Abstract
The export of nutrients from source organs to parts of the body where they are required (e.g. sink organs) is a fundamental biological process. Export of amino acids, one of the most abundant nitrogen species in plant long-distance transport tissues (i.e. xylem and phloem), is an essential process for the proper distribution of nitrogen in the plant. Physiological studies have detected the presence of multiple amino acid export systems in plant cell membranes. Yet, surprisingly little is known about the molecular identity of amino acid exporters, partially due to the technical difficulties hampering the identification of exporter proteins. In this short review, we will summarize our current knowledge about amino acid export systems in plants. Several studies have described plant amino acid transporters capable of bi-directional, facilitative transport, reminiscent of activities identified by earlier physiological studies. Moreover, recent expansion in the number of available amino acid transporter sequences have revealed evolutionary relationships between amino acid exporters from other organisms with a number of uncharacterized plant proteins, some of which might also function as amino acid exporters. In addition, genes that may regulate export of amino acids have been discovered. Studies of these putative transporter and regulator proteins may help in understanding the elusive molecular mechanisms of amino acid export in plants.
Keywords: Nitrogen, amino acids, membrane transport
INTRODUCTION
Transfer of nutrients between organs is of fundamental importance in multi-cellular organisms for the proper supply of nutrients and removal of unwanted products. In addition to being central metabolites, amino acids are the most abundant nitrogen species in animal serum and long-distance transport systems of plants (i.e. xylem and phloem) and are hence considered the main nitrogen carriers in both kingdoms.
In plants, inorganic nitrogen (i.e. NO3– and NH4+) taken up by roots is incorporated into glutamine and glutamate (primary nitrogen assimilation), which is used to synthesize other amino acids and nitrogenous compounds by transamination. This process happens either in root or shoot tissue, depending on factors such as the molecular species of nitrogen taken up and the carbon/nitrogen balance of the plant (Andrews, 1986; Kruse et al., 2002; Marschner, 1995). Once synthesized, amino acids are delivered to the so-called sink organs (developing roots and leaves, flowers, and seeds) that are largely dependent on reduced nitrogen supplied by the long-distance transport systems of the plant (Pate, 1973; Pate et al., 1981).
Amino acid transfer between organs through xylem and phloem is critical for optimizing nitrogen allocation in the plant to the growth conditions or developmental stage (Tegeder and Rentsch, 2010). In Ricinus, half of the amino acids delivered to the roots via the phloem are eventually relocated into the ascending xylem sap, indicating a significant amount of phloem-to-xylem transport (Schobert and Komor, 1987). Amino acids delivered to developing fruits are predominantly supplied by the phloem, the content of which becomes enriched in amino acids as it moves towards fruits, revealing active xylem-to-phloem transfer (Jeschke and Hartung, 2000). Distribution and recycling of amino acids through the xylem and phloem ensures optimal nitrogen allocation between organs (Figure 1). Amino acid transfer is also very active at the cellular level. Many amino acids are synthesized in the chloroplast and transported into the cytosol for protein synthesis and secondary metabolite production, or transported and stored in the vacuole. Distribution among the organelles is unequal: the highest amino acid content is found in the cytosol and lowest in the vacuole, suggesting directional amino acid transfer across the membrane rather than simple equilibrative diffusion (Leidreiter et al., 1995; Riens et al., 1991; Winter et al., 1993).
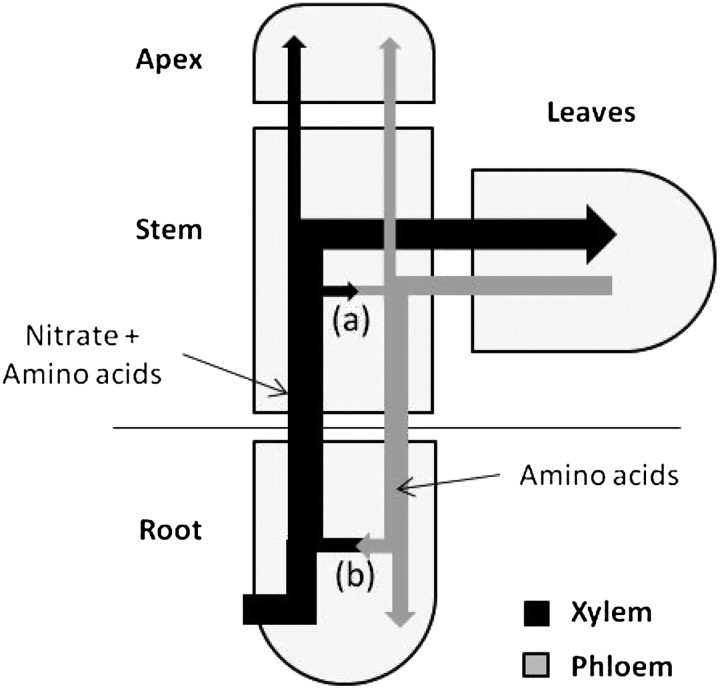
Figure 1.
Nitrogen Cycling between Root and Shoots.
Arrows represent the fluxes of nitrogen uptake, transport, and utilization in whole plants of Ricinus communis. Part of the taken up nitrate is reduced in the root as amino acids and is transported to the leaves and shoot apex by the xylem (black arrow). The remaining nitrate is reduced in the leaves, and exported to the root and the shoot apex by the phloem (gray arrow). (A) Xylem to phoem and (B) phloem to xylem transfers occur during the transport in the root and the stem, leading to amino acid cycling. Adapted from Jeschke and Hartung (Jeschke and Hartung 2000).
The proper distribution of amino acids as described above requires amino acids be imported and exported in several locations. Amino acids synthesized in roots or acquired from the descending phloem are exported across the plasma membrane of root cells to the xylem sap, which is part of the apoplasm. Depending on the plant species, phloem can be loaded with amino acids symplasmically or apoplasmically. In the latter case, no symplasmic connection between the leaf parenchyma cells and the phloem exists and amino acids need to be first exported to the leaf apoplasm before being imported into the phloem at the level of the companion cell (Lalonde et al., 2003, 2004). In the reproductive organs, the embryo is symplasmically disconnected from the maternal tissues of the seed coat. Here, again, amino acids need to be exported from the seed coat cells to the apoplasm and taken up by the embryo cells (deJong et al., 1997; Zhang et al., 2007). Plant roots release a large variety of compounds into the rhizosphere, including amino acids (Bertin et al., 2003; Walker et al., 2003). Nodule-forming symbiotic bacteria have recently been shown to depend on plants for branched chain amino acids while supplying other amino acids to their hosts (Lodwig et al., 2003; Prell et al., 2009). The presence of the efflux of amino acids into the rhizosphere or to the nodule shows that most root cells, just like seed coat, or leaf xylem and phloem parenchyma, are capable of amino acid export.
In the past two decades, multiple mechanisms of amino acid import in plant have been discovered. The establishment of effective heterologous expression systems, notably Saccharomyces cerevisiae mutants auxotrophic for amino acids as the sole nitrogen source, enabled isolation of transporters that restore amino acid import into the cells with an unprecedented efficiency. Since most of these importers turned out to be electrogenic H+-symporters, the mode of transport could be characterized using voltage-clamp techniques in Xenopus oocytes. In contrast, elucidation of the molecular mechanisms responsible for cellular amino acid export (or solute export in general) is lagging behind, primarily due to the lack of an efficient method for the identification of proteins with export activity. Furthermore, both earlier physiological studies and recent molecular evidence suggest that some plant amino acid exporters mediate bi-directional, facilitated diffusions (see below), which limits the use of voltage-clamp techniques, and obscures the results from radio-tracer experiments.
