Abstract
Cationic antimicrobial peptides and polymyxins are a group of naturally occurring antibiotics that can also possess immunomodulatory activities. They are considered a new source of antibiotics for treating infections by bacteria that are resistant to conventional antibiotics. Members of the genus Burkholderia, which includes various human pathogens, are inherently resistant to antimicrobial peptides. The resistance is several orders of magnitude higher than that of other Gram-negative bacteria such as Escherichia coli, Salmonella enterica, or Pseudomonas aeruginosa. This review summarizes our current understanding of antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. These bacteria possess major and minor resistance mechanisms that will be described in detail. Recent studies have revealed that many other emerging Gram-negative opportunistic pathogens may also be inherently resistant to antimicrobial peptides and polymyxins and we propose that Burkholderia sp. are a model system to investigate the molecular basis of the resistance in extremely resistant bacteria. Understanding resistance in these types of bacteria will be important if antimicrobial peptides come to be used regularly for the treatment of infections by susceptible bacteria because this may lead to increased resistance in the species that are currently susceptible and may also open up new niches for opportunistic pathogens with high inherent resistance.
Keywords: Burkholderia, antimicrobial peptides, polymyxin, outer membrane, lipopolysaccharide, antibiotics, bacterial resistance mechanisms
Introduction
As the resistance of pathogenic bacteria to conventional antibiotics is on the rise, new sources of antibiotics are being sought including cationic antimicrobial peptides and polymyxins (Hancock and Sahl, 2006). These compounds are naturally occurring and are produced by most forms of life from microbes to mammals (Yeaman and Yount, 2007). They can act as antibiotics against bacteria but also have a wide range of important immunomodulatory activities in vivo (Lai and Gallo, 2009). Antimicrobial peptides represent a potential new source of antibiotics for treatment of various bacterial infections, including opportunistic infections in cystic fibrosis (CF) patients (Zhang et al., 2005), and are being used to as a last resort to treat some infections by multidrug resistant bacteria (Zavascki et al., 2007). Antimicrobial peptides cause disruption of the cytoplasmic membrane of bacteria (inner membrane in Gram-negative bacteria), but can also inhibit intracellular processes such as nucleic acid, DNA, or protein synthesis (Brogden, 2005). Resistance mechanisms have been characterized in various Gram-negative bacteria, including Escherichia coli, Salmonella enterica, and Pseudomonas aeruginosa. Many resistance mechanisms in Gram-negative bacteria involve alterations of the lipopolysaccharide (LPS) molecule of the outer membrane, which reduce the access of antimicrobial peptides and polymyxins to the inner membrane (Vaara et al., 1981; Gunn et al., 1998; Guo et al., 1998; Tamayo et al., 2005). Many Gram-positive bacteria display analogous alterations to their lipoteichoic and wall teichoic acids resulting in reduced access of these compounds to their cytoplasmic membranes (Peschel et al., 1999; Kovács et al., 2006; Giaouris et al., 2008).
A group of extremely antimicrobial peptide resistant Gram-negative bacteria exist and the best characterized of these organisms are species of the genus Burkholderia. This review summarizes the current state of knowledge concerning antimicrobial peptide and polymyxin resistance among the Burkholderia and discusses how the treatment of infections caused by sensitive bacteria with antimicrobial peptides or polymyxins could impact infections by resistant Gram-negative bacteria.
The Genus Burkholderia: Pathogens Amongst the Weeds
The genus Burkholderia comprises over 40 species of bacteria, most of which have been isolated from soil or fresh water. Members of this genus are symbionts of plants, fungi, and insects and pathogens of plants, fungi, and non-human mammals (for an excellent review on the distribution of Burkholderia bacteria within the environment see Compant et al., 2008). Within this genus are a group of at least 17 species, collectively known as the Burkholderia cepacia complex, that cause chronic opportunistic infections particularly in patients with CF or chronic granulomatous disease (Mahenthiralingam et al., 2005; Vanlaere et al., 2009). The genus also includes two species that cause acute infections in humans: B. pseudomallei, the causative agent of melioidosis (Cheng and Currie, 2005), and B. mallei, a facultative intracellular bacterium that causes glanders (Gregory et al., 2007). Acute and opportunistic infections by other members of the genus have also been reported (Gerrits et al., 2005; Sudo et al., 2005; Glass et al., 2006; Weinberg et al., 2007; Lestin et al., 2008; Deris et al., 2010).
Within the natural environment abundant opportunities arise during which Burkholderia bacteria may be exposed to antimicrobial peptides, polymyxins, and other similar compounds; this exposure is likely to have contributed a selective pressure for the evolution of high levels of resistance in these organisms. The rhizosphere is home to many microorganisms that produce antibiotics and many antibiotics have been directly detected from rhizosphere soil (Berg et al., 2005). These include bacteria such as Paenibacillus polymyxa, which contains the genetic material required to make polymyxins, a lantibiotic, bacitracin, and other antibiotics (Ma et al., 2011). Burkholderia sp. also form symbiotic and/or pathogenic interactions with insects and plants (Compant et al., 2008). These hosts also produce antimicrobial peptides (García-Olmedo et al., 1998; Bulet and Stocklin, 2005) which Burkholderia bacteria are likely exposed to at some point in their symbiosis or pathogenesis.
Antimicrobial Peptide and Polymyxin Resistance in the Genus Burkholderia
Members of the genus Burkholderia are inherently resistant to many antimicrobial agents (Mahenthiralingam et al., 2005); they have been shown to grow on penicillin as a sole carbon source (Vermis et al., 2003) and survive in solutions of chlorhexidine used for disinfection of hospital equipment (Heo et al., 2008). For the purpose of this review we considered Gram-negative bacteria to be extremely resistant to antimicrobial peptides and polymyxins if minimum inhibitory concentration (MIC) values of 500 μg/ml or more have been reported. Numerous species of Burkholderia bacteria fall into this category (Table 1). For comparison, reported MIC values for sensitive Gram-negative bacteria such as E. coli, S. enterica, and P. aeruginosa are several orders of magnitude lower than those for Burkholderia sp. (Table 1).
Table 1.
