ABSTRACT
Microbes use directed motility to colonize harsh and dynamic environments. We discovered that Helicobacter pylori strains establish bacterial colonies deep in the gastric glands and identified a novel protein, ChePep, necessary to colonize this niche. ChePep is preferentially localized to the flagellar pole. Although mutants lacking ChePep have normal flagellar ultrastructure and are motile, they have a slight defect in swarming ability. By tracking the movement of single bacteria, we found that ∆ChePep mutants cannot control the rotation of their flagella and swim with abnormally frequent reversals. These mutants even sustain bursts of movement backwards with the flagella pulling the bacteria. Genetic analysis of the chemotaxis signaling pathway shows that ChePep regulates flagellar rotation through the chemotaxis system. By examining H. pylori within a microscopic pH gradient, we determined that ChePep is critical for regulating chemotactic behavior. The chePep gene is unique to the Epsilonproteobacteria but is found throughout this diverse group. We expressed ChePep from other members of the Epsilonproteobacteria, including the zoonotic pathogen Campylobacter jejuni and the deep sea hydrothermal vent inhabitant Caminibacter mediatlanticus, in H. pylori and found that ChePep is functionally conserved across this class. ChePep represents a new family of chemotaxis regulators unique to the Epsilonproteobacteria and illustrates the different strategies that microbes have evolved to control motility.
IMPORTANCE
Helicobacter pylori strains infect half of all humans worldwide and contribute to the development of peptic ulcers and gastric cancer. H. pylori cannot survive within the acidic lumen of the stomach and uses flagella to actively swim to and colonize the protective mucus and epithelium. The chemotaxis system allows H. pylori to navigate by regulating the rotation of its flagella. We identified a new protein, ChePep, which controls chemotaxis in H. pylori. ChePep mutants fail to colonize the gastric glands of mice and are completely outcompeted by normal H. pylori. Genes encoding ChePep are found only in the class Epsilonproteobacteria, which includes the human pathogen Campylobacter jejuni and environmental microbes like the deep-sea hydrothermal vent colonizer Caminibacter mediatlanticus, and we show that ChePep function is conserved in this class. Our study identifies a new colonization factor in H. pylori and also provides insight into the control and evolution of bacterial chemotaxis.
Introduction
The Epsilonproteobacteria are an ancient class of bacteria that dominate extreme environments, such as deep sea hydrothermal vents, sulfidic caves, and the human stomach (1). Some, like Campylobacter jejuni and Helicobacter pylori, are also major human pathogens (2, 3). To avoid harmful chemicals and access beneficial conditions, many Epsilonproteobacteria utilize chemotaxis to control the direction of their motility in response to their environment. For example, H. pylori uses chemotaxis to avoid the acidic conditions of the gastric lumen and colonize a narrow niche of buffered mucus overlaying the stomach epithelium as well as the surface of epithelial cells (4).
Bacteria rotate flagella to generate propulsion, but also modulate the speed and direction of flagellar rotation to steer their overall trajectory (5, 6). The chemotaxis system couples control of flagellar rotation with environmental sensing to enable bacteria to move towards attractants and away from repellents (7). The role of chemotaxis in the Epsilonproteobacteria has mostly been investigated as a colonization factor for pathogenic bacteria, but many environmental members of this class also possess chemotaxis homologs, indicating that this process is broadly utilized (8, 9).
The chemotaxis signaling system in the Epsilonproteobacteria is a slightly modified histidine-aspartate phosphorelay (HAP) system and is similar to the basic chemotaxis signaling system conserved across all bacteria (7, 10, 11). Extensive study of bacteria like Escherichia coli and Bacillus subtilis has provided a useful framework for understanding the chemotaxis system in Epsilonproteobacteria, as all previously identified chemotaxis genes in Epsilonproteobacteria are homologous to genes initially described in E. coli and B. subtilis (12–14). Therefore, we were surprised to identify a previously uncharacterized family of proteins that control chemotaxis in Epsilonproteobacteria, which we term “ChePep.”
By studying the pathogenic member of the Epsilonproteobacteria, H. pylori, which is associated with peptic ulcers and gastric cancer (15, 16), we show that ChePep controls flagellar switching in a chemotaxis signaling protein-dependent manner. When examining gastric tissue from mice infected with H. pylori, we find colonization of the gastric antral glands requires ChePep and that ChePep confers a significant overall advantage to colonizing the stomach. Finally, we show that ChePep function is conserved across the Epsilonproteobacteria as we can complement H. pylori lacking its endogenous ChePep with orthologs from the zoonotic pathogen Campylobacter jejuni and the deep sea hydrothermal vent inhabitant Caminibacter mediatlanticus.
RESULTS
H. pylori ChePep localizes to the bacterial poles and is part of a novel family of proteins unique to the Epsilonproteobacteria.
We identified ChePep because of our interest in Helicobacter pylori colonization of the epithelial cell surface. H. pylori cells preferentially adhere to the cell surface near the epithelial junctions and grow into cell-associated microcolonies at these sites (17, 18). We have shown that H. pylori cells modify their attachment sites by recruiting junction-associated proteins near the sites of bacterial attachment (19). While investigating host proteins associated with the adhered bacteria, we noticed that an affinity-purified antibody against the junctional protein Par-3 recognized not just the tight junctions, but also an H. pylori protein.
Immunoprecipitation of the unknown H. pylori protein with the anti-Par-3 antibody yielded a single polypeptide that runs at 98 kDa on SDS-PAGE (see Fig. S1A in the supplemental material). Using mass spectrometry, we identified this protein as “poly-E-rich protein” (HPG27_303), a protein of unknown function annotated for its preponderance of glutamic acid residues. The predicted amino acid sequence of poly-E-rich protein has no homology to Par-3, but shares an antigenic site, as anti-Par-3 antibody recognizes recombinant poly-E-rich protein expressed in E. coli (Fig. S1B).
