Abstract
A simian immunodeficiency virus (SIV) vaccine coexpressing granulocyte-macrophage colony stimulating factor (GM-CSF) prevented infection in 71% of macaques that received 12 rectal challenges. The SIVsmE660 challenge had the tropism of incident human immunodeficiency virus (HIV) infections and a similar genetic distance from the SIV239 vaccine as intraclade HIV isolates. The heterologous prime-boost vaccine regimen used recombinant DNA for priming and recombinant modified vaccinia Ankara for boosting. Co-expression of GM-CSF in the DNA prime enhanced the avidity of elicited immunoglobulin G for SIV envelope glycoproteins, the titers of neutralizing antibody for easy-to-neutralize SIV isolates, and antibody-dependent cellular cytotoxicity. Impressively, the co-expressed GM-CSF increased vaccine-induced prevention of infection from 25% in the non–GM-CSF co-expressing vaccine group to 71% in the GM-CSF co-expressing vaccine group. The prevention of infection showed a strong correlation with the avidity of the elicited Env-specific antibody for the Env of the SIVsmE660 challenge virus (r = 0.9; P < .0001).
A community-based efficacy trial in Thailand provided the first evidence that a human immunodeficiency virus (HIV)/AIDS vaccine could prevent infection. In this trial, 31% of the participants were protected by a vaccine that elicited both antibody and T cells [1]. Here, we tested the ability of a SIVmac239 (SIV239)–based vaccine that induces both antibody and T cells to prevent infection by a heterologous SIVsmE660 (SIVE660) challenge. The vaccine consisted of a recombinant DNA used to prime immune responses and a recombinant modified vaccinia Ankara (MVA) used to boost responses. Both the DNA and MVA components of the vaccine expressed the 3 major proteins of immunodeficiency viruses (Gag, Pol, and Env) and produced noninfectious virus like particles (VLPs).
The simian immunodeficiency virus (SIV) vaccine was tested in the presence and absence of granulocyte-macrophage colony stimulating factor (GM-CSF) co-expressed with the SIV immunogens. GM-CSF is a critical factor for the development and differentiation of dendritic cells [2] and a favored adjuvant for microbial and cancer vaccines [3]. Provenge, an approved cancer vaccine, uses GM-CSF as a fusion protein [4]. Studies involving cancer models have shown that GM-CSF expressed in cells has higher immune-stimulatory activity than when it is administered as a recombinant protein and that overly high expression of GM-CSF elicits suppressive immune responses [5–7].
GM-CSF was co-expressed in the DNA vaccine to imprint effects of the co-expressed GM-CSF on the immune response [8]. In our prior macaque studies, co-expression of GM-CSF enhanced the avidity of the elicited Env-specific serum immunoglobulin (Ig) G [9, 10], increased titers of neutralizing antibody for easy to neutralize “tier 1” isolates of HIV-1 [11], and increased the production of virus-specific IgA in rectal secretions [9] but did not augment CD4+ and CD8+ T-cell responses. The enhanced antibody responses—in particular, the avidity of the Env-specific IgG—has correlated with 100–10,000-fold reductions in peak viremia after high-dose rectal challenges with chimeras of simian and human immunodeficiency viruses (SHIV) and SIV [9, 10, 12]. The GM-CSF adjuvant has also improved control of the transient re-emergence of SHIV during the chronic phase of vaccine-mediated control [11].
Although our prior preclinical vaccine trials delivered high-dose rectal challenges, which infect all animals at the first exposure, this study was designed using repeated moderate-dose rectal challenges to better mimic human exposures [13, 14]. Also, to better represent human exposures, the study included the use of a challenge virus that was heterologous to the immunogen. Specifically, SIVmac239 sequences were used in the vaccine, and SIVsmE660—a virus 91% related in Gag and 83% related in Env—was used for the challenge [15, 16]. This level of variation is comparable to that observed between clade B isolates in the current pandemic [15, 16]. Our primary objectives were to test the effect of the immunizations on the number of challenges to infection and, for animals that became infected, to test the effect of the immunizations on control of post-challenge virus replication. A secondary objective was to identify potential correlates for protection.
METHODS
Vaccines
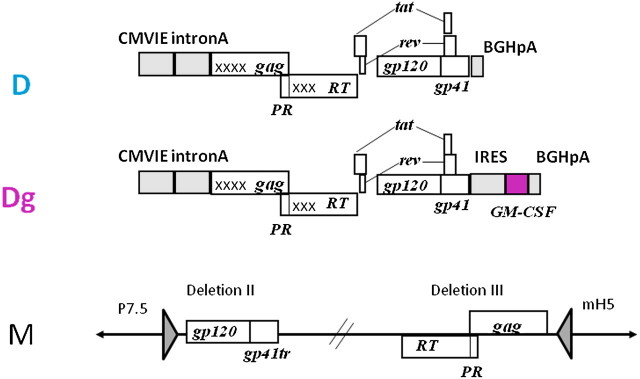
The GM-CSF co-expressing DNA vaccine was constructed by inserting rhesus macaque GM-CSF sequences into the pGA1/SIV239 DNA plasmid (termed D) that expresses SIV239 Gag, PR, RT, Env, Tat, and Rev to create the GM-CSF co-expressing plasmid (termed Dg) (Figure 1). The DNA vaccines express multiple SIV proteins from a single RNA by subgenomic splicing and frameshifting. GM-CSF is expressed by the same mRNA as Env using the encephalomyocarditis virus internal ribosome entry site. Dg expressed ∼200 ng of GM-CSF per 106 transiently transfected 293T cells, a level of expression that is associated with enhanced immune responses for cellular cancer vaccines [5]. A single recombinant MVA (previously designated DR1 or MVASIVgpe and designated M here) expressed Gag, Pol, and Env but did not co-express GM-CSF [17] (Figure 1). The MVA vaccine encodes gag and RT sequences in deletion III and env sequences in deletion II of MVA. The MVA vaccine expressed VLP whereas the over-expressed Gag in the DNA vaccine formed intracellular aggregates as well as VLP. The DNA vaccine expressed the complete gp160 form of Env and the MVA vaccine encoded a gp150 form, which was truncated to remove 146 amino acids at the C-terminus of the gp41 subunit to enhance expression on the plasma membrane of infected cells and stabilize the insert. [18]. Both vaccines expressed membrane bound trimeric forms of the envelope glycoprotein.
