Abstract
Recent developments in reagent design can address problems in single cells that were not previously approachable. We have attempted to foresee what will become possible, and the sorts of biological problems that become tractable with these novel reagents. We have focused on the novel fluorescent proteins that allow convenient multiplexing, and provide for a time-dependent analysis of events in single cells. Methods for fluorescently labeling specific molecules, including endogenously expressed proteins and mRNA have progressed and are now commonly used in a variety of organisms. Finally, sensitive microscopic methods have become more routine practice. This article emphasizes that the time is right to coordinate these approaches for a new initiative on single cell imaging of biological molecules.
Introduction
Recent advances in fluorescent probes with red-shifted spectra resulted in creation of novel red fluorescent proteins (RFPs) and RFP-based biosensors with enhanced spectral and biochemical characteristics. Reduced autofluorescence, low light scattering, and minimal absorbance at the longer wavelengths make RFPs superior probes for cell, tissue, and whole-body imaging [1]. Moreover, introduction of novel RFPs enables multi-color labeling, intravital imaging, super-resolution microscopy, and provides new pairs for FRET techniques. In this review we focus on novel monomeric RFPs and their application for studying gene expression, nuclear localization, and dynamics using advanced imaging. For properties and applications of green fluorescent proteins (GFPs) and other blue, cyan and yellow fluorescent proteins (FPs) we refer to recent reviews [2,3].
Modern red fluorescent proteins
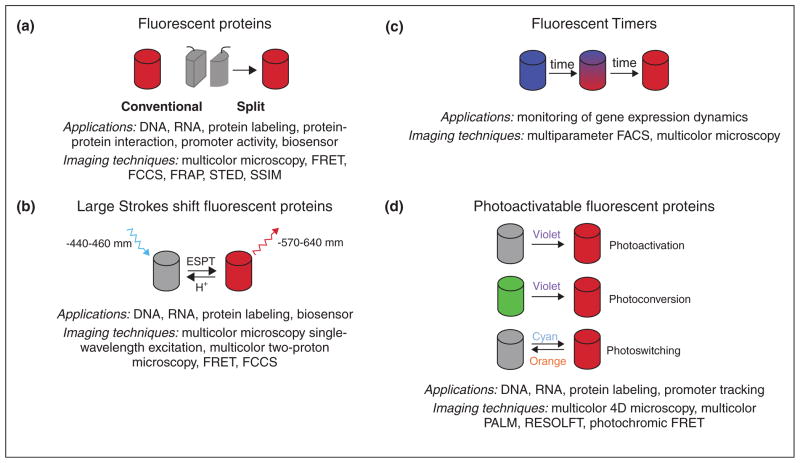
Modern RFPs, with emission maxima exceeding 560 nm, can be divided into five main groups: conventional and split orange, red and far-red FPs, RFPs with a large Stokes shift (LSS-RFPs), fluorescent timers (FTs), and photo-activatable RFPs (PA-RFPs) (Figure 1). We list the currently recommended FPs of each class and their key spectroscopic properties in Table 1.
Figure 1.
Major groups of RFPs, their photophysical properties, and potential applications are shown. (a) Conventional and split RFPs. Two non-fluorescent fragments of split FP when brought together form a complete FP barrel. (b) Large Stokes shift RFPs. Excited state proton transfer was shown to be responsible for large Stokes shift. (c) Fluorescent timers. (d) Three types of photoactivatable RFPs. Dark-to-red PAFPs irreversably convert from non-fluorescent state to the fluorescent state under violet light (photoactivation). Green-to-red PAFPs irreversably convert from green fluorescent state to red fluorescent state under violet light (photoconvertion). Red-to-dark photoswitchable FPs reversably convert from non-fluorescent state to the fluorescent state under different lights (photoswitching).
Table 1.