The identification of amino acid exporters is essential in understanding how amino acid cycling is achieved in plants, and how these processes are regulated. In addition, it has previously been demonstrated that the supply of amino acids has fundamental effects on organs such as developing seeds (Lohaus and Moellers, 2000; Lohaus et al., 1995; Riens et al., 1991). Previous studies aiming at modifying amino acid content and composition in the consumed organs depended largely on activations of metabolic enzymes with varying degrees of success (Frankard et al., 1992; Shaul and Galili, 1992; Ufaz and Galili, 2008; Zhu and Galili, 2003). Manipulations of transport activities might pose an alternative approach to achieve the same goal (Koch et al., 2003). Therefore, a better understanding of the export process would provide valuable target genes for manipulating and improving the yield and quality of proteins in grains.
In this review, we will summarize our knowledge on amino acid export in plants, obtained from physiological experiments and computational analyses enabled by the major expansion in sequences of amino acid transporter homologs in a myriad of organisms. The combination of bioinformatics and technological advances in the detection of amino acids would certainly lead to the identification of the ‘missing link’ in amino acid transport—cellular export mechanisms of these important compounds.
AMINO ACID EXPORT ACROSS THE PLASMA MEMBRANE
Membrane transporters are often categorized as: importers (catalyzing the transport into the cell or a sub-cellular compartment), exporters (catalyzing transport out of the cell or a sub-cellular compartment), and bi-directional transporters (mediating both export and import, usually a facilitator). The terms ‘import’ and ‘export’ are somewhat arbitrary: for example, the transport of solute from the vacuole into the cytosol can be seen as either ‘export’ of solute from the vacuole to the cytosol, or ‘import’ from the vacuole into the cytosol. In this review, ‘export’ is defined as the movement of solute from the cytosol to either the apoplasmic space or into an intracellular organelle such as the vacuole, and ‘import’ will be used to describe transport in the opposite direction. To further avoid confusion, the direction of the transport will be stated whenever appropriate. The net transport of solutes across membranes results from the summation of both import and export activities. Since the terms used in the literature to define transport vary greatly, import activity minus export will be defined in this review as ‘uptake’ and export activity minus import as ‘efflux’. Strictly speaking, determining individual contributions of import and export to the net transport is nearly impossible. However, in experimental situations in which transport activities in the other direction are expected to be minimal, such as heterologous expression systems with limited endogenous transport activities, it is reasonable to approximate import or export activities by measuring uptake or efflux of a compound (Nasholm et al., 2009).
For cellular import mechanisms, the amounts of solutes taken up can be measured using isotope-labeled compounds, which can be quantified using scintillation counting or mass spectrometry. In this way, the functional properties of transporters, such as apparent Km, Vmax, and mode of transport can be determined (Gu et al., 2007; Kinraide, 1981; Kinraide et al., 1984; Persson and Nasholm, 2001; Schobert and Komor, 1987). On the other hand, measurement of efflux has been more challenging. Since plant cells exhibit generally a high amino acid import activity, amino acid re-uptake needs to be taken into account in the estimation of efflux. This problem can at least be partially addressed by labeling either the plant or external medium with an isotope (i.e. 15N) and following the exchange of two isotopes between the plant and external medium over the course of an experiment (Lesuffleur and Cliquet, 2010; Lesuffleur et al., 2007; Phillips et al., 2004). While this method is effective in determining contributions of import and export to the net uptake/efflux, only amino acids that are excreted in relatively large amounts can be reliably detected. Alternatively, plants cells can be pre-loaded with radio-labeled amino acids and excretion into the medium can be estimated by measuring the amount of released radioactivity (Pratelli et al., 2010; Schobert and Komor, 1987). This method allows the net amounts of excreted compound to be detected with high sensitivity, but conclusions drawn from such data can be erroneous if the radioisotope is released as another compound due to intracellular metabolism. Because of such limitations, export of amino acids remains less explored at the physiological level than amino acid import (Nasholm et al., 2009). A small number of physiological studies have been conducted to characterize the nature of amino acid efflux from plant cells. Although amino acid efflux is expected to occur in multiple tissues such as leaf and root xylem parenchyma (see above), seed coat and root have been the main focus of such studies due to the ease of access.
Assimilates are continuously transferred from maternal tissues (i.e. integuments, the future seed coat) to the endosperm and embryo during seed development. Since embryonic tissues and endosperm are symplasmically isolated from the maternal tissues, assimilates are released from the seed coat to the apoplasmic compartment and actively taken up by the cells of the developing seed (Zhang et al., 2007). The development of the ‘empty-seed-coat’ technique, in which the embryo from a developing seed is removed and replaced by an appropriate solution, enabled studies of assimilate effluxes from the seed coat (deJong et al., 1997; Lanfermeijer et al., 1992; Wolswinkel and Deruiter, 1985). Analysis of the composition of the solution after several hours showed that glutamine, alanine, and threonine accounted for about 55% of the amino acids excreted from the seed coat of Vicia faba and Pisum sativum (Lanfermeijer et al., 1992; Wolswinkel and Deruiter, 1985). Amino acid flux from the seed coat was found to be four to five orders of magnitude greater than simple diffusion through the lipid bilayer (deJong et al., 1997), suggesting that export is transporter-mediated. To determine the mechanism of export, deJong et al. (1997) investigated the effect of the pH gradient across the membrane and chemical inhibitors that either modify extracellular sulfhydryl groups (p-chloromercuribenzenesulfonic acid, PCMBS) or dissipate the proton gradients (cyanide-m-chlorophenylhydrazone, CCCP). The authors found that amino acid export from the seed coat is decreased by the action of PCMBS, suggesting that amino acid efflux is transporter-mediated. Insensitivity to pH changes or CCCP suggests that export is not energized by the proton gradient. Likewise, amino acid import into the seed coat was found to be insensitive to the proton-gradient and driven only by the amino acid gradient—characteristics reminiscent of the export mechanism (deJong et al., 1997; van Dongen et al., 2001). The combination of these results suggests that amino acid export from the seed coat is mediated by a bi-directional facilitator.
The root system of plants not only import water and nutrients from the soil solution, but also releases low- and high-molecular-weight compounds to the rhizosphere. Excretion of solutes from the root system would fulfill several purposes, such as controlling the quantity and quality of the microbial community, altering the physical and chemical properties of the soil and inhibiting the growth of competing plants (Bertin et al., 2003; Walker et al., 2003). Amino acids have been shown to be part of the numerous compounds released by plant roots (Kraffczyk et al., 1984; Phillips et al., 2004). At the same time, amino acid uptake systems are expressed in roots and involved in the acquisition of external amino acids for nitrogen nutrition (Jones et al., 2005; Nasholm et al., 2009). Consequently, the net flux of amino acids is determined by the activities of both export and import (Jones and Darrah, 1994; Jones et al., 2005; Schobert and Komor, 1987). It has been shown that net amino acid efflux occurs for some amino acids (e.g. glycine and serine (Lesuffleur et al., 2007)). Net efflux was observed when roots were treated with inhibitors targeting the proton gradient, abolishing the activity of the proton-coupled importers (Jones and Darrah, 1994; Rroco et al., 2002). Amino acid release rate was also found to be enhanced by the application of microbial products (e.g. zearalenone), suggesting further that export is transporter-mediated (Phillips et al., 2004).