Further bioinformatic analysis of poly-E-rich protein revealed homologous genes throughout the Epsilonproteobacteria, but none from any other bacterial classes (see Fig. S1C in the supplemental material). Yet despite conserved synteny in all epsilonproteobacterial genomes, poly-E-rich protein homologs are differentially annotated in this class, with many homologs being referred to as hypothetical proteins. To unify these homologs under a shared name and reflect their proposed function, we will refer to all poly-E-rich protein homologs as “ChePep.”
Although ChePep homologs vary in predicted molecular weight and amino acid sequence, they all contain a conserved amino-terminal region with a putative response regulator motif present in histidine-aspartate phosphorelay (HAP) systems (see Fig. S1D in the supplemental material) (20). The presence of a putative response regulator suggested the hypothesis that ChePep homologs function with a HAP system.
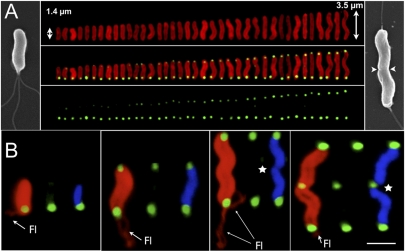
Bacteria utilize HAP systems to sense and respond to a wide variety of environmental changes (21). One of the best understood HAP systems is the chemotaxis system, which controls movement of bacteria in relation to changing environmental chemical concentrations (7). Many chemotaxis proteins concentrate at the poles of bacteria (22–24), so to determine ChePep localization, we raised antibodies specific to H. pylori ChePep. Antibodies made against recombinant ChePep detect a single polypeptide band by immunoblotting bacterial lysates, and ChePep localizes exclusively to the poles of H. pylori (see Fig. S2B in the supplemental material). By analyzing bacterial populations at different stages in the cell cycle, we noticed that immunofluorescence intensity of ChePep varies at one bacterial pole. We therefore double labeled bacteria with both anti-H. pylori and anti-ChePep antibodies and aligned them by their length. This revealed that ChePep immunofluorescence signal is relatively constant at one bacterial pole, while signal at the other pole increases as the bacteria elongate during the cell cycle (Fig. 1A). Quantification of anti-ChePep fluorescence intensity at the distal pole shows a strong positive correlation between ChePep levels at this pole and bacterial length (Fig. S2A). This observation suggests that ChePep levels remain relatively constant at the old bacterial pole, while increasing in concentration at the new pole as bacteria elongate. ChePep intensity reaches equivalent levels at both poles just prior to cell division (Fig. 1A). Also at the time of cell division, we observed that H. pylori can have flagella at both poles, which upon septation will yield two daughter cells with a bundle of flagella at the old bacterial pole (Fig. 1A). This suggested that ChePep is preferentially concentrated at the flagellar pole of H. pylori. To test this we performed higher-resolution confocal microscopy with three-dimensional (3D) reconstruction of H. pylori under conditions where the flagella, ChePep, and chromosome can be simultaneously visualized (Fig. 1B). This analysis confirmed that in the shortest bacteria, with no evidence of chromosomal segregation, the highest concentration of ChePep is at the flagellar pole. The immunofluorescence intensity of ChePep at the distal pole increases as the bacteria elongate, although ChePep accumulates at this pole prior to the appearance of flagella (Fig. 1B). Once ChePep reaches equal concentration at each pole, we observe chromosomal segregation, indicating these bacteria are preparing for imminent septation (Fig. 1B).
FIG 1 .
ChePep localizes to the flagellar poles. (A) H. pylori cells at different stages of the cell cycle were stained with antibodies to H. pylori (red) and ChePep (green) and aligned according to their length to analyze ChePep expression and localization in individual bacteria. Scanning electron microscopy (SEM) illustrates the location of flagella in a young (left) versus septating (right; arrowheads indicate septation cleft) H. pylori cell. (For the quantitative relationship between bacterial length and ChePep intensity, see Fig. S2A in the supplemental material.) (B) Confocal 3D reconstructions of H. pylori cells at different stages of growth stained with anti-ChePep (green), anti-H. pylori (red), and DAPI for DNA (blue). Fl, flagella. Stars indicate chromosomal segregation. Scale bar, 1 µm in B.
Besides their role in motility, polar proteins in bacteria function in cell division and chemotaxis (25). Therefore, we constructed a chePep isogenic deletion mutant (∆ChePep) (see Fig. S2B in the supplemental material) to test for defects in growth kinetics, flagellar structure, and motility. ∆ChePep mutants have identical in vitro growth curves to the wild type (WT) (Fig. S2C). At the ultrastructural level, we found no differences in the number of flagella, flagellar location, or flagellar morphology of ∆ChePep mutants (Fig. S2E). In soft agar motility assays, we noted that the ∆ChePep mutant is motile; however, it forms significantly smaller swarming halos than those formed by the WT (77% ± 5% the diameter of the WT) (Fig. S2D).
Taken together, this slight defect in ∆ChePep mutant swarming ability combined with ChePep’s polar localization and putative amino-terminal response regulator motif suggested that it could be part of an environmental response system such as chemotaxis. We were, therefore, particularly interested in examining the role of ChePep in colonization of the stomach and whether it is important for H. pylori interactions with the epithelial surface.
ChePep is required to colonize the gastric glands and confers an advantage for in vivo colonization.