Properties of the modern monomeric red fluorescent proteins
| Protein | Exmax, nm | Emmax, nm | ε, M−1·CM−1 | QY | Brightnessa | pKa | Additional parameter | Ref | |
|---|---|---|---|---|---|---|---|---|---|
| Red fluorescent proteins | |||||||||
| Timeb, h | |||||||||
| mKOκ | 551 | 563 | 105,000 | 0.61 | 64 | 4.2 | 1.8 | 4 | |
| mOrange2 | 549 | 565 | 58,000 | 0.60 | 35 | 6.5 | 4.5 | 6 | |
| TagRFP-T | 555 | 584 | 81,000 | 0.41 | 33 | 4.6 | 1.7 | 6 | |
| mRuby | 558 | 605 | 112,000 | 0.35 | 39 | 5 | 2.8 | 5 | |
| LSSmKate2 | 460 | 605 | 26,000 | 0.17 | 4.5 | 2.7 | 2.5 | 12 | |
| mLumin | 587 | 621 | 70,000 | 0.46 | 32 | 4.7 | 1.3 | 8 | |
| mKate2 | 588 | 633 | 62,500 | 0.40 | 25 | 5.4 | <0.33 | 7 | |
| mNeptune | 600 | 650 | 67,000 | 0.20 | 13 | 5.4 | ND | 10 | |
| TagRFP657 | 611 | 657 | 34,000 | 0.10 | 3.4 | 5.0 | 2.0 | 9 | |
| Fluorescent times | |||||||||
| Timec, h | |||||||||
| Slow-FT | 402 | 465 | 33,400 | 0.35 | 12 | 2.6 | 9.8 | 14 | |
| 583 | 604 | 84,200 | 0.05 | 4 | 4.6 | 28 | |||
| Medium-FT | 401 | 464 | 44,800 | 0.41 | 18 | 2.7 | 1.2 | 14 | |
| 579 | 600 | 73,100 | 0.08 | 6 | 4.7 | 3.9 | |||
| Fast-FT | 403 | 466 | 49,700 | 0.30 | 15 | 2.8 | 0.25 | 14 | |
| 583 | 606 | 75,300 | 0.09 | 7 | 4.1 | 7.1 | |||
| mK-GO | 500 | 509 | 35,900 | ND | ND | 6.0 | 10 | 15 | |
| 548 | 561 | 42,000 | ND | ND | 4.8 | ||||
| Photoactivatable red fluorescent proteins | |||||||||
| Conditiond | |||||||||
| PAmCherry | 564 | 594 | 18,000 | 0.46 | 8 | 6.3 | Violet | 16 | |
| PATagRFP | 562 | 595 | 66,000 | 0.38 | 25 | 5.3 | Violet | 17 | |
| Dendra2 | 490 | 507 | 45,000 | 0.50 | 22 | 6.6 | Matures to green | 18 | |
| 553 | 573 | 35,000 | 0.55 | 19 | 6.9 | Violet | |||
| mEos2 | 506 | 519 | 56,000 | 0.84 | 47 | 5.6 | Matures to green | 19 | |
| 573 | 584 | 46,000 | 0.66 | 30 | 6.4 | Violet | |||
| mKikGR | 505 | 515 | 49,000 | 0.69 | 34 | ND | Matures to green | 20 | |
| 580 | 591 | 28,000 | 0.63 | 18 | ND | Violet | |||
| mIrisFP | 486 | 516 | 47,000 | 0.54 | 25 | 5.4 | Violet | 21 | |
| 546 | 578 | 33,000 | 0.59 | 19 | 7.6 | Violet | Cyan | ||
| rsTagRFP | 440 | 585 | 15,300 | 0.001 | 0.02 | ND | Orange | 22 | |
| 567 | 585 | 36,800 | 0.11 | 4 | 6.6 | Blue | |||
Exmax is the excitation maximum. Emmax is the emission maximum. ε is the molar extinction coefficient. QY is the quantum yield.
Fluorescent protein brightness is determined as a product of quantum yield and molar extinction coefficient, divided by 1000.
Maturation half time.
Characteristic time for the color transition.
Condition for the chromophore formation: spontaneous maturation or photoactivation (PAmCherry, PATagRFP), photoconversion (Dendra2, mEos2, mKikGR, mIrisFP), or photoswitching (mIrisFP, rsTagRFP).
Conventional red fluorescent proteins
Orange and red FPs
The palette of conventional RFPs have been enriched by the number of enhanced monomeric orange FPs (OFPs) and RFPs for DNA, RNA, and protein labeling in living cells. The novel orange mKOκ [4] and red mRuby [5] FPs are the brightest among the currently available monomeric FPs. High extinction coefficients, pH-stability and extended Stokes shift (47 nm) in case of mRuby make these RFPs attractive as FRET acceptors for yellow donors. However, mKOκ and mRuby are less photostable than mCherry under arc lamp illumination. TagRFP-T and mOrange2, which preserve spectral properties of their precursors TagRFP and mOrange, are attractive for long-term imaging owing to their photostability both under arc lamp and laser illumination, [6]. The improved version of mKate, mKate2, combines brightness and photostability with rapid maturation [7]. Transgenic expression of mKate2 in Xenopus embryos revealed reduced cytotoxicity even at high concentration in the cells. Another mKate derivative, split-mLumin, is a novel red bimolecular fluorescent complementation system that shows improved performance in mammalian cells at 37 °C [8].