Figure 1.
Schematics of SIV239 DNA and recombinant modified vaccinia Ankara (MVA) vaccines. Transcriptional control elements are shaded. For the DNA vaccines, transcription is initiated by the cytomegalovirus immediate early promoter (CMVIE) including intron A and terminated by the bovine growth hormone polyadenylation sequence (BGHpA). For the MVA vaccine, transcription is under the control of the p7.5 (env) and mH5 (gag-pol) promoters. gag, Pr, RT, tat, rev, and env are sequences encoding the group-specific antigens, protease, reverse transcriptase, transcriptional activator, regulatory protein, and envelope glycoprotein, respectively, of SIV239. Xs indicate inactivating point mutations in reverse transcriptase and packaging sequences in gag. D, SIV239 DNA vaccine; Dg, GM-CSF co-expressing SIV239 DNA vaccine; M, SIV239 MVA vaccine.
Study Design
Animal studies were conducted at the Yerkes National Primate Research Center (Atlanta, GA) and were approved by the Emory University Animal Care and Use Committee (Atlanta). Young, adult, male rhesus macaques were prescreened to preclude the use of animals with the Mamu-A*01 histocompatibility type and to limit the use of animals with Mamu-B*08 and B*17 types to 1 per group, because these histocompatability types are correlated with enhanced control of SIV infection [16]. Animals were randomized to adjuvanted and nonadjuvanted vaccine groups of 8 each. Three mg of the DNA vaccine was administered at weeks 0 and 8, and 1 × 108 plaque forming units of the MVA vaccine was administered at weeks 16 and 24. All vaccinations were delivered intramuscularly by needle injection. The control group, added at the time of challenge, consisted of 9 young adult, male animals that were also selected to be Mamu A*01, B*08 and B*17 negative.
Twelve weekly intrarectal challenge were administered starting 6 months after the final MVA immunization using 5000 50% tissue culture infective dose (1.8 × 107 copies of viral RNA) of the SIVsmE660-Hirsch2000 stock [14, 19]. In 3 independent trials, this dose infected ∼30% of nonvacinated animals at each exposure independent of Mamu type, sex, age, and institutional environment (data not shown; B. Felber and G. Pavlakis, personal communication). Before challenge, 1 animal in the GM-CSF-adjuvanted group was euthanized because of self-mutilation. Throughout the study, hematologic and clinical chemistry testing were performed to assess any potential toxicological effects associated with the use of the GM-CSF. TRIM5 genotype was determined by sequence analysis of polymerase chain reaction fragments representing the TRIM5 TFP, CYPA, and Q alleles, as described elsewhere [20].
Antibody Assays
Titers of Env-specific IgG in serum and Env-specific IgA in rectal secretions collected with Weck-Cel sponges were determined using SIV239 VLP or rgp130mac251 (Immunodiagnostics) as a source of Env antigen in assays for IgG and IgA, respectively [9]. To be significant, Env-specific IgA had to be both 3.4-fold greater than the preimmune value and 0.145, the mean value plus 3 standard deviations for naive macaques. Avidity indices, or the fraction of retained antibody after a 1.5-M NaSCN wash times 100, were determined using duplicate enzyme-linked immunosorbent assays (ELISAs) [9]. SIV239 Env captured from VLP produced by transient transfection of 293 T cells and SIVE660 ENV captured from the challenge stock after 1 round of amplification on rhesus peripheral blood mononuclear cells (PBMCs) were used as antigen substrates. Pooled serum specimens from vaccinated rhesus were used as a reference standard in each assay. This sample had a mean avidity index of 38 and a standard deviation of 3. Neutralization assays were conducted using HIV pseudovirions, with Envs representing isolates from the genetically diverse SIVE660 stock and a luciferase reporter gene assay in TZM-bl cells [21]. Assays for antibody-dependent cellular cytotoxicity (ADCC) were conducted by adapting a previously published method [22]. In brief, recombinant SIVmac239 gp120 (Immune Tech) was used to coat CEM.NKRCCR5 cells as targets, and leukopheresis samples from an uninfected human healthy donor were used as effectors at an effector-to-target ratio of 30:1. The target cells were preloaded with a substrate that undergoes fluorescence after cleavage with granzymeB. After 1 hour of incubation at 37°C, the percentage of target cells that had received granzyme B from the effector cells and scored as fluorescence positive were reported as the percentage of Granzyme B activity. A serum dilution is considered positive if the percentage of Granzyme B is >9% after subtraction of the the percentage of Granzyme B for effector and target cells incubated without serum.
Cellular Immune Assays
Cellular immune assays and breadth of responses were conducted using pools of peptides (15 mers overlapping by 11) matched to the SIV239 immunogen for stimulation of PBMCs [9]. Responding cells were measured with use of intracellular cytokine staining (ICS). Positive results were at least twice the background level and >0.01%. Breadth of response was tested using 13 Gag and 11 Env peptide pools. Boolean analysis was performed to measure polyfunctionality [23]. Proliferation was tested using loss of carboxyfluorscein succinmidyl ester staining [24].
Statistical Analysis
Statistical analyses were conducted using Prism (Graphpad Software) and Spotfire S-PLUS 8.1 (TIBCO).
RESULTS
Rhesus macaques received DNA vaccines at weeks 0 and 8 and the MVA vaccine at weeks 16 and 24 in regimens referred to as DDMM (unadjuvanted DNA) and DgDgMM (Gm-CSF-adjuvanted DNA). The co-expression of GM-CSF in the DNA immunogen did not cause changes in hematology or blood chemistries or elicit detectable antibody to GM-CSF (data not shown).