A recent study focused on the specificity and mechanism of root exudation of amino acids. For this purpose, Lesuffleur and Cliquet (2010) incubated plant roots in a medium containing 15N-labeled amino acid and measured the variation in the 14N/15N ratio in the roots and the medium to assess both efflux and uptake. While application of 10 μM CCCP and vanadate, an inhibitor of ATP hydrolysis, led to reductions in Gly uptake, efflux was hardly affected by the same treatments (Lesuffleur and Cliquet, 2010). These experiments suggest that, similarly to export from seed coat, amino acid export from root cells is transporter-mediated and not energized by the proton gradient across the membrane.
Comparison of the amino acid composition of the seed coat and root tissues with their respective exudates showed that the composition of amino acids in cells does not match the composition of the exported amino acids (Lesuffleur and Cliquet, 2010; Lesuffleur et al., 2007; Wolswinkel and Deruiter, 1985). These differences suggest that the amino acid export systems in seed coat and root are capable of exporting a broad range of amino acids but show some selectivity for certain amino acids, analogous to the amino acid import systems characterized so far (Rentsch et al., 2007; Tegeder and Rentsch, 2010). On the other hand, the composition of the leaf apoplasm was found to be very similar to the composition of the cytosolic compartment, suggesting that the amino acid export mechanism of the leaf parenchyma cells might be non-selective (Lohaus et al., 1995) or mediated by transporters displaying different selectivity.
In summary, studies of amino acid efflux from seed coat, root, and leaf cells suggest that export is mediated by transporters with limited selectivity. The transport systems expressed in seed coats and roots require no proton gradient, allowing transfer of substrate downstream of the electrochemical gradient, and at least in seed coat, the system also seems to allow bi-directional transport.
EXPORT OF AMINO ACIDS FROM THE CYTOSOL TO THE VACUOLE
Many transporter families include members that are targeted to the vacuolar membrane as well as the plasma membrane: for example, aquaporins (Maurel et al., 2008), Ca2+-ATPases (Kabala and Klobus, 2005), potassium channels (Dunkel et al., 2008), Na+/H+ exchangers (NHXs) (Apse and Blumwald, 2007), and sucrose transporters (Endler et al., 2006; Reinders et al., 2008). The presence of transporters with the same transport mechanism on both plasma and vacuolar membranes would mean that the substrate for exporters can either be excreted into the apoplasm or transported into the vacuole (i.e. ‘internal excretion’, Martinoia et al., 1993). Amino acid transporters mediating transport into the vacuole are most likely evolutionally related to plasma membrane exporters. Therefore, considering transport processes into the vacuole is relevant in investigating the mechanisms of cellular solute export. Transporters isolated so far that mediate amino acid flux across the membranes of mitochondria and chloroplasts are phylogenetically different from plasma membrane or vacuolar membrane transporters (Jack et al., 2000; Pudelski et al., 2010), and therefore will not be discussed in this review.
Plant vacuoles fulfill essential roles in cell expansion, storage of proteins, nutrients and defense compounds, degradation of proteins and compounds, regulation of turgor pressure, and are involved in metabolite partitioning with the cytosol (Martinoia et al., 2007; Mueller et al., 2007). Analysis of the composition of isolated vacuoles (Dietz et al., 1990) or estimation of the composition of leaf compartments using non-aqueous partitioning (Leidreiter et al., 1995; Riens et al., 1991; Winter et al., 1993) has shown that amino acid composition in the vacuole differs significantly from that in the cytosol, indicating that tonoplast transporters are selective and directional. Study of amino acid transport across the tonoplast using isolated vacuoles suggested that multiple mechanisms are responsible for amino acid transport. Transport of neutral amino acids (Ala, Leu, Gln, Gly) was found to be insensitive to the proton gradient (Dietz et al., 1990; Goerlach and Willmshoff, 1992; Martinoia et al., 1991). Transport of these amino acids was stimulated by the application of ATP and its non-hydrolysable analog, but not MgATP, indicating that the transport is stimulated, but not energized, by ATP (Dietz et al., 1990; Goerlach and Willmshoff, 1992). Interestingly, transport of amino acids from the vacuole to the cytosol was also stimulated by ATP and its non-hydrolysable analog, suggesting that tonoplastic amino acid transport is mediated by bi-directional transporters (Dietz et al., 1989; Thume and Dietz, 1991). Transport analysis of reconstituted amino acid transporters into liposomes recapitulated ATP dependence, inhibition by amino acids, and the bi-directionality of the transport (Thume and Dietz, 1991). On the other hand, transport of phenylalanine into the vacuole was found to be dependent on the proton gradient, and inhibited by other aromatic amino acids but not alanine or valine, suggesting the existence of an independent transport system for aromatic amino acids (Homeyer et al., 1989). Likewise, the existence of a permease specific for positively charged amino acids (Arg and Lys) has been proposed (Martinoia et al., 1991). From these studies, it was concluded at least three independent transport systems for neutral, aromatic, and basic amino acids are present in plant vacuolar membranes (Homeyer et al., 1989; Homeyer and Schultz, 1988; Martinoia et al., 1991), but the molecular identity of the corresponding proteins is still unknown.
MOLECULAR MECHANISMS OF AMINO ACID EXPORT
Currently, over two dozen transporter families are known to include transporters for amino acids and their derivatives (Saier, 2000; Saier et al., 2009), and the number of family members continues to grow as the number of sequenced genomes expands. Phylogenetic analyses have revealed that while some amino acid transporter families are specific to a single domain or even a single kingdom, many are conserved between more evolutionally distant organisms (Supplemental Table 1 and Supplemental Figures 1–3). Also, even though the members of one family can have diverse substrates, these substrates rarely correspond to structurally divergent compounds (i.e. sugars versus amino acids). Therefore, it is likely that uncharacterized plant proteins evolutionally related to amino acid exporters from other kingdoms would share functional similarities with these proteins. Here, we will summarize our knowledge on superfamilies and families that include plant members and contain amino acid exporters characterized in other organisms. For a more comprehensive review of amino acid transporters in all kingdoms, readers are referred to other excellent reviews (Burkovski and Kramer, 2002; Saier, 2000; Wipf et al., 2002). Although there is little information about amino acid exporters in plants, proteins from the families listed below can be considered candidates for amino acid exporter mechanisms in plants.
Plant transporters that mediate energy-coupled amino acid uptake into the cytosol have been reviewed extensively (Rentsch et al., 2007; Tegeder and Rentsch, 2010; Wipf et al., 2002), and therefore will not be covered by this review. Likewise, there has been a marked progress in the identification and characterization of exporters for the important phytohormone auxin, which can be considered an amino acid derivative. Transporters that mediate export of auxin have been reviewed recently, and will not be covered in this review (Geisler and Murphy, 2006; Grunewald and Friml, 2010; Zazimalova et al., 2010).
THE AMINO ACID-POLYAMINE-ORGANOCATION (APC) SUPERFAMILY
Members of the APC superfamily are ubiquitous, found in all three domains of life (Supplemental Figures 1 and 3B). According to the transporter classification (TC) system developed by Saier and collaborators, the APC superfamily includes five families, four of which mediate amino acid transport (Chang et al., 2004): the Amino acid-Polyamine-Organocation (APC) family; the Amino Acid/Auxin Permease (AAAP) family; the Alanine or Glycine: Cation Symporter (AGCS) family; the Cation-Chloride Cotransporter (CCC) family; and the Hydroxy/Aromatic Amino Acid Permease (HAAAP) family.
The APC family, the founding member of the APC superfamily, is of particular interest in terms of amino acid export. Most of the members of the APC family mediate transport of amino acids and their derivatives, with a few notable exceptions (Saier, 2000). Transporters belonging to the APC family characterized so far mediate solute–cation symport, solute–solute antiport, or facilitated diffusion (Jack et al., 2000; Verrey et al., 2004). Plants have known homologs in three subfamilies belonging to the APC family: the Cationic Amino Acid Transporters (CATs), the Amino Acid/Choline Transporters (ACTs), and the Polyamine H+-Symporters (PHSs).