Far-red FPs
The development of monomeric RFPs with emission beyond 650 nm has recently been achieved. Far-red FPs can be preferable for labeling cellular proteins in strong autofluorescence conditions and for multicolor imaging with OFPs. The TagRFP657 protein, characterized by absorption/emission at 611/657 nm, exhibits low cytotoxicity, high pH-stability and photostability and can be efficiently excited by the standard 633–640 nm red lasers [9]. mNeptune, exhibiting absorption/emission at 600/650 nm, outperforms TagRFP657 in brightness in mammalian cells [10].
Large Stokes shift fluorescent proteins
Recently, several orange and red FPs with large Stokes shifts (LSS; a difference between excitation and emission maxima more than 100 nm) have been developed on the basis of conventional RFPs [11•]. An excited-state proton transfer (ESPT) occurring upon excitation of a neutral chromophore was shown to be responsible for the LSS observed in these proteins (Figure 1b). The LSS-RFPs are beneficial for imaging under autofluorescence conditions since autofluorescence has a shorter Stokes shift. Moreover, LSS-FPs can be efficiently used with regular FPs for multicolor imaging with a single excitation wavelength and as an additional red color for conventional RFPs. LSSmKate2, optimized for expression in mammalian cells, is recommended owing to its photostability, pH insensitivity and excellent fusion property [12].
Fluorescent timers (FT)
A fluorescent timer changes its color with time owing to a chemical conversion of its chromophore (Figure 1c) [13]. The predictable time course of fluorescence transition allows a quantitative analysis of temporal and spatial molecular events based on the ratio between fluorescence intensities of the two forms. The first monomeric FTs that exhibited distinctive fast, medium, and slow blue-to-red chromophore maturation rates (from around 10 min to 28 h) were developed on the basis of mCherry [14]. The blue and red forms of FTs are bright either alone in protein fusions or together with green FPs for multicolor microscopy. However, noticeable blue-to-red photoactivation of FTs under intense illumination by blue light may complicate their long-term imaging, but still allows efficient application for flow cytometry. Another monomeric FT named Kusabira Green Orange (mK-GO) changes fluorescence from green to orange. The ratio of orange per green fluorescence determined by in vitro translation linearly increased and reached a plateau at approximately 10 h [15].
Photoactivatable red fluorescent proteins (PARFP)
PARFPs change fluorescent properties upon irradiation with a certain wavelength. All PARFPs can be divided into the three main groups by color transitions upon illumination (Figure 1d).
Dark-to-red photoactivatable FPs
PAmCherrys [16] and PATagRFP [17••] are non-fluorescent in the dark (non-activated) state, but easily undergo irreversible activation under violet light irradiation of relatively low intensity. High photoactivation contrast and photostable red forms make long-term visualization of the activated proteins possible. However, PATagRFP significantly outperforms PAmCherry in pH stability, brightness, and photostability (Table 1).
Green-to-red photoswitchable FPs
All members of this group initially mature to a green-emitting state, which can be irreversably photoconverted into the red fluorescent form upon violet light illumination. The most promising variants of green-to-red PAFPs, which are Dendra2 [18], mEos2 [19], and mKikGR [20], are characterized by high brightness and photostabilities of both fluorescent forms, efficient maturation at 37 °C. Additionally, excellent performance in difficult fusions has already allowed their succesful application for a variety of cell biology problems. It was shown that mKikGR can be also activated by soft radiation of IR laser. A remarkable protein mIrisFP combines properties of photoactivatable and photoswitchable FPs [21]. It undergoes irreversible photoactivation from green to red fluorescent form under violet light, moreover the green and red fluorescent forms can be reversibly switched between dark and fluorescent states by light.
Reversibly photoswitchable RFPs
A small class of photoswitchable RFPs is represented by rsTagRFP [22••]. Initially rsTagRFP matures to a red fluorescent form. However, illumination with blue and yellow light switches the protein into a red fluorescent state or nonfluorescent state, respectively. Switching can be repeated hundreds of times reaching a 20-fold ratio of fluorescence intensities. Thus, rsTagRFP spectral properties are beneficial for sensitive imaging of the switched form.
