Abstract
A number of compounds from different chemical classes are known to bind competitively to the ATP-pocket of Hsp90 and inhibit its chaperone function. The natural product geldanamycin was the first reported inhibitor of Hsp90 and since then synthetic inhibitors from purine, isoxazole and indazol-4-one chemical classes have been discovered and are currently or soon to be in clinical trials for the treatment of cancer. In spite of a similar binding mode to Hsp90, distinct biological profiles were demonstrated amongst these molecules, both in vitro and in vivo. To better understand the molecular basis for these dissimilarities, we report here the synthesis of chemical tools for three Hsp90 inhibitor classes. These agents will be useful for probing tumor-by-tumor the Hsp90 complexes isolated by specific inhibitors. Such information will lead to better understanding of tumor specific molecular markers to aid in their clinical development. It will also help to elucidate the molecular basis for the biological differences observed among Hsp90 inhibitors.
Keywords: Heat shock protein 90, PU-H71, NVP-AUY922, SNX-2112, Cancer, Docking
1. Introduction
Heat shock protein 90 (Hsp90) is a molecular chaperone with key roles in folding and maintaining the conformational integrity of its client proteins through its ATPase activity.1, 2 Because many of these proteins (i.e. Her2, Raf-1, Akt, Cdk4, Polo-1 kinase, cMet, mutant B-Raf, mutant p53, AR, ER, Bcr-Abl, HIF-1 alpha, hTERT) are associated with signaling pathways involved in cell regulation, it plays an important role in maintaining the transformed phenotype.3 As such, Hsp90 has become one of the highly pursued molecular targets in cancer therapy, with efforts in the development of Hsp90-inhibitory agents in neurodegenerative diseases and pathogenic resistance (i.e. viral, fungal, bacterial) closely following behind.3, 4 A search on esp@cenet patent database returned over 700 hits in September 2010, with several applications claiming distinct compositions of matter that interact with Hsp90.
At least five distinct Hsp90 inhibitor chemotypes are currently in clinical studies, with many others following in late-stage IND evaluation or different stages of pre-clinical evaluation.4, 5 While the interest in Hsp90 inhibitors has spread fast, knowledge on the target has however, lagged behind. It is known that while all current Hsp90 inhibitors in advanced stage (clinical or late-stage IND) inhibit Hsp90 by binding to its N-terminal site regulatory pocket, evidence suggests important distinctions in the in vitro and in vivo behavior of these agents.6–9 Among most critical differences are the kinetics of client protein modulation and in vivo pharmacodynamic profiles which both can have a determining effect on the clinical efficacy and the therapeutic window of the Hsp90-agents.6–9
In addition, with the advancement of Hsp90 inhibitors into several cancers, and potentially other diseases, it will be important to identify tumor- and disease-specific Hsp90 clients and implement these in the clinic as potential molecular markers to provide proof that the target, Hsp90, is inhibited. Identification of proper clinical biomarkers has become of utmost importance. The cost of successful anticancer drug development to the stage of approval has escalated recently to more than $800 million, and having a properly chosen pharmacodynamic disease biomarker can increase the efficacy of the process. In particular, it can be a useful indicator of drug activity and accelerate the ability to make go-no go decisions early in the clinical process.10
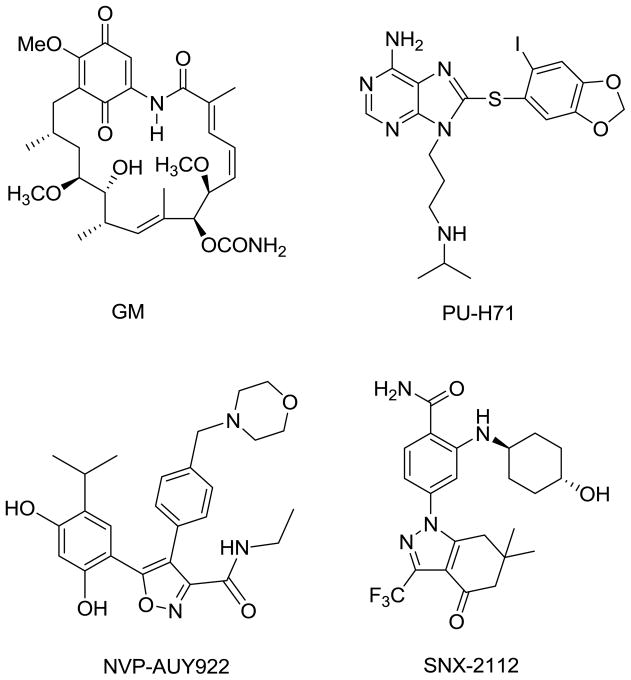
Most molecules currently known to inhibit Hsp90 function (Figure 1) mimic or adopt scaffolds based on those of geldanamycin11 (GM, ansamycin class), PU-H7112 (purine class), NVP-AUY9227 (isoxazole class) and SNX-211213 (indazol-4-one class). Geldanamycin was the first identified Hsp90 inhibitor and as such has served a central role in the study of Hsp90 biology. Originally, it was believed to inhibit Src kinase directly, but by using geldanamycin covalently bound to Affi-Gel® 10 solid support, it was later shown to bind to Hsp90 and inhibit heterocomplex formation with Src.11 Biotinylated geldanamycin was also synthesized.14
Figure 1.
Structures of Hsp90 inhibitors.
Here, we report on the design and synthesis of molecules based on purine, isoxazole and indazol-4-one chemical classes attached to Affi-Gel® 10 beads and on the synthesis of a biotinylated purine compound. These are chemical tools to investigate and understand the molecular basis for the distinct behavior of Hsp90 inhibitors. They can be also used to better understand Hsp90 tumor biology by examining bound client proteins and co-chaperones. Understanding the tumor specific clients of Hsp90 most likely to be modulated by each Hsp90 inhibitor could lead to a better choice of pharmacodynamic markers, and thus a better clinical design. Not lastly, understanding the molecular differences among these Hsp90 inhibitors could result in identifying characteristics that could lead to the design of an Hsp90 inhibitor with most favorable clinical profile.
2. Design of Hsp90 probes and precursor evaluation
The attachment of small molecules to a solid support is a very useful method to probe their target and the target’s interacting partners. Indeed, as mentioned above, geldanamycin attached to solid support enabled for the identification of Hsp90 as its target.11 Perhaps the most crucial aspects in designing such chemical probes are determining the appropriate site for attachment of the small molecule ligand, and designing an appropriate linker between the molecule and the solid support. Our strategy to design Hsp90 chemical probes entails several steps. First, in order to validate the optimal linker length and its site of attachment to the Hsp90 ligand, the linker-modified ligand was docked onto an appropriate X-ray crystal structure of Hsp90α. Second, the linker-modified ligand was evaluated in a fluorescent polarization (FP) assay that measures competitive binding to Hsp90 derived from a cancer cell extract. This assay uses Cy3b-labeled geldanamycin as the FP-optimized Hsp90 ligand.15 These steps are important to ensure that the solid-support immobilized molecules maintain a strong affinity for Hsp90. Finally, the linker-modified small molecule was attached to the solid support, and its interaction with Hsp90 was validated by incubation with an Hsp90-containing cell extract.
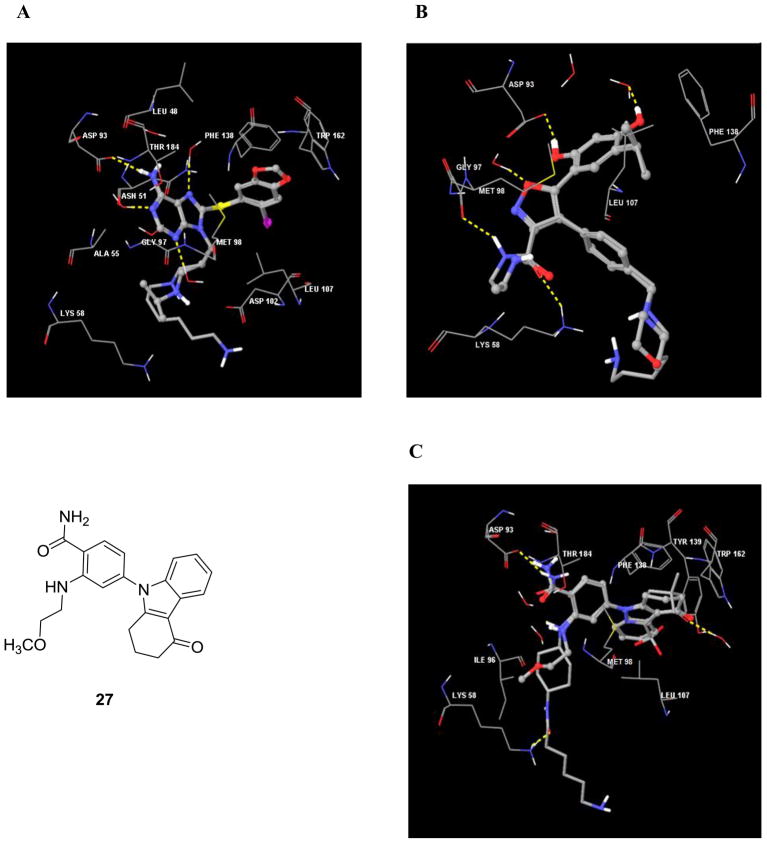
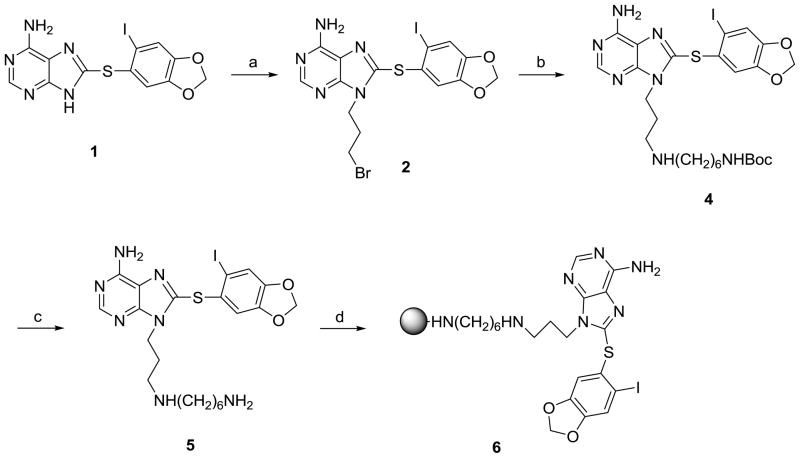
We chose Affi-Gel® 10 (BioRad) for ligand attachment. These agarose beads have an N-hydroxysuccinimide ester at the end of a 10C spacer arm, and in consequence, each linker was designed to contain a distal amine functionality. The site of linker attachment to PU-H71 was aided by the co-crystal structure of it bound to the N-terminal domain of human Hsp90α (PDB ID: 2FWZ). This structure shows that the purine’s N9 amine makes no direct contact with the protein and is directed towards solvent (Figure 2A).16 As well, a previous SAR indicated that this is an attractive site since it was previously used for the introduction of water solubilizing groups.12 Compound 5 (PU-H71-C6 linker) was designed and docked onto the Hsp90 active site (Figure 2A). All the interactions of PU-H71 were preserved, and the computer model clearly showed that the linker oriented towards the solvent exposed region. Therefore, compound 5 was synthesized as the immediate precursor for attachment to solid support (see Chemistry, Scheme 1). In the FP assay, 5 retained affinity for Hsp90 (IC50 = 19.8 nM compared to 22.4 nM for PU-H71, Table 1) which then enabled us to move forward with confidence towards the synthesis of solid support immobilized PU-H71 probe (6) by attachment to Affi-Gel® 10 (Scheme 1).
