrel proteins have been identified (3, 4, 15) as well as the signals required for the proper sorting and assembly of β-barrel proteins (12, 23). Recent findings imply that it is the β-barrel structure that is sufficient for a protein to be targeted to and imported into mitochondria, as well as that the β-sorting signal is universally recognized in bacteria and mitochondria (18, 19). Our data suggest, however, that these findings are only partially applicable to human cells.
We overexpressed several bacterial β-barrel proteins in human cells. Of the limited number of proteins tested, only those from Neisseria sp. located exclusively to mitochondria. All of the others could be found mainly as cytosolic aggregates. These results can be interpreted in two ways. The first possibility is that cytosolic chaperones of human cells are not capable of maintaining all β-barrel proteins in an import-competent state long enough for them to be taken up by mitochondria. The other option is that the central entry point for mitochondrial proteins, the TOM complex, exercises certain selectivity toward foreign β-barrel proteins. Therefore, only certain β-barrel structures will be recognized and imported by human mitochondria. It is possible that by lowering the rate of overexpression some import of these bacterial β-barrel proteins still might occur because both chaperones and the TOM complex would be better able to handle lower amounts of the foreign substrate. However, in microscopy studies we observed that even in the cells where there was less of the overexpressed protein, no mitochondrial targeting occurred. Both interpretations would imply that there are similarities in structures or sequences among those β-barrel proteins imported by mitochondria, whereas these similarities are not present in cytosol-remaining β-barrel proteins. However, alignment of sequences of proteins used in this study does not point to a specific signal that would direct some, but not other β-barrel proteins to the TOM complex. One indication, however, is the observation that mitochondrial localizing of FLAG-Omp85Ngo-12 was impaired (supplemental Fig. 3), pointing to a possible targeting role of the β-sorting signal. However, considering that β-signals are conserved among members of the YaeT/BamA/Omp85 family of proteins, and not all of them localize to mitochondria, it is possible that other regions of these proteins are also important for proper targeting to mitochondria in human cells.
Our results further demonstrate that, upon import into mitochondria, gonococcal Omp85, but not PorB, is assembled into complexes in the OMM. PorBNgo, as shown previously (20), avoids recognition by the SAM complex. The protein then most likely remains in the intermembrane space, and the reported IMM localization (20) is probably due to mislocalization and not specific IMM targeting. On the other hand, Omp85Ngo is recognized by both the TOM and the SAM complex. In addition to possible differences between C-terminal β-targeting signals of PorBNgo and Omp85Ngo, it is also likely that upstream residues in other β-strands play a role in recognition of these proteins by the SAM complex, similar to what we have shown for endogenous mitochondrial porin VDAC1 (13).
The complexes of Omp85 we detected by BN-PAGE and Western blotting are of a similar size but not identical to oligomeric complexes of E. coli Omp85 after refolding (23) and of N. meningitidis Omp85 after overexpression in yeast (19). After purification of Omp85Ngo complexes from human mitochondria, we could detect no other proteins present in significant amounts (data not shown). Omp85Ngo complexes remained unchanged after Sam50 depletion, and we could not immunoprecipitate FLAG-Omp85Ngo together with Sam50 or Metaxins (Fig. 5, E and G). Therefore, it is highly likely that Omp85Ngo in human mitochondria also forms homo-oligomeric complexes.
When Omp85Ngo and PorBNgo were overexpressed simultaneously, we could see specific protein complexes being formed by PorBNgo. Omp85Ngo was, therefore, capable of assembling PorBNgo in mitochondria. PorB proteins from other species of Neisseria could also be assembled by Omp85Ngo. The efficiency depended on the similarity of these proteins with the original substrate PorBNgo. This is the first time that a bacterial Sam50 homolog was shown to be able to function in mitochondria in folding and assembly of other bacterial β-barrel proteins. Previously, Robert et al. could not detect any membrane insertion of E. coli PhoE and LamB pores that were added to planar lipid bilayers containing reconstituted E. coli Omp85 (23). They concluded that this was the consequence of the absence of accessory lipoproteins known to function together with Omp85 in the membrane insertion and assembly of bacterial β-barrel proteins (15). Our results, on the other hand, imply that Omp85Ngo is capable of functioning without accessory lipoproteins in the OMM to promote insertion of bacterial β-barrel proteins. Considering that Omp85Ngo could not substitute for the lack of its mitochondrial homolog Sam50, it is obvious, however, that endogenous mitochondrial β-barrel proteins are not assembled by Omp85Ngo into the outer membrane of human mitochondria, although Omp85 seems to be able to assemble a β-barrel protein from fungal mitochondria into bacterial outer membrane (18). The ability of Omp85 to function without accessory lipoproteins in the assembly of β-barrel proteins might have been its original quality. Accessory proteins, such as lipoproteins present in bacteria (15, 17) or additional components of the SAM complex (3, 7–9, 11) have possibly been acquired later during evolution, to enhance efficiency and specificity of these complexes.
It seems that the specialization of mitochondria and their import and assembly machineries have continued, leading to the differences we observe between yeast and human mitochondria in regard to recognition and membrane integration of bacterial β-barrel proteins. Identifying the source of these differences, as well as further characterizing the signals that direct β-barrel proteins to mitochondria, is the next step that will contribute to our knowledge about the biogenesis of β-barrel proteins in mitochondria.