We have briefly described enhanced versions of FPs from each group that can be used to study problems in cell biology. Following are some applications of these novel RFPs to study gene expression, nuclear localization, and dynamics using advanced imaging techniques.
Microscopy techniques utilizing fluorescent proteins
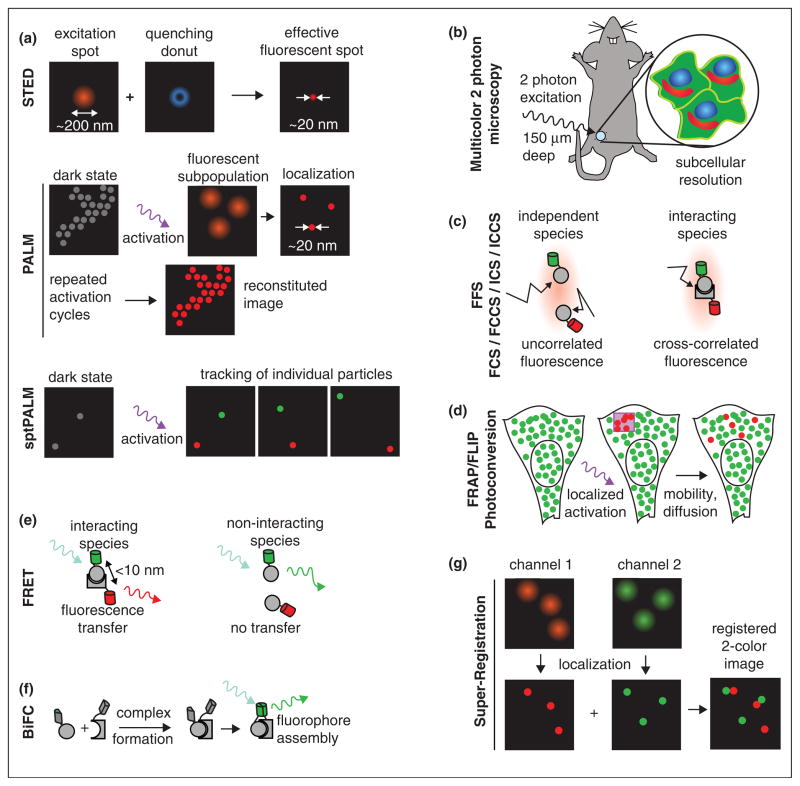
A variety of microscopy/spectroscopy techniques have been developed in the past decades, which are briefly summarized in Figure 2. Together with FPs, these methods provide key information about cellular function that is otherwise unattainable.
Figure 2.
Advanced microscopy and spectroscopy techniques for imaging gene expression, nuclear localization, and dynamics. (a) Super-resolution microscopy: The first class of super-resolution microscopy exploits the nonlinear optics to reduce the illumination spot size in technique such as stimulated emission depletion (STED) microscopy, reversible saturable optical fluorescence transition (RESOLFT) microscopy [47], and saturated structured illumination microscopy (SSIM) [48]. The second class involves repeated activation and bleaching of sparsely selected fluorescent molecule and subsequently accurate localization to build up the high resolution images, such as photoactivation localization microscopy (PALM) and its close variants STORM and fPALM [25•]. Single particle tracking PALM (sptPALM) allows tracking of high density molecules in live cell [29]. (b) MPM: Multiphoton microscopy [49] offers attractive feature over traditional confocal and widefield microscopy for live cell and thick tissue imaging for its increased penetration depth owing to less light scattering, reduced autofluorescence and photobleaching, minimal absorbance of hemoglobin and skin melanin at the longer wavelengths, and its optical sectioning effect. Development of RFPs with large Stokes shift and far-red spectrum enables multicolor in vivo MPM with subcellular resolution [12]. (c) FFS: Fluorescence fluctuation spectroscopy includes a variety of techniques that utilize the fluctuating fluorescence signal when molecules randomly diffuse through a subfemtoliter observation volume created by confocal or two-photon microscope. Fluorescence correlation spectroscopy (FCS) and fluorescence cross-correlation spectroscopy (FCCS) [31,50] exploit the temporal decay of correlation/crosscorrelation of the signal to extract the concentration, mobility, and the interaction information. Brightness analysis studies the amplitude of the fluctuation and provides stoichiometry and affinity information of interactions [33]. Image correlation spectroscopy (ICS) and cross correlation spectroscopy (ICCS) [32] measures spatially fluctuating signal from raster-scan laser confocal/two-photon microscopy. They are powerful tools to measure the clustering and dynamics of membrane proteins and receptors. (d) FRAP, FLIP, Photoactivation, Photoconversion: Molecules in a region of interest are optically highlighted by photobleaching or photoactivation [28]. As the highlighted molecule exchanges with the surrounding unhighlighted ones owing to diffusion and binding, the fluorescence in the ROI is monitored to obtain the kinetic information about mobility and interaction. (e) FRET: Fluorescence resonance energy transfer measures the effect of excited-state energy transfer from donor to an adjacent acceptor protein. FRET provides evidence for direct interaction since the energy transfer occurs only when donor and acceptor are within 10 nm of each other. Compared with FFS, FRET is independent of the mobility of the molecule