Figure 2.
A) Interactions of Hsp90α (PDB ID: 2FWZ) with PU-H71 (ball and stick model) and compound 5 (tube model). B) Interactions of Hsp90α (PDB ID: 2VCI) with NVP-AUY922 (ball and stick model) and compound 10 (tube model). C) Interactions of Hsp90α (PDB ID: 3D0B) with compound 27 (ball and stick model) and compound 20 (tube model). Hydrogen bonds are shown as dotted yellow lines and important active site amino acid residues and water molecules are represented as sticks.
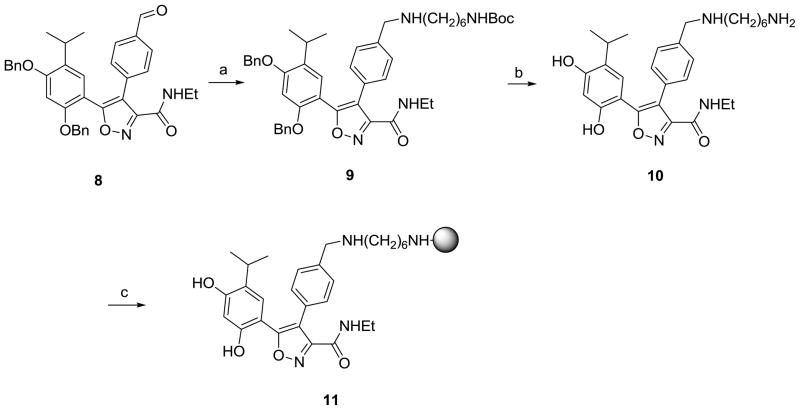
Scheme 1.
Synthesis of PU-H71 beads (6).
Reagents and conditions: (a) Cs2C03, 1,3-dibromopropane, DMF, rt; (b) NH2(CH2)6NHBoc (3), DMF, rt, 24h; (c) TFA, CH2C12, rt; (d) Affigel-10, DIEA, DMAP, DMF.
Table 1.
Binding affinity for Hsp90 from SKBr3 cellular extracts.
| Compound | IC50 (nM) |
|---|---|
| GM | 15.4 |
| PU-H71 | 22.4 |
| 5 | 19.8 |
| 7 | 67.1 |
| NVP-AUY922 | 4.1 |
| 10 | 7.0 |
| SNX-2112 | 15.1 |
| 18 | 210.1 |
| 20 | 24.7 |
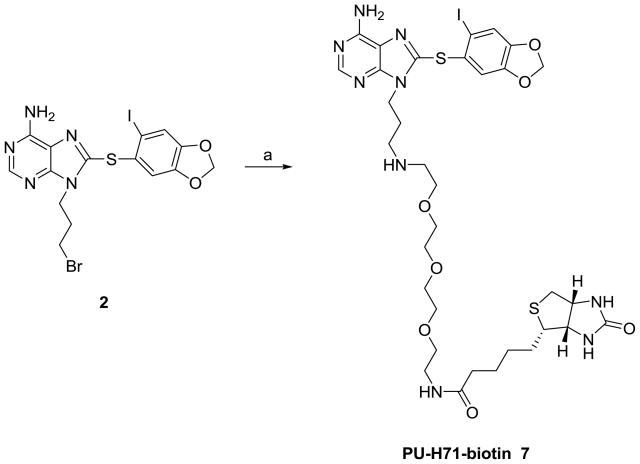
We also designed a biotinylated derivative of PU-H71. One advantage of the biotinylated agent over the solid supported agents is that they can be used to probe binding directly in cells or in vivo systems. The ligand-Hsp90 complexes can then be captured on biotin-binding avidin or streptavidin containing beads. Typically this process reduces the unspecific binding associated with chemical precipitation from cellular extracts. Alternatively, for in vivo experiments, the presence of active sites (in this case Hsp90), can be detected in specific tissues (i.e. tumor mass in cancer) by the use of a labeled-streptavidin conjugate (i.e. FITC-streptavidin). Biotinylated PU-H71 (7) was obtained by reaction of 2 with biotinyl-3,6,9-trioxaundecanediamine (EZ-Link® Amine-PEO3-Biotin) (Scheme 2). 7 retained affinity for Hsp90 (IC50 = 67.1 nM) and contains an exposed biotin capable of interacting with streptavidin for affinity purification.
Scheme 2.
Synthesis of PU-H71-biotin (7).
Reagents and conditions: (a) EZ-Link® Amine-PE03-Biotin, DMF, rt.
From the available co-crystal structure of NVP-AUY922 with Hsp90α (PDB ID: 2VCI, Figure 2B) and co-crystal structures of related 3,4-diarylpyrazoles with Hsp90α, as well as from SAR, it was evident that there was a considerable degree of tolerance for substituents at the para-position of the 4-aryl ring.7, 17–19 Because the 4-aryl substituent is largely directed towards solvent and substitution at the para-position seems to have little impact on binding affinity, we decided to attach the molecule to solid support at this position. In order to enable attachment, the morpholine group was changed to the 1,6-diaminohexyl group to give 10 as the immediate precursor for attachment to solid support. Docking 10 onto the active site (Figure 2B) shows that it maintains all of the interactions of NVP-AUY922 and that the linker orients towards the solvent exposed region. When 10 was tested in the binding assay it also retained affinity (IC50 = 7.0 nM compared to 4.1 nM for NVP-AUY922, Table 1) and was subsequently used for attachment to solid support (see Chemistry, Scheme 3).
Scheme 3.
Synthesis of NVP-AUY922 beads (11).
Reagents and conditions: (a) NH2(CH2)6NHBoc (3), NaCNBH3, AcOH, MeOH, rt; (b) BC13, CH2C12, rt; (c) Affigel-10, DIEA, DMAP, DMF.
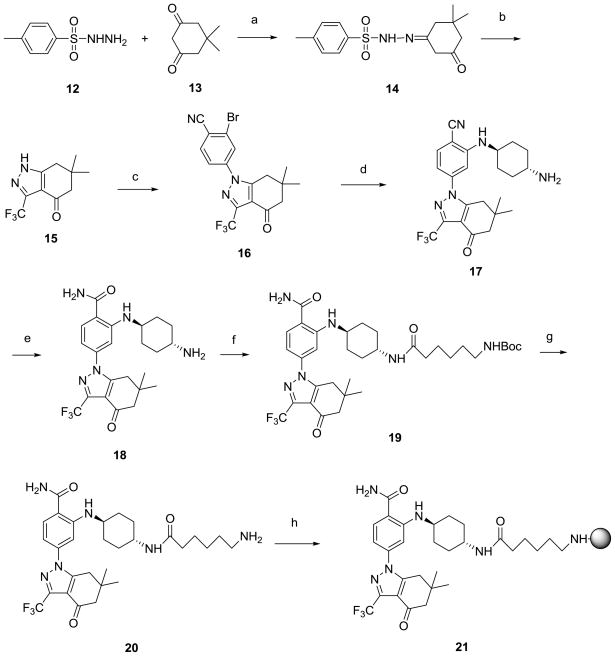
Although a co-crystal structure of SNX-2112 with Hsp90 is not publicly available, that of a related tetrahydro-4H-carbazol-4-one (27) bound to Hsp90α (PDB ID: 3D0B, Figure 2C) is.20 This, along with the reported SAR for 27 suggests linker attachment to the hydroxyl of the trans-4-aminocylohexanol substituent. Direct attachment of 6-amino-caproic acid via an ester linkage was not considered desirable because of the potential instability of such bonds in lysate mixtures due to omnipresent esterases. Therefore, the hydroxyl was substituted with amino to give the trans-1,4-diaminocylohexane derivative 18 (Scheme 4). Such a change resulted in nearly a 14-fold loss in potency as compared to SNX-2112 (Table 1). 6-(Boc-amino)caproic acid was attached to 18 and following deprotection, 20 was obtained as the immediate precursor for attachment to beads (see Chemistry, Scheme 4). Docking suggested that 20 interacts similarly to 27 (Figure 2C) and that the linker orients towards the solvent exposed region. 20 was determined to have good affinity for Hsp90 (IC50 = 24.7 nM compared to 15.1 nM for SNX-2112 and 210.1 nM for 18, Table 1) and to have regained almost all of the affinity lost by 18. The difference in activity between 18 and both 20 and SNX-2112 is well explained by our binding model, as compounds 20 (-C=O, Figure 2C) and SNX-2112 (-OH, Figure not shown) form a hydrogen bond with the side-chain amino of Lys 58. 18 contains a strongly basic amino group and is incapable of forming a hydrogen bond with Lys 58 side chain (NH2, Figure not shown). This is in good agreement with the observation of Huang et al13 that basic amines at this position are disfavored. The amide bond of 20 converts the basic amino of 18 into a non-basic amide group capable of acting as an H-bond acceptor to Lys 58, similarly to the hydroxyl of SNX-2112.
Scheme 4.
Synthesis of SNX-2112 beads (21).
Reagents and conditions: (a) p -toluene sulfonic acid, toluene, reflux, 1.5 h; (b) trifluoroacetic anhydride, Et3N, THF, 55°C, 3 h, then methanol/NaOH rt, 3 h; (c) 2-bromo-4-fluorobenzonitrile, NaH, DMF, 90°C, 5.5 h.; (d) trans-l,4-diaminocyclohexane, NaOtBu, Pd2(dba)3, DavePhos, DME, 50°C, overnight; (e) DMSO, EtOH, NaOH, H2O2, rt, 3 h.; (f) 6-(BOC-amino)caproic acid, EDCI, DMAP, rt, 2 h; (g) TFA, CH2C12, rt; (h) Affigel-10, DIEA, DMAP, DMF.
3. Chemistry
Synthesis of PU-H71 beads (6) is shown in Scheme 1 and commences with the 9-alkylation of 8-arylsulfanylpurine (1)12 with 1,3-dibromopropane to afford 2 in 35% yield. The low yield obtained in the formation of 2 can be primarily attributed to unavoidable competing 3-alkylation. Five equivalents of 1,3-dibromopropane were used to ensure complete reaction of 1 and to limit other undesirable side-reactions, such as dimerization, which may also contribute to the low yield. 2 was reacted with tert-butyl 6-aminohexylcarbamate (3) to give the Boc-protected amino purine 4 in 90% yield. Deprotection with TFA followed by reaction with Affi-Gel® 10 resulted in 6. Biotinylated PU-H71 (7) was also synthesized by reacting 2 with EZ-Link® Amine-PEO3-Biotin (Scheme 2).