To study ChePep’s role in H. pylori infection of the stomach, we orally infected mice with the WT, ∆ChePep mutant, or a 1:1 ratio of both bacteria. Two weeks later, we harvested the stomachs to determine the numbers of H. pylori colony-forming units (CFU). Mice singly colonized by the ∆ChePep strain had slightly lower, but not significantly different, CFU counts than mice singly colonized by the WT (Fig. 2A). Yet, when mice were coinfected with the WT and ∆ChePep mutant in a 1:1 ratio, the WT colonized at normal levels, but the ∆ChePep mutant was undetectable (Fig. 2B). To determine whether ChePep confers a colonization advantage in multiple strains of H. pylori, we also constructed ChePep mutants of strain SS2000 (26) and infected mice both singly and in competition. Similar to the previous results with strain SS1, we found that the SS2000 ∆ChePep mutant colonizes mice comparably to the WT in single infections, but ∆ChePep cells are completely undetectable in competition infections (see Fig. S3A and S3B in the supplemental material).
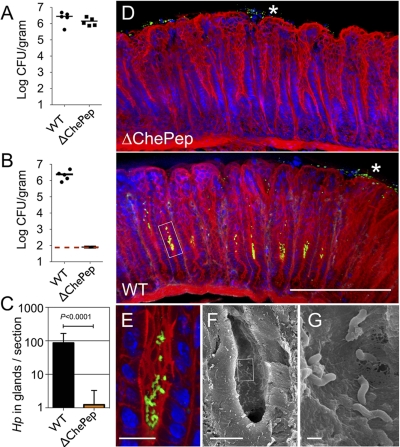
FIG 2 .
ChePep is essential for colonizing the antral gastric glands and confers a significant advantage in colonization of the stomach. (A) Colony-forming units (CFU) of H. pylori in the stomachs of mice colonized with either the WT or ∆ChePep mutant for 2 weeks. Each marker represents an individual mouse. (B) CFU counts from mice coinfected with both the WT and ∆ChePep mutant in a 1:1 ratio for 2 weeks. The dashed red line indicates the limit of detection. P < 0.0001. (C) Volumetric analysis of bacteria colonizing the antral glands calculated from 100-µm-thick sections imaged by 3D confocal microscopy from single infections of either the WT or ∆ChePep mutant. The average number of H. pylori cells within the gastric glands per section is plotted. Data from 20 sections from three mice infected with the WT and 26 sections from three mice infected with the ∆ChePep mutant were compared. P < 0.0001. (D) 3D confocal microscopy of murine stomachs infected with either the WT or the ∆ChePep mutant. F-actin is stained with phallodin (red), and nuclei (blue) and H. pylori cells (green) are immunolabeled. Asterisks indicate H. pylori cells in the surface mucus of the stomach, while a box highlights bacterial colonies in mid-glands. (E) Magnified view of the area boxed in panel D. (F) SEM of WT-infected gland. (G) Magnified view of area boxed in panel F. Scale bars represent 100 µm in panel D, 10 µm in panels E and F, and 1 µm in G. P values are from the two-tailed Student t test.
To investigate whether ChePep is involved not only in the establishment of infection but also in persistence, we asked whether WT can displace a previously established infection with the ∆ChePep strain. We infected mice first with the ∆ChePep strain for 2 weeks and then subsequently reinfected these mice with the WT. After a further 4 weeks of coinfection, the WT reached normal single infection levels, but ∆ChePep strain colonization declined to 0.03% of equivalent ∆ChePep strain single infections (see Fig. S3C in the supplemental material). These in vivo experiments show that ChePep confers a significant colonization advantage in the murine stomach for both the establishment and persistence of infection.
Chemotaxis was shown to be important for H. pylori colonization of the stomach and to facilitate interactions between H. pylori and the gastric epithelium (27). In particular, inactivation of chemotaxis through mutation of the genes that encode the conserved signal transduction proteins CheW, CheA, and CheY results in competition defects in mixed infections similar to what we observe between the WT and ∆ChePep strains (28). CheW, CheA, and CheY mutants colonize mouse stomachs in single infections, but they are significantly outcompeted when coinfected with the WT (28). These experiments thus support a possible role for ChePep in chemotaxis.
Given the colonization disadvantage of ∆ChePep in competition experiments, we suspected that the mutant could be defective in colonizing a particular niche within the stomach. The WT strain of SS1 mainly colonizes the gastric antrum of the C57BL/6 mice (29). Consequently, we were interested in determining whether single infections of the ∆ChePep mutant and WT showed differences in their spatial distribution within this region of the stomach. Typically H. pylori infection is visualized using histology, mainly to confirm the presence of H. pylori and assess the associated inflammation (30, 31). Routine histological examination relies on random sampling of the tissue sectioned in 5- to 10-µm samples. Unless laborious serial sectioning is performed, comparison and quantification of the anatomical localization of H. pylori are limited by this technique. To overcome this limitation, we adapted methods of three-dimensional (3D) confocal microscopy of whole tissues (32, 33) and analyzed 100- to 200-µm sections of infected gastric tissue at a time. This allowed us to reconstruct complete infected gastric glands in three dimensions to compare and quantify the anatomical localization of WT versus the ∆ChePep mutant (Fig. 2). Both WT and ∆ChePep strains were readily identified in the mucus layer overlying the stomach surface (Fig. 2D, asterisks). Additionally, using 3D confocal reconstruction and scanning electron microscopy (SEM), we were able to document that WT cells also form discrete clusters of bacteria adhered to the epithelial surface in the mid-glandular zone of the gastric antrum (Fig. 2D to G). These WT bacteria within the glands resemble clonal microcolonies observed growing on epithelial surfaces in vitro (17) since they form discrete clusters of tightly packed spiral bacteria (Fig. 2E and F). Interestingly, we found that ∆ChePep cells are essentially missing from this glandular region and are mainly found in the overlying surface mucus layer (Fig. 2D, top). We quantified the numbers of H. pylori cells colonizing the gastric glands by volumetric image analysis of the bacterial colonies within the antral glands from 3D confocal reconstructions. We found an average of 86.2 WT cells per section (n = 20), while the ∆ChePep strain had only 1.6 bacteria per section (n = 27) (Fig. 2C). These findings show that ChePep is essential for colonizing the glands in the antrum of the stomach.