under investigation. FRET can be measured simply by acceptor bleaching or ratiometric imaging. However ratiometric imaging is not appropriate for general purpose protein interaction assays since it depends on relative concentration of donor and acceptor. Fluorescence lifetime imaging microscopy (FLIM) based FRET assay is not limited by this and is commonly applied to detect protein interactions. (f) BiFC [38]: In bimolecular fluorescence complementation experiment, an FP is split into two segments and fused to two interacting molecules. The two segments remain dark until the interacting partners bring them together and form a complete FP. However, owing to the maturation of fluorophore, there is delay between the interaction and the appearance of fluorescence. In certain scenario, the formation of bimolecular complex is irreversible, which complicates the physiological process understudy. BiFC has been successfully applied to study protein–protein interaction. (g) Super-registration microscopy: Imaging two interacting molecules in different color with high spatial and temporal resolution is challenging. The super-registration microscopy [26] exploits a natural cellular marker to register positions in different detection channels beyond the diffraction limit. It has been applied to detect a single mRNA particle passing through a single nuclear pore. Currently, the technique is limited to the case that the cellular marker is relative immobile during the time of imaging.
Measuring molecular localization
The localization of molecules within the cell can be followed by 4D microscopy. Time-lapse imaging of FP labeled proteins or mRNAs provides information on their localization and translocation in living cells [23,24]. A new set of methods, termed super-resolution microscopy, have broken the diffraction limit of conventional light microscopy (Figure 2a) [25•]. One form of super-resolution imaging, photoactivation localization microscopy (PALM), involves repeatedly activation and bleaching of sparsely selected fluorescent molecules followed by accurate localization. Introduction of novel photoactivatable RFPs enables multicolor PALM of fixed and living cells [16]. Super-registration microscopy allows co-registration of two spectrally distinct molecules with 20 ms temporal and 26 nm spatial precision in live cells by exploiting a natural cellular marker such as a nuclear pore [26]. A transition from cellular imaging to tissue imaging also become possible with intravital multiphoton microscopy with subcellular resolution [12].
Measuring molecular mobility
The mobility of molecules can be measured by highlighting a subset of molecules in a small region of interest. This group of techniques includes fluorescence recovery after photobleaching (FRAP), its variation fluorescence loss in photobleaching (FLIP) and reversibly or irreversibly photoactivation of FPs [27,28]. Photoactivation overcomes some limitation of FRAP and FLIP, such as phototoxicity and complex photophysics of some FPs. It enables tracking fast protein movement [28] or even dual-color single particle tracking PALM in live cells [17••,29]. With proper mathematical modeling, these measurements also yield information about binding with the subcellular structure [30•]. Alternative approaches to measure mobility include fluorescence correlation spectroscopy (FCS) [31] and image correlation spectroscopy (ICS) [32]. FCS is able to measure fast dynamics ranging from submicrosecond to second in a specific location. ICS is especially suitable for slower events such as receptors moving on the plasma membrane.