Synthesis of NVP-AUY922 beads (11) from aldehyde 87 is shown in Scheme 3. 9 was obtained from the reductive amination of 8 with 3 in 75% yield with no detectable loss of the Boc group. In a single step, both the Boc and benzyl protecting groups were removed with BCl3 to give isoxazole 10 in 78% yield, which was then reacted with Affi-Gel® 10 to give 11.
Synthesis of SNX-2112 beads (21) is shown in Scheme 4, and while compounds 17 and 18 are referred to in the patent literature,21, 22 neither is adequately characterized, nor are their syntheses fully described. Therefore, we feel that it is worth describing the synthesis in detail. Tosylhydrazone 14 was obtained in 89% yield from the condensation of tosyl hydrazide (12) with dimedone (13). The one-pot conversion of 14 to tetrahydroindazolone 15 occurs following base promoted cyclocondensation of the intermediate trifluoroacyl derivative generated by treatment with trifluroacetic anhydride in 55% yield. 15 was reacted with 2-bromo-4-fluorobenzonitrile in DMF to give 16 in 91% yield. It is interesting to note the regioselectivity of this reaction as arylation occurs selectively at N1. In computational studies of indazol-4-ones similar to 15, both 1H and 2H-tautomers are known to exist in equilibrium, however, because of its higher dipole moment the 1H tautomer is favored in polar solvents.23 The amination of 16 with trans-1,4-diaminocyclohexane was accomplished under Buchwald conditions24 using tris(dibenzylideneacetone)dipalladium [Pd2(dba)3] and 2-dicyclohexylphosphino-2′-(N,N-dimethylamino)biphenyl (DavePhos) to give nitrile 17 (24%) along with amide 18 (17%) for a combined yield of 41%. Following complete hydrolysis of 17, 18 was coupled to 6-(Boc-amino)caproic acid with EDCI/DMAP to give 19 in 91% yield. Following deprotection, 20 was obtained which was then reacted with Affi-Gel® 10 to give 21.
Several methods were employed to measure the progress of the reactions for the synthesis of the final probes. UV monitoring of the liquid was used by measuring a decrease in λmax for each compound. In general, it was observed that that there was no further decrease in the λmax after 1.5 h, indicating completion of the reaction. TLC was employed as a crude measure of the progress of the reaction whereas LC-MS monitoring of the liquid was used to confirm complete reaction. While on TLC the spot would not disappear since excess compound was used (1.2 eq.), a clear decrease in intensity indicated progress of the reaction.
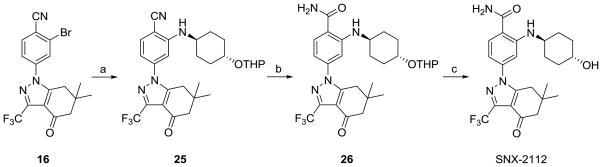
The synthesis and full characterization of the Hsp90 inhibitors PU-H7112 and NVP-AUY9227 have been reported elsewhere. SNX-2112 had previously been mentioned in the patent literature,21, 22 and only recently has it been fully characterized and its synthesis adequately described.13 At the time this research project began specific details on its synthesis were lacking. Additionally, we had difficulty reproducing the amination of 16 with trans-4-aminocyclohexanol under conditions reported for similar compounds [Pd(OAc)2, DPPF, NaOtBu, toluene, 120°C, microwave]. In our hands, only trace amounts of product were detected at best. Changing catalyst to PdCl2, Pd(PPh3)4 or Pd2(dba)3 or solvent to DMF or 1,2-dimethoxyethane (DME) or base to K3PO4 did not result in any improvement. Therefore, we modified this step and were able to couple 16 to trans-4-aminocyclohexanol tetrahydropyranyl ether (24) under Buchwald conditions24 using Pd2(dba)3 and DavePhos in DME to give nitrile 25 (28%) along with amide 26 (17%) for a combined yield of 45% (Scheme 5). These were the conditions used to couple 16 to trans-1,4-diaminocyclohexane, and similarly some of 25 was hydrolysed to 26 during the course of the reaction. Because for our purpose it was unnecessary, we did not optimize this reaction for 25. We surmised that a major hindrance to the reaction was the low solubility of trans-4-aminocyclohexanol in toluene and that using the THP protected alcohol 24 at the very least increased solubility. SNX-2112 was obtained and fully characterized (1H, 13C-NMR, MS) following removal of the THP group from 26.
Scheme 5.
Synthesis of SNX-2112.
Reagents and conditions: (a) O-THP-trans-cylohexanolamine (24), NaOtBu, Pd2(dba)3, DavePhos, DME, 60°C, 3.5 h; (b) DMSO, EtOH, 5N NaOH, H2O2, rt, 4 h; (c) PPTS, EtOH, 65°C, 17 h.
4. Biological evaluation
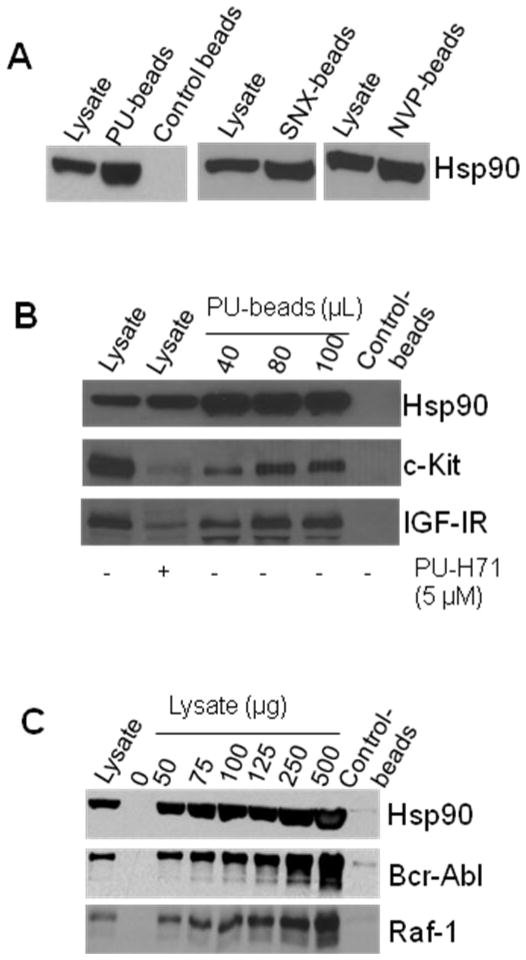
Next, we investigated whether the synthesized beads retained interaction with tumor Hsp90. Agarose beads covalently attached to either of PU-H71, NVP-AUY922, SNX-2112 or 2-methoxyethylamine (PU-, NVP-, SNX-, control-beads, respectively), were incubated with K562 chronic myeloid leukemia (CML) or MDA-MB-468 breast cancer cell extracts. As seen in Figure 3A, the Hsp90 inhibitor, but not the control-beads, efficiently isolated Hsp90 in the cancer cell lysates. Control beads contain an Hsp90 inactive chemical (2-methoxyethylamine) conjugated to Affi-Gel® 10 (see Experimental) providing an experimental control for potential unspecific binding of the solid-support to proteins in cell extracts.
Figure 3.
A) Hsp90 in K562 extracts (250 μg) was isolated by precipitation with PU-, SNX- and NVP-beads or Control-beads (80 μL). Control beads contain 2-methoxyethylamine, an Hsp90-inert molecule. Proteins in pull-downs were analyzed by Western blot. B) In MDA-MB-468 cell extracts (300 μg), PU-beads isolate Hsp90 in complex with its onco-client proteins, c-Kit and IGF-IR. To evaluate the effect of PU-H71 on the steady-state levels of Hsp90 onco-client proteins, cells were treated for 24 h with PU-H71 (5 μM). C) In K562 cell extracts, PU-beads (40 μL) isolate Hsp90 in complex with the Raf-1 and Bcr-Abl onco-proteins. Lysate = endogenous protein content; PU- and Control-beads indicate proteins isolated on the particular beads. The data are consistent with those obtained from multiple repeat experiments (n ≥ 2).
Further, to probe the ability of these chemical tools to isolate genuine Hsp90 client proteins in tumor cells, we incubated PU-H71 attached to solid support (6) with cancer cell extracts. We were able to demonstrate dose-dependent isolation of Hsp90/c-Kit and Hsp90/IGF-IR complexes in MDA-MB-468 cells (Figure 3B) and of Hsp90/Bcr-Abl and Hsp90/Raf-1 complexes in K562 cells (Figure 3C). These are Hsp90-dependent onco-proteins with important roles in driving the transformed phenotype in triple-negative breast cancers and CML, respectively.3, 25, 26 In accord with an Hsp90 mediated regulation of c-Kit and IGF-IR, treatment of MDA-MB-468 cells with PU-H71 led to a reduction in the steady-state levels of these proteins (Figure 3B, compare Lysate, - and + PU-H71). Using the PU-beads (6), we were recently able to isolate and identify novel Hsp90 clients, such as the transcriptional repressor BCL-6 in diffuse large B-cell lymphoma27 and JAK2 in mutant JAK2 driven myeloproliferative disorders.28 We were also able to identify Hsp90 onco-clients specific to triple-negative breast cancer.9 In addition to shedding light on the mechanisms of action of Hsp90 in these tumors, the identified proteins are important tumor-specific onco-clients and will be introduced as biomarkers in monitoring the clinical efficacy of PU-H71 and Hsp90 inhibitors in these cancers during clinical studies.
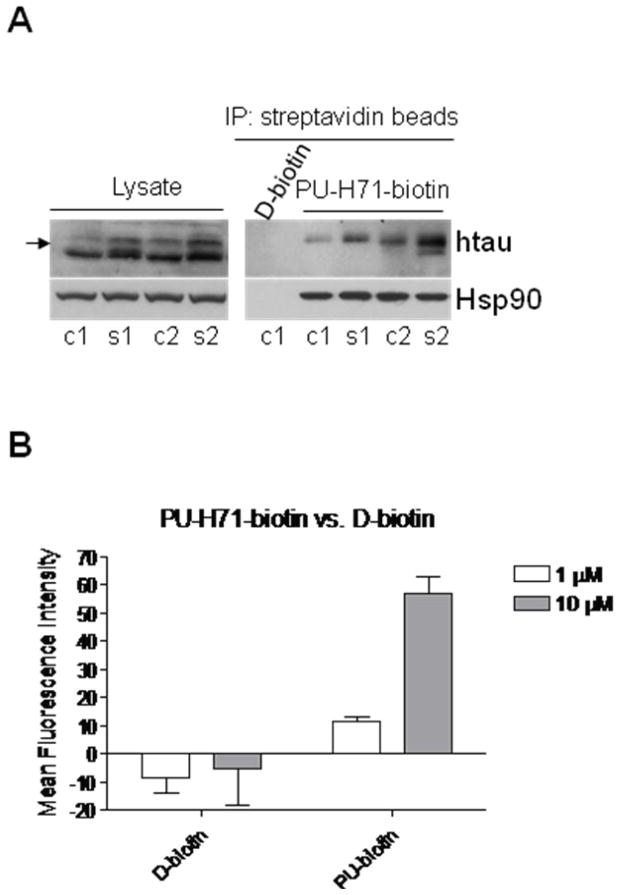
Similar experiments were possible with PU-H71-biotin (7), which selectively isolated Hsp90 in complex with mutant tau species in brain homogenates obtained from Alzheimer’s disease transgenic mice (Figure 4A). The protein tau when of aberrant activity, such as in Alzheimer’s disease and in a cluster of tauopathies termed “frontotemporal dementia and parkinsonism linked to chromosome 17”, is regulated by Hsp90.29–31
Figure 4.