Plant members of CATs were initially identified through their ability to mediate amino acid uptake in a heterologous expression system (Frommer et al., 1995). Amino acid uptake by AtCAT1, AtCAT5, and AtCAT6 were shown to be proton-gradient-dependent, suggestive of proton-coupled transport (Frommer et al., 1995; Hammes et al., 2006; Rentsch et al., 2007; Su et al., 2004). In contrast, AtCAT8 might mediate proton-independent, non-energized transport of amino acids (Yang et al., 2010), possibly mediating efflux through facilitated diffusion. Interestingly, proteomics studies identified four members of the AtCAT group (AtCAT2, 4, 8, and 9) on the tonoplast (Jaquinod et al., 2007), and localization of AtCAT2 and AtCAT8 on the tonoplast has been experimentally confirmed (Su et al., 2004; Yang et al., 2010). AtCAT8 is thus a good candidate for the previously observed amino acid transport across the tonoplast, but whether it mediates bi-directional transport has still to be demonstrated.
Transporters that belong to the Amino Acid/Choline Transporters (ACTs) are found in bacteria, fungi, yeasts, and plants. Only one member of this family has been characterized so far in plants: Bidirectional Amino acid Transport (BAT)1 from Arabidopsis increased the accumulation of Arg and Ala, but decreased the accumulation of Lys and Glu. From these results, it was suggested that BAT1 is a bi-directional amino acid transporter, possibly an exporter for certain amino acids (Dundar and Bush, 2009). Here, again, the transport mechanism of BAT1 still remains to be investigated.
Transporters that belong to the PHS family are not well characterized. A transporter belonging to this family, LmPOT1 from Leishmania major, is a proton–polyamine symporter localized on the plasma membrane. Neither the substrates nor transport mechanism have been characterized for plant members (LATs) of the PHS family (Wipf et al., 2002).
The AAAP family, which includes many previously characterized proton-symporters for amino acids, contains sub-families whose members mediate transport of amino acids into the vacuole (Rentsch et al., 2007; Tegeder and Rentsch, 2010; Wipf et al., 2002). Plant proteins responsible for amino acid export have not yet been identified in this family. Saccharomyces cerevisiae Amino Acid Vacuolar Transporters (AVTs) of the AAAP family encode proteins targeted to the tonoplast and involved in amino acid transport (Chahomchuen et al., 2009; Chardwiriyapreecha et al., 2008; Russnak et al., 2001; Shimazu et al., 2005). The AVTs are of particular interest since many plant transporters belong to the same sub-family, including Aromatic and Neutral Transporters (ANTs). From these groups, only ANT1 has been functionally characterized, and shown to import neutral, aromatic amino acids and arginine into yeast (Chen et al., 2001). Unfortunately, neither the sub-cellular localization of ANT1, nor its functional properties have been experimentally determined. Proteomics approaches have found three VAATs (see Supplemental Table 2) and two presently uncharacterized putative amino acid transporters (AT2G40420 and AT3G30390; T1 and T3, Supplemental Table 2) on the Arabidopsis tonoplast (SUBA database: http://suba.plantenergy.uwa.edu.au/ (Heazlewood et al., 2007)), suggesting that some of these transporters could be the long-sought amino acid transporter of the tonoplast, but it remains to be shown if they mediate bi-directional transport across the tonoplast.
THE DRUG/METABOLITE TRANSPORTER (DMT) SUPERFAMILY
The Drug/Metabolite Transporter (DMT) superfamily consists of multiple families (currently the number of families recognized by the TC in this superfamily is 28, www.tcdb.org) that transport a variety of solutes including sugars, nucleobases, nucleotide-sugars, and amino acids (Jack et al., 2001). Transport mechanisms of this superfamily include solute–solute antiport, solute–cation symport, and solute–cation antiport.
Among the families that belong to the DMT superfamily, nine families include plant members: the Plant Drug/metabolite exporters (P-DMEs), triose phosphate transporters (TPTs), the CMP-Sialate:CMP Antiporters (CSAs), the UDP-glucose transporters (UGAs), the GDP-mannose:GMP antiporters (GMAs), the plant organocation permeases (POPs), the Nucleobase Uptake Transporters (NBUTs), the Thiamine Pyrophosphate Transporters (TPPTs), and the NIPA Mg2+ Uptake Permeases (NIPAs). The P-DMEs are of particular interest as potential amino acid transport systems, since they are more closely related to the Drug/metabolite exporter (DMEs) family than any other family. The DME family includes bacterial and archaeal proteins that mediate the efflux of amino acids and amines (Dassler et al., 2000; Livshits et al., 2003; Paul et al., 2000). Members of the P-DME family are poorly characterized. One member from Medicago truncatula, Nodulin 21, has been identified as a nodulation-specific protein (Delauney et al., 1990), but no function has been assigned to this gene. Recently, a member of this family, named Walls Are Thin (WAT1), was isolated as an Arabidopsis homolog of a gene expressed at the onset of secondary cell wall formation in Zinnia elegans tracheary element cultures (Ranocha et al., 2010). T-DNA insertion mutants in WAT1 led to a pleiotropic phenotype, including reduced deposition of secondary cell walls and changes in the content of aromatic amino acids. The mutants showed increased sensitivity to a toxic analog of tryptophan, 5-Methyltryptophan (5-Me-Trp), which could be explained by a modification in the activity of the aromatic biosynthesis pathway (Ranocha et al., 2010). Alternatively, since WAT1 localizes to the tonoplast, this result could suggest that WAT1 sequesters 5-Me-Trp through transporting it from the cytosol into the vacuole. Since vacuolar transport is often mediated by H+-solute antiporters, a plasma membrane protein with a similar mechanism can be expected to mediate solute export from the cell (see above). Therefore, it is possible that some members of P-DMEs function as plasma membrane exporters.
OTHER SUPERFAMILIES OF AMINO ACID EXPORTERS
Other superfamilies of transporters that (1) include amino acid exporters from other organisms and (2) include plant members are summarized below. A large part of plant members belonging to these superfamilies remains uncharacterized, and whether some of them also mediate amino acid export remains to be seen.
ATP Binding Cassette (ABC) Transporter Superfamily
All ABC transporters possess a monophyletic ATP-hydrolyzing constituent that energizes the vectorial transport of substrates. Forty-eight families of ABC transporters, including both prokaryotic and eukaryotic uptake and efflux systems, have been annotated so far (Saier, 2000). The substrate of ABC transporters are diverse, including sugars, amino acids, peptides, nucleotides, lipids, alkaloids, xenobiotic drugs, and glucans (Saier, 2000; Wang et al., 2009). ABCC family (also known as the Drug Conjugate Transporters) can transport amino acid derivatives in eukaryotes. Plant members of the ABCC family include transporters that recognize diverse substrates including glutathione-conjugates, chlorophyll catabolites, inositol phosphate, and folate (reviewed in Rea, 2007; Wanke and Kolukisaoglu, 2010). Interestingly, T-DNA insertion in AtMRP2, a gene encoding a vacuolar transporter of glutathione conjugates and possibly chlorophyll catabolites, causes alternation in amino acids secreted from roots (Badri et al., 2008). Similarly, expression of Cystic Fibrosis Transmembrane Conductance Regulator (CFTR), an ABCC transporter from mammals, has been documented to have an effect on amino acid efflux (Rotoli et al., 1996). In neither case is the exact mechanism that leads to the change in amino acid efflux known.