Because of the ∆ChePep strain’s failure to colonize the antral glands (27), ChePep localization to the flagellar pole, the presence of a putative CheY response regulator motif, and the ∆ChePep mutant’s slight defect in swarm plates, we hypothesized that ChePep could be involved in chemotaxis or motility.
ChePep controls H. pylori swimming direction and switching of flagellar rotation.
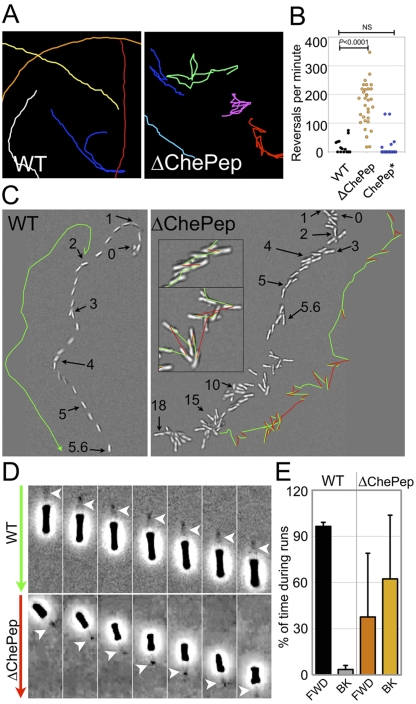
To examine the swimming behavior of both the WT and ∆ChePep mutant, we used video microscopy and single particle tracking of individual bacteria grown in broth. WT bacteria generally swim in smooth paths that are straight or curved and reverse their swimming direction at an average rate of 14.8 reversals/min (Fig. 3A and 3B). However, ∆ChePep bacteria swim with a dramatically different pattern, exemplified by frequent changes in swimming direction. The average reversal frequency of ∆ChePep is over an order of magnitude greater than that of the WT (average of 162.7 reversals/min), and the frequent ∆ChePep strain reversals are interspersed with only occasional bursts of straight swimming (Fig. 3A and 3B; see Movie S1 in the supplemental material).
FIG 3 .
ChePep reduces switching of the flagellar rotational direction. (A) Magnified tracings of both WT and ∆ChePep cell swimming behavior (see Movie S1 in the supplemental material). (B) Quantification of reversals per minute of the WT, ∆ChePep, and ChePep* (∆ChePep complemented in trans) strains. (C) DIC video microscopy of WT and ∆ChePep cell swimming (see Movie S2 in the supplemental material). The position of a single swimming bacterium over time is shown for the WT versus the ∆ChePep mutant. The position at a particular time (in seconds) is marked with arrows, and the swimming path is marked in green in one direction and red when the bacteria reverse swimming direction. Inset images of ∆ChePep cell movement at higher magnification are also shown. (D) High-magnification phase-contrast video microscopy shows a ∆ChePep mutant swimming backwards with the flagella “pulling” (see Movie S3 in the supplemental material). The arrows on the left indicate the direction that the bacteria are swimming. (Green indicates swimming forward, and red indicates swimming backward.) The position of the flagella is marked with an arrowhead in each panel. (E) Quantification of the percentage of time that WT and ∆ChePep cells swim forward (FWD) with the flagella “pushing” or backwards (BK) with the flagella “pulling.” n = 8 for each strain. P values are from the two-tailed Student t test.
To characterize the behavior of ∆ChePep mutants at higher spatial resolution, we used differential interference contrast (DIC) imaging and found that the reversal events are not due to tumbling, as seen in peritrichously flagellated bacteria (34). Instead, we observed that ∆ChePep cells rapidly reverse swimming direction while maintaining the orientation of the bacterial body (Fig. 3C; see Movie S2 in the supplemental material). This suggests that the flagella can support swimming in either direction by propelling the bacteria forward or pulling the bacteria in reverse. We hypothesized that these reversals may represent changes in the direction of flagellar rotation.
To test this hypothesis, we filmed the swimming bacteria by high-magnification phase-contrast video microscopy at a rate of 30 frames per s, where we could see the localization of the flagellar tuft at one pole of the bacteria while recording their swimming direction. We found that indeed H. pylori can reverse swimming direction with the flagella propelling the bacteria either forward (“pushing”) or in reverse (“pulling”) (Fig. 3D; see Movie S3 in the supplemental material). Under normal conditions, WT bacteria mostly swim forward, with the flagella pushing the bacteria. In contrast, ∆ChePep bacteria spend a significant percentage of time swimming in reverse (Fig. 3E). This pattern of behavior by ∆ChePep cells indicates that ChePep reduces switching of flagellar rotation, and without ChePep, the flagella sustain periods of reverse rotation that enable backwards runs.
In other bacteria, including H. pylori, mutations within the chemotaxis signaling system result in loss of turning responses and straight swimming trajectories (35, 36). However, in other Proteobacteria, mutations in chemotaxis regulators such as CheZ and CheB result in hyperreversal or increased tumbling behavior, reminiscent of ∆ChePep mutants (6, 35, 37). Therefore, our results suggest that ChePep could affect flagellar switching through regulation of the chemotaxis system.
ChePep controls flagellar rotation through the chemotaxis system.