A) Hsp90-containing protein complexes from the brains of JNPL3 mice, an Alzheimer’s disease transgenic mouse model, isolated through chemical precipitation with beads containing a streptavidin-immobilized PU-H71-biotin construct or control streptavidin-immobilized D-biotin. Aberrant tau species are indicated by arrow. c1, c2 and s1, s2, cortical and subcortical brain homogenates, respectively, extracted from 6-month-old female JNPL3 mice (Right). Western blot analysis of brain lysate protein content (Left). B) Cell surface Hsp90 in MV4-11 leukemia cells as detected by PU-H71-biotin. The data are consistent with those obtained from multiple repeat experiments (n ≥ 2).
It is important to note that previous attempts to isolate Hsp90/client protein complexes using a solid-support immobilized GM were of little success.32 In that case, the proteins bound to Hsp90 were washed away during the preparative steps. To prevent the loss of Hsp90-interacting proteins, the authors had to subject the cancer cell extracts to cross-linking with DSP, a homobifunctional amino-reactive DTT-reversible cross-linker, suggesting that unlike PU-H71, GM is unable to stabilize Hsp90/client protein interactions. We observed a similar profile when using beads with GM directly covalently attached to the Affi-Gel® 10 resin (Chiosis et al, unpublished results). Crystallographic and biochemical investigations suggest that GM preferentially interacts with Hsp90 in an apo, open-conformation, that is unfavorable for certain client protein binding33–35 providing a potential explanation for the limited ability of GM-beads to capture Hsp90/client protein complexes. It is currently unknown what Hsp90 conformations are preferred by the other Hsp90 chemotypes, but with the NVP- and SNX-beads also available, as reported here, similar evaluations are now possible, leading to a better understanding of the interaction of these agents with Hsp90, and of the biological significance of these interactions.
In another application of the chemical tools designed here, we show that PU-H71-biotin (7) can also be used to specifically detect Hsp90 when expressed on the cell surface (Figure 4B). Hsp90, which is mainly a cytosolic protein, has been reported in certain cases to translocate to the cell surface. In breast cancer for example, membrane Hsp90 is involved in aiding cancer cell invasion.36 Specific detection of the membrane Hsp90 in live cells is possible by the use of PU-H71-biotin (7) because, while the biotin conjugated Hsp90 inhibitor may potentially enter the cell, the streptavidin conjugate used to detect the biotin, is cell impermeable. Figure 4B shows that PU-H71-biotin but not D-biotin can detect Hsp90 expression on the surface of leukemia cells.
5. Conclusions
In conclusion, we have prepared useful chemical tools based on three different Hsp90 inhibitors, each of a different chemotype. These were prepared either by attachment onto solid support, such as PU-H71 (purine), NVP-AUY922 (isoxazole) and SNX-2112 (indazol-4-one)-beads, or by biotinylation (PU-H71-biotin). The utility of these probes was demonstrated by their ability to efficiently isolate Hsp90 and, in the case of PU-H71 beads (6), isolate Hsp90 onco-protein containing complexes from cancer cell extracts. Available co-crystal structures and SAR were utilized in their design, and docking to the appropriate X-ray crystal structure of Hsp90α used to validate the site of attachment of the linker. These are important chemical tools in efforts towards better understanding Hsp90 biology and towards designing Hsp90 inhibitors with most favorable clinical profile.
6. Experimental
6.1. General
1H and 13C NMR spectra were recorded on a Bruker 500 MHz instrument. Chemical shifts were reported in δ values in ppm downfield from TMS as the internal standard. 1H data were reported as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, br = broad, m = multiplet), coupling constant (Hz), integration. 13C chemical shifts were reported in δ values in ppm downfield from TMS as the internal standard. Low resolution mass spectra were obtained on a Waters Acquity Ultra Performance LC with electrospray ionization and SQ detector. High-performance liquid chromatography analyses were performed on a Waters Autopurification system with PDA, MicroMass ZQ and ELSD detector and a reversed phase column (Waters X-Bridge C18, 4.6 × 150 mm, 5 μm) using a gradient of (a) H2O + 0.1% TFA and (b) CH3CN + 0.1% TFA, 5 to 95% b over 10 minutes at 1.2 mL/min. Column chromatography was performed using 230–400 mesh silica gel (EMD). All reactions were performed under argon protection. Affi-Gel® 10 beads were purchased from Bio-Rad (Hercules, CA). EZ-Link® Amine-PEO3-Biotin was purchased from Pierce (Rockford, Il). PU-H7112 and NVP-AUY9227 were synthesized according to previously published methods. GM was purchased from Aldrich.
6.2. Synthesis
6.2.1. 9-(3-Bromopropyl)-8-(6-iodobenzo[d][1,3]dioxol-5-ylthio)-9H-purin-6-amine (2)
112 (0.500 g, 1.21 mmol) was dissolved in DMF (15 mL). Cs2CO3 (0.434 g, 1.33 mmol) and 1,3-dibromopropane (1.22 g, 0.617 mL, 6.05 mmol) were added and the mixture was stirred at rt for 45 minutes. Then additional Cs2CO3 (0.079 g, 0.242 mmol) was added and the mixture was stirred for 45 minutes. Solvent was removed under reduced pressure and the resulting residue was chromatographed (CH2Cl2:MeOH:AcOH, 120:1:0.5 to 80:1:0.5) to give 0.226 g (35%) of 2 as a white solid. 1H NMR (CDCl3/MeOH-d4) δ 8.24 (s, 1H), 7.38 (s, 1H), 7.03 (s, 1H), 6.05 (s, 2H), 4.37 (t, J = 7.1 Hz, 2H), 3.45 (t, J = 6.6 Hz, 2H), 2.41 (m, 2H); MS (ESI): m/z 534.0/536.0 [M+H]+.
6.2.2. tert-Butyl 6-aminohexylcarbamate (3)37
1,6-diaminohexane (10 g, 0.086 mol) and Et3N (13.05 g, 18.13 mL, 0.129 mol) were suspended in CH2Cl2 (300 mL). A solution of di-tert-butyl dicarbonate (9.39 g, 0.043 mol) in CH2Cl2 (100 mL) was added dropwise over 90 minutes at rt and stirring continued for 18 h. The reaction mixture was added to a seperatory funnel and washed with water (100 mL), brine (100 mL), dried over Na2SO4 and concentrated under reduced pressure. The resulting residue was chromatographed [CH2Cl2:MeOH-NH3 (7N), 70:1 to 20:1] to give 7.1 g (76%) of 3. 1H NMR (CDCl3) δ 4.50 (br s, 1H), 3.11 (br s, 2H), 2.68 (t, J = 6.6 Hz, 2H), 1.44 (s, 13H), 1.33 (s, 4H); MS (ESI): m/z 217.2 [M+H]+.
6.2.3. tert-Butyl 6-(3-(6-amino-8-(6-iodobenzo[d][1,3]dioxol-5-ylthio)-9H-purin-9-yl)propylamino)hexylcarbamate (4)
2 (0.226 g, 0.423 mmol) and 3 (0.915 g, 4.23 mmol) in DMF (7 mL) was stirred at rt for 24 h. The reaction mixture was concentrated and the residue chromatographed [CHCl3:MeOH:MeOH-NH3 (7N), 100:7:3] to give 0.255 g (90%) of 4. 1H NMR (CDCl3) δ 8.32 (s, 1H), 7.31 (s, 1H), 6.89 (s, 1H), 5.99 (s, 2H), 5.55 (br s, 2H), 4.57 (br s, 1H), 4.30 (t, J = 7.0 Hz, 2H), 3.10 (m, 2H), 2.58 (t, J = 6.7 Hz, 2H), 2.52 (t, J = 7.2 Hz, 2H), 1.99 (m, 2H), 1.44 (s, 13H), 1.30 (s, 4H); 13C NMR (125 MHz, CDCl3) δ 156.0, 154.7, 153.0, 151.6, 149.2, 149.0, 146.3, 127.9, 120.1, 119.2, 112.4, 102.3, 91.3, 79.0, 49.8, 46.5, 41.8, 40.5, 31.4, 29.98, 29.95, 28.4, 27.0, 26.7; HRMS (ESI) m/z [M+H]+ calcd. for C26H37IN7O4S, 670.1673; found 670.1670; HPLC: tR = 7.02 min.
6.2.4. N1-(3-(6-Amino-8-(6-iodobenzo[d][1,3]dioxol-5-ylthio)-9H-purin-9-yl)propyl)hexane-1,6-diamine (5)
4 (0.310 g, 0.463 mmol) was dissolved in 15 mL of CH2Cl2:TFA (4:1) and the solution was stirred at rt for 45 min. Solvent was removed under reduced pressure and the residue chromatographed [CH2Cl2:MeOH-NH3 (7N), 20:1 to 10:1] to give 0.37 g of a white solid. This was dissolved in water (45 mL) and solid Na2CO3 added until pH~12. This was extracted with CH2Cl2 (4 × 50 mL) and the combined organic layers were washed with water (50 mL), dried over Na2SO4, filtered and concentrated under reduced pressure to give 0.200 g (76%) of 5. 1H NMR (CDCl3) δ 8.33 (s, 1H), 7.31 (s, 1H), 6.89 (s, 1H), 5.99 (s, 2H), 5.52 (br s, 2H), 4.30 (t, J = 6.3 Hz, 2H), 2.68 (t, J = 7.0 Hz, 2H), 2.59 (t, J = 6.3 Hz, 2H), 2.53 (t, J = 7.1 Hz, 2H), 1.99 (m, 2H), 1.44 (s, 4H), 1.28 (s, 4H); 13C NMR (125 MHz, CDCl3/MeOH-d4) δ 154.5, 152.6, 151.5, 150.0, 149.6, 147.7, 125.9, 119.7, 119.6, 113.9, 102.8, 94.2, 49.7, 46.2, 41.61, 41.59, 32.9, 29.7, 29.5, 27.3, 26.9; HRMS (ESI) m/z [M+H]+ calcd. for C21H29IN7O2S, 570.1148; found 570.1124; HPLC: tR = 5.68 min.