Antimicrobial peptide and polymyxin resistance values for many of the studies discussed in the text.
| Species/strain | Assay | Compound | Value | References |
|---|---|---|---|---|
| B. pseudomallei 1026b | MIC | Polymyxin B | 128 mg/ml | Burtnick and Woods (1999) |
| Colistin | 128 mg/ml | |||
| B. cepacia ATCC 17770 | MIC | Human β-defensin-3 | 6.6 μg/ml | García et al. (2001) |
| B. vietnamiensis (11/11 strains) | MIC | Bactenecin 2a | > 128 μg/ml | Jassem et al. (2011) |
| LL-37 | > 128 μg/ml | |||
| B. vietnamiensis (10/11 strains) | MIC | Polymyxin B | > 75 μg/ml | |
| B. vietnamiensis FC0656 | MIC | Polymyxin B | 37.5 μg/ml | |
| MIC50 | Polymyxin B | > 1024 μg/ml | Loutet et al. (2006, 2009) | |
| Melittin | > 512 μg/ml | |||
| B. cenocepacia K56-2 | Inhibition on solid media | Human neutrophil peptide-1 | > 100 μg/ml | |
| B. cenocepacia J2315 | MIC50 | Polymyxin B | > 512 μg/ml | |
| 18 Strains from five species: | MIC50 | Polymyxin B | > 256 μg/ml | Thwaite et al. (2009) |
| B. cepacia | Calcitermin | > 256 μg/ml | ||
| B. multivorans | Cecropin A | > 256 μg/ml | ||
| B. cenocepacia | Granulysin | > 256 μg/ml | ||
| B. stabilis | LL-37 | > 256 μg/ml | ||
| B. vietnamiensis | MUC-7 | > 256 μg/ml | ||
| P113 | > 256 μg/ml | |||
| Ovine polyaspartic acid | > 256 μg/ml | |||
| Magainin II | 8 to > 256 μg/ml | |||
| S. enterica (Typhimurium) 1408s | MIC | Polymyxin B | 1.9 μg/ml | Gunn et al. (1998) |
| P. aeruginosa H103 | MIC | Polymyxin B | 8 μg/ml | McPhee et al. (2003) |
| E. coli SC9251 | MIC | Polymyxin B | 0.5 μg/ml | Vaara and Vaara (1994) |
Antimicrobial peptide and polymyxin resistance has been investigated in various different Burkholderia sp. and a variety of major and minor determinants of antimicrobial peptide and polymyxin resistance have been proposed for members of the genus Burkholderia (Figure 1). An early study in B. cepacia (formerly Pseudomonas cepacia) showed that unlike P. aeruginosa, polymyxin B bound poorly to whole cells of B. cepacia and was unable to permeabilize the outer membrane of B. cepacia to a second molecule (1-N-phenylnaphthylamine; Moore and Hancock, 1986); these observations have since been confirmed in B. cenocepacia and B. pseudomallei (Burtnick and Woods, 1999; Ortega et al., 2009). These initial stages in the action of polymyxin B are known as the self-promoted uptake pathway, a process by which a cationic antibiotic displaces divalent cations that bridge LPS molecules and promote the uptake of other molecules across the outer membrane by decreasing the permeability of the membrane (Hancock and Wong, 1984). Moore and Hancock (1986) concluded that unique properties of the B. cepacia outer membrane blocked the self-promoted uptake of polymyxin B and that this explained, at least in part, the high levels of resistance seen in the organism.
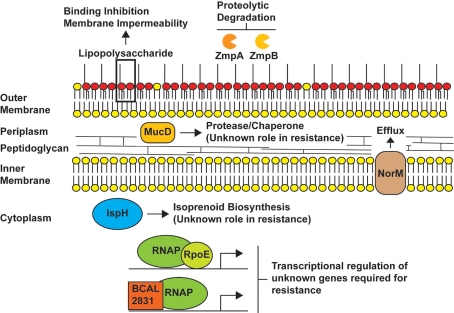
Figure 1.
Proposed antimicrobial peptide and/or polymyxin resistance mechanisms in Burkholderia species. The model depicts LPS as providing a major barrier to binding of peptides (Loutet et al., 2011) and the possible role of the metalloproteases ZmpA and ZmpB in degrading antimicrobial peptides. The periplasmic protein involved in resistance by an unknown mechanism is MucD, which is homologous to members of the HtrA protease family. In our model, IspH, an enzyme involved in the synthesis of isoprenoids could play a role in antimicrobial peptide resistance by introducing modifications of the lipid composition of the inner membrane, while the efflux pump NorM could mediate the efflux of peptides that cross the inner membrane and gain access to the cytoplasm. The regulators RpoE and BCAL2831 participate in global regulatory responses leading to an increase in antimicrobial peptide resistance. The genes regulated by RpoE and BCAL2831 and involved in these responses have not been identified, but it is likely RpoE regulates mucD since the genes encoding both proteins are co-transcribed (Flannagan and Valvano, 2008).
Burtnick and Woods (1999) used a random screen to identify determinants of antimicrobial peptide resistance in B. pseudomallei. These authors reported that B. pseudomallei had the ability to grow in concentrations of polymyxin B as high at 128 mg/ml and obtained polymyxin B-sensitive transposon mutants in three genes. One gene (waaF) encodes a protein required for LPS core oligosaccharide biosynthesis. The others encoded a predicted enzyme with UDP-glucose dehydrogenase activity and an enzyme called IspH (formerly LytB), which at the time was thought to be involved in the bacterial stringent response (Gustafson et al., 1993) but has since been shown to be required for the synthesis of isoprenoids (Rohdich et al., 2002). All of these pathways are also involved in antimicrobial peptide and polymyxin resistance in B. cenocepacia (Loutet et al., 2006, 2009, 2011; Ortega et al., 2009).