In bacteria such as E. coli, switching the direction of flagellar rotation requires altering the interaction between the flagellar stator and the flagellar switch complex (38). This interaction is controlled by the chemotaxis system through the interaction of phosphorylated CheY with the flagellar switch complex protein, FliM (39, 40). H. pylori possesses homologs of flagellar switch proteins, and it was shown in vitro that only phosphorylated H. pylori CheY interacts with FliM (41). Therefore, we hypothesized that the underlying mechanism of increased flagellar switching in the ∆ChePep mutant could be due to either a direct interaction between ChePep and the flagellar switch complex or, alternatively, ChePep regulation of the interaction of the chemotaxis system with the flagellar switch complex.
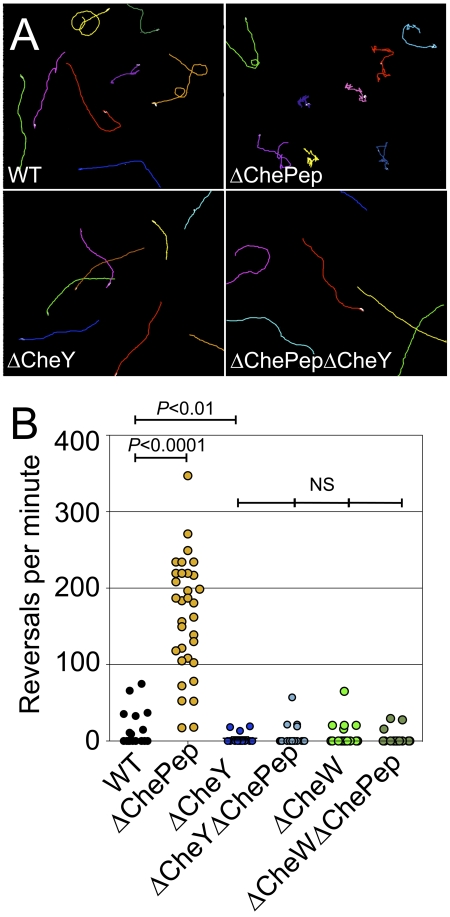
To distinguish between these two hypotheses, we created a chePep cheY double mutant. If ChePep functioned directly on the flagellar motor, then the hyperswitching phenotype of the ∆ChePep mutant would be independent of chemotaxis and would persist in the absence of CheY. If, however, ChePep controls flagellar rotation by regulating interaction of the chemotaxis system with the flagella, then loss of CheY would be epistatic over the ∆ChePep phenotype. Quantitative behavioral analysis of these double mutants revealed that loss of CheY completely abolishes the hyperreversal phenotype of the ∆ChePep mutant as the ∆ChePep ∆CheY mutant reverses at a rate of only 5.8 reversals/min, while the ∆ChePep mutant reverses at a frequency of 162.7 reversals/min (Fig. 4A and B). Importantly, comparison of ∆CheY ∆ChePep mutant’s reversal frequency to that of the ∆CheY mutant shows no significant difference, indicating that the ∆CheY mutation is entirely epistatic over ∆ChePep (Fig. 4A and B). To further verify that ChePep acts through the chemotaxis signaling pathway and to test if this action requires phosphorylation of CheY, we constructed a ChePep CheW double mutant. CheW is an upstream adapter protein that facilitates interactions between transmembrane methyl-accepting chemotaxis protein (MCP) receptors and the chemotaxis pathway, and it is necessary for phosphate transfer to CheY (7). As with the ∆ChePep ∆CheY mutant, the hyperreversal phenotype of the ∆ChePep mutant is completely abolished in a ∆CheW background, and there is no significant difference between the ∆ChePep ∆CheW and ∆CheW strains (Fig. 4B). In the absence of a functional chemotaxis system, the ∆ChePep mutant no longer exhibits hyperswitching of the flagella. Taken together, these data suggest that ChePep controls H. pylori flagellar rotation by regulating the interaction of the chemotaxis system with the flagellar complex.
FIG 4 .
ChePep controls flagellar switching through the chemotaxis system. (A) Motility tracings of the WT, the ∆ChePep and ∆CheY single mutants, and the ∆ChePep ∆CheY double mutant strain. (B) Quantification of reversals per minute of the WT and ∆ChePep, ∆CheY, ∆CheY ∆ChePep, ∆CheW, and ∆CheW ∆ChePep mutants. P values are from the two-tailed Student t test.
ChePep regulates H. pylori chemotactic responses.
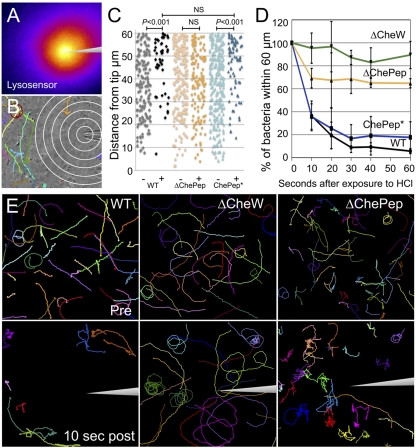
Based on the genetic and behavioral data presented above, we hypothesized that in ∆ChePep mutants, the chemotaxis signaling pathway remains intact but overactive. Therefore, the ∆ChePep strain should have deficient but not null chemotactic responses. To functionally test the role of ChePep in H. pylori chemotaxis, we developed an assay to analyze the swimming response of individual H. pylori cells within a microscopic pH gradient. Acid was shown to be a potent chemorepellent for H. pylori in vitro (42), and the pH gradient of the gastric mucus is thought to be the primary chemical cue used by H. pylori for orientation in the stomach (4). We utilized a micropipette to create a point source of acid by releasing a small flow (0.3 to 0.8 pmol/min) of hydrochloric acid (HCl). We verified the presence of a stable pH gradient within the viewing field of the video microscope by the differential fluorescence of a pH-sensitive dye (Fig. 5A; see Fig. S4 in the supplemental material). By monitoring pH-dependent fluorescence over time, we confirmed that the distribution of the microscopic pH gradient remained constant (measured for over 5 min) (Fig. S4). The device was controlled with a micromanipulator, which allowed the micropipette to be precisely inserted or completely removed from H. pylori cultures while continuously tracking the bacteria through video phase-contrast microscopy (Fig. 5B; see Movie S4 in the supplemental material). To ensure that the pH gradient was not bactericidal, nor that the flow physically displaced H. pylori, we verified that a chemotaxis null mutant, the ∆CheW strain, continued to swim near the needle tip (Movie S4).