6.2.5. PU-H71-Affi-Gel 10 beads (6)
4 (0.301 g, 0.45 mmol) was dissolved in 15 mL of CH2Cl2:TFA (4:1) and the solution was stirred at rt for 45 min. Solvent was removed under reduced pressure and the residue dried under high vacuum overnight. This was dissolved in DMF (12 mL) and added to 25 mL of Affi-Gel 10 beads (prewashed, 3 × 50 mL DMF) in a solid phase peptide synthesis vessel. 225 μL of N,N-diisopropylethylamine and several crystals of DMAP were added and this was shaken at rt for 2.5 h. Then 2-methoxyethylamine (0.085 g, 97 μl, 1.13 mmol) was added and shaking was continued for 30 minutes. Then the solvent was removed and the beads washed for 10 minutes each time with CH2Cl2:Et3N (9:1, 4 × 50 mL), DMF (3 × 50 mL), Felts buffer (3 × 50 mL) and i-PrOH (3 × 50 mL). The beads 6 were stored in i-PrOH (beads: i-PrOH (1:2), v/v) at −80°C.
6.2.6. PU-H71-biotin (7)
2 (4.2 mg, 0.0086 mmol) and EZ-Link® Amine-PEO3-Biotin (5.4 mg, 0.0129 mmol) in DMF (0.2 mL) was stirred at rt for 24 h. The reaction mixture was concentrated and the residue chromatographed [CHCl3:MeOH-NH3 (7N), 5:1] to give 1.1 mg (16%) of 7. 1H NMR (CDCl3) δ 8.30 (s, 1H), 8.10 (s, 1H), 7.31 (s, 1H), 6.87 (s, 1H), 6.73 (br s, 1H), 6.36 (br s, 1H), 6.16 (br s, 2H), 6.00 (s, 2H), 4.52 (m, 1H), 4.28–4.37 (m, 3H), 3.58–3.77 (m, 10H), 3.55 (m, 2H), 3.43 (m, 2H), 3.16 (m, 1H), 2.92 (m, 1H), 2.80 (m, 2H), 2.72 (m, 1H), 2.66 (m, 2H), 2.17 (t, J = 7.0 Hz, 2H), 2.04 (m, 2H), 1.35–1.80 (m, 6H); MS (ESI): m/z 872.2 [M+H]+.
6.2.7. tert-Butyl 6-(4-(5-(2,4-bis(benzyloxy)-5-isopropylphenyl)-3-(ethylcarbamoyl)isoxazol-4-yl)benzylamino)hexylcarbamate (9)
AcOH (0.26 g, 0.25 mL, 4.35 mmol) was added to a mixture of 87 (0.5 g, 0.87 mmol), 3 (0.56 g, 2.61 mmol), NaCNBH3 (0.11 g, 1.74 mmol), CH2Cl2 (21 mL) and 3 Å molecular sieves (3 g). The reaction mixture was stirred for 1 h at rt. It was then concentrated under reduced pressure and chromatographed [CH2Cl2:MeOH-NH3 (7N), 100:1 to 60:1] to give 0.50 g (75%) of 9. 1H NMR (CDCl3) δ 7.19–7.40 (m, 12H), 7.12–7.15 (m, 2H), 7.08 (s, 1H), 6.45 (s, 1H), 4.97 (s, 2H), 4.81 (s, 2H), 3.75 (s, 2H), 3.22 (m, 2H), 3.10 (m, 3H), 2.60 (t, J = 7.1 Hz, 2H), 1.41–1.52 (m, 13H), 1.28–1.35 (m, 4H), 1.21 (t, J = 7.2 Hz, 3H), 1.04 (d, J = 6.9 Hz, 6H); MS (ESI): m/z 775.3 [M+H]+.
6.2.8. 4-(4-((6-Aminohexylamino)methyl)phenyl)-5-(2,4-dihydroxy-5-isopropylphenyl)-N-ethylisoxazole-3-carboxamide (10)
To a solution of 9 (0.5 g, 0.646 mmol) in CH2Cl2 (20 mL) was added a solution of BCl3 (1.8 mL, 1.87 mmol, 1.0 M in CH2Cl2) and this was stirred at rt for 10 h. Saturated NaHCO3 was added and CH2Cl2 was evaporated under reduced pressure. The water was carefully decanted and the remaining yellow precipitate was washed a few times with EtOAc and CH2Cl2 to give 0.248 g (78%) of 10. 1H NMR (CDCl3/MeOH-d4) δ 7.32 (d, J = 8.1 Hz, 2H), 7.24 (d, J = 8.1 Hz, 2H), 6.94 (s, 1H), 6.25 (s, 1H), 3.74, (s, 2H), 3.41 (q, J = 7.3 Hz, 2H), 3.08 (m, 1H), 2.65 (t, J = 7.1 Hz, 2H), 2.60 (t, J = 7.1 Hz, 2H), 1.40–1.56 (m, 4H), 1.28–1.35 (m, 4H), 1.21 (t, J = 7.3 Hz, 3H), 1.01 (d, J = 6.9 Hz, 6H); 13C NMR (125 MHz, CDCl3/MeOH-d4) δ 168.4, 161.6, 158.4, 157.6, 155.2, 139.0, 130.5, 129.5, 128.71, 128.69, 127.6, 116.0, 105.9, 103.6, 53.7, 49.2, 41.8, 35.0, 32.7, 29.8, 27.6, 27.2, 26.4, 22.8, 14.5; HRMS (ESI) m/z [M+H]+ calcd. for C28H39N4O4, 495.2971; found 495.2986; HPLC: tR = 6.57 min.
6.2.9. NVP-AUY922-Affi-Gel 10 beads (11)
10 (46.4 mg, 0.094 mmol) was dissolved in DMF (2 mL) and added to 5 mL of Affi-Gel 10 beads (prewashed, 3 × 8 mL DMF) in a solid phase peptide synthesis vessel. 45 μl of N,N-diisopropylethylamine and several crystals of DMAP were added and this was shaken at rt for 2.5 h. Then 2-methoxyethylamine (17.7 mg, 21 μl, 0.235 mmol) was added and shaking was continued for 30 minutes. Then the solvent was removed and the beads washed for 10 minutes each time with CH2Cl2 (3 × 8 mL), DMF (3 × 8 mL), Felts buffer (3 × 8 mL) and i-PrOH (3 × 8 mL). The beads 11 were stored in i-PrOH (beads: i-PrOH, (1:2), v/v) at −80°C.
6.2.10. N′-(3,3-Dimethyl-5-oxocyclohexylidene)-4-methylbenzenesulfonohydrazide (14)38
10.00 g (71.4 mmol) of dimedone (13), 13.8 g (74.2 mmol) of tosyl hydrazide (12) and p-toluene sulfonic acid (0.140 g, 0.736 mmol) were suspended in toluene (600 mL) and this was refluxed with stirring for 1.5 h. While still hot, the reaction mixture was filtered and the solid was washed with toluene (4 × 100 mL), ice-cold ethyl acetate (2 × 200 mL) and hexane (2 × 200 mL) and dried to give 19.58 g (89%) of 14 as a solid. TLC (100% EtOAc) Rf = 0.23; 1H NMR (DMSO-d6) δ 9.76 (s, 1H), 8.65 (br s, 1H), 7.69 (d, J = 8.2 Hz, 2H), 7.41 (d, J = 8.1 Hz, 2H), 5.05 (s, 1H), 2.39 (s, 3H), 2.07 (s, 2H), 1.92 (s, 2H), 0.90 (s, 6H); MS (ESI): m/z 309.0 [M+H]+.
6.2.11. 6,6-Dimethyl-3-(trifluoromethyl)-6,7-dihydro-1H-indazol-4(5H)-one (15)
To 5.0 g (16.2 mmol) of 14 in THF (90 mL) and Et3N (30 mL) was added trifluoroacetic anhydride (3.4 g, 2.25 mL, 16.2 mmol) in one portion. The resulting red solution was heated at 55°C for 3 h. After cooling to rt, methanol (8 mL) and 1M NaOH (8 mL) were added and the solution was stirred for 3 h at rt. The reaction mixture was diluted with 25 mL of saturated NH4Cl, poured into a seperatory funnel and extracted with EtOAc (3 × 50 mL). The combined organic layers were washed with brine (3 × 50 mL), dried over Na2SO4 and concentrated under reduced pressure to give a red oily residue which was chromatographed (hexane:EtOAc, 80:20 to 60:40) to give 2.08 g (55%) of 15 as an orange solid. TLC (hexane:EtOAc, 6:4) Rf = 0.37; 1H NMR (CDCl3) δ 2.80 (s, 2H), 2.46 (s, 2H), 1.16 (s, 6H); MS (ESI): m/z 231.0 [M-H]−.
6.2.12. 2-Bromo-4-(6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)benzonitrile (16)
To a mixture of 15 (0.100 g, 0.43 mmol) and NaH (15.5 mg, 0.65 mmol) in DMF (8 mL) was added 2-bromo-4-fluorobenzonitrile (86 mg, 0.43 mmol) and heated at 90°C for 5 h. The reaction mixture was concentrated under reduced pressure and the residue chromatographed (hexane:EtOAc, 10:1 to 10:2) to give 0.162 g (91%) of 16 as a white solid. 1H NMR (CDCl3) δ 7.97 (d, J = 2.1 Hz, 1H), 7.85 (d, J = 8.4 Hz, 1H), 7.63 (dd, J = 8.4, 2.1 Hz, 1H), 2.89 (s, 2H), 2.51 (s, 2H), 1.16 (s, 6H); MS (ESI): m/z 410.0/412.0 [M-H]−.
6.2.13. 2-(trans-4-Aminocyclohexylamino)-4-(6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)benzonitrile (17)
A mixture of 16 (0.200 g, 0.485 mmol), NaOtBu (93.3 mg, 0.9704 mmol), Pd2(dba)3 (88.8 mg, 0.097 mmol) and DavePhos (38 mg, 0.097 mmol) in 1,2-dimethoxyethane (15 mL) was degassed and flushed with argon several times. trans-1,4-Diaminocyclohexane (0.166 g, 1.456 mmol) was added and the flask was again degassed and flushed with argon before heating the reaction mixture at 50°C overnight. The reaction mixture was concentrated under reduced pressure and the residue purified by preparatory TLC (CH2Cl2:MeOH-NH3 (7N), 10:1) to give 52.5 mg (24%) of 17. Additionally, 38.5 mg (17%) of amide 18 was isolated for a total yield of 41%. 1H NMR (CDCl3) δ 7.51 (d, J = 8.3 Hz, 1H), 6.81 (d, J = 1.8 Hz, 1H), 6.70 (dd, J = 8.3, 1.8 Hz, 1H), 4.64 (d, J = 7.6 Hz, 1H), 3.38 (m, 1H), 2.84 (s, 2H), 2.81 (m, 1H), 2.49 (s, 2H), 2.15 (d, J = 11.2 Hz, 2H), 1.99 (d, J = 11.0 Hz, 2H), 1.25–1.37 (m, 4H), 1.14 (s, 6H); MS (ESI): m/z 446.3 [M+H]+.