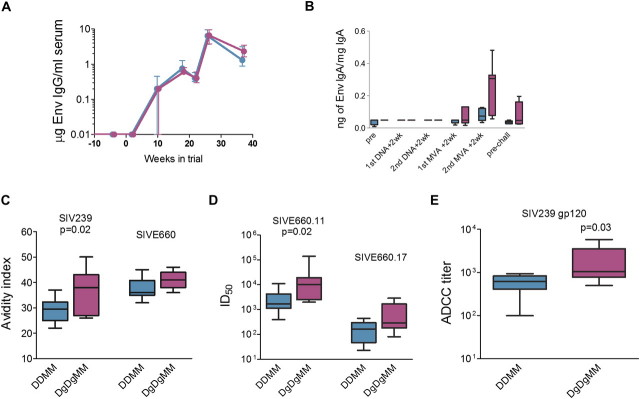
The DDMM and DgDgMM regimens elicited similar temporal patterns and magnitudes of Env-specific serum IgG but different patterns of Env-specific IgA in rectal secretions (Figure 2A and 2B). In both groups, the IgG responses increased after the MVA boosts and had decreased to ∼20% of peak values by the time of challenge. IgA, measured as a specific activity (as nanograms of Env IgA per micrograms of total IgA) was detected after the first MVA boost and increased in both frequency of detection and height after the second MVA boost. At the time of challenge, IgA titers had contracted by ∼50%. At peak IgA responses, Env-specific IgA was detected in 57% of the animals in the DgDgMM group, as opposed to 12% of the animals in the DDMM group. The specific activity of Env IgA in secretions was greater than that in blood, indicating that the rectal IgA had originated from local mucosal synthesis.
Figure 2.
Humoral immune responses elicited by the granulocyte-macrophage colony stimulating factor (GM-CSF)–adjuvanted and nonadjuvanted DNA/ modified vaccinia Ankara (MVA) vaccines. DNA priming immunizations were administered at weeks 0 and 8, and MVA booster immunizations were administered at weeks 16 and 24. A, Env-specific immunoglobulin (Ig) G responses measured in serum at before immunization and at weeks 2, 10, 18, 21, 26, and 37 in the trial. Micrograms of IgG are estimated relative to a standard curve of rhesus IgG. Values are medians ± interquartile ranges. B, Tukey plots presenting Env-specific IgA responses in rectal secretions before immunization, 2 weeks after the indicated immunizations, and before challenge. IgA is presented as Env-specific IgA divided by total IgA. C, Avidity indices for elicited IgG for the SIV239 Env of the immunogen and the SIVE660 Env of the challenge measured at 2 weeks after the second MVA immunization. Avidity indices increased with time in the trial and further increased after infection. D, Neutralization titers for pseudotypes with 2 Envs molecularly cloned from the genetically diverse SIVE660 stock. Titers for SIVE660.11 were determined 2 weeks after the second MVA boost and, for SIVE660.17, at 13 weeks after the second MVA boost. Differences in overall titers for the 2 tested isolates reflect differences in the susceptibility of different isolates from the E660 quasi-species to neutralization and differences in the timing of assays. Titers are the reciprocal for the dilution of serum achieving an inhibitory dose of 50% (ID50) in the TZM-bl assay. E, Antibody-dependent cellular cytotoxicity titers for SIVmac239 gp120 coated CEM.NKRCCR5 cells at 2 weeks after the second MVA boost. In panels C–E, box plots present median and 25th and 75th percentiles for responses. Target Envs and the significance for differences between the DDMM and DgDgMM regimens are indicated above the box plots. Statistical comparisons were made using a 2-sided Wilcoxon rank-sum test. Data for the unadjuvanted DNA (DDMM) regimen are presented in turquoise, and those for the Gm-CSF-adjuvanted DNA (DgDgMM) regimen are presented in fuchsia.
Additional analysis of the elicited IgG response revealed that the Env-specific IgG elicited by the DgDgMM regimen was qualitatively different from that elicited by the DDMM regimen. Co-expression of GM-CSF in the immunogen increased the avidity of the Env-specific IgG response (Figure 2C). This enhancement was significant for the SIV239 Env of the immunogen and showed a trend for the SIVE660 Env of the challenge virus. Consistent with the higher avidity, the Env-specific antibody in the GM-CSF–adjuvanted group had higher neutralizing activity and higher ADCC activity (Figure 2 D and E) [25]. Titers of neutralizing activity were increased for 2 easy- to-neutralize isolates derived from the genetically diverse SIVE660 challenge stock (SIV660.11 and SIVE660.17) and achieved significance for SIVE660.11. Neutralizing antibody for a more difficult-to-neutralize isolate, SIVE660.CR54, was less than the level of detection in the TZM-bl assay (data not shown). ADCC activity was also tested, and serum samples from animals in both the DDMM and DgDgMM groups contained antibodies capable of mediating ADCC activity. Serum specimens from animals in the DgDgMM group had significantly higher ADCC activity than did the DDMM serum specimens (Figure 2E).
In contrast to the antibody responses, the T-cell responses did not vary significantly between the 2 vaccine groups. T-cell responses were characterized for their magnitude, breadth, and cytokine co-expression using ICS and PBMC stimulated with peptide pools representing SIV239 Gag and Env (Figure 3). ICS scored patterns of expression of interferon (IFN)-γ, interleukin (IL)-2, and tumor necrosis factor (TNF)-α. Both vaccine regimens elicited similar temporal magnitudes of CD4+ and CD8+ T-cell responses (Figure 3A, B), similar breadths of responses (Figure 3C, D), similar patterns of polyfunctionality (Figure 3E, F) and similar proliferative responses (data not shown). In both groups, CD4+ T-cell responses peaked at median magnitudes of 1.6% of total CD4+ T cells; and CD8+ T-cell responses, at median magnitudes of 0.1% of total CD8+ T cells. In terms of polyfunctionality, 55% of the nonadjuvanted CD4+ T-cell response and 40% of the GM-CSF–adjuvanted CD4+ T-cell response coproduced IFN-γ, IL-2, and TNF-α. For CD8+ T cells, 14% of the nonadjuvanted and 12% of the GM-CSF–adjuvanted response were triple producers.