FIG 5 .
ChePep regulates H. pylori chemotaxis response in a pH gradient. (A) A microscopic pH gradient generated by a microinjection needle is visualized by Lysosensor dye fluorescence (pseudocolor image) (see Fig. S4 in the supplemental material for details). (B) A sample frame from a movie of WT bacteria and their motility tracings after 20 s in the pH gradient is shown. Rings are 10 µm apart. (C) The distance of each H. pylori cell to the acid point source is plotted before (−) or 10 s after (+) exposure to the pH gradient; ChePep* is ∆ChePep complemented in trans. NS, no statistical difference. (D) Plot of the percentage of H. pylori cells remaining within 60 µm of the micropipette tip over time. One hundred percent is defined as the number of bacteria within this radius directly before addition of acid. Error bars indicate standard deviations from the mean of five independent movies. Results for the ∆ChePep mutant and WT strains are significantly different (P < 0.0001, F test). (E) Bacterial motility paths of WT versus different mutants 2 s before (upper panels) and after 10 s in the pH gradient (bottom panels) (see Movie S5 in the supplemental material).
We tracked the swimming paths of individual bacteria before and after exposure to the pH gradient and monitored their behavioral responses (Fig. 5E; see Movie S5 in the supplemental material). Before exposure to acid, the bacteria swim throughout the field of view without directional preference. The WT and ∆CheW mutants swim straight or with curved arcs and ∆ChePep mutant cells swim with a visibly higher reversal frequency. Within a second of exposure to acid, WT cells respond by increasing their reversal frequency and begin redistributing away from the needle tip (Fig. 5E; Movie S5). ∆ChePep cells respond to the acid gradient by further increasing their reversal rate, but have a less efficient escape response since more bacteria remain within the field and close to the acid point source (Fig. 5E; Movie S5).
To quantify the response to the pH gradient, we measured the distance of individual bacteria from the acid point source. At 10 s after introduction of the pH gradient, WT cells redistribute away from the needle tip in a graded fashion, while ∆ChePep cells display no significant redistribution (Fig. 5C). The defective response of the ∆ChePep mutant is rescued by complementation in trans with its own promoter (ChePep*) (Fig. 5C). To determine whether this failure in orientation also results in a deficient escape response, we tracked how many H. pylori cells escape from a 60-µm radius around the acid point source over 60 s. Within 10 s, only 31% of WT cells remain within this radius (Fig. 5D). In contrast, ∆ChePep cells have an attenuated response, as 70% of ∆ChePep cells remain within a 60-µm radius in an identical gradient (Fig. 5D). The nonchemotactic ∆CheW mutant had no escape response (Fig. 5D). Observations at later time points showed that approximately 70% of ∆ChePep cells persisted within a 60-µm radius, while WT cells continued to clear the area until approximately 30 s after acid exposure, by which time they had achieved a steady state of 10% remaining bacteria in a 60-µm radius (Fig. 5D). Our data indicate that the ∆ChePep strain is impaired in chemotaxis, but unlike the ∆CheW strain, it is not a complete chemotaxis null mutant. Its swimming phenotype is the opposite of straight-swimming ∆CheW and other chemotaxis null mutants, suggesting that either excess CheY-phosphate is created or that the phosphate is not removed from CheY in the ∆ChePep mutant. This result is consistent with the proposed role of ChePep as a novel chemotaxis regulator in H. pylori. Examination of other Epsilonproteobacteria revealed the presence of chePep homologs throughout this class of diverse bacteria, and so we wondered whether ChePep function is a broadly conserved mechanism of chemotaxis regulation.
ChePep is functionally conserved in the Epsilonproteobacteria.
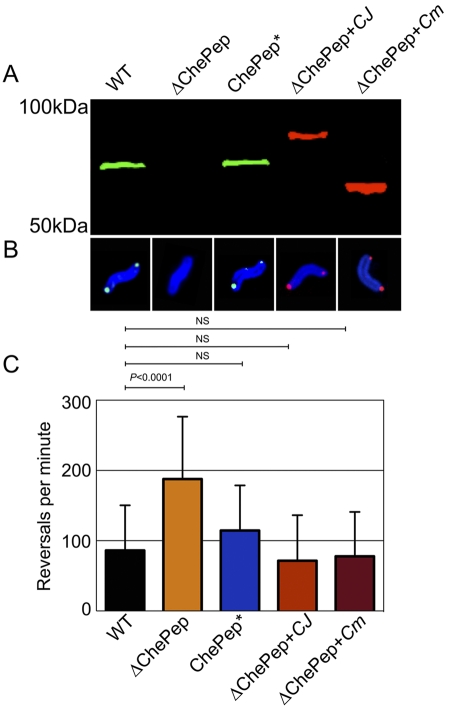
To test whether distantly related ChePep proteins are functionally conserved, we cloned chePep homologs from other Epsilonproteobacteria and complemented the ∆ChePep H. pylori mutant with these genes. The ChePep homologs chosen were from the pathogen Campylobacter jejuni, which is a major cause of food poisoning in humans (2), and the environmental bacterium Caminibacter mediatlanticus, which was isolated from a hydrothermal vent at the Mid-Atlantic Ridge (43). An immunoblot verified WT levels of ChePep when the endogenous promoter was used in complementing ∆ChePep with H. pylori ChePep (Fig. 6A). We confirmed expression of C. jejuni and C. mediatlanticus ChePep by immunoblotting for a C-terminal tag that was added to each foreign ChePep (Fig. 6A). The sizes and sequences of ChePep varied considerably amongst the three different bacteria, with the chePep genes from C. jejuni, C. mediatlanticus, and H. pylori having 30% conserved sequence identity (Fig. 6A; see Fig. S1C in the supplemental material). Despite the differences in ChePep size and sequence, C. jejuni and C. mediatlanticus ChePep proteins concentrate at the poles in H. pylori (Fig. 6B) and complement the hyperreversal phenotype of the ∆ChePep mutant (Fig. 6C). This provides functional evidence that, despite the large evolutionary distance between H. pylori, C. jejuni, and C. mediatlanticus, ChePep is conserved across a diverse set of Epsilonproteobacteria.