6.2.14. 2-(trans-4-Aminocyclohexylamino)-4-(6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)benzamide (18)
A solution of 17 (80 mg, 0.18 mmol) in DMSO (147 μl), EtOH (590 μl), 5N NaOH (75 μl) and H2O2 (88 μl) was stirred at rt for 3 h. The reaction mixture was concentrated under reduced pressure and the residue purified by preparatory TLC [CH2Cl2:MeOH-NH3 (7N), 10:1] to give 64.3 mg (78%) of 18. 1H NMR (CDCl3) δ 8.06 (d, J = 7.5 Hz, 1H), 7.49 (d, J = 8.4 Hz, 1H), 6.74 (d, J = 1.9 Hz, 1H), 6.62 (dd, J = 8.4, 2.0 Hz, 1H), 5.60 (br s, 2H), 3.29 (m, 1H), 2.85 (s, 2H), 2.77 (m, 1H), 2.49 (s, 2H), 2.13 (d, J = 11.9 Hz, 2H), 1.95 (d, J = 11.8 Hz, 2H), l.20–1.42 (m, 4H), 1.14 (s, 6H); MS (ESI): m/z 464.4 [M+H]+; HPLC: tR = 7.05 min.
6.2.15. tert-Butyl 6-(trans-4-(2-carbamoyl-5-(6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)phenylamino)cyclohexylamino)-6-oxohexylcarbamate (19)
To a mixture of 18 (30 mg, 0.0647 mmol) in CH2Cl2 (1 ml) was added 6-(Boc-amino)caproic acid (29.9 mg, 0.1294 mmol), EDCI (24.8 mg, 0.1294 mmol) and DMAP (0.8 mg, 0.00647 mmol). The reaction mixture was stirred at rt for 2 h then concentrated under reduced pressure and the residue purified by preparatory TLC [hexane:CH2Cl2:EtOAc:MeOH-NH3 (7N), 2:2:1:0.5] to give 40 mg (91%) of 19. 1H NMR (CDCl3/MeOH-d4) δ 7.63 (d, J = 8.4 Hz, 1H), 6.75 (d, J = 1.7 Hz, 1H), 6.61 (dd, J = 8.4, 2.0 Hz, 1H), 3.75 (m, 1H), 3.31 (m, 1H), 3.06 (t, J = 7.0 Hz, 2H), 2.88 (s, 2H), 2.50 (s, 2H), 2.15 (m, 4H), 2.03 (d, J = 11.5 Hz, 2H), 1.62 (m, 2H), l.25–1.50 (m, 17H), 1.14 (s, 6H); 13C NMR (125 MHz, CDCl3/MeOH-d4) δ 191.5, 174.1, 172.3, 157.2, 151.5, 150.3, 141.5, 140.6 (q, J = 39.4 Hz), 130.8, 120.7 (q, J = 268.0 Hz), 116.2, 114.2, 109.5, 107.3, 79.5, 52.5, 50.7, 48.0, 40.4, 37.3, 36.4, 36.0, 31.6, 31.3, 29.6, 28.5, 28.3, 25.7, 25.4; HRMS (ESI) m/z [M+Na]+ calcd. for C34H47F3N6O5Na, 699.3458; found 699.3472; HPLC: tR = 9.10 min.
6.2.16. 2-(trans-4-(6-Aminohexanamido)cyclohexylamino)-4-(6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)benzamide (20)
19 (33 mg, 0.049 mmol) was dissolved in 1 mL of CH2Cl2:TFA (4:1) and the solution was stirred at rt for 45 min. Solvent was removed under reduced pressure and the residue purified by preparatory TLC [CH2Cl2:MeOH-NH3 (7N), 6:1] to give 24 mg (86%) of 20. 1H NMR (CDCl3/MeOH-d4) δ 7.69 (d, J = 8.4 Hz, 1H), 6.78 (d, J = 1.9 Hz, 1H), 6.64 (dd, J = 8.4, 1.9 Hz, 1H), 3.74 (m, 1H), 3.36 (m, 1H), 2.92 (t, J = 7.5 Hz, 2H), 2.91 (s, 2H), 2.51 (s, 2H), 2.23 (t, J = 7.3 Hz, 2H), 2.18 (d, J = 10.2 Hz, 2H), 2.00 (d, J = 9.1 Hz, 2H), 1.61–1.75 (m, 4H), l.34–1.50 (m, 6H), 1.15 (s, 6H); 13C NMR (125 MHz, MeOH-d4) δ 191.2, 173.6, 172.2, 151.8, 149.7, 141.2, 139.6 (q, J = 39.5 Hz), 130.3, 120.5 (q, J = 267.5 Hz), 115.5, 114.1, 109.0, 106.8, 51.6, 50.0, 47.8, 39.0, 36.3, 35.2, 35.1, 31.0, 30.5, 26.8, 26.7, 25.4, 24.8; HRMS (ESI) m/z [M+H]+ calcd. for C29H40F3N6O3, 577.3114; found 577.3126; HPLC: tR = 7.23 min.
6.2.17. SNX-2112-Affi-Gel 10 beads (21)
19 (67 mg, 0.0992 mmol) was dissolved in 3.5 mL of CH2Cl2:TFA (4:1) and the solution was stirred at rt for 20 min. Solvent was removed under reduced pressure and the residue dried under high vacuum for two hours. This was dissolved in DMF (2 mL) and added to 5 mL of Affi-Gel 10 beads (prewashed, 3 × 8 mL DMF) in a solid phase peptide synthesis vessel. 45 μl of N,N-diisopropylethylamine and several crystals of DMAP were added and this was shaken at rt for 2.5 h. Then 2-methoxyethylamine (18.6 mg, 22 μl, 0.248 mmol) was added and shaking was continued for 30 minutes. Then the solvent was removed and the beads washed for 10 minutes each time with CH2Cl2 (3 × 8 mL), DMF (3 × 8 mL) and i-PrOH (3 × 8 mL). The beads 21 were stored in i-PrOH (beads: i-PrOH, (1:2), v/v) at −80°C.
6.2.18. N-Fmoc-trans-4-aminocyclohexanol (22)39
To a solution of trans-4-aminocyclohexanol hydrochloride (2.0 g, 13.2 mmol) in dioxane:water (26:6.5 mL) was added Et3N (1.08 g, 1.49 mL, 10.7 mmol) and this was stirred for 10 min. Then Fmoc-OSu (3.00 g, 8.91 mmol) was added over five minutes and the resulting suspension was stirred at rt for 2 h. The reaction mixture was concentrated to ~5 mL, then some CH2Cl2 was added. This was filtered and the solid was washed with H2O (4 × 40 mL) then dried to give 2.85 g (95%) of 22 as a white solid. Additional 0.100 g (3%) of 22 was obtained by extracting the filtrate with CH2Cl2 (2 × 100 mL), drying over Na2SO4, filtering and removing solvent for a combined yield of 98%. TLC (hexane:EtOAc, 20:80) Rf = 0.42; 1H NMR (CDCl3) δ 7.77 (d, J = 7.5 Hz, 2H), 7.58 (d, J = 7.4 Hz, 2H), 7.40 (t, J = 7.4 Hz, 2H), 7.31 (t, J = 7.4 Hz, 2H), 4.54 (br s, 1H), 4.40 (d, J = 5.6 Hz, 2H), 4.21 (t, J = 5.6 Hz, 1H), 3.61 (m, 1H), 3.48 (m, 1H), 1.9–2.1 (m, 4H), 1.32–1.48 (m, 2H), 1.15–1.29 (m, 2H); MS (ESI): m/z 338.0 [M+H]+.
6.2.19. N-Fmoc-trans-4-aminocyclohexanol tetrahydropyranyl ether (23)
1.03 g (3.05 mmol) of 22 and 0.998 g (1.08 mL, 11.86 mmol) of 3,4-dihydro-2H-pyran (DHP) was suspended in dioxane (10 mL). Pyridinium p-toluenesulfonate (0.153 g, 0.61 mmol) was added and the suspension stirred at rt. After 1 hr additional DHP (1.08 mL, 11.86 mmol) and dioxane (10 mL) were added and stirring continued. After 9 h additional DHP (1.08 mL, 11.86 mmol) was added and stirring continued overnight. The resulting solution was concentrated and the residue chromatographed (hexane:EtOAc, 75:25 to 65:35) to give 1.28 g (100%) of 23 as a white solid. TLC (hexane:EtOAc, 70:30) Rf = 0.26; 1H NMR (CDCl3) δ 7.77 (d, J = 7.5 Hz, 2H), 7.58 (d, J = 7.5 Hz, 2H), 7.40 (t, J = 7.4 Hz, 2H), 7.31 (dt, J = 7.5, 1.1 Hz, 2H), 4.70 (m, 1H), 4.56 (m, 1H), 4.40 (d, J = 6.0 Hz, 2H), 4.21 (t, J = 6.1 Hz, 1H), 3.90 (m, 1H), 3.58 (m, 1H), 3.45–3.53 (m, 2H), 1.10–2.09 (m, 14H); MS (ESI): m/z 422.3 [M+H]+.
6.2.20. trans-4-Aminocylohexanol tetrahydropyranyl ether (24)
1.28 g (3.0 mmol) of 23 was dissolved in CH2Cl2 (20 mL) and piperidine (2 mL) was added and the solution stirred at rt for 5 h. Solvent was removed and the residue was purified by chromatography [CH2Cl2:MeOH-NH3 (7N), 80:1 to 30:1] to give 0.574 g (96%) of 24 as an oily residue which slowly crystallized. 1H NMR (CDCl3) δ 4.70 (m, 1H), 3.91 (m, 1H), 3.58 (m, 1H), 3.49 (m, 1H), 2.69 (m, 1H), 1.07–2.05 (m, 14H); MS (ESI): m/z 200.2 [M+H]+.