Detecting molecular interactions
An effective way to measure protein–protein interactions in living cells is fluorescence resonance energy transfer (FRET) (reviewed in [33]). Intensity-based ratiometric FRET imaging is easy to implement and widely used to measure fast signaling events of biosensors. Using recently developed photoswitchable rsTagRFP as acceptor and YFP as donor, FRET can be turned on and off, offering an internal control for photochromic FRET (pcFRET) [22••]. FRET, although powerful, suffers from the high false-negative rate to measure protein–protein interactions since it is distance dependent. An alternative approach that is not limited by distance is fluorescence fluctuation spectroscopy (FFS). Brightness analysis in FFS provides straightforward measurements of protein homo-oligomerization [34]. By labeling proteins with different colors, FCCS and ICCS are able to detect interacting species [32,35]. The recently developed hetero-species partition analysis (HSP) utilizes dual-color brightness to measure stoichiometry as well as generate binding curves in living cells [36]. Large Stokes shift proteins provide unique advantages for multicolor FFS experiments since they allow efficient excitation of multiple fluorophores with a single wavelength, eliminating the complications of overlapping lasers and FRET between protein pairs [37•]. Bimolecular fluorescence complementation (BiFC) [38] represents one of the newly developed approaches for visualizing protein–protein interactions. The recently introduced split-mLu-min [8] allows simultaneously three-color imaging with a Cerulean and Venus based BiFC system in a single cell.
Imaging gene expression
Gene expression in eukaryotic cells involves many steps and numerous components (Figure 2) [39••,40]. Biochemical studies have identified most players and detailed the enzymatic nature of the process. Various hues of FPs allow multicolor labeling of DNA, RNA, and protein factors involved in gene expression. In addition, novel spectral properties such as photoswitching or fluorescent timers open the way for pulse-chase experiments at a single cell level. Currently it is possible to image three red colors (simultaneous imaging mOrange2 and TagRFP657, and asynchronous imaging of LSS-mKate2). Combination of RFPs with conventional blue/green and large Stokes shift GFPs could image as many as six colors in a single cell.
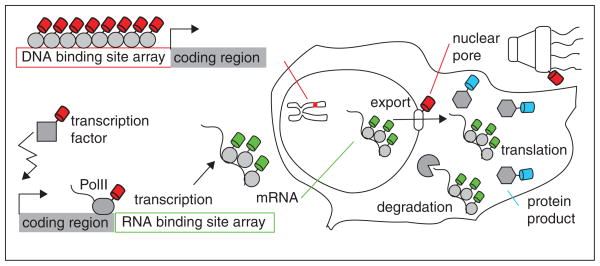
In Figure 3, we have shown schematically the process of gene expression and how each step can be visualized. First, a specific gene locus on a chromosome can be tagged with DNA binding protein fused to FP by inserting recombinant DNA sequences carrying specific binding sites (such as Lac operator/repressor). Additionally, multiple mRNAs can be visualized in a single cell by incorporating a specific sequence recognized by an RNA binding protein labeled by FPs [41,42]. When the gene is transcribed, multiple nascent transcripts accumulate and illuminate the transcription site.
Figure 3.
The gene expression in eukaryotic cells involves many steps and numerous components. First transcription requires close cooperation between transcription factor, corregulator, mediator, chromatin remodeler, histone covalent modifier, and basal transcription machinery. After transcription, the mRNA is again subjected to post-transcription modification, export, localization, translation, and degradation. Each individual step can be visualized by tagging corresponding factors with different FPs. Quantitative microscopy techniques allow one to extract dynamic information as reviewed in the text.
In order to investigate mechanistic details, various factors that participate or regulate transcription can be labeled. Nuclear receptors (NR) are transcription factors that regulate gene expression in a ligand-dependent manner. Binding of agonist ligand triggers conformation changes of NR that leads to the recruitment of coactivators. Dual-color FFS has been successfully applied to study the concentration, mobility, and interactions of NR and its interaction with coactivators [36]. The transcription dynamics are measured by applying FRAP or photoactivation to the transcription site. In this way, the residence time of various factors and dynamics of RNA polymerase has been measured [43•,44]. It reveals surprisingly dynamic behavior and short binding times for most factors at the transcription site except the polymerase, which elongates the transcript. Novel photoswitching FPs will allow us to follow transcription initiation, elongation, and termination at the same time. MS2 labeled mRNA was tracked in the nucleus and showed that Brownian diffusion dictates the transport [45]. By labeling the nuclear pore complex and applying super-registration microscopy, we and others have observed mRNA going through a single nuclear pore [26,46]. Finally, the mRNA reaches cytoplasm and is translated. Fluorescent protein is commonly used as reporter for gene activity. For example, gene product tagged with fluorescent timers enables monitoring gene expression by conventional microscopy or flow cytometry [4,14].