Figure 3.
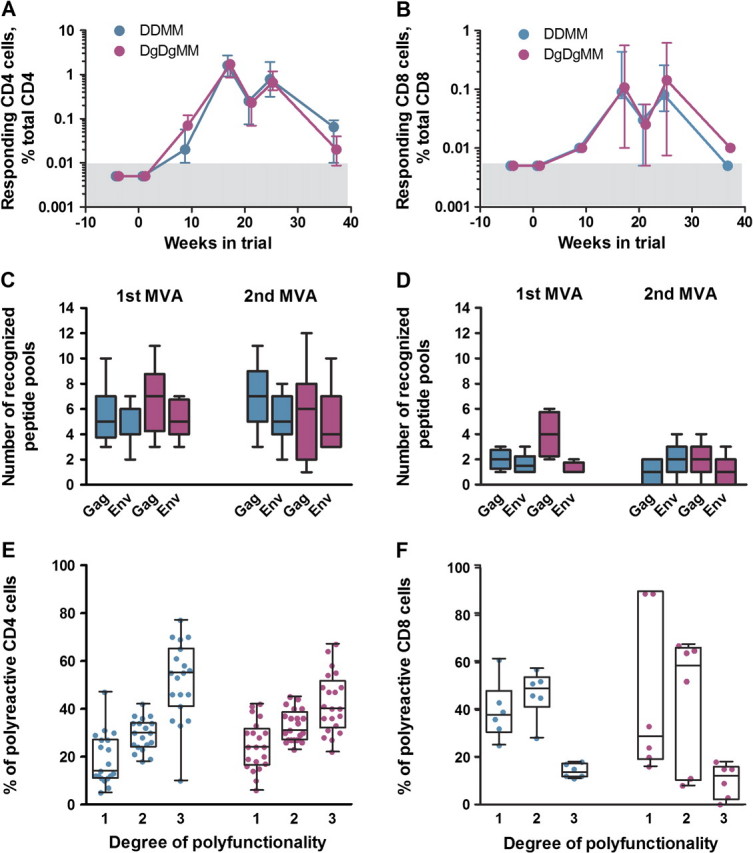
Cellular immune responses elicited by the granulocyte-macrophage colony stimulating factor (GM-CSF)–adjuvanted and nonadjuvanted DNA/ modified vaccinia Ankara (MVA) vaccines. DNA priming immunizations were administered at weeks 0 and 8, and MVA booster immunizations were administered at weeks 16 and 24. Shown are Vaccine-elicited CD4+ T cell responses (A) and CD8+ T-cell responses (B) before immunization and at weeks 2, 10, 17, 21, 25, and 37 in the trial. Responses are shown as interferon (IFN)–γ secreting cells measured by intracellular cytokine staining (ICS) after Gag and Env peptide stimulation of peripheral blood mononuclear cells (PBMCs). Gray boxes represent the background for detection. Also shown are the breadth of vaccine-elicited IFN-γ secreting CD4+ T-cell responses (C) and CD8+ T-cell responses (D) measured by ICS of PBMCs stimulated with 13 Gag and 11 Env peptide pools at 1 week after the first and second MVA immunizations. E and F, Polyfunctionality for cytokine production by elicited CD4+ and CD8+ T-cell responses at 1 week after the second MVA immunization. Boolean analyses were used to determine the frequencies of IFN-γ–, interleukin-2–, and tumor necrosis factor–α–producing cells responding to Gag and Env. Only those responses that were ≥0.07% of total cytokine-positive cells were considered for analysis. The box plots present the median and interquartile ranges for the percentage of responding cells (as a proportion of total cytokine positive cells) producing 1, 2, or 3 cytokines. Patterns of cytokine production for individual subsets of single or double producers were overall similar (data not shown). Data for the unadjuvanted DNA (DDMM) regimen are presented in turquoise, and those for the Gm-CSF-adjuvanted DNA (DgDgMM) regimen are presented in fuchsia.
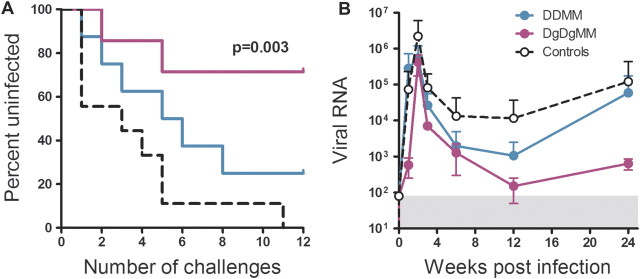
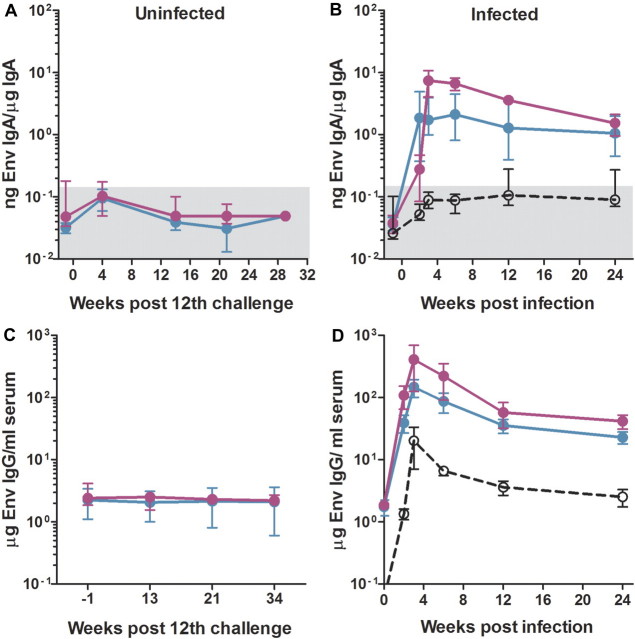
The repeated rectal challenge was initiated 6 months after the last MVA vaccination, a time when vaccine-elicited responses had contracted into memory. Infection was delayed in both the DDMM and DgDgMM vaccine groups, with the DgDgMM group resisting infection at a highly significant level (Figure 4A). Five (71%) of the 7 animals in the DgDgMM group were protected against the 12 challenges, whereas only 2 (25%) of 8 in the DDMM group were protected. All but 1 of the 9 control animals were infected by the fifth challenge, and the remaining animal was infected at the 11th challenge. The difference in protection for the GM-CSF–adjuvanted and unvaccinated group was highly significant (P = .003, by Mantel Cox test). Differences between the adjuvanted and nonadjuvanted groups and between the nonadjuvanted and unvaccinated groups showed trends that did not achieve significance in our group sizes. Temporal levels of post-challenge viremia did not show differences between groups but did show good control in the 2 GM-CSF-adjuvanted animals that became infected (Figure 4b). In both vaccine groups, the prevention of infection was complete. No evidence of viral replication or evolving SIV-specific immune responses were found during the 12 months after the last challenge. This includes the lack of anamnestic Env-specific IgA responses in rectal secretions (Figure 5A), anamnestic Env-specific IgG responses in blood (Figure 5C), and responding T cells for Nef—a protein present in the challenge virus but not the vaccine. This is in strong contrast to the vaccinated and infected animals, in which strong anamnestic IgA responses in rectal secretions (Figure 5B), anamnestic IgG responses in blood (Figure 5D), and responding T cells to Nef (data not shown) were clearly evident.