FIG 6 .
ChePep is functionally conserved throughout the Epsilonproteobacteria. (A) Immunoblot of the WT and ∆ChePep, ChePep* (∆ChePep complemented with H. pylori ChePep), ∆ChePep + CJ (∆ChePep complemented with FLAG-tagged C. jejuni ChePep), and ∆ChePep + Cm (∆ChePep complemented with FLAG-tagged C. mediatlanticus ChePep) strains. The blot is probed with anti-H. pylori ChePep (green) and anti-FLAG (red). (B) Immunofluorescence of H. pylori stained with anti-H. pylori (blue), anti-ChePep (green), and anti-FLAG (red). (C) Reversals per minute of the WT and ∆ChePep, ChePep*, ∆ChePep + Cj, and ∆ChePep + Cm strains. P values are from the two-tailed Student t test. NS, no statistical difference.
DISCUSSION
Chemotaxis is estimated to be present in over half of all microbes (10) and is arguably the best understood signaling system in biology. Despite the wealth of information on chemotaxis, it is becoming increasingly apparent that microbes have evolved diverse mechanisms to control their motility and regulate chemotactic responses to the environment (11). We have discovered a novel family of chemotaxis regulator proteins in the Epsilonproteobacteria called “ChePep.” We show that ChePep preferentially localizes to the flagellar pole of H. pylori, yet ChePep mutants have normal flagella and are motile. These mutants have aberrant flagellar rotation, but genetic experiments show that ChePep does not directly control flagellar switching, since it requires the presence of a functional chemotaxis signaling system. This suggests that ChePep functions as a chemotaxis regulator. Analysis of H. pylori chemotactic behavior in a pH gradient is consistent with this hypothesis. ChePep is also required for normal colonization of the stomach, especially in competition with the WT, and is essential for colonization of the epithelial surface inside the antral glands. Despite substantial variation between ChePep orthologs, we demonstrate that ChePep is functionally conserved in Epsilonproteobacteria as different as the deep sea colonizing organism C. mediatlanticus and the human pathogens H. pylori and C. jejuni.
When H. pylori cells are exposed to a chemorepellent, like acid, they increase their turning frequency because of a predicted increase in phosphate signaling in the chemotaxis system. ∆ChePep mutants can respond to acid by increasing their turning frequency; however, they constantly switch flagellar rotation even under standard culture conditions. This suggests that ∆ChePep mutants have abnormally high levels of phosphorylated CheY protein. In some enteric bacteria, similar hypertumbling phenotypes are attained by mutating chemotaxis regulators such as CheZ and CheB, which normally function to reduce the phosphorylation state of CheY (6, 35, 37).
In comparison to E. coli and B. subtilis, our understanding of the negative regulation of phospho-CheY by H. pylori and the Epsilonproteobacteria is far less understood. The initial analysis of Epsilonproteobacteria genomes concluded that this class lacked a CheZ homolog (44, 45), but more recent reports have identified a remote homolog to CheZ, termed CheZHP (13). Though CheZHP has very limited homology to the CheZ of E. coli, it conserves the active site residues analogous to E. coli Asp143 and Gln147 and has in vitro CheY phosphatase activity (46). Further biochemical investigation of CheZHP demonstrated phosphatase activity not only with CheY but also with CheA and CheV2, thereby indicating that CheZHP may have a more varied and complex function than E. coli CheZ (46). The identification of ChePep suggests that H. pylori and the Epsilonproteobacteria have evolved an additional mechanism of chemotaxis regulation. Further examination of the relationship between ChePep and CheZHP may provide insight into the unique mechanism of chemotaxis regulation in the Epsilonproteobacteria.
With the exception of the putative amino-terminal response regulator domain, ChePep has no homology to other chemotaxis proteins. ChePep orthologs are some of the most negatively charged proteins in Epsilonproteobacteria (47, 48), with an average cumulative composition of 29.6% glutamic and aspartic acids. In H. pylori, ChePep is composed of 23% glutamic acid and 5% aspartic acid, while in C. jejuni, ChePep contains 15.3% glutamic acid and 15.7% aspartic acid. Despite considerable sequence variation between ChePep orthologs, including the content of glutamic and aspartic acids, ChePep proteins from C. jejuni and C. mediatlanticus are able to complement H. pylori ∆ChePep. This suggests that ChePep requires an abundance of negatively charged amino acids for its function rather than a specific amino acid sequence in these domains. High net charge is typical of many natively unfolded proteins, and their intrinsic lack of structure can facilitate functional flexibility (49). A total of 77.4% of H. pylori ChePep is predicted to be natively disordered, and it is tempting to speculate that this may explain the complementation of H. pylori ∆ChePep with such disparate ChePep orthologs. Natively unfolded proteins are relatively uncommon in bacteria, but they are estimated to be abundant in eukaryotic signaling systems (50). Precisely how the negatively charged and predicted disordered region of ChePep contributes to the mechanistic function of regulating chemotaxis signaling requires further investigation.