The Major Facilitator Superfamily (MFS)
The major facilitator superfamily (MFS) is the largest family of transporters that includes members from bacteria, archaea, and eukarya. In bacteria, MFS transporters are one of the major mechanisms for the extrusion of diverse cytotoxic chemicals such as antibiotics (Fluman and Bibi, 2009). Substrates of MFS members include sugars, polyols, drugs, neurotransmitters, peptides, and inorganic anions and amino acids (Fluman and Bibi, 2009). MFS members that transport amino acids include the yeast Vacuolar Basic Amino Acid Transporters (V-BAAT) (Chardwiriyapreecha et al., 2008; Shimazu et al., 2005), the low affinity aromatic amino acid (Tyr, Trp, Phe) transporter, TAT1 (Kim et al., 2001), the human L-Amino Acid Transporters LAT3 and 4 (Babu et al., 2003; Bodoy et al., 2005), Proteobacterial Intraphagosomal Amino Acid Transporters (Pht) (Sauer et al., 2005), and Acids and Quinidine Resistant 1(AQR1) in yeasts (Velasco et al., 2003, 2004). Among these transporters, LAT3, LAT4, and TAT1 mediate facilitated diffusion and therefore can function as amino acid exporters. AQR1, which shares the highest sequence similarity with H+ antiporters, is likely to mediate amino acid excretion by either H+–amino acid exchanger activity or vesicular loading and subsequent exocytosis (Velasco et al., 2004).
REGULATION OF AMINO ACID EXPORT
Since amino acids are the major form of nitrogen transported between organs, it is not surprising that the quantity and composition of extracellular amino acids are subject to environmental and developmental changes such as light intensity (Lam et al., 1995), seasonal changes (Couturier et al., 2010; Sagisaka, 1974), and plant–microbe interaction (Phillips et al., 2004; Rico and Preston, 2008). Most likely, these changes involve regulations of multiple enzymes and transporters. Enzymes involved in nitrogen assimilation and amino acid biosynthesis are regulated by environmental conditions such as nitrogen availability, biotic and abiotic stresses, and carbon/nitrogen balance (Hsieh et al., 1996; Lam et al., 1995; Nunes-Nesi et al., 2010; Oliveira et al., 2001; Scheible et al., 2004; Tzin and Galili, 2010). Likewise, some amino acid importers are regulated by addition of external nitrate (Hirner et al., 2006; Liu and Bush, 2006).
Because the molecular identities of amino acid exporters are largely unknown, the regulation of amino acid export is similarly obscure. However, a mutant that might be deficient in the regulation of amino acid export has been discovered. The gdu1-1D mutant that overexpresses GDU1, a single transmembrane protein with no similarity to previously characterized proteins, secretes large amounts of Gln from the hydathodes (Pilot et al., 2004). This mutant displays a large increase of free amino acids, especially glutamine, in the xylem sap and apoplasmic fluid, suggesting that GDU1 is involved in the regulation of amino acid export. These results corroborate previous studies that found over-representation of Gln secreted in the xylem sap and seed coat exudates (Lanfermeijer et al., 1992; Pilot et al., 2004; Wolswinkel and Deruiter, 1985). Moreover, the export process stimulated in gdu1-1D is passive, specific for amino acids, and shows a weak preference for Gln over the other amino acids (Pratelli et al., 2010)—characteristics that were observed for amino acid export systems in previous physiological studies (deJong et al., 1997; Lanfermeijer et al., 1992; Wolswinkel and Deruiter, 1985).
The mechanism of how GDU1 regulates the export of amino acids is currently unknown. Because GDU1 is a single transmembrane protein, it is unlikely that the GDU1 itself constitutes an amino acid exporter (Pilot et al., 2004). Similarly to the small subunit of the heteromeric amino acid exchanger (4F2hc or rBAT) from mammals (Chillaron et al., 2001), GDU1 might be a part of a transporter complex. Alternatively, GDU1 might be a chaperone that is necessary for the correct localization and/or function of an amino acid exporter. In a curious analogy, mutation in a single transmembrane protein, Tie-dyed 1, results in defective sucrose export from the source leaves in maize (Ma et al., 2009). The search for interacting partners for membrane proteins (Lalonde et al., 2008, 2010; Obrdlik et al., 2004) may help in discovering the mechanisms through which GDU1 controls the amino acid export process.
CONCLUSION
Export of solutes from cells is one of the least studied areas in transport physiology. Although cellular export of amino acids is essential for the proper distribution of nitrogen in plants, surprisingly little is known at the molecular level. Yet, physiological studies clearly indicate that amino acid export is a process mediated by multiple transporters, some of which are predicted to be previously uncharacterized bi-directional facilitators. Deep phylogenetic analysis between transporter proteins from all domains of life, enabled by the rapid expansion in the number of sequenced genomes, suggests that plant genomes encode multiple proteins that share homology to amino acid exporter proteins from other organisms. These proteins, most of which are uncharacterized, are likely candidates of amino acid exporters in plants. Novel methods that can detect metabolite export with higher throughput might help the discovery of amino acid exporters in plants (Okumoto et al., 2008). Another exciting area is the regulation of amino acid export. In addition to further studies of mechanisms through which GDU1 controls amino acid export, efforts such as the high-throughput metabolite analysis of large number of T-DNA mutants (Ajjawi et al., 2010; Chen et al., 2010) may lead to the identification of additional players in the regulation of amino acid export processes.
FUNDING
The work in the author's lab was supported by National Institutes of Health (1R21NS064412-01) and Jeffress Memorial Trust (J-908).