In numerous Gram-negative bacteria, the sugar 4-amino-4-deoxy-l-arabinose (Ara4N) is synthesized and then added to the lipid A component of LPS to increase antimicrobial peptide and polymyxin resistance (Nummila et al., 1995; Gunn et al., 1998; Ernst et al., 1999; Winfield et al., 2005). However, in most Gram-negative bacteria synthesis of this sugar is dispensable under most growth conditions and the sugar is not normally found in the lipid A molecule. Many studies have shown that Burkholderia sp. produce Ara4N constitutively as part of their LPS molecule, in some cases placing the sugar in both the lipid A and core oligosaccharide portions of the molecules (Isshiki et al., 1998; Gronow et al., 2003; Silipo et al., 2005, 2007; Brett et al., 2007; Novem et al., 2009; Ortega et al., 2009). The constitutive presence of this sugar in the LPS molecule was suggested to be a major determinant of antimicrobial peptide resistance in Burkholderia sp. (Cox and Wilkinson, 1991; Vinion-Dubiel and Goldberg, 2003). Initial attempts in our laboratory to disrupt the genes required for the synthesis of Ara4N in B. cenocepacia failed, which led to the hypothesis that this sugar was essential for B. cenocepacia viability. Ortega et al. (2007) constructed conditional mutants in this pathway and showed that both the synthesis of Ara4N and the transfer of Ara4N to lipid A are essential. Depletion of proteins from this pathway in conditional mutants resulted in morphological changes such as accumulation of membranous material inside the cell, empty sacculi, and division septa defects, decreased viability as measured by live/dead staining, and increased sensitivity to detergents (Ortega et al., 2007). The morphological changes are reminiscent of those observed in mutants of the essential Lpt pathway, responsible for the translocation of LPS from the inner membrane to the outer membrane (Sperandeo et al., 2007, 2008). The first step in the synthesis of Ara4N is the conversion of UDP-glucose to UDP-glucuronic acid, a reaction catalyzed by the protein UDP-glucose dehydrogenase (Ugd; Raetz and Whitfield, 2002). Loutet et al. (2009) showed that the combination of two ugd genes, ugdBCAL2946 and ugdBCAM0855, is essential for the viability of B. cenocepacia, while the most highly expressed gene, ugdBCAL2946, is required for polymyxin B resistance. As a component of the Ara4N synthesis pathway, ugd genes have often been implicated in antimicrobial peptide resistance in other organisms (Burtnick and Woods, 1999; Mouslim and Groisman, 2003; Hung et al., 2007). Although this study links a gene in the Ara4N synthesis pathway with a predicted role in polymyxin B resistance, the content of Ara4N in the LPS molecule of this mutant has not yet been determined (Loutet et al., 2009). To date it has not been possible to assess the precise contribution of Ara4N to antimicrobial peptide resistance in B. cenocepacia because due to their essential nature no mutant strain completely lacking Ara4N in the LPS molecule has been reported in the literature.
A second region of the LPS molecule, the core oligosaccharide, also contributes to antimicrobial peptide resistance in B. cenocepacia. Progressive truncation of the LPS core oligosaccharide results in increasing sensitivity of B. cenocepacia to polymyxin B and increasing binding of polymyxin B to whole cells (Ortega et al., 2009). A severe LPS core oligosaccharide truncation leads to the loss of the sugar l-glycero-d-manno-heptose and all sugars added after these residues. This “deep-rough” truncation typically causes pleiotropic effects in Gram-negative bacteria (Raetz and Whitfield, 2002; Valvano et al., 2002). A heptoseless LPS mutant of B. cenocepacia displayed increased sensitivity to numerous antimicrobial agents, and in particular to polymyxin B and to structurally unrelated antimicrobial peptides such as melittin and human neutrophil peptide-1 (Loutet et al., 2006). The mutant strain used in this study was unable to synthesize l-glycero-d-manno-heptose and these results were later confirmed in a B. cenocepacia mutant strain able to synthesize this sugar but unable to transfer it to the LPS molecule (Ortega et al., 2009).
The other major determinant of resistance described in B. cenocepacia is the alternative sigma factor RpoE (Loutet et al., 2011). This alternative sigma factor controls the expression of a regulon of genes that encode proteins required for bacteria to respond to extracytoplasmic stress (Raina et al., 1995). In B. cenocepacia, RpoE plays a significant role in polymyxin B resistance (but not melittin resistance; Loutet et al., 2011). It is also required for polymyxin B resistance at 37°C but not at 30°C (Flannagan and Valvano, 2008; Loutet et al., 2011). These results support the notion that an extremely resistant organism can have resistance determinants that are required for some compounds but not others and only under certain conditions.
Three studies have proposed additional minor determinants of antimicrobial peptide resistance in Burkholderia sp. First, Fehlner-Gardiner and Valvano (2002) demonstrated that in B. vietnamiensis, NorM, a member of the multidrug and toxic compound extrusion family of efflux systems (Kuroda and Tsuchiya, 2009), contributed to polymyxin B resistance, but only in the presence of exogenously added tetracycline. In the absence of tetracycline, the efflux pump played little part in polymyxin B resistance. Second, Kooi and Sokol (2009) demonstrated that two secreted zinc metalloproteases of B. cenocepacia (ZmpA and ZmpB) could degrade various antimicrobial peptides in vitro but that B. cenocepacia mutant strains lacking either ZmpA, ZmpB, or both did not exhibit increases in susceptibility to antimicrobial peptides other than a small increase in mutants lacking ZmpB to protamine. Third, we recently demonstrated that determinants of antimicrobial peptide resistance could be identified in a deep-rough LPS B. cenocepacia mutant (which lacks one of the major determinants of antimicrobial peptide resistance). These determinants, which included a two-component response regulator (BCAL2831), a periplasmic protease (MucD), and one of two IspH proteins, play little or no detectable role in a strain with its LPS molecule intact (Loutet et al., 2011).
Together, these studies provide evidence that extreme resistance of Burkholderia sp. to antimicrobial peptides is likely multifactorial (Figure 1) and consists of major and minor determinants of resistance. This led us to propose a two-tier model of antimicrobial peptide resistance in B. cenocepacia (Loutet et al., 2011). In the first tier of this model, we hypothesize that the outer membrane impermeability barrier and poor peptide binding maintained, at least in part, by unique aspects of the LPS molecule provides the majority of the resistance seen in this organism in conjunction with major peptide- or condition-specific resistance mechanisms (such as the requirement for RpoE for polymyxin B resistance at 37°C). In the second tier of the model, we have proposed that various other resistance mechanisms exist, each playing a small role in resistance but as a whole contributing substantially to the high total resistance of the organism. These mechanisms may be difficult to identify experimentally in the presence of the strong outer membrane impermeability barrier and poor peptide binding mediated by LPS.