Figure 4.
Co-expressed granulocyte-macrophage colony stimulating factor enhances protection against infection from 25% to 71%. A, Kaplan-Meier curve for number of challenges to infection. P = .003 is the significance for the difference in number of challenges to infection between the GM-CSF-adjuvanted DNA (DgDgMM) and unvaccinated group (by log-rank Mandel-Cox test). B, Temporal post challenge viremia in animals that became infected. Infection dates are adjusted, with week 1 being the first week an infection was detected. Data are presented as means ± 1 standard deviation to show the differences in overall levels of viremia in the groups. The gray box represents the background for detection of viral RNA in the quantitative real-time polymerase chain reaction test. Data for the unadjuvanted DNA (DDMM) regimen are presented in turquoise, and those for the DgDgMM regimen are presented in fuchsia.
Figure 5.
Absence of anamnestic antibody responses in repeatedly challenged animals that did not become infected. A, Absence of a detectable anamnestic Env-specific immunoglobulin (Ig) A response in uninfected rhesus macaques at various weeks after the last challenge. B, Strong anamestic IgA responses for Env in vaccinated animals that became infected. C, Absence of a detectable anamnestic IgG response for Env in uninfected rhesus macaques at various weeks after the last challenge. D, Strong anamnestic IgG responses for Env in vaccinated animals that became infected. Data are presented as medians ± interquartile ranges. The gray boxes represent backgrounds for detection. Data for the unadjuvanted DNA (DDMM) regimen are presented in turquoise, and those for the Gm-CSF-adjuvanted DNA (DgDgMM) regimen are presented in fuchsia.
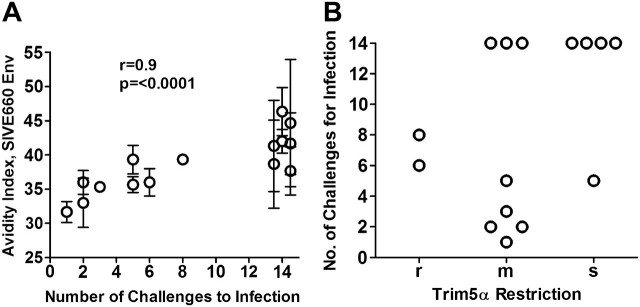
The avidity of the Env-specific IgG for the SIVE660 Env of the challenge virus correlated with the number of challenges to infection (r = .9; P <.0001, by Spearman test) (Figure 6A). In contrast, titers of neutralizing and ADCC antibody activities and the presence of anti-Env IgA, which were higher in the GM-CSF-adjuvanted group, did not correlate with protection. Elicited T-cell responses also did not correlate with the number of challenges to infection. The correlation with avidity was specific for the Env of the challenge virus and was not observed for the Env of the SIV239 immunogen.
Figure 6.
Avidity of the vaccine-elicited immunoglobulin (Ig) G for the Env of the challenge virus correlates with protection. A, Significant correlation between avidity of the elicited IgG for the SIVE660 Env of the challenge virus and the number of challenges to infection. Data are presented as the mean ± 1 standard deviation for 3 independent assays. Animals that did not become infected by the 12 challenges are plotted at 14 challenges. Correlations were done using the 2-sided Spearman rank order statistical analysis. B, The TRIM5α genotype of vaccinated rhesus macaques does not restrict the number of challenges to infection r, restrictive TRIM5α genotype (homozygous or heterozygous for TRIM5αTFP or CYPA); s, susceptible genotype (homozygous for TRIM5αQ); m, moderately susceptible (heterozygous for a restrictive and permissive allele). Animals that were not infected by the 12 challenges are plotted at challenge 14.
To test whether TRIM5α, an innate restriction factor that is polymorphic in rhesus macaques, might have played a role in our findings, rhesus macaques were typed for TRIM5α. The results of these analyses revealed no correlation in the vaccinated animals between the number of challenges to infection and the presence of restrictive (r), moderately restrictive (m), or susceptible (s) TRIM5α genotypes (Figure 6C). Of the 7 protected animals, 4 had the susceptible TRIM5α genotype, 3 had a moderately susceptible genotype, and none had the restrictive genotype. Thus no evidence could be found that TRIM5α restricted infection in the vaccinated and protected animals.
DISCUSSION
The co-expressed GM-CSF in the DNA prime for an MVA boost achieved highly significant protection against a repeated rectal challenge, whereas the vaccine without the co-expressed GM-CSF showed only a trend toward prevention of infection. In the presence of the co-expressed GM-CSF, 71% of the vaccinated animals were protected against 12 repeated rectal challenges; however, in the absence of the co-expressed GM-CSF, only 25% of the group was protected. These results suggest that targeting low levels of GM-CSF expression to the site of DNA immunization can serve as a strong adjuvant for preventing immunodeficiency virus infections.