Supplementary Material
Acknowledgments
We thank Dr. G. Krohne and his team from the Central Dept. for Electron Microscopy, University of Würzburg, for the help with transmission electron microscopy and Dr. C. Meisinger for critically reading the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grants KO3882/1-1 (to V. K.-P.) and RU 631/7-1 (to T. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods and Figs. S1–S4.
- OMM
- outer mitochondrial membrane
- BAM
- β-barrel assembly machinery
- BN
- blue native
- IMM
- inner mitochondrial membrane
- SAM
- sorting and assembly machinery
- TOB
- topogenesis of outer membrane β-barrel proteins
- TOM
- translocase of outer membrane
- VDAC
- voltage-dependent anion-selective channel.
REFERENCES
- 1. Tommassen J. (2010) Microbiology 156, 2587–2596 [DOI] [PubMed] [Google Scholar]
- 2. Wimley W. C. (2003) Curr. Opin. Struct. Biol. 13, 404–411 [DOI] [PubMed] [Google Scholar]
- 3. Wiedemann N., Kozjak V., Chacinska A., Schönfisch B., Rospert S., Ryan M. T., Pfanner N., Meisinger C. (2003) Nature 424, 565–571 [DOI] [PubMed] [Google Scholar]
- 4. Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W. (2003) Nature 426, 862–866 [DOI] [PubMed] [Google Scholar]
- 5. Kozjak V., Wiedemann N., Milenkovic D., Lohaus C., Meyer H. E., Guiard B., Meisinger C., Pfanner N. (2003) J. Biol. Chem. 278, 48520–48523 [DOI] [PubMed] [Google Scholar]
- 6. Gentle I., Gabriel K., Beech P., Waller R., Lithgow T. (2004) J. Cell Biol. 164, 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Milenkovic D., Kozjak V., Wiedemann N., Lohaus C., Meyer H. E., Guiard B., Pfanner N., Meisinger C. (2004) J. Biol. Chem. 279, 22781–22785 [DOI] [PubMed] [Google Scholar]
- 8. Waizenegger T., Habib S. J., Lech M., Mokranjac D., Paschen S. A., Hell K., Neupert W., Rapaport D. (2004) EMBO Rep. 5, 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ishikawa D., Yamamoto H., Tamura Y., Moritoh K., Endo T. (2004) J. Cell Biol. 166, 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gratzer S., Lithgow T., Bauer R. E., Lamping E., Paltauf F., Kohlwein S. D., Haucke V., Junne T., Schatz G., Horst M. (1995) J. Cell Biol. 129, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kozjak-Pavlovic V., Ross K., Benlasfer N., Kimmig S., Karlas A., Rudel T. (2007) EMBO Rep. 8, 576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kutik S., Stojanovski D., Becker L., Becker T., Meinecke M., Krüger V., Prinz C., Meisinger C., Guiard B., Wagner R., Pfanner N., Wiedemann N. (2008) Cell 132, 1011–1024 [DOI] [PubMed] [Google Scholar]
- 13. Kozjak-Pavlovic V., Ross K., Götz M., Goosmann C., Rudel T. (2010) J. Mol. Biol. 397, 219–232 [DOI] [PubMed] [Google Scholar]
- 14. Bos M. P., Robert V., Tommassen J. (2007) Annu. Rev. Microbiol. 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 15. Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., Kahne D. (2005) Cell 121, 235–245 [DOI] [PubMed] [Google Scholar]
- 16. Voulhoux R., Bos M. P., Geurtsen J., Mols M., Tommassen J. (2003) Science 299, 262–265 [DOI] [PubMed] [Google Scholar]
- 17. Volokhina E. B., Beckers F., Tommassen J., Bos M. P. (2009) J. Bacteriol. 191, 7074–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walther D. M., Bos M. P., Rapaport D., Tommassen J. (2010) Mol. Biol. Evol. 27, 887–895 [DOI] [PubMed] [Google Scholar]
- 19. Walther D. M., Papic D., Bos M. P., Tommassen J., Rapaport D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kozjak-Pavlovic V., Dian-Lothrop E. A., Meinecke M., Kepp O., Ross K., Rajalingam K., Harsman A., Hauf E., Brinkmann V., Günther D., Herrmann I., Hurwitz R., Rassow J., Wagner R., Rudel T. (2009) PLoS Pathog. 5, e1000629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Müller A., Rassow J., Grimm J., Machuy N., Meyer T. F., Rudel T. (2002) EMBO J. 21, 1916–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Humphries A. D., Streimann I. C., Stojanovski D., Johnston A. J., Yano M., Hoogenraad N. J., Ryan M. T. (2005) J. Biol. Chem. 280, 11535–11543 [DOI] [PubMed] [Google Scholar]
- 23. Robert V., Volokhina E. B., Senf F., Bos M. P., Van Gelder P., Tommassen J. (2006) PLoS Biol. 4, e377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Endo T., Yamano K. (2010) Biochim. Biophys. Acta 1803, 706–714 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.