There are Exceptions to Every Rule
Of course, there are exceptions to the idea that Burkholderia bacteria are universally resistant to antimicrobial peptides and polymyxins (Table 1). In a study using numerous isolates (both clinical and environmental) of B. vietnamiensis, Jassem et al. (2011) found that while a large majority of the isolates were resistant to all of the peptides tested, the growth of one environmental isolate (strain FC0656) was inhibited by 37.5 μg/ml of polymyxin B. Thwaite et al. (2009) found that of eight antimicrobial peptides tested, seven failed to inhibit the growth of five different Burkholderia sp. at up to 256 μg/ml of antimicrobial peptide. However one, magainin II, could inhibit the growth of many different Burkholderia strains at concentrations ranging from as low as 8 to 256 μg/ml. Additionally, the growth of one strain of B. cepacia was inhibited by a relatively low concentration (6.6 μg/ml) of human β-defensin-3 (García et al., 2001).
Emerging Opportunists may be Extremely Resistant to Antimicrobial Peptides
There is evidence that various emerging opportunistic pathogens may be extremely resistant to antimicrobial peptides. Coenye et al. (2002) analyzed a collection of unusual bacteria isolated from CF patients and misidentified at hospitals as members of the Bcc. The authors of this study found various different isolates, most of which had never or only very rarely been reported as isolated from CF patients. One reason many of these isolates were misidentified as members of the Bcc was their ability to grow on B. cepacia selective agar (BCSA), a media formulated for the selection of members of the Bcc. BCSA contains approximately 100 μg/ml of polymyxin B, which typically allows for the selective isolation of Bcc bacteria from other opportunistic pathogens of CF patients (Henry et al., 1997). Non-Bcc isolates from CF patients that were able to grow on BCSA included Ralstonia mannitolilytica, R. gilardii, Pandoraea sp., Acinetobacter baumannii, Chromobacterium violaceum, P. huttiensis, Xanthomonas hyacinthi, and Inquilinus limosus (Coenye et al., 2002). Since this study, a second report detailed pulmonary infections in 28 CF patients with Herbaspirillum sp., most of which also grew on BCSA and were misidentified as Bcc bacteria (Spilker et al., 2008). These results show that various organisms, phenotypically similar to B. cenocepacia and resistant to polymyxin B, have the ability to infect patients with CF. Finally, treatment of susceptible P. aeruginosa (MIC values between 1.6 and 6.1 μg/ml) infections in CF patients with colistin (polymyxin E) has lead to the isolation of colistin-resistant P. aeruginosa isolates (MIC values between 100 and 400 μg/ml; Johansen et al., 2008).
Future Directions of Study
There are multiple experimental lines to continue in the investigation of antimicrobial peptide and polymyxin resistance in Burkholderia sp. It is important to determine if Burkholderia bacteria make a transcriptional response to antimicrobial peptides. Since two regulatory systems, RpoE and a two-component system (BCAL2830/BCAL2831), have been implicated in the resistance of B. cenocepacia (Loutet et al., 2011) it seems likely that some transcriptional response occurs upon exposure to antimicrobial peptides. Genes whose transcription change upon treatment with antimicrobial peptides, as well as those controlled by RpoE and BCAL2830/BCAL2831, could prove to be important players in resistance and provide additional insight into mechanisms of resistance in these bacteria. The genes regulated by RpoE and BCAL2830/BCAL2831 have not been identified, though mucD is likely regulated by RpoE, since it is in the rpoE operon (Flannagan and Valvano, 2008), in other organisms rpoE gene expression is autoregulated (Raina et al., 1995), and preliminary experiments suggest that this is also the case in B. cenocepacia (El-Halfawy and Valvano, unpublished).
Understanding the essential nature of Ara4N substitutions of the LPS molecule in Burkholderia will likely provide significant insight into the unique properties of the outer membrane of these bacteria that set them apart from other Gram-negative bacteria. Compounds to disrupt the Ara4N pathway are being developed as potential therapeutics to administer with antimicrobial peptides or polymyxins (Kline et al., 2008). Such compounds might be very powerful anti-Burkholderia agents due to the essential nature of this pathway.
Since so many pathways have been implicated in the resistance of Burkholderia to antimicrobial peptides and polymyxins, another avenue to explore that could lead to the development of an anti-Burkholderia therapeutic is to identify compounds that synergize with these antibiotics against Burkholderia. We have recently begun to test this concept with a small library of naturally occurring compounds derived from marine life and have identified some potential lead molecules that increase polymyxin B sensitivity (Loutet and Valvano, unpublished). Other groups are screening compound libraries to find inhibitors of specific pathways (De Leon et al., 2006), such as the synthesis of l-glycero-d-manno-heptose sugars required for the LPS core oligosaccharide of Gram-negative bacteria (Valvano et al., 2002) and that have been shown to be required for antimicrobial peptide resistance and in vivo survival of B. cenocepacia (Loutet et al., 2006). All of these avenues of future research could potentially lead to a therapeutic strategy where a patient with a Burkholderia infection would receive a therapeutically applied antimicrobial peptide and a second drug that potentiates the activity of the peptide by reducing bacterial resistance to it.
Concluding Remarks
Much progress has been made to identify antimicrobial peptide resistance mechanisms in many different Gram-negative and Gram-positive bacteria (Yeaman and Yount, 2003). These resistance mechanisms include: changes to the cytoplasmic membrane in both Gram-negative and Gram-positive bacteria (Dorrer and Teuber, 1977; Peschel et al., 2001), alterations to teichoic acids in Gram-positive bacteria (Peschel et al., 1999) and LPS in Gram-negative bacteria (Vaara et al., 1981), efflux pumps (Shafer et al., 1998), proteases (Stumpe et al., 1998; Guina et al., 2000; Caldas et al., 2002; Belas et al., 2004), exopolysaccharides (Benincasa et al., 2009; Foschiatti et al., 2009), capsule polysaccharides (Campos et al., 2004; Spinosa et al., 2007; Llobet et al., 2008), modification of intracellular antimicrobial peptide targets (Vizán et al., 1991), and the coordination of resistance mechanisms through transcriptional regulation (Gunn and Miller, 1996; Guo et al., 1997; Humphreys et al., 1999; McPhee et al., 2003; Winfield et al., 2005; Kraus et al., 2008).