6.2.21. 4-(6,6-Dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)-2-(trans-4-(tetrahydro-2H-pyran-2-yloxy)cyclohexylamino)benzonitrile (25)
A mixture of 16 (0.270 g, 0.655 mmol), NaOtBu (0.126 g, 1.31 mmol), Pd2(dba)3 (0.120 g, 0.131 mmol) and DavePhos (0.051 g, 0.131 mmol) in 1,2-dimethoxyethane (20 mL) was degassed and flushed with argon several times. 24 (0.390 g, 1.97 mmol) was added and the flask was again degassed and flushed with argon before heating the reaction mixture at 60°C for 3.5 h. The reaction mixture was concentrated under reduced pressure and the residue purified by preparatory TLC [hexane:CH2Cl2:EtOAc:MeOH-NH3 (7N), 7:6:3:1.5] to give 97.9 mg (28%) of 25. Additionally, 60.5 mg (17%) of amide 26 was isolated for a total yield of 45%. 1H NMR (CDCl3) δ 7.52 (d, J = 8.3 Hz, 1H), 6.80 (d, J = 1.7 Hz, 1H), 6.72 (dd, J = 8.3, 1.8 Hz, 1H), 4.72 (m, 1H), 4.67 (d, J = 7.6 Hz, 1H), 3.91 (m, 1H), 3.68 (m, 1H), 3.50 (m, 1H), 3.40 (m, 1H), 2.84 (s, 2H), 2.49 (s, 2H), 2.06–2.21 (m, 4H), 1.30–1.90 (m, 10H), 1.14 (s, 6H); MS (ESI): m/z 529.4 [M-H]−.
6.2.22. 4-(6,6-Dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)-2-(trans-4-(tetrahydro-2H-pyran-2-yloxy)cyclohexylamino)benzamide (26)
A solution of 25 (120 mg, 0.2264 mmol) in DMSO (220 μl), EtOH (885 μl), 5N NaOH (112 μl) and H2O2 (132 μl) was stirred at rt for 4 h. Then 30 mL of brine was added and this was extracted with EtOAc (5 × 15 mL), dried over Na2SO4, filtered and concentrated under reduced pressure. The residue was purified by preparatory TLC [hexane:CH2Cl2:EtOAc:MeOH-NH3 (7N), 7:6:3:1.5] to give 102 mg (82%) of 26. 1H NMR (CDCl3) δ 8.13 (d, J = 7.4 Hz, 1H), 7.50 (d, J = 8.4 Hz, 1H), 6.74 (d, J = 1.9 Hz, 1H), 6.63 (dd, J = 8.4, 2.0 Hz, 1H), 5.68 (br s, 2H), 4.72 (m, 1H), 3.91 (m, 1H), 3.70 (m, 1H), 3.50 (m, 1H), 3.34 (m, 1H), 2.85 (s, 2H), 2.49 (s, 2H), 2.05–2.19 (m, 4H), 1.33–1.88 (m, 10H), 1.14 (s, 6H); MS (ESI): m/z 547.4 [M-H]−.
6.2.23. 4-(6,6-Dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)-2-(trans-4-hydroxycyclohexylamino)benzamide (SNX-2112)
26 (140 mg, 0.255 mmol) and pyridinium p-toluenesulfonate (6.4 mg, 0.0255 mmol) in EtOH (4.5 mL) was heated at 65°C for 17 h. The reaction mixture was concentrated under reduced pressure and the residue purified by preparatory TLC [hexane:CH2Cl2:EtOAc:MeOH-NH3 (7N), 2:2:1:0.5] to give 101 mg (85%) of SNX-2112. 1H NMR (CDCl3) δ 8.10 (d, J = 7.4 Hz, 1H), 7.52 (d, J = 8.4 Hz, 1H), 6.75 (d, J = 1.3 Hz, 1H), 6.60 (dd, J = 8.4, 1.6 Hz, 1H), 5.97 (br s, 2H), 3.73 (m, 1H), 3.35 (m, 1H), 2.85 (s, 2H), 2.48 (s, 2H), 2.14 (d, J = 11.8 Hz, 2H), 2.04 (d, J = 11.1 Hz, 2H), 1.33–1.52 (m, 4H), 1.13 (s, 6H); 13C NMR (125 MHz, CDCl3/MeOH-d4) δ 191.0, 171.9, 151.0, 150.0, 141.3, 140.3 (q, J = 39.6 Hz), 130.4, 120.3 (q, J = 270.2 Hz), 115.9, 113.7, 109.2, 107.1, 69.1, 52.1, 50.2, 40.1, 37.0, 35.6, 33.1, 30.2, 28.0; MS (ESI): m/z 463.3 [M-H] −, 465.3 [M+H]+; HPLC: tR = 7.97 min.
6.2.24. Preparation of control beads
DMF (8.5 mL) was added to 20 mL of Affi-Gel 10 beads (prewashed, 3 × 40 mL DMF) in a solid phase peptide synthesis vessel. 2-Methoxyethylamine (113 mg, 129 μL, 1.5 mmol) and several crystals of DMAP were added and this was shaken at rt for 2.5 h. Then the solvent was removed and the beads washed for 10 minutes each time with CH2Cl2 (4 × 35 mL), DMF (3 × 35 mL), Felts buffer (2 × 35 mL) and i-PrOH (4 × 35 mL). The beads were stored in i-PrOH (beads: i-PrOH (1:2), v/v) at −80°C.
6.3. Competition assay
For the competition studies, fluorescence polarization (FP) assays were performed as previously reported.15 Briefly, FP measurements were performed on an Analyst GT instrument (Molecular Devices, Sunnyvale, CA). Measurements were taken in black 96-well microtiter plates (Corning # 3650) where both the excitation and the emission occurred from the top of the wells. A stock of 10 μM GM-cy3B was prepared in DMSO and diluted with Felts buffer (20 mM Hepes (K), pH 7.3, 50 mM KCl, 2 mM DTT, 5 mM MgCl2, 20 mM Na2MoO4, and 0.01% NP40 with 0.1 mg/mL BGG). To each 96-well were added 6 nM fluorescent GM (GM-cy3B), 3 μg SKBr3 lysate (total protein), and tested inhibitor (initial stock in DMSO) in a final volume of 100 μL HFB buffer. Drugs were added in triplicate wells. For each assay, background wells (buffer only), tracer controls (free, fluorescent GM only) and bound GM controls (fluorescent GM in the presence of SKBr3 lysate) were included on each assay plate. GM was used as positive control. The assay plate was incubated on a shaker at 4°C for 24 h and the FP values in mP were measured. The fraction of tracer bound to Hsp90 was correlated to the mP value and plotted against values of competitor concentrations. The inhibitor concentration at which 50% of bound GM was displaced was obtained by fitting the data. All experimental data were analyzed using SOFTmax Pro 4.3.1 and plotted using Prism 4.0 (Graphpad Software Inc., San Diego, CA).
6.4. Chemical Precipitation, Western blotting and Flow Cytometry
The leukemia cell lines K562 and MV4-11 and the breast cancer cell line MDA-MB-468 were obtained from the American Type Culture Collection. Cells were cultured in RPMI (K562), in Iscove’s modified Dulbecco’s media (MV4-11) or in DME/F12 (MDA-MB-468) supplemented with 10% FBS, 1% L-glutamine, 1% penicillin and streptomycin, and maintained in a humidified atmosphere of 5% CO2 at 37°C. Cells were lysed by collecting them in Felts buffer (HEPES 20 mM, KCl 50 mM, MgCl2 5 mM, NP40 0.01%, freshly prepared Na2MoO4 20 mM, pH 7.2–7.3) with added 10 μg/μL of protease inhibitors (leupeptin and aprotinin), followed by three successive freeze (in dry ice) and thaw steps. Total protein concentration was determined using the BCA kit (Pierce) according to the manufacturer’s instructions.
Hsp90 inhibitor beads or control beads containing an Hsp90 inactive chemical (2-methoxyethylamine) conjugated to agarose beads were washed three times in lysis buffer. The bead conjugates (80 μL or as indicated) were then incubated overnight at 4ºC with cell lysates (250 μg), and the volume was adjusted to 200–300 μL with lysis buffer. Following incubation, bead conjugates were washed 5 times with the lysis buffer and analyzed by Western blot, as indicated below.
For treatment with PU-H71, cells were grown to 60–70% confluence and treated with inhibitor (5 μM) for 24h. Protein lysates were prepared in 50 mM Tris pH 7.4, 150 mM NaCl and 1% NP-40 lysis buffer.
For Western blotting, protein lysates (10–50 μg) were electrophoretically resolved by SDS/PAGE, transferred to nitrocellulose membrane and probed with a primary antibody against Hsp90 (1:2000, SMC-107A/B, StressMarq), anti-IGF-IR (1:1000, 3027, Cell Signaling) and antic-Kit (1:200, 612318, BD Transduction Laboratories). The membranes were then incubated with a 1:3000 dilution of a corresponding horseradish peroxidase conjugated secondary antibody. Detection was performed using the ECL-Enhanced Chemiluminescence Detection System (Amersham Biosciences) according to manufacturer’s instructions.
To detect the binding of PU-H71 to cell surface Hsp90, MV4-11 cells at 500,000 cells/ml were incubated with the indicated concentrations of PU-H71-biotin or D-biotin as control for 2 h at 37°C followed by staining of phycoerythrin (PE) conjugated streptavidin (SA) (BD Biosciences) in FACS buffer (PBS + 0.5% FBS) at 4°C for 30 min. Cells were then analyzed using the BD-LSRII flow cytometer. Mean fluorescence intensity (MFI) was used to calculate the binding of PU-H71-biotin to cells and values were normalized to the MFI of untreated cells stained with SA-PE.
6.5. Docking
Molecular docking computations were carried out on a HP workstation xw8200 with the Ubuntu 8.10 operating system using Glide 5.0.40 The coordinates for the Hsp90α complexes with bound inhibitor PU-H71 (PDB ID: 2FWZ), NVP-AUY922 (PDB ID: 2VCI) and 27 (PDB ID: 3D0B) were downloaded from the RCSB Protein Data Bank. For docking experiments, compounds PU-H71, NVP-AUY922, 5, 10, 20 and 27 were constructed using the fragment dictionary of Maestro 8.5 and geometry-optimized using the Optimized Potentials for Liquid Simulations-All Atom (OPLS-AA) force field41 with the steepest descent followed by truncated Newton conjugate gradient protocol as implemented in Macromodel 9.6, and were further subjected to ligand preparation using default parameters of LigPrep 2.2 utility provided by Schrödinger LLC. Each protein was optimized for subsequent grid generation and docking using the Protein Preparation Wizard provided by Schrödinger LLC. Using this tool, hydrogen atoms were added to the proteins, bond orders were assigned, water molecules of crystallization not deemed to be important for ligand binding were removed, and the entire protein was minimized. Partial atomic charges for the protein were assigned according to the OPLS-2005 force field. Next, grids were prepared using the Receptor Grid Generation tool in Glide. With the respective bound inhibitor in place, the centroid of the workspace ligand was chosen to define the grid box. The option to dock ligands similar in size to the workspace ligand was selected for determining grid sizing.