Supplementary Material
References
- Ajjawi I, Lu Y, Savage LJ, Bell SM, Last RL. Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project. Plant Physiol. 2010;152:529–540. doi: 10.1104/pp.109.148494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews M. The partitioning of nitrate assimilation between the root and shoot of higher plants. Plant Cell Environ. 1986;9:511–519. [Google Scholar]
- Apse MP, Blumwald E. Na+ transport in plants. FEBS Lett. 2007;581:2247–2254. doi: 10.1016/j.febslet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Babu E, et al. Identification of a novel system L amino acid transporter structurally distinct from heterodimeric amino acid transporters. J. Biol. Chem. 2003;278:43838–43845. doi: 10.1074/jbc.M305221200. [DOI] [PubMed] [Google Scholar]
- Badri DV, et al. Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP-binding cassette transporter mutants. Plant Physiol. 2008;146:762–771. doi: 10.1104/pp.107.109587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin C, Yang XH, Weston LA. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil. 2003;256:67–83. [Google Scholar]
- Bodoy S, Martin L, Zorzano A, Palacin M, Estevez R. Identification of LAT4, a novel amino acid transporter with system L activity. J. Biol. Chem. 2005;280:12002–12011. doi: 10.1074/jbc.M408638200. [DOI] [PubMed] [Google Scholar]
- Burkovski A, Kramer R. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Applied Microbiol. Biotechnol. 2002;58:265–274. doi: 10.1007/s00253-001-0869-4. [DOI] [PubMed] [Google Scholar]
- Chahomchuen T, Hondo K, Ohsaki M, Sekito T, Kakinuma Y. Evidence for Avt6 as a vacuolar exporter of acidic amino acids in Saccharomyces cerevisiae cells. J. General Applied Microbiol. 2009;55:409–417. doi: 10.2323/jgam.55.409. [DOI] [PubMed] [Google Scholar]
- Chang AB, Lin R, Studley WK, Tran CV, Saier MH. Phylogeny as a guide to structure and function of membrane transport proteins (Review) Molecular Membrane Biol. 2004;21:171–181. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- Chardwiriyapreecha S, et al. Identification of the fnx1+ and fnx2+ genes for vacuolar amino acid transporters in Schizosaccharomyces pombe. FEBS Lett. 2008;582:2225–2230. doi: 10.1016/j.febslet.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Chen LQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LS, Ortiz-Lopez A, Jung A, Bush DR. ANT1, an aromatic and neutral amino acid transporter in Arabidopsis. Plant Physiol. 2001;125:1813–1820. doi: 10.1104/pp.125.4.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillaron J, Roca R, Valencia A, Zorzano A, Palacin M. Heteromeric amino acid transporters: biochemistry, genetics, and physiology. Amer. J. Physiol.—Renal Physiol. 2001;281:F995–F1018. doi: 10.1152/ajprenal.2001.281.6.F995. [DOI] [PubMed] [Google Scholar]
- Couturier J, Doidy J, Guinet F, Wipf D, Blaudez D, Chalot M. Glutamine, arginine and the amino acid transporter Pt-CAT11 play important roles during senescence in poplar. Ann. Bot. 2010;105:1159–1169. doi: 10.1093/aob/mcq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassler T, Maier T, Winterhalter C, Bock A. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol. Microbiol. 2000;36:1101–1112. doi: 10.1046/j.1365-2958.2000.01924.x. [DOI] [PubMed] [Google Scholar]
- deJong A, KoerselmanKooij J, Schuurmans J, Borstlap A. The mechanism of amino acid efflux from seed coats of developing pea seeds as revealed by uptake experiments. Plant Physiol. 1997;114:731–736. doi: 10.1104/pp.114.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauney AJ, Cheon CI, Snyder PJ, Verma DPS. A nodule-specific sequence encoding a methionine-rich polypeptide, nodulin-21. Plant Mol. Biol. 1990;14:449–451. doi: 10.1007/BF00028782. [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Jager R, Kaiser G, Martinoia E. Amino acid transport across the tonoplast of vacuoles isolated from barley mesophyll protoplasts: uptake of alanine, leucine, and glutamine. Plant Physiol. 1990;92:123–129. doi: 10.1104/pp.92.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Martinoia E, Heber U. Mobilisation of vacuolar amino acids in leaf cells as affected by ATP and the level of cytosolic amino acids: ATP regulates but appears not to energize vacuolar amino-acid release. Biochim. Biophys. Acta. 1989;984:57–62. [Google Scholar]
- Dundar E, Bush DR. BAT1, a bidirectional amino acid transporter in Arabidopsis. Planta. 2009;229:1047–1056. doi: 10.1007/s00425-009-0892-8. [DOI] [PubMed] [Google Scholar]
- Dunkel M, Latz A, Schumacher K, Wuller T, Becker D, Hedrich R. Targeting of vacuolar membrane localized members of the TPK channel family. Mol. Plant. 2008;1:938–949. doi: 10.1093/mp/ssn064. [DOI] [PubMed] [Google Scholar]
- Endler A, et al. Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 2006;141:196–207. doi: 10.1104/pp.106.079533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluman N, Bibi E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim. Biophys. Acta—Proteins and Proteomics. 2009;1794:738–747. doi: 10.1016/j.bbapap.2008.11.020. [DOI] [PubMed] [Google Scholar]
- Frankard V, Ghislain M, Jacobs M. Two feedback-insensitive enzymes of the aspartate pathway in Nicotiana sylvestris. Plant Physiol. 1992;99:1285–1293. doi: 10.1104/pp.99.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Unseld M, Ninnemann O. Seed and vascular expression of a high-affinity transporter for cationic amino acids in Arabidopsis. Proc. Natl Acad. Sci. U S A. 1995;92:12036–12040. doi: 10.1073/pnas.92.26.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett. 2006;580:1094–1102. doi: 10.1016/j.febslet.2005.11.054. [DOI] [PubMed] [Google Scholar]
- Goerlach J, Willmshoff I. Glycine uptake into barley mesophyll vacuoles is regulated but not energized by ATP. Plant Physiol. 1992;99:134–139. doi: 10.1104/pp.99.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Friml J. The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 2010;29:2700–2714. doi: 10.1038/emboj.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL. LC–MS/MS assay for protein amino acids and metabolically related compounds for large-scale screening of metabolic phenotypes. Analyt. Chem. 2007;79:8067–8075. doi: 10.1021/ac070938b. [DOI] [PubMed] [Google Scholar]
- Hammes UZ, Nielsen E, Honaas LA, Taylor CG, Schachtman DP. AtCAT6, a sink-tissue-localized transporter for essential amino acids in Arabidopsis. Plant J. 2006;48:414–426. doi: 10.1111/j.1365-313X.2006.02880.x. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis subcellular database. Nucleic Acids Res. 2007;35:D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirner A, et al. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell. 2006;18:1931–1946. doi: 10.1105/tpc.106.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homeyer U, Schultz G. Transport of phenylalanine into vacuoles isolated from barley mesophyll protoplasts. Planta. 1988;176:378–382. doi: 10.1007/BF00395418. [DOI] [PubMed] [Google Scholar]
- Homeyer U, Litek K, Huchzermeyer B, Schultz G. Uptake of phenylalanine into isolated barley vacuoles is driven by both tonoplast adenosine triphosphatase and pyrophosphatase: evidence for a hydrophobic l-amino acid carrier system. Plant Physiol. 1989;89:1388–1393. doi: 10.1104/pp.89.4.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Lam HM, Coruzzi G. Metabolic regulation of nitrogen assimilatory genes in Arabidopsis. Plant Physiol. 1996;111:615–615. [Google Scholar]
- Jack DL, Paulsen IT, Saier MH. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiol. 2000;146(Pt 8):1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- Jack DL, Yang NM, Saier MH. The drug/metabolite transporter superfamily. Euro. J. Biochem. 2001;268:3620–3639. doi: 10.1046/j.1432-1327.2001.02265.x. [DOI] [PubMed] [Google Scholar]
- Jaquinod M, et al. A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteom. 2007;6:394–412. doi: 10.1074/mcp.M600250-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke WD, Hartung W. Root-shoot interactions in mineral nutrition. Plant Soil. 2000;226:57–69. [Google Scholar]