Members of the genus Burkholderia are the best studied among the highly antimicrobial peptide resistant species of Gram-negative bacteria, and a series of major and minor resistance determinants have been proposed for these bacteria (Figure 1). Studies in the Burkholderia could provide a model for the other extremely resistant Gram-negative bacteria. It is important to understand resistance in these organisms because many of these bacteria are opportunistic human pathogens that establish chronic infections. In this context, using antimicrobial peptides and polymyxins to treat infections by susceptible bacteria (such as P. aeruginosa infections in the CF lung) may give rise to subsequent infections by opportunistic pathogens that are resistant to these compounds.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank past and present members of the Valvano laboratory for many discussions about B. cenocepacia, lipopolysaccharide, antibiotic resistance, and other relevant topics. Slade A. Loutet was supported by a Cystic Fibrosis Canada studentship and a portion of this work appeared in his Ph.D. thesis. Work on B. cenocepacia in Miguel A. Valvano’s lab is supported by operating grants from Cystic Fibrosis Canada, the Canadian Institutes of Health Research Special Initiative on Novel Antimicrobials, and the B. cepacia microarray initiative from the U.S. Cystic Fibrosis Foundation. Miguel A. Valvano holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis and the Cystic Fibrosis Canada Zeller’s Senior Researcher Award.
References
- Belas R., Manos J., Suvanasuthi R. (2004). Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect. Immun. 72, 5159–5167 10.1128/IAI.72.9.5159-5167.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benincasa M., Mattiuzzo M., Herasimenka Y., Cescutti P., Rizzo R., Gennaro R. (2009). Activity of antimicrobial peptides in the presence of polysaccharides produced by pulmonary pathogens. J. Pept. Sci. 15, 595–600 10.1002/psc.1142 [DOI] [PubMed] [Google Scholar]
- Berg G., Eberl L., Hartmann A. (2005). The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7, 1673–1685 10.1111/j.1462-2920.2005.00891.x [DOI] [PubMed] [Google Scholar]
- Brett P. J., Burtnick M. N., Snyder D. S., Shannon J. G., Azadi P., Gherardini F. C. (2007). Burkholderia mallei expresses a unique lipopolysaccharide mixture that is a potent activator of human Toll-like receptor 4 complexes. Mol. Microbiol. 63, 379–390 10.1111/j.1365-2958.2006.05519.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- Bulet P., Stocklin R. (2005). Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept. Lett. 12, 3–11 10.2174/0929866053406011 [DOI] [PubMed] [Google Scholar]
- Burtnick M. N., Woods D. E. (1999). Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob. Agents Chemother. 43, 2648–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas C., Cherqui A., Pereira A., Simões N. (2002). Purification and characterization of an extracellular protease from Xenorhabdus nematophila involved in insect immunosuppression. Appl. Environ. Microbiol. 68, 1297–1304 10.1128/AEM.68.3.1297-1304.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M. A., Vargas M. A., Regueiro V., Llompart C. M., Albertí S., Bengoechea J. A. (2004). Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 72, 7107–7114 10.1128/IAI.72.12.7107-7114.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. C., Currie B. J. (2005). Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18, 383–416 10.1128/CMR.18.2.383-416.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T., Goris J., Spilker T., Vandamme P., LiPuma J. J. (2002). Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J. Clin. Microbiol. 40, 2062–2069 10.1128/JCM.40.9.3300-3307.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S., Nowak J., Coenye T., Clément C., Barka E. A. (2008). Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626 10.1111/j.1574-6976.2008.00113.x [DOI] [PubMed] [Google Scholar]
- Cox A. D., Wilkinson S. G. (1991). Ionizing groups in lipopolysaccharides of Pseudomonas cepacia in relation to antibiotic resistance. Mol. Microbiol. 5, 641–646 10.1111/j.1365-2958.1991.tb00775.x [DOI] [PubMed] [Google Scholar]
- De Leon G. P., Elowe N. H., Koteva K. P., Valvano M. A., Wright G. D. (2006). An in vitro screen of bacterial lipopolysaccharide biosynthetic enzymes identifies an inhibitor of ADP-heptose biosynthesis. Chem. Biol. 13, 437–441 10.1016/j.chembiol.2006.02.010 [DOI] [PubMed] [Google Scholar]
- Deris Z. Z., Van Rostenberghe H., Habsah H., Noraida R., Tan G. C., Chan Y. Y., Rosliza A. R., Ravichandran M. (2010). First isolation of Burkholderia tropica from a neonatal patient successfully treated with imipenem. Int. J. Infect. Dis. 14, e73–e74 10.1016/j.ijid.2009.11.019 [DOI] [PubMed] [Google Scholar]
- Dorrer E., Teuber M. (1977). Induction of polymyxin resistance in Pseudomonas fluorescens by phosphate limitation. Arch. Microbiol. 114, 87–89 10.1007/BF00429636 [DOI] [PubMed] [Google Scholar]
- Ernst R. K., Yi E. C., Guo L., Lim K. B., Burns J. L., Hackett M., Miller S. I. (1999). Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286, 1561–1565 10.1126/science.286.5444.1561 [DOI] [PubMed] [Google Scholar]
- Fehlner-Gardiner C. C., Valvano M. A. (2002). Cloning and characterization of the Burkholderia vietnamiensis norM gene encoding a multi-drug efflux protein. FEMS Microbiol. Lett. 215, 279–283 10.1111/j.1574-6968.2002.tb11403.x [DOI] [PubMed] [Google Scholar]
- Flannagan R. S., Valvano M. A. (2008). Burkholderia cenocepacia requires RpoE for growth under stress conditions and delay of phagolysosomal fusion in macrophages. Microbiology 154, 643–653 10.1099/mic.0.2007/013714-0 [DOI] [PubMed] [Google Scholar]