Conclusions
We are entering a new era of designing probes. These probes have the essential features required for live imaging in cells and tissues: low autofluorescence in the emission spectrum, non-toxic excitation wavelengths amenable to intravital imaging, and timer aspects for following molecules as a function of the biological processes that govern them. The novel reagents can provide a mix-and-match smorgasbord for an increasing complexity of biological processes to investigate. For instance, illuminating with a single excitation wavelength can now provide four colors of labeled species. The future is bright for researchers searching for biological gold under this rainbow.
Acknowledgments
This work was supported by the grants from the National Institutes of Health, GM084364, GM086217, GM057071, and GM080264 (to R.H.S) and GM073913 (to V.V.V.). T.L. is supported by a Human Frontier Science Program Long-Term Fellowship.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Konig K. Multiphoton microscopy in life sciences. J Microsc. 2000;200:83–104. doi: 10.1046/j.1365-2818.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- 2.Stepanenko OV, Verkhusha VV, Kuznetsova IM, Uversky VN, Turoverov KK. Fluorescent proteins as biomarkers and biosensors: throwing color lights on molecular and cellular processes. Curr Protein Pept Sci. 2008;9:338–369. doi: 10.2174/138920308785132668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 4.Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat Methods. 2008;5:683–685. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- 5.Kredel S, Oswald F, Nienhaus K, Deuschle K, Rocker C, Wolff M, Heilker R, Nienhaus GU, Wiedenmann J. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS ONE. 2009;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu J, Zhang Z, Zheng Y, Yang J, Qin L, Lu J, Huang Z-L, Zeng S, Luo Q. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions. Biosens Bioelectron. 2009;25:234–239. doi: 10.1016/j.bios.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Morozova KS, Piatkevich KD, Gould TJ, Zhang J, Bewersdorf J, Verkhusha VV. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys J. 2010;99:L13–15. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin MZ, McKeown MR, Ng HL, Aguilera TA, Shaner NC, Campbell RE, Adams SR, Gross LA, Ma W, Alber T, et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Piatkevich KD, Malashkevich VN, Almo SC, Verkhusha VV. Engineering ESPT pathways based on structural analysis of LSSmKate red fluorescent proteins with large Stokes shift. J Am Chem Soc. 2010;132:10762–10770. doi: 10.1021/ja101974k. The paper describes a structure-based rational design strategy to develop RFPs with large Stokes shifts. The strategy was successfully applied to RFPs of the different genetic background and yielded a number of FP variants exhibited LSS fluorescence emission in a wide range of wavelengths from 560 to 640 nm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piatkevich KD, Hulit J, Subach OM, Wu B, Abdulla A, Segall JE, Verkhusha VV. Monomeric red fluorescent proteins with a large Stokes shift. Proc Natl Acad Sci USA. 2010;107:5369–5374. doi: 10.1073/pnas.0914365107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pletnev S, Subach FV, Dauter Z, Wlodawer A, Verkhusha VV. Understanding blue-to-red conversion in monomeric fluorescent timers and hydrolytic degradation of their chromophores. J Am Chem Soc. 2010;132:2243–2253. doi: 10.1021/ja908418r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subach FV, Subach OM, Gundorov IS, Morozova KS, Piatkevich KD, Cuervo AM, Verkhusha VV. Monomeric fluorescent timers that change color from blue to red report on cellular trafficking. Nat Chem Biol. 2009;5:118–126. doi: 10.1038/nchembio.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuboi T, Kitaguchi T, Karasawa S, Fukuda M, Miyawaki A. Age-dependent preferential dense-core vesicle exocytosis in neuroendocrine cells revealed by newly developed monomeric fluorescent timer protein. Mol Biol Cell. 2010;21:87–94. doi: 10.1091/mbc.E09-08-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Subach FV, Patterson GH, Renz M, Lippincott-Schwartz J, Verkhusha VV. Bright monomeric photoactivatable red fluorescent protein for two-color super-resolution sptPALM of live cells. J Am Chem Soc. 2010;132:6481–6491. doi: 10.1021/ja100906g. The authors applied structure-based rational and random mutagenesis to the TagRFP protein in order to convert it into a bright, irreversibly photoactivatable red FP, named PATagRFP. Compared to other PARFPs, it has higher brightness and photoactivation contrast, faster maturation, better pH-stability, and better photostability; additionally it lacks a green emission state observed in green-to-redPAFPs. These properties make PATagRFP an excellent protein tag for both conventional diffraction-limited microscopy and multicolor super-resolution imaging techniques. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 19.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habuchi S, Tsutsui H, Kochaniak AB, Miyawaki A, van Oijen AM. mKikGR, a monomeric photoswitchable fluorescent protein. PLoS ONE. 2008;3:e3944. doi: 10.1371/journal.pone.0003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs J, Bohme S, Oswald F, Hedde PN, Krause M, Wiedenmann J, Nienhaus GU. A photoactivatable marker protein for pulse-chase imaging with superresolution. Nat Methods. 2010;7:627–630. doi: 10.1038/nmeth.1477. [DOI] [PubMed] [Google Scholar]