- Jones DL, Darrah PR. Amino-acid influx at the soil-root interface of Zea mays L. and its implications in the rhizosphere. Plant Soil. 1994;163:1–12. [Google Scholar]
- Jones DL, Shannon D, Junvee-Fortune T, Farrarc JF. Plant capture of free amino acids is maximized under high soil amino acid concentrations. Soil Biol. Biochem. 2005;37:179–181. [Google Scholar]
- Kabala K, Klobus GY. Plant Ca2+-ATPases. Acta Physiolog. Plantarum. 2005;27:559–574. [Google Scholar]
- Kim DK, Kanai Y, Chairoungdua A, Matsuo H, Cha SH, Endou H. Expression cloning of a Na+-independent aromatic amino acid transporter with structural similarity to H+/monocarboxylate transporters. J. Biol. Chem. 2001;276:17221–17228. doi: 10.1074/jbc.M009462200. [DOI] [PubMed] [Google Scholar]
- Kinraide TB. Interamino acid inhibition of transport in higher plants: evidence for two transport channels with ascertainable affinities for amino acids. Plant Physiol. 1981;68:1327–1333. doi: 10.1104/pp.68.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinraide TB, Newman IA, Etherton B. A quantitative simulation model for H+-amino acid cotransport to interpret the effects of amino acids on membrane potential and extracellular pH. Plant Physiol. 1984;76:806–813. doi: 10.1104/pp.76.3.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W, et al. Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J. 2003;33:211–220. doi: 10.1046/j.1365-313x.2003.01618.x. [DOI] [PubMed] [Google Scholar]
- Kraffczyk I, Trolldenier G, Beringer H. Soluble root exudates of maize: influence of potassium supply and rhizosphere microorganisms. Soil Biol. Biochem. 1984;16:315–322. [Google Scholar]
- Kruse J, et al. Elevated pCO2 favours nitrate reduction in the roots of wild-type tobacco (Nicotiana tabacum cv. Gat.) and significantly alters N-metabolism in transformants lacking functional nitrate reductase in the roots. J. Exp. Bot. 2002;53:2351–2367. doi: 10.1093/jxb/erf094. [DOI] [PubMed] [Google Scholar]
- Lalonde S, Ehrhardt DW, Loque D, Chen J, Rhee SY, Frommer WB. Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J. 2008;53:610–635. doi: 10.1111/j.1365-313X.2007.03332.x. [DOI] [PubMed] [Google Scholar]
- Lalonde S, et al. A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Frontier Plant Physiol. 2010;1:1–14. doi: 10.3389/fphys.2010.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Tegeder M, Throne-Holst M, Frommer WB, Patrick JW. Phloem loading and unloading of sugars and amino acids. Plant Cell Environ. 2003;26:37–56. [Google Scholar]
- Lalonde S, Wipf D, Frommer WB. Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Ann. Rev. Plant Biol. 2004;55:341–372. doi: 10.1146/annurev.arplant.55.031903.141758. [DOI] [PubMed] [Google Scholar]
- Lam HM, et al. Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell. 1995;7:887–898. doi: 10.1105/tpc.7.7.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfermeijer FC, Vanoene MA, Borstlap AC. Compartmental analysis of amino-acid release from attached and detached pea seed coats. Planta. 1992;187:75–82. doi: 10.1007/BF00201626. [DOI] [PubMed] [Google Scholar]
- Leidreiter K, Kruse A, Heineke D, Robinson DG, Heldt HW. Subcellular volumes and metabolite concentrations in potato (Solanum tuberosum cv. Desiree) leaves. Botan. Acta. 1995;108:439–444. [Google Scholar]
- Lesuffleur F, Cliquet JB. Characterisation of root amino acid exudation in white clover (Trifolium repens L.) Plant Soil. 2010;333:191–201. [Google Scholar]
- Lesuffleur F, Paynel F, Bataille MP, Le Deunff E, Cliquet JB. Root amino acid exudation: measurement of high efflux rates of glycine and serine from six different plant species. Plant Soil. 2007;294:235–246. [Google Scholar]
- Liu X, Bush DR. Expression and transcriptional regulation of amino acid transporters in plants. Amino Acids. 2006;30:113–120. doi: 10.1007/s00726-005-0248-z. [DOI] [PubMed] [Google Scholar]
- Livshits VA, Zakataeva NP, Aleshin VV, Vitushkina MV. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 2003;154:123–135. doi: 10.1016/S0923-2508(03)00036-6. [DOI] [PubMed] [Google Scholar]
- Lodwig EM, et al. Amino-acid cycling drives nitrogen fixation in the legume–Rhizobium symbiosis. Nature. 2003;422:722–726. doi: 10.1038/nature01527. [DOI] [PubMed] [Google Scholar]
- Lohaus G, Moellers C. Phloem transport of amino acids in two Brassica napus L. genotypes and one B-carinata genotype in relation to their seed protein content. Planta. 2000;211:833–840. doi: 10.1007/s004250000349. [DOI] [PubMed] [Google Scholar]
- Lohaus G, Winter H, Riens B, Heldt HW. Further studies of the phloem loading process in leaves of barley and spinach: the comparison of metabolite concentrations in the apoplastic compartment with those in the cytosolic compartment and in the sieve tubes. Botan. Acta. 1995;108:270–275. [Google Scholar]
- Ma Y, Slewinski TL, Baker RF, Braun DM. Tie-dyed1 encodes a novel, phloem-expressed transmembrane protein that functions in carbohydrate partitioning. Plant Physiol. 2009;149:181–194. doi: 10.1104/pp.108.130971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. London and San Diego: Academic Press; 1995. [Google Scholar]
- Martinoia E, Grill E, Tommasini R, Kreuz K, Amrhein N. ATP-dependent glutathione S-conjugate ‘export’ pump in the vacuolar membrane of plants. Nature. 1993;364:247–249. [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE. Vacuolar transporters and their essential role in plant metabolism. J. Exper. Bot. 2007;58:83–102. doi: 10.1093/jxb/erl183. [DOI] [PubMed] [Google Scholar]
- Martinoia E, Thume M, Vogt E, Rentsch D, Dietz KJ. Transport of arginine and aspartic acid into isolated barley mesophyll vacuoles. Plant Physiol. 1991;97:644–650. doi: 10.1104/pp.97.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Verdoucq L, Luu DT, Santoni V. Plant aquaporins: membrane channels with multiple integrated functions. Ann. Rev. Plant Biol. 2008;59:595–624. doi: 10.1146/annurev.arplant.59.032607.092734. [DOI] [PubMed] [Google Scholar]
- Mueller J, Mettbach U, Menzel D, Samaj J. Molecular dissection of endosomal compartments in plants. Plant Physiol. 2007;145:293–304. doi: 10.1104/pp.107.102863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasholm T, Kielland K, Ganeteg U. Uptake of organic nitrogen by plants. New Phytologist. 2009;182:31–48. doi: 10.1111/j.1469-8137.2008.02751.x. [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant. 2010;3:973–996. doi: 10.1093/mp/ssq049. [DOI] [PubMed] [Google Scholar]
- Obrdlik P, et al. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl Acad. Sci. U S A. 2004;101:12242–12247. doi: 10.1073/pnas.0404467101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S, Takanaga H, Frommer WB. Quantitative imaging for discovery and assembly of the metabo-regulome. New Phytologist. 2008;180:271–295. doi: 10.1111/j.1469-8137.2008.02611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira IC, et al. Metabolite and light regulation of metabolism in plants: lessons from the study of a single biochemical pathway. Brazilian J. Med. Biol. Res. 2001;34:567–575. doi: 10.1590/s0100-879x2001000500003. [DOI] [PubMed] [Google Scholar]
- Pate JS. Uptake, assimilation and transport of nitrogen compounds by plants. Soil Biol. Biochem. 1973;5:109–119. [Google Scholar]
- Pate JS, Atkins CA, Herridge DF, Layzell DB. Synthesis, storage, and utilization of amino compounds in white lupin (Lupinus albus L.) Plant Physiol. 1981;67:37–42. doi: 10.1104/pp.67.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul L, Ferguson DJ, Krzycki JA. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 2000;182:2520–2529. doi: 10.1128/jb.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson J, Nasholm T. A GC–MS method for determination of amino acid uptake by plants. Physiol. Plant. 2001;113:352–358. doi: 10.1034/j.1399-3054.2001.1130308.x. [DOI] [PubMed] [Google Scholar]
- Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teuber LR. Microbial products trigger amino acid exudation from plant roots. Plant Physiol. 2004;136:2887–2894. doi: 10.1104/pp.104.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot G, et al. Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from Hydathodes of Arabidopsis leaves. Plant Cell. 2004;16:1827–1840. doi: 10.1105/tpc.021642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Voll LM, Horst RJ, Frommer WB, Pilot G. Stimulation of non-selective amino acid export by Glutamine Dumper proteins. Plant Physiol. 2010;152:762–773. doi: 10.1104/pp.109.151746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prell J, White JP, Bourdes A, Bunnewell S, Bongaerts RJ, Poole PS. Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc. Natl Acad. Sci. U S A. 2009;106:12477–12482. doi: 10.1073/pnas.0903653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudelski B, Kraus S, Soll J, Philippar K. The plant PRAT proteins: preprotein and amino acid transport in mitochondria and chloroplasts. Plant Biol. 2010;12:42–55. doi: 10.1111/j.1438-8677.2010.00357.x. [DOI] [PubMed] [Google Scholar]
- Ranocha P, et al. Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 2010;63:469–483. doi: 10.1111/j.1365-313X.2010.04256.x. [DOI] [PubMed] [Google Scholar]
- Rea PA. Plant ATP-binding cassette transporters. Ann. Rev. Plant Biol. 2007;58:347–375. doi: 10.1146/annurev.arplant.57.032905.105406. [DOI] [PubMed] [Google Scholar]
- Reinders A, Sivitz AB, Starker CG, Gantt JS, Ward JM. Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lotus japonicus. Plant Mol. Biol. 2008;68:289–299. doi: 10.1007/s11103-008-9370-0. [DOI] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M. Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 2007;581:2281–2289. doi: 10.1016/j.febslet.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Rico A, Preston GM. Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol. Plant Microbe Interact. 2008;21:269–282. doi: 10.1094/MPMI-21-2-0269. [DOI] [PubMed] [Google Scholar]
- Riens B, Lohaus G, Heineke D, Heldt HW. Amino acid and sucrose content determined in the cytosolic, chloroplastic, and vacuolar compartments and in the phloem sap of spinach leaves. Plant Physiol. 1991;97:227–233. doi: 10.1104/pp.97.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotoli BM, Bussolati O, Cabrini G, Gazzola GC. Changes in neutral amino acid efflux and membrane potential associated with the expression of GFTR protein. Amino Acids. 1996;11:247–255. doi: 10.1007/BF00813863. [DOI] [PubMed] [Google Scholar]
- Rroco E, Kosegarten H, Mengel K. Importance of plasmalemma H+-ATPase activity for N losses from intact roots of spring wheat (Triticum aestivum L.) Euro. J. Agron. 2002;16:187–196. [Google Scholar]
- Russnak R, Konczal D, McIntire SL. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J. Biol. Chem. 2001;276:23849–23857. doi: 10.1074/jbc.M008028200. [DOI] [PubMed] [Google Scholar]
- Sagisaka S. Effect of low temperature on amino acid metabolism in wintering poplar. Plant Physiol. 1974;53:319–322. doi: 10.1104/pp.53.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology. 2000;146:1775–1795. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
- Saier MH, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Bachman MA, Swanson MS. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl Acad. Sci. U S A. 2005;102:9924–9929. doi: 10.1073/pnas.0502767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert C, Komor E. Amino acid uptake by Ricinus communis roots: characterization and physiological significance. Plant Cell Environ. 1987;10:493–500. [Google Scholar]
- Shaul O, Galili G. Threonine overproduction in transgenic tobacco plants expressing a mutant desensitized aspartate kinase of Escherichia coli. Plant Physiol. 1992;100:1157–1163. doi: 10.1104/pp.100.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu M, Sekito T, Akiyama K, Ohsumi Y, Kakinuma Y. A family of basic amino acid transporters of the vacuolar membrane from Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:4851–4857. doi: 10.1074/jbc.M412617200. [DOI] [PubMed] [Google Scholar]
- Su YH, Frommer WB, Ludewig U. Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiol. 2004;136:3104–3113. doi: 10.1104/pp.104.045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Rentsch D. Uptake and partitioning of amino acids and peptides. Mol. Plant. 2010;3:997–1011. doi: 10.1093/mp/ssq047. [DOI] [PubMed] [Google Scholar]
- Thume M, Dietz KJ. Reconstitution of the tonoplast amino-acid carrier into liposomes: evidence for an ATP-regulated carrier in different species. Planta. 1991;185:569–575. doi: 10.1007/BF00202968. [DOI] [PubMed] [Google Scholar]
- Tzin V, Galili G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant. 2010;3:956–952. doi: 10.1093/mp/ssq048. [DOI] [PubMed] [Google Scholar]
- Ufaz S, Galili G. Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol. 2008;147:954–961. doi: 10.1104/pp.108.118091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Laan RGW, Wouterlood M, Borstlap AC. Electrodiffusional uptake of organic cations by pea seed coats: further evidence for poorly selective pores in the plasma membrane of seed coat parenchyma cells. Plant Physiol. 2001;126:1688–1697. doi: 10.1104/pp.126.4.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco I, Calderon IL, Andre B. The AQR1 transporter mediates amino acid excretion in Saccharomyces cerevisiae. Yeast. 2003;20:S237. doi: 10.1128/EC.3.6.1492-1503.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco I, Tenreiro S, Calderon IL, Andre B. Saccharomyces cerevisiae Aqr1 is an internal-membrane transporter involved in excretion of amino acids. Eukaryot. Cell. 2004;3:1492–1503. doi: 10.1128/EC.3.6.1492-1503.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrey F, Closs EI, Wagner CA, Palacin M, Endou H, Kanai Y. CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch. 2004;447:532–542. doi: 10.1007/s00424-003-1086-z. [DOI] [PubMed] [Google Scholar]
- Walker TS, Bais HP, Grotewold E, Vivanco JM. Root exudation and rhizosphere biology. Plant Physiol. 2003;132:44–51. doi: 10.1104/pp.102.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Dukarevich M, Sun EI, Yen MR, Saier MH. Membrane porters of ATP-binding cassette transport systems are polyphyletic. J. Membrane Biol. 2009;231:1–10. doi: 10.1007/s00232-009-9200-6. [DOI] [PubMed] [Google Scholar]
- Wanke D, Kolukisaoglu HU. An update on the ABCC transporter family in plants: many genes, many proteins, but how many functions? Plant Biol. 2010;12:15–25. doi: 10.1111/j.1438-8677.2010.00380.x. [DOI] [PubMed] [Google Scholar]
- Winter H, Robinson DG, Heldt HW. Subcellular volumes and metabolite concentrations in barley leaves. Planta. 1993;191:180–190. [Google Scholar]
- Wipf D, Ludewig U, Tegeder M, Rentsch D, Koch W, Frommer WB. Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem. Sci. 2002;27:139–147. doi: 10.1016/s0968-0004(01)02054-0. [DOI] [PubMed] [Google Scholar]
- Wolswinkel P, Deruiter H. Amino acid release from the seed coat of developing seeds of Vicia faba and Pisum sativum. Annals Bot. 1985;55:283–287. [Google Scholar]
- Yang HY, Bogner M, Stierhof YD, Ludewig U. H+-independent glutamine transport in plant root tips. Plos One. 2010;5:e8197. doi: 10.1371/journal.pone.0008917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazimalova E, Murphy AS, Yang HB, Hoyerova K, Hosek P. Auxin transporters: why so many? Cold Spring Harbor Perspect. Biol. 2010;2:a001552. doi: 10.1101/cshperspect.a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WH, Zhou YC, Dibley KE, Tyerman SD, Furbank RT, Patrick JW. Nutrient loading of developing seeds. Func. Plant Biol. 2007;34:314–331. doi: 10.1071/FP06271. [DOI] [PubMed] [Google Scholar]
- Zhu X, Galili G. Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell. 2003;15:845–853. doi: 10.1105/tpc.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.