- Foschiatti M., Cescutti P., Tossi A., Rizzo R. (2009). Inhibition of cathelicidin activity by bacterial exopolysaccharides. Mol. Microbiol. 72, 1137–1146 10.1111/j.1365-2958.2009.06707.x [DOI] [PubMed] [Google Scholar]
- García J. R., Jaumann F., Schulz S., Krause A., Rodríguez-Jiménez J., Forssmann U., Adermann K., Klüver E., Vogelmeier C., Becker D., Hedrich R., Forssmann W. G., Bals R. (2001). Identification of a novel, multifunctional β-defensin (human β-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. 306, 257–264 [DOI] [PubMed] [Google Scholar]
- García-Olmedo F., Molina A., Alamillo J. M., Rodríguez-Palenzuéla P. (1998). Plant defense peptides. Biopolymers (Pept. Sci.) 47, 479–491 [DOI] [PubMed] [Google Scholar]
- Gerrits G. P., Klaasen C., Coenye T., Vandamme P., Meis J. F. (2005). Burkholderia fungorum septicemia. Emerg. Infect. Dis. 11, 1115–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaouris E., Briandet R., Meyrand M., Courtin P., Chapot-Chartier M.-P. (2008). Variations in the degree of D-alanylation of teichoic acids in Lactococcus lactis alter resistance to cationic antimicrobials but have no effect on bacterial surface hydrophobicity and charge. Appl. Environ. Microbiol. 74, 4764–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass M. B., Gee J. E., Steigerwalt A. G., Cavuoti D., Barton T., Hardy R. D., Godoy D., Spratt B. G., Clark T. A., Wilkins P. P. (2006). Pneumonia and septicemia caused by Burkholderia thailandensis in the United States. J. Clin. Microbiol. 44, 4601–4604 10.1128/AEM.00078-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory C. W., Estes D. M., Alfredo G. T. (2007). Glanders: off to the races with Burkholderia mallei. FEMS Microbiol. Lett. 277, 115–122 10.1128/JCM.01585-06 [DOI] [PubMed] [Google Scholar]
- Gronow S., Noah C., Blumenthal A., Lindner B., Brade H. (2003). Construction of a deep-rough mutant of Burkholderia cepacia ATCC 25416 and characterization of its chemical and biological properties. J. Biol. Chem. 278, 1647–1655 10.1111/j.1574-6968.2007.00949.x [DOI] [PubMed] [Google Scholar]
- Guina T., Yi E. C., Wang H., Hackett M., Miller S. I. (2000). A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182, 4077–4086 10.1074/jbc.M206942200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn J. S., Lim K. B., Krueger J., Kim K., Guo L., Hackett M., Miller S. I. (1998). PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27, 1171–1182 10.1128/JB.182.14.4077-4086.2000 [DOI] [PubMed] [Google Scholar]
- Gunn J. S., Miller S. I. (1996). PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178, 6857–6864 10.1046/j.1365-2958.1998.00757.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Lim K. B., Gunn J. S., Bainbridge B., Darveau R. P., Hackett M., Miller S. I. (1997). Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276, 250–253 [DOI] [PubMed] [Google Scholar]
- Guo L., Lim K. B., Poduje C. M., Daniel M., Gunn J. S., Hackett M., Miller S. I. (1998). Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95, 189–198 10.1126/science.276.5313.795 [DOI] [PubMed] [Google Scholar]
- Gustafson C. E., Kaul S., Ishiguro E. E. (1993). Identification of the Escherichia coli lytB gene, which is involved in penicillin tolerance and control of the stringent response. J. Bacteriol. 175, 1203–1205 10.1016/S0092-8674(00)81750-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Wong P. G. (1984). Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 26, 48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E. W., Sahl H.-G. (2006). Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557 [DOI] [PubMed] [Google Scholar]
- Henry D. A., Campbell M. E., LiPuma J. J., Speert D. P. (1997). Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J. Clin. Microbiol. 35, 614–619 10.1038/nbt1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo S. T., Kim S. J., Jeong Y. G., Bae I. G., Jin J. S., Lee J. C. (2008). Hospital outbreak of Burkholderia stabilis bacteraemia related to contaminated chlorhexidine in haematological malignancy patients with indwelling catheters. J. Hosp. Infect. 70, 241–245 [DOI] [PubMed] [Google Scholar]
- Humphreys S., Stevenson A., Bacon A., Weinhardt A. B., Roberts M. (1999). The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67, 1560–1568 10.1016/j.jhin.2008.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung R. J., Chien H. S., Lin R. Z., Lin C. T., Vatsyayan J., Peng H. L., Chang H. Y. (2007). Comparative analysis of two UDP-glucose dehydrogenases in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 282, 17738–17748 [DOI] [PubMed] [Google Scholar]
- Isshiki Y., Kawahara K., Zähringer U. (1998). Isolation and characterisation of disodium (4-amino-4- deoxy-β-l-arabinopyranosyl)-(1-8)-(d-glycero-α-d-talo-oct-2-ulopyranosylona te)-(2-4)-(methyl 3- deoxy-d-manno-oct-2-ulopyranos- id)onate from the lipopolysaccharide of Burkholderia cepacia. Carbohydr. Res. 313, 21–27 10.1074/jbc.M702319200 [DOI] [PubMed] [Google Scholar]
- Jassem A. N., Zlosnik J. E. A., Henry D. A., Hancock R. E. W., Ernst R. K., Speert D. P. (2011). In vitro susceptibility of Burkholderia vietnamiensis to aminoglycosides. Antimicrob. Agents Chemother. 55, 2256–2264 10.1016/S0008-6215(98)00179-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen H. K., Moskowitz S. M., Ciofu O., Pressler T., HØiby N. (2008). Spread of colistin resistant non-mucoid Pseudomonas aeruginosa among chronically infected Danish cystic fibrosis patients. J. Cyst. Fibros. 7, 391–397 10.1128/AAC.01434-10 [DOI] [PubMed] [Google Scholar]