- 22••.Subach FV, Zhang L, Gadella TW, Gurskaya NG, Lukyanov KA, Verkhusha VV. Red fluorescent protein with reversibly photoswitchable absorbance for photochromic FRET. Chem Biol. 2010;17:745–755. doi: 10.1016/j.chembiol.2010.05.022. Compared to the available monomeric red rsFP, the rsTagRFP protein has a substantially larger brightness and the higher contrast that is independent of the switching cycle. Moreover, the rsTagRFP photocon-version is accompanied by dramatic absorbance changes that have allowed for its application in pcFRET with EYFP as a donor. The rsTagRFP-EYFP pair demonstrated a high potential of the pcFRET imaging to study subcellular protein interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 25••.Patterson G, Davidson M, Manley S, Lippincott-Schwartz J. Superresolution imaging using single-molecule localization. Annu Rev Phys Chem. 2010;61:345–367. doi: 10.1146/annurev.physchem.012809.103444. This paper provides excellent review about the recent progress of photoactivation based super-resolution microscopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grunwald D, Singer RH. In vivo imaging of labelled endogenous [bgr]-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker SM, Buckheit RW, 3rd, Falk MM. Green-to-red photoconvertible fluorescent proteins: tracking cell and protein dynamics on standard wide-field mercury arc-based microscopes. BMC Cell Biol. 2010;11:15. doi: 10.1186/1471-2121-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chudakov DM, Chepurnykh TV, Belousov VV, Lukyanov S, Lukyanov KA. Fast and precise protein tracking using repeated reversible photoactivation. Traffic. 2006;7:1304–1310. doi: 10.1111/j.1600-0854.2006.00468.x. [DOI] [PubMed] [Google Scholar]
- 29.Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 30•.Mueller F, Mazza D, Stasevich TJ, McNally JG. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.03.002. The authors review the state of the art of the FRAP technique and its quantitative analysis. Importantly, they reconcile previous results and provide technical guidelines that yield robust interpretations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magde D, Elson E, Webb WW. Thermodynamics fluctuations in a reacting system: measurement by fluorescence correlation spectroscopy. Phys Rev Lett. 1972;29:705–708. [Google Scholar]
- 32.Kolin D, Wiseman P. Cell Biochemistry and Biophysics. Humana Press Inc; 2007. Advances in image correlation spectroscopy: measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells; pp. 141–164. [DOI] [PubMed] [Google Scholar]
- 33.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Wei LN, Muller JD. Probing protein oligomerization in living cells with fluorescence fluctuation spectroscopy. Proc Natl Acad Sci USA. 2003;100:15492–15497. doi: 10.1073/pnas.2533045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bacia K, Schwille P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat Protoc. 2007;2:2842–2856. doi: 10.1038/nprot.2007.410. [DOI] [PubMed] [Google Scholar]
- 36.Wu B, Chen Y, Müller JD. Heterospecies partition analysis reveals binding curve and stoichiometry of protein interactions in living cells. Proc Natl Acad Sci. 2010;107:4117–4122. doi: 10.1073/pnas.0905670107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Wu B, Chen Y, Müller JD. Fluorescence fluctuation spectroscopy of mCherry in living cells. Biophys J. 2009;96:2391–2404. doi: 10.1016/j.bpj.2008.12.3902. This paper demonstrates the necessity of detailed characterization of FP for proper interpretation of quantitative spectroscopic experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev. 2009;38:2876–2886. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, et al. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. This review summarizes the quantitative approaches to image gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 42.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 43•.McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. This paper demonstrates an excellent example of combining quantitative microscopy technique with fluorescent protein to reveal the dynamic behavior of transcription factors. [DOI] [PubMed] [Google Scholar]
- 44.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 47.Hell SW. Toward fluorescence nanoscopy. Nat Biotechnol. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 48.Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MGL. Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 50.Schwille P, Meyer-Almes FJ, Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]