A strong correlate for the prevention of infection was the avidity of the vaccine-elicited Env specific IgG. Animals that had avidity indices of ≥40 were not infected, whereas those with indices <40 showed a strong correlation between their avidity index and the number of challenges required for infection. Given these results, we hypothesize that antibody elicited by trimeric membrane-bound Env can recognize Env on virions and infected cells and that, if this antibody has sufficient avidity, it can initiate Fc-mediated mechanisms of protection such as complement (C)-mediated lysis, opsonization, ADCC, and antibody dependent cell-mediated virus inhibition (ADCVI) [25, 26]. The Fc region of the antibody can also bind to cervical mucus, providing an antibody trap for viral infections [27].
In studies of rhesus with our HIV vaccines, we have shown that the Env-specific antibody elicited by our clade B vaccine has broad avidity for incident clade B but not incident clade C isolates; moreover, antibody elicited by our clade C vaccine has broad avidity for the Envs of incident clade C but not incident clade B isolates[10]. Thus, we suggest that high-avidity antibody can have broad intraclade activity. This suggestion is consistent with studies of complement and Fc-mediated mechanisms of antibody-mediated protection that show patient serum having good breadth for mediating these activities against patient isolates (for review, see [26]).
Co-expression of GM-CSF in the DNA vaccine augmented avidity for both the Env of the SIV239 immunogen and the Env of the SIVE660 challenge. However, to observe the correlation between avidity and number of challenges to infection, avidity needed to be measured for the SIVE660 Env of the challenge stock. The SIV239 Env could elicit protective avidity for the SIVE660 Env, but the targets for this protection needed to be assessed using the challenge Env. These results indicate that there are multiple conserved targets for high avidity antibody on Env and suggest that each isolate will display different constellations of conserved targets.
In contrast to antibody responses, for which 3 of the 5 measured features were significantly enhanced by the co-expressed GM-CSF, none of the features we measured for T-cell responses were changed. This could reflect our assays testing for type 1 T-cell help and not the follicular CD4+ T-cell help that supports the maturation of antibody responses in germinal centers or the type 2 T-cell help favored by GM-CSF–stimulated dendritic cells [28–30]. GM-CSF stimulates the expansion and differentiation of myeloid dendritic cells, which display the receptor for GM-CSF [31]. Myeloid dendritic cells preferentially migrate to the marginal zone of lymph nodes where germinal centers for the maturation of B cells undergo formation [32]. The GM-CSF–stimulated myeloid dendritic cells produce IL-6, an important cytokine for the formation of germinal centers and the growth and differentiation of B cells in germinal centers [33, 34]. GM-CSF–stimulated myeloid dendritic cells favor the elicitation of type 2 T-cell help [29, 30]—a type of help that does not display the CCR5 chemokine receptor that is used as a co-receptor by HIV [35]. Thus, the GM-CSF adjuvant may facilitate prevention of infection by eliciting types of T-cell help that do not seed mucosal surfaces with preferred targets for infection [36]
The strong correlation between the avidity of vaccine-elicited IgG and the number of challenges to infection is the first demonstration that avidity can provide a serological correlate for prevention (not just control) of infection by an immunodeficiency virus. This demonstration introduces a new concept for HIV vaccine development: nonneutralizing but tightly binding antibody can mediate prevention of a mucosal infection. The ability to elicit broadly neutralizing antibody has eluded vaccine developers and is rare in natural infections [37–40]. In contrast, binding antibody for the native form of Env is elicited in virtually all infections. Thus, vaccines that elicit high-avidity binding antibody for the native form of Env may be able to provide a protective humoral component for mucosal infections.
Prior examples of vaccines for which the avidity of an antibody response was found to be important for protection include the conjugate vaccines. These vaccines convert T-cell–independent to T-cell–dependent immunogens and allow antibody stimulated by polysaccharides to undergo affinity maturation in children aged <2 years. For example, the avidity of the antibody responses elicited by vaccines for Haemophilus influenzae type B [41] and Streptococcus pneumononiae (pneumococcus) [42] are key to their protective activities. Failed measles and respiratory syncytial viral vaccines elicit nonprotective low-avidity antibody [43, 44]. The measurement of avidity for HIV-1 immunogens may be of particular importance because of the slow maturation of antibody to the highly glycosylated Env [45].
The achievement of 71% prevention of infection against 12 repeated rectal challenges by a heterologous SIV is a milestone for the preclinical development of HIV/AIDS vaccines. The previous benchmark for success has been live, attenuated vaccines—vaccines that are generally considered to be too risky for general use because of their ability to revert to virulence and recombine with challenge viruses [46]. In contrast to the 71% protection achieved here (5 or 7 animals), a live, attenuated SIV239 vaccine protected only 3 (37%) of 8 animals subjected to 10 repeated SIVE660 challenges [47]. A cytomegalovirus-vectored SIV239 vaccine has induced immune responses that controlled infection in 4 (33%) of 12 rhesus macaques subjected to repeated challenges with the homologous SIV239 virus, but it failed to induce responses that prevented infection [48].
In summary, our results show a GM-CSF co-expressing DNA prime for a MVA boost eliciting immune responses that prevented infection in 71% of macaques receiving 12 repeated intrarectal challenges with doses of a heterologous SIV that are transmitted 30–300 times more frequently than HIV-1 during human heterosexual intercourse [49]. The SIVE660 challenge had the same tropism as typical HIV infections [50] and a similar genetic distance from the SIV239 vaccine strain as HIV-1 clade-specific vaccines have for within-clade isolates [15, 16]. Provocatively, a nonneutralizing serological marker, avidity of the elicited IgG for the Env of the challenge virus, was identified as a correlate for prevention of infection.