- Kline T., Trent M. S., Stead C. M., Lee M. S., Sousa M. C., Felise H. B., Nguyen H. V., Miller S. I. (2008). Synthesis of and evaluation of lipid A modification by 4-substituted 4-deoxy arabinose analogs as potential inhibitors of bacterial polymyxin resistance. Bioorg. Med. Chem. Lett. 18, 1507–1510 10.1016/S1569-1993(08)60146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooi C., Sokol P. A. (2009). Burkholderia cenocepacia zinc metalloproteases influence resistance to antimicrobial peptides. Microbiology 155, 2818–2825 10.1016/j.bmcl.2007.12.061 [DOI] [PubMed] [Google Scholar]
- Kovács M., Halfmann A., Fedtke I., Heintz M., Peschel A., Vollmer W., Hakenbeck R., Brückner R. (2006). A functional dlt operon, encoding proteins required for incorporation of D-alanine in teichoic acids in Gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188, 5797–5805 10.1099/mic.0.028969-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus D., Herbert S., Kristian S., Khosravi A., Nizet V., Götz F., Peschel A. (2008). The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol. 8, 85–89 10.1128/JB.00336-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda T., Tsuchiya T. (2009). Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 1794, 763–768 10.1186/1471-2180-8-85 [DOI] [PubMed] [Google Scholar]
- Lai Y., Gallo R. L. (2009). AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestin F., Kraak R., Podbielski A. (2008). Two cases of keratitis and corneal ulcers caused by Burkholderia gladioli. J. Clin. Microbiol. 46, 2445–2449 10.1016/j.it.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llobet E., Tomás J. M., Bengoechea J. A. (2008). Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology 154, 3877–3886 10.1128/JCM.02442-07 [DOI] [PubMed] [Google Scholar]
- Loutet S. A., Bartholdson S. J., Govan J. R., Campopiano D. J., Valvano M. A. (2009). Contributions of two UDP-glucose dehydrogenases to viability and polymyxin B resistance of Burkholderia cenocepacia. Microbiology 155, 2029–2039 10.1099/mic.0.2008/022301-0 [DOI] [PubMed] [Google Scholar]
- Loutet S. A., Flannagan R. S., Kooi C., Sokol P. A., Valvano M. A. (2006). A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J. Bacteriol. 188, 2073–2080 10.1099/mic.0.027607-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet S. A., Mussen L. E., Flannagan R. S., Valvano M. A. (2011). A two-tier model of polymyxin B resistance in Burkholderia cenocepacia. Environ. Microbiol. Rep. 3, 278–285 10.1128/JB.188.6.2073-2080.2006 [DOI] [PubMed] [Google Scholar]
- Ma M., Wang C., Ding Y., Li L., Shen D., Jiang X., Guan D., Cao F., Chen H., Feng R., Wang X., Ge Y., Yao L., Bing X., Yang X., Li J., Du B. (2011). Complete genome sequence of Paenibacillus polymyxa SC2, a strain of plant growth-promoting rhizobacterium with broad-spectrum antimicrobial activity. J. Bacteriol. 193, 311–312 10.1111/j.1758-2229.2010.00222.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E., Urban T. A., Goldberg J. B. (2005). The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3, 144–156 10.1128/JB.01234-10 [DOI] [PubMed] [Google Scholar]
- McPhee J. B., Lewenza S., Hancock R. E. (2003). Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50, 205–217 10.1038/nrmicro1085 [DOI] [PubMed] [Google Scholar]
- Moore R. A., Hancock R. E. (1986). Involvement of outer membrane of Pseudomonas cepacia in aminoglycoside and polymyxin resistance. Antimicrob. Agents Chemother. 30, 923–926 10.1046/j.1365-2958.2003.03673.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouslim C., Groisman E. A. (2003). Control of the Salmonella ugd gene by three two-component regulatory systems. Mol. Microbiol. 47, 335–344 [DOI] [PubMed] [Google Scholar]
- Novem V., Shui G., Wang D., Bendt A. K., Sim S. H., Liu Y., Thong T. W., Sivalingam S. P., Ooi E. E., Wenk M. R., Tan G. (2009). Structural and biological diversity of lipopolysaccharides from Burkholderia pseudomallei and Burkholderia thailandensis. Clin. Vaccine Immunol. 16, 1420–1428 10.1046/j.1365-2958.2003.03318.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nummila K., Kilpeläinen I., Zähringer U., Vaara M., Helander I. M. (1995). Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16, 271–278 10.1128/CVI.00472-08 [DOI] [PubMed] [Google Scholar]
- Ortega X., Silipo A., Saldías M. S., Bates C. C., Molinaro A., Valvano M. A. (2009). Biosynthesis and structure of the Burkholderia cenocepacia K56-2 lipopolysaccharide core oligosaccharide: truncation of the core oligosaccharide leads to increased binding and sensitivity to polymyxin B. J. Biol. Chem. 284, 21738–21751 10.1111/j.1365-2958.1995.tb02299.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega X. P., Cardona S. T., Brown A. R., Loutet S. A., Flannagan R. S., Campopiano D. J., Govan J. R., Valvano M. A. (2007). A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J. Bacteriol. 189, 3639–3644 10.1074/jbc.M109.008532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A., Jack R. W., Otto M., Collins L. V., Staubitz P., Nicholson G., Kalbacher H., Nieuwenhuizen W. F., Jung G., Tarkowski A., van Kessel K. P. M., van Strijp J. A. G. (2001). Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor Mprf is based on modification of membrane lipids with L-lysine. J. Exp. Med. 193, 1067–1076 10.1128/JB.00153-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A., Otto M., Jack R. W., Kalbacher H., Jung G., Götz F. (1999). Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410 10.1084/jem.193.9.1067 [DOI] [PubMed] [Google Scholar]