Next, the extra precision (XP) Glide docking method was used to flexibly dock compounds PU-H71 and 5 (to 2FWZ), NVP-AUY922 and 10 (to 2VCI), and 20 and 27 (to 3D0B) into their respective binding site. Although details on the methodology used by Glide are described elsewhere,42–44 a short description about parameters used is provided below. The default setting of scale factor for van der Waals radii was applied to those atoms with absolute partial charges less than or equal to 0.15 (scale factor of 0.8) and 0.25 (scale factor of 1.0) electrons for ligand and protein, respectively. No constraints were defined for the docking runs. Upon completion of each docking calculation, at most 100 poses per ligand were allowed to generate. The top-scored docking pose based on the Glide scoring function45 was used for our analysis. In order to validate the XP Glide docking procedure the crystallographic bound inhibitor (PU-H71 or NVP-AUY922 or 27) was extracted from the binding site and re-docked into its respective binding site. There was excellent agreement between the localization of the inhibitor upon docking and the crystal structure as evident from the 0.098 Å (2FWZ), 0.313 Å (2VCI) and 0.149 Å (3D0B) root mean square deviations. Thus, the present study suggests the high docking reliability of Glide in reproducing the experimentally observed binding mode for Hsp90 inhibitors and the parameter set for the Glide docking reasonably reproduces the X-ray structure.
Acknowledgments
This work was supported in part by W.H. Goodwin and A. Goodwin and the Commonwealth Cancer Foundation for Research, The Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center (MSKCC), the Translational and Integrative Medicine Research Fund of MSKCC, SPORE Pilot Award and Research & Therapeutics Program in Prostate Cancer, 1R01CA155226-01, U01AG032969-01A1, R21AG028811, 3P30CA008748-44S2, Leukemia and Lymphoma Society LLS#6114-10, the Byrne Fund, the Geoffrey Beene Cancer Research Center of MSKCC (G.C.), Hirshberg Foundation for Pancreatic Cancer Research (G.C.), grant UL1RR024996 of the Clinical and Translational Science Center at Weill Cornell Medical College and Susan G. Komen for the Cure (G.C. and T.T.). We also thank Dr. George Sukenick and Dr. Hui Liu of the NMR Analytical Core Facility at MSKCC for expert mass spectral analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. EMBO. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obermann WMJ, Sondermann H, Russo AA, Pavletich NP, Hartl FU. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitesell L, Lindquist SL. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 4.Taldone T, Sun W, Chiosis G. Bioorg Med Chem. 2009;17:2225–2235. doi: 10.1016/j.bmc.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janin Y. Drug Discov Today. 2010;15:342–353. doi: 10.1016/j.drudis.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Solit DB, Zheng FF, Drobnjak M, Munster PN, Higgins B, Verbel D, Heller G, Tong W, Cardon-Cardo C, Agus DB, Scher HI, Rosen N. Clin Cancer Res. 2002;8:986–993. [PubMed] [Google Scholar]
- 7.Brough PA, Aherne W, Barril X, Borgognoni J, Boxall K, Cansfield JE, Cheung KMJ, Collins I, Davies NGM, Drysdale MJ, Dymock B, Eccles SA, Finch H, Fink A, Hayes A, Howes R, Hubbard RE, James K, Jordan AM, Lockie A, Martins V, Massey A, Matthews TP, McDonald E, Northfield CJ, Pearl LH, Prodromou C, Ray S, Raynaud FI, Roughley SD, Sharp SY, Surgenor A, Walmsley DL, Webb P, Wood M, Workman P, Wright L. J Med Chem. 2008;51:196–218. doi: 10.1021/jm701018h. [DOI] [PubMed] [Google Scholar]
- 8.Chandarlapaty S, Sawai A, Ye Q, Scott A, Silinski M, Huang K, Fadden P, Partdrige J, Hall S, Steed P, Norton L, Rosen N, Solit DB. Clin Cancer Res. 2008;14:240–248. doi: 10.1158/1078-0432.CCR-07-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldas-Lopes E, Cerchietti L, Ahn JH, Clement CC, Robles AI, Rodina A, Moulick K, Taldone T, Gozman A, Guo Y, Wu N, de Stanchina E, White J, Gross SS, Ma Y, Varticovski L, Melnick A, Chiosis G. Proc Natl Acad Sci USA. 2009;106:8368–8373. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carden CP, Sarker D, Postel-Vinay S, Yap TA, Attard G, Banerji U, Garrett MD, Thomas GV, Workman P, Kaye SB, de Bono JS. Drug Discov Today. 2010;15:88–97. doi: 10.1016/j.drudis.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He H, Zatorska D, Kim J, Aguirre J, Llauger L, She Y, Wu N, Immormino RM, Gewirth DT, Chiosis G. J Med Chem. 2006;49:381–390. doi: 10.1021/jm0508078. [DOI] [PubMed] [Google Scholar]
- 13.Huang KH, Veal JM, Fadden RP, Rice JW, Eaves J, Strachan JP, Barabasz AF, Foley BE, Barta TE, Ma W, Silinski MA, Hu M, Partridge JM, Scott A, DuBois LG, Freed T, Steed PM, Ommen AJ, Smith ED, Hughes PF, Woodward AR, Hanson GJ, McCall WS, Markworth CJ, Hinkley L, Jenks M, Geng L, Lewis M, Otto J, Pronk B, Verleysen K, Hall SE. J Med Chem. 2009;52:4288–4305. doi: 10.1021/jm900230j. [DOI] [PubMed] [Google Scholar]
- 14.Clevenger RC, Raibel JM, Peck AM, Blagg BSJ. J Org Chem. 2004;69:4375–4380. doi: 10.1021/jo049848m. [DOI] [PubMed] [Google Scholar]
- 15.Du Y, Moulick K, Rodina A, Aguirre J, Felts S, Dingledine R, Fu H, Chiosis G. J Biomol Screen. 2007;12:915–924. doi: 10.1177/1087057107306067. [DOI] [PubMed] [Google Scholar]
- 16.Immormino RM, Kang Y, Chiosis G, Gewirth DT. J Med Chem. 2006;49:4953–4960. doi: 10.1021/jm060297x. [DOI] [PubMed] [Google Scholar]
- 17.Cheung KM, Matthews TP, James K, Rowlands MG, Boxall KJ, Sharp SY, Maloney A, Roe SM, Prodromou C, Pearl LH, Aherne GW, McDonald E, Workman P. Bioorg Med Chem Lett. 2005;15:3338–3343. doi: 10.1016/j.bmcl.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 18.Dymock BW, Barril X, Brough PA, Cansfield JE, Massey A, McDonald E, Hubbard RE, Surgenor A, Roughley SD, Webb P, Workman P, Wright L, Drysdale MJ. J Med Chem. 2005;48:4212–4215. doi: 10.1021/jm050355z. [DOI] [PubMed] [Google Scholar]
- 19.Barril X, Beswick MC, Collier A, Drysdale MJ, Dymock BW, Fink A, Grant K, Howes R, Jordan AM, Massey A, Surgenor A, Wayne J, Workman P, Wright L. Bioorg Med Chem Lett. 2006;16:2543–2548. doi: 10.1016/j.bmcl.2006.01.099. [DOI] [PubMed] [Google Scholar]
- 20.Barta TE, Veal JM, Rice JW, Partridge JM, Fadden RP, Ma W, Jenks M, Geng L, Hanson GJ, Huang KH, Barabasz AF, Foley BE, Otto J, Hall SE. Bioorg Med Chem Lett. 2008;18:3517–3521. doi: 10.1016/j.bmcl.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 21.Serenex, Huang K, Ommen AJ, Barta TE, Hughes PF, Veal JM, Ma W, Smith ED, Woodward AR, McCall WS. 2008130879A2. WO. 2008
- 22.Serenex, Huang K, Ommen AJ, Barta TE, Hughes PF, Veal JM, Ma W, Smith ED, Woodward AR, McCall WS. 20080269193A1. US. 2008
- 23.Claramunt RM, López C, Pérez-Medina C, Pinilla E, Torres MR, Elguero J. Tetrahedron. 2006;62:11704–11713. [Google Scholar]
- 24.Old DW, Wolfe JP, Buchwald SL. J Am Chem Soc. 1998;120:9722–9723. [Google Scholar]
- 25.Hurvitz SA, Finn RS. Future Oncol. 2009;5:1015–1025. doi: 10.2217/fon.09.68. [DOI] [PubMed] [Google Scholar]
- 26.Law JH, Habibi G, Hu K, Masoudi H, Wang MY, Stratford AL, Park E, Gee JM, Finlay P, Jones HE, Nicholson RI, Carboni J, Gottardis M, Pollak M, Dunn SE. Cancer Res. 2008;68:10238–10246. doi: 10.1158/0008-5472.CAN-08-2755. [DOI] [PubMed] [Google Scholar]
- 27.Cerchietti LC, Lopes EC, Yang SN, Hatzi K, Bunting KL, Tsikitas LA, Mallik A, Robles AI, Walling J, Varticovski L, Shaknovich R, Bhalla KN, Chiosis G, Melnick A. Nat Med. 2009;15:1369–1376. doi: 10.1038/nm.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marubayashi S, Koppikar P, Taldone T, Abdel-Wahab O, West N, Bhagwat N, Lopes-Vazquez EC, Ross KN, Gönen M, Gozman A, Ahn J, Rodina A, Ouerfelli O, Yang G, Hedvat C, Bradner JE, Chiosis G, Levine RL. J Clin Invest. 2010;120:3578–3593. doi: 10.1172/JCI42442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo W, Dou F, Rodina A, Chip S, Kim J, Zhao Q, Moulick K, Aguirre J, Wu N, Greengard P, Chiosis G. Proc Natl Acad Sci USA. 2007;104:9511–9518. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W, Rodina A, Chiosis G. BMC Neurosci. 2008;9(Suppl 2):S7. doi: 10.1186/1471-2202-9-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo W, Sun W, Taldone T, Rodina A, Chiosis G. Mol Neurodegener. 2010;5:24. doi: 10.1186/1750-1326-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsaytler PA, Krijgsveld J, Goerdayal SS, Rüdiger S, Egmond MR. Cell Stress Chaperones. 2009;14:629–638. doi: 10.1007/s12192-009-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH. J Med Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 34.Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 35.Nishiya Y, Shibata K, Saito S, Yano K, Oneyama C, Nakano H, Sharma SV. Anal Biochem. 2009;385:314–320. doi: 10.1016/j.ab.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 36.Sidera K, Patsavoudi E. Cell Cycle. 2008;7:1564–1568. doi: 10.4161/cc.7.11.6054. [DOI] [PubMed] [Google Scholar]
- 37.Hansen JB, Nielsen MC, Ehrbar U, Buchardt O. Synthesis. 1982:404–405. [Google Scholar]
- 38.Hiegel GA, Burk P. J Org Chem. 1973;38:3637–3639. [Google Scholar]
- 39.Crestey F, Ottesen LK, Jaroszewski JW, Franzyk H. Tetrahedron Lett. 2008;49:5890–5893. [Google Scholar]
- 40.Schrodinger LLC. New York: [Google Scholar]
- 41.Jorgensen WL, Maxwell D, Tirado-Rives J. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 42.Patel PD, Patel MR, Kaushik-Basu N, Talele TT. J Chem Inf Model. 2008;48:42–55. doi: 10.1021/ci700266z. [DOI] [PubMed] [Google Scholar]
- 43.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 44.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 45.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. J Comput-Aided Mol Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]