Funding
This research was supported by an Integrated Preclinical/Clinical AIDS Vaccine Development program project 5U19 AI074073 (HLR), NIH grants AI30034 (DCM) and AI083118 (WEJ), the Emory University CFAR P30 AI050409 and by NCRR/NIH base grants P30 RR00165(YNPRC) and P30 RR00168 (NEPRC)
Acknowledgments
We are indebted to Dr Tianwei Yu for help with statistical analyses and to Drs B. Felber and G. Pavlakis for sharing data from SIVsmE660 challenge studies at the Frederick Cancer Research Center. We thank Robert Wilson for assistance with rectal IgA assays; Ken Rogers for assays for GM-CSF and production of SIVE660 virus for enzyme-linked immunosorbent assays; Jeffrey Americo for preparation of the MVA vaccine; Celia Labranche, William Rothwell, Corrine Rose, and Cindi Emmerson for assistance with neutralization assays; Craig Auden, Jen Morgan, and Laura Hall and the genetics core of the New England Primate Research Center (NEPRC) for Trim5 genotyping; the Emory Center for AIDS Research (CFAR) for viral load measurements, and the Yerkes Pathology Laboratory for hematology analyses. We are indebted to Susan Reuland for administrative assistance.
References
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–0. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 3.Borrello I, Pardoll D. GM-CSF-based cellular vaccines: a review of the clinical experience. Cytokine Growth Factor Rev. 2002;13:185–93. doi: 10.1016/s1359-6101(01)00034-x. [DOI] [PubMed] [Google Scholar]
- 4.Small EJ, Fong L. Developing immunotherapy as legitimate therapy for patients with prostate cancer. J Clin Oncol. 2010;28:1085–7. doi: 10.1200/JCO.2009.26.3483. [DOI] [PubMed] [Google Scholar]
- 5.Serafini P, Carbley R, Noonan KA, Tan G, Bronte V, Borrello I. High-dose granulocyte-macrophage colony-stimulating factor-producing vaccines impair the immune response through the recruitment of myeloid suppressor cells. Cancer Res. 2004;64:6337–43. doi: 10.1158/0008-5472.CAN-04-0757. [DOI] [PubMed] [Google Scholar]
- 6.Filipazzi P, Valenti R, Huber V, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. 2007;25:2546–3. doi: 10.1200/JCO.2006.08.5829. [DOI] [PubMed] [Google Scholar]
- 7.Parmiani G, Castelli C, Pilla L, Santinami M, Colombo MP, Rivoltini L. Opposite immune functions of GM-CSF administered as vaccine adjuvant in cancer patients. Ann Oncol. 2007;18:226–32. doi: 10.1093/annonc/mdl158. [DOI] [PubMed] [Google Scholar]
- 8.Feltquate DM, Heaney S, Webster RG, Robinson HL. Different T helper cell types and antibody isotypes generated by saline and gene gun DNA immunization. J Immunol. 1997;158:2278–84. [PubMed] [Google Scholar]
- 9.Lai L, Vodros D, Kozlowski PA, et al. GM-CSF DNA: an adjuvant for higher avidity IgG, rectal IgA, and increased protection against the acute phase of a SHIV-89.6P challenge by a DNA/MVA immunodeficiency virus vaccine. Virology. 2007;369:153–67. doi: 10.1016/j.virol.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Lai L, Amara RR, et al. Preclinical studies of human immunodeficiency virus/AIDS vaccines: inverse correlation between avidity of anti-Env antibodies and peak postchallenge viremia. J Virol. 2009;83:4102–11. doi: 10.1128/JVI.02173-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson HL, Montefiori DC, Villinger F, et al. Studies on GM-CSF DNA as an adjuvant for neutralizing Ab elicited by a DNA/MVA immunodeficiency virus vaccine. Virology. 2006;352:285–94. doi: 10.1016/j.virol.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Kannanganat S, Nigam P, Velu V, et al. Preexisting vaccinia virus immunity decreases SIV-specific cellular immunity but does not diminish humoral immunity and efficacy of a DNA/MVA vaccine. J Immunol. 2010;185:7262–73. doi: 10.4049/jimmunol.1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDermott AB, Mitchen J, Piaskowski S, et al. Repeated low-dose mucosal simian immunodeficiency virus SIVmac239 challenge results in the same viral and immunological kinetics as high-dose challenge: a model for the evaluation of vaccine efficacy in nonhuman primates. J Virol. 2004;78:3140–4. doi: 10.1128/JVI.78.6.3140-3144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keele BF, Li H, Learn GH, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med. 2009;206:1117–34. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh WW, Jaru-Ampornpan P, Nevidomskyte D, et al. Partial protection of simian immunodeficiency virus (SIV)-infected rhesus monkeys against superinfection with a heterologous SIV isolate. J Virol. 2009;83:2686–96. doi: 10.1128/JVI.02237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds MR, Weiler AM, Weisgrau KL, et al. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J Exp Med. 2008;205:2537–0. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Rompay KK, Greenier JL, Cole KS, et al. Immunization of newborn rhesus macaques with simian immunodeficiency virus (SIV) vaccines prolongs survival after oral challenge with virulent SIVmac251. J Virol. 2003;77:179–90. doi: 10.1128/JVI.77.1.179-190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt LS, Belyakov IM, Earl PL, Berzofsky JA, Moss B. Enhanced cell surface expression, immunogenicity and genetic stability resulting from a spontaneous truncation of HIV Env expressed by a recombinant MVA. Virology. 2008;372:260–72. doi: 10.1016/j.virol.2007.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amara RR, Villinger F, Altman JD, et al. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science. 2001;292:69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- 20.Kirmaier A, Wu F, R.M. N, et al. TRIM5. suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV and SHIV in a luciferase reporter gene assay. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, Coico R, editors. Current Protocols in Immunology. New York: John Wiley and Sons; 2004. [DOI] [PubMed] [Google Scholar]
- 22.Packard BZ, Telford WG, Komoriya A, Henkart PA. Granzyme B activity in target cells detects attack by cytotoxic lymphocytes. J Immunol. 2007;179:3812–20. doi: 10.4049/jimmunol.179.6.3812. [DOI] [PubMed] [Google Scholar]