- Raetz C. R., Whitfield C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 10.1074/jbc.274.13.8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina S., Missiakis D., Georgopoulos C. (1995). The rpoE gene encoding the σE σ24 heat shock sigma factor of Escherichia coli. EMBO J. 14, 1043–1055 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F., Hecht S., Gärtner K., Adam P., Krieger C., Amslinger S., Arigoni D., Bacher A., Eisenreich W. (2002). Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. U.S.A. 99, 1158–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer W. M., Qu X., Waring A. J., Lehrer R. I. (1998). Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U.S.A. 95, 1829–1833 10.1073/pnas.032658999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A., Molinaro A., Cescutti P., Bedini E., Rizzo R., Parrilli M., Lanzetta R. (2005). Complete structural characterization of the lipid A fraction of a clinical strain of B. cepacia genomovar I lipopolysaccharide. Glycobiology 15, 561–570 10.1073/pnas.95.4.1829 [DOI] [PubMed] [Google Scholar]
- Silipo A., Molinaro A., Ieran‘ T., De Soyza A., Sturiale L., Garozzo D., Aldridge C., Corris P. A., Anjam Khan C. M., Lanzetta R., Parrilli M. (2007). The complete structure and pro-inflammatory activity of the lipooligosaccharide of the highly epidemic and virulent Gram-negative bacterium Burkholderia cenocepacia ET-12 (Strain J2315). Chem. Eur. J. 13, 3501–3511 10.1093/glycob/cwi029 [DOI] [PubMed] [Google Scholar]
- Sperandeo P., Cescutti R., Villa R., Di Benedetto C., Candia D., Deho G., Polissi A. (2007). Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189, 244–253 10.1002/chem.200601406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandeo P., Lau F. K., Carpentieri A., De Castro C., Molinaro A., Deho G., Silhavy T. J., Polissi A. (2008). Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 190, 4460–4469 10.1128/JB.01126-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilker T., Uluer A. Z., Marty F. M., Yeh W. W., Levison J. H., Vandamme P., LiPuma J. J. (2008). Recovery of Herbaspirillum species from persons with cystic fibrosis. J. Clin. Microbiol. 46, 2774–2777 10.1128/JB.00270-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinosa M. R., Progida C., Talà A., Cogli L., Alifano P., Bucci C. (2007). The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect. Immun. 75, 3594–3603 10.1128/JCM.00460-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpe S., Schmid R., Stephens D. L., Georgiou G., Bakker E. P. (1998). Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J. Bacteriol. 180, 4002–4006 10.1128/IAI.01945-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo H., Hisada Y., Ito M., Kotaki H., Minami A. (2005). Burkholderia pickettii spondylitis. Spinal Cord 43, 499–502 [DOI] [PubMed] [Google Scholar]
- Tamayo R., Choudhury B., Septer A., Merighi M., Carlson R., Gunn J. S. (2005). Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar typhimurium lipopolysaccharide core. J. Bacteriol. 187, 3391–3399 10.1038/sj.sc.3101740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thwaite J. E., Humphrey S., Fox M. A., Savage V. L., Laws T. R., Ulaeto D. O., Titball R. W., Atkins H. S. (2009). The cationic peptide magainin II is antimicrobial for Burkholderia cepacia-complex strains. J. Med. Microbiol. 58, 923–929 10.1128/JB.187.10.3391-3399.2005 [DOI] [PubMed] [Google Scholar]
- Vaara M., Vaara T. (1994). Ability of cecropin B to penetrate the enterobacterial outer membrane. Antimicrob. Agents Chemother. 38, 2498–2501 10.1099/jmm.0.008128-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M., Vaara T., Jensen M., Helander I., Nurminen M., Rietschel E. T., Mäkelä P. H. (1981). Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 129, 145–149 [DOI] [PubMed] [Google Scholar]
- Valvano M. A., Messner P., Kosma P. (2002). Novel pathways for biosynthesis of nucleotide-activated glycero-manno-heptose precursors of bacterial glycoproteins and cell surface polysaccharides. Microbiology 148, 1979–1989 10.1016/0014-5793(81)80777-6 [DOI] [PubMed] [Google Scholar]
- Vanlaere E., Baldwin A., Gevers D., Henry D., De Brandt E., Lipuma J. J., Mahenthiralingam E., Speert D. P., Dowson C., Vandamme P. (2009). Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59, 102–111 [DOI] [PubMed] [Google Scholar]
- Vermis K., Vandamme P. A. R., Nelis H. J. (2003). Burkholderia cepacia complex genomovars: utilization of carbon sources, susceptibility to antimicrobial agents and growth on selective media. J. Appl. Microbiol. 95, 1191–1199 10.1099/ijs.0.001123-0 [DOI] [PubMed] [Google Scholar]
- Vinion-Dubiel A. D., Goldberg J. B. (2003). Lipopolysaccharide of Burkholderia cepacia complex. J. Endotoxin Res. 9, 201–213 10.1046/j.1365-2672.2003.02054.x [DOI] [PubMed] [Google Scholar]
- Vizán J. L., Hernandez-Chico C., del Castillo I., Moreno F. (1991). The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J. 10, 467–476 10.1179/096805103225001404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J. B., Alexander B. D., Majure J. M., Williams L. W., Kim J. Y., Vandamme P., LiPuma J. J. (2007). Burkholderia glumae infection in an infant with chronic granulomatous disease. J. Clin. Microbiol. 45, 662–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield M. D., Latifi T., Groisman E. A. (2005). Transcriptional regulation of the 4-Amino-4-deoxy-L-arabinose biosynthetic genes in Yersinia pestis. J. Biol. Chem. 280, 14765–14772 10.1128/JCM.02058-06 [DOI] [PubMed] [Google Scholar]
- Yeaman M. R., Yount N. Y. (2003). Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55 10.1074/jbc.M413900200 [DOI] [PubMed] [Google Scholar]
- Yeaman M. R., Yount N. Y. (2007). Unifying themes in host defence effector polypeptides. Nat. Rev. Microbiol. 5, 727–740 10.1124/pr.55.1.2 [DOI] [PubMed] [Google Scholar]
- Zavascki A. P., Goldani L. Z., Li J., Nation R. L. (2007). Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60, 1206–1215 10.1038/nrmicro1744 [DOI] [PubMed] [Google Scholar]
- Zhang L., Parente J., Harris S. M., Woods D. E., Hancock R. E. W., Falla T. J. (2005). Antimicrobial peptide therapeutics for cystic fibrosis. Antimicrob. Agents Chemother. 49, 2921–2927 10.1093/jac/dkm357 [DOI] [PMC free article] [PubMed] [Google Scholar]