- 23.Kannanganat S, Ibegbu C, Chennareddi L, Robinson HL, Amara RR. Multiple-cytokine-producing antiviral CD4 T cells are functionally superior to single-cytokine-producing cells. J Virol. 2007;81:8468–76. doi: 10.1128/JVI.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velu V, Kannanganat S, Ibegbu C, et al. Elevated expression levels of inhibitory receptor programmed death 1 on simian immunodeficiency virus-specific CD8 T cells during chronic infection but not after vaccination. J Virol. 2007;81:5819–28. doi: 10.1128/JVI.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao P, Zhao J, Patterson LJ, et al. Multiple vaccine-elicited nonneutralizing antienvelope antibody activities contribute to protective efficacy by reducing both acute and chronic viremia following simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J Virol. 2010;84:7161–73. doi: 10.1128/JVI.00410-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber M, Trkola A. Humoral immunity to HIV-1: neutralization and beyond. J Intern Med. 2007;262:5–25. doi: 10.1111/j.1365-2796.2007.01819.x. [DOI] [PubMed] [Google Scholar]
- 27.Hope T, Carias A, Rothwangl K, Veazey R. Global HIV Vaccine Enterprise NY. Abstract Book, AIDS Vaccine 2010. September 28--October 1, 2010, Atlanta, Georgia: Defining. the interaction of HIV with the mucosal barriers to gain insights into the mechanisms of sexual transmission [abstract S04.01] 39. [Google Scholar]
- 28.Poholek AC, Hansen K, Hernandez SG, et al. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 2010;185:313–26. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faith A, McDonald J, Peek E, et al. Functional plasticity of human respiratory tract dendritic cells: GM-CSF enhances T(H)2 development. J Allergy Clin Immunol. 2005;116:1136–43. doi: 10.1016/j.jaci.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Stumbles PA, Thomas JA, Pimm CL, et al. Resting respiratory tract dendritic cells preferentially stimulate T helper cell type 2 (Th2) responses and require obligatory cytokine signals for induction of Th1 immunity. J Exp Med. 1998;188:2019–31. doi: 10.1084/jem.188.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rissoan MC, Soumelis V, Kadowaki N, et al. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283:1183–6. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- 32.Pulendran B, Smith JL, Caspary G, et al. Distinct dendritic cell subsets differentially regulate the class of immune response in vivo. Proc Natl Acad Sci U S A. 1999;96:1036–41. doi: 10.1073/pnas.96.3.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopf M, Herren S, Wiles MV, Pepys MB, Kosco-Vilbois MH. Interleukin 6 influences germinal center development and antibody production via a contribution of C3 complement component. J Exp Med. 1998;188:1895–06. doi: 10.1084/jem.188.10.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burdin N, Galibert L, Garrone P, Durand I, Banchereau J, Rousset F. Inability to produce IL-6 is a functional feature of human germinal center B lymphocytes. J Immunol. 1996;156:4107–3. [PubMed] [Google Scholar]
- 35.Loetscher P, Uguccioni M, Bordoli L, et al. CCR5 is characteristic of Th1 lymphocytes. Nature. 1998;391:344–5. doi: 10.1038/34814. [DOI] [PubMed] [Google Scholar]
- 36.Douek DC, Brenchley JM, Betts MR, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–8. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 37.Binley JM, Lybarger EA, Crooks ET, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–8. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doria-Rose NA, Klein RM, Manion MM, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol. 2009;83:188–99. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Svehla K, Louder MK, et al. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol. 2009;83:1045–59. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sather DN, Armann J, Ching LK, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83:757–69. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlesinger Y, Granoff DM. Avidity and bactericidal activity of antibody elicited by different Haemophilus influenzae type b conjugate vaccines. The Vaccine Study Group. JAMA. 1992;267:1489–94. [PubMed] [Google Scholar]
- 42.Anttila M, Eskola J, Ahman H, Kayhty H. Avidity of IgG for Streptococcus pneumoniae type 6B and 23F polysaccharides in infants primed with pneumococcal conjugates and boosted with polysaccharide or conjugate vaccines. J Infect Dis. 1998;177:1614–21. doi: 10.1086/515298. [DOI] [PubMed] [Google Scholar]
- 43.Polack FP, Hoffman SJ, Crujeiras G, Griffin DE. A role for nonprotective complement-fixing antibodies with low avidity for measles virus in atypical measles. Nat Med. 2003;9:1209–13. doi: 10.1038/nm918. [DOI] [PubMed] [Google Scholar]
- 44.Delgado MF, Coviello S, Monsalvo AC, et al. Lack of antibody affinity maturation due to poor Toll-like receptor stimulation leads to enhanced respiratory syncytial virus disease. Nat Med. 2009;15:34–41. doi: 10.1038/nm.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parekh BS, Pau CP, Kennedy MS, Dobbs TL, McDougal JS. Assessment of antibody assays for identifying and distinguishing recent from long-term HIV type 1 infection. AIDS Res Hum Retroviruses. 2001;17:137–46. doi: 10.1089/08892220150217229. [DOI] [PubMed] [Google Scholar]
- 46.Ruprecht RM, Baba TW, Liska V. Attenuated HIV vaccine: caveats [letter; comment] Comment on: Science 1995 Nov 10;270(5238):988-91. Science. 1996;271:1790–2. [PubMed] [Google Scholar]
- 47.Reynolds MR, Weiler AM, Piaskowski SM, et al. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J Virol. 2010;84:9190–9. doi: 10.1128/JVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen SG, Vieville C, Whizin N, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293–9. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Royce RA, Sena A, Cates WJ, Cohen MS. Current concepts: sexual transmission of HIV. New Engl J Med. 1997;336:1072–8. doi: 10.1056/NEJM199704103361507. [DOI] [PubMed] [Google Scholar]
- 50.Margolis L, Shattock R. Selective transmission of CCR5-utilizing HIV-1: the ‘gatekeeper' problem resolved? Nat Rev Microbiol. 2006;4:312–7. doi: 10.1038/nrmicro1387. [DOI] [PubMed] [Google Scholar]