In the context of H. pylori pathogenesis, ChePep is important in allowing the bacteria to colonize a specialized niche on the surface of the epithelium. H. pylori cells primarily colonize a 30-µm band of buffered mucus that extends above the cell surface (4) and also directly attach to and grow on epithelial surfaces (17). H. pylori uses both motility and chemotaxis to reach this habitat and maintain colonization of the host (27, 51, 52). There are several gradients that extend from the epithelial surface to the lumen of the stomach, but experimental evidence suggests the pH gradient is critical for H. pylori in vivo spatial orientation (4). Chemotaxis has also been reported to be important for facilitating adhesion to the gastric epithelium and contributes to host inflammation (27, 53). Our work expands on this theme by documenting that H. pylori forms microcolonies directly adhered to the epithelium within the mid-glands of the stomach. ChePep mutants are absent from this niche, indicating that chemotaxis is necessary to locate or persist within the mid-glands. This tropism for the mid-glandular zone is particularly intriguing because of the gastric progenitor cells that are known to reside within this approximate zone (54). A direct association between H. pylori and the gastric progenitor cells could have implications for the increased gastric cancer associated with H. pylori infection (15), so evaluation of ChePep’s role in animal models of carcinogenesis will be important for future studies.
Besides H. pylori, the class of Epsilonproteobacteria contains additional pathogens, including C. jejuni, which is a major source of diarrheal illness in humans (2). Motility and chemotaxis were identified as important colonization factors in C. jejuni (55, 56) and influence bacterial virulence in the ferret model (57). The ability to complement H. pylori ∆ChePep with ChePep from C. jejuni suggests that ChePep function is conserved in C. jejuni. ChePep may also significantly contribute to colonization, access to specialized niches, and disease outcome of C. jejuni and other pathogenic Epsilonproteobacteria.
Finally, the discovery of ChePep has implications for the evolution of bacterial chemotaxis. Flagellated bacteria generally share the same basic components of the chemotaxis signaling pathway (7, 10, 11), and all previously identified chemotaxis genes in H. pylori have homologs in other classes of Proteobacteria (12, 44, 46, 58). However, ChePep is unique to the Epsilonproteobacteria. The extensive distribution of chePep homologs throughout the Epsilonproteobacteria combined with its restriction to this class suggests that ChePep evolved within an ancient ancestral member of the Epsilonproteobacteria as a unique mechanism for controlling an otherwise conserved chemotaxis system.
MATERIALS AND METHODS
H. pylori strains and bacterial culture.
The SS1 strain (29) of H. pylori was used throughout this study, with the exception of Fig. S3A and S3B in the supplemental material, where we used another mouse-colonizing strain, SS2000 (26), and Fig. 6, where we used the strain G27MA (19) because it is more readily genetically modified by natural transformation. H. pylori strains were either grown on Columbia blood agar plates or shaking brucella broth cultures under standard conditions (59). The ∆ChePep, ∆CheW, ∆CheY, ∆ChePep ∆CheY, ∆ChePep ∆CheW, and ∆FliF isogenic mutants were constructed by a PCR-based method (60), and mutants were verified by sequencing (sequences and primers available upon request). ∆ChePep was complemented in trans by insertion of a construct into the rdxA locus comprising 320 bp upstream of chePep (containing the chePep promoter), the chePep open reading frame (ORF) (from strain G27MA), and the aphA gene (conferring kanamycin resistance). Complementation of ∆ChePep with ChePep from Caminibacter mediatlanticus and Campylobacter jejuni was performed similarly, except the chePep ORF from H. pylori was replaced with the chePep ORF from either C. mediatlanticus or C. jejuni, both with a 3×FLAG tag (Sigma) encoded at the C terminus. Genomic DNA from C. mediatlanticus was kindly provided by Constantino Vetriani (Rutgers University). C. jejuni 81-176 was kindly provided by Victor DiRita (University of Michigan).
pH gradient generation and single-particle tracking of swimming bacteria.
H. pylori cultures were grown to mid-log phase and placed into a glass-bottom 35-mm dish (MatTek) on a Zeiss Axiovert-35 inverted microscope equipped with phase-contrast optics and a heated stage. A Hammamatsu C2400 video charge-coupled-device (CCD) camera was used to record via an Argus-20 image processor onto Quicktime at 30 frames per s. A Femtotip II microinjection micropipette (Eppendorf) containing 1 N hydrochloric acid (HCl) was inserted into or removed from the viewing field using a micromanipulator (Eppendorf 5171). To create a point source of acid, a constant flow through the tip was controlled with an Eppendorf transjector 5246. Liquid flow and acid concentration were empirically adjusted to avoid physically dispersing the bacteria while obtaining a chemotactic response within the viewing field. Typical flow settings were 0.3 to 0.8 pmol of HCl per min, measured by inserting the needle into a known volume of water and measuring the pH change with a microelectrode after 2 h of constant flow. The pH gradient was local since there was no detectable change in the overall pH of the bacterial cultures during the tracking experiments. To confirm the presence of a stable microscopic pH gradient, a pH-sensitive dye, Lysosensor green DND-189 (Molecular Probes), was added to water at a concentration of 100 nM and imaged by epifluorescence. To test for chemotactic changes in bacterial spatial distribution, the position of individual motile bacteria was recorded before and 10 s after introduction of acid. The distance of each bacterium to the micropipette tip was plotted. Statistical significance of differences between the pre- and postacid distributions was assessed by the Mann-Whitney test. The H. pylori chemotactic escape response to a pH gradient was quantified by counting the number of motile bacteria within 60 µm of the micropipette tip every 10 s over a time course of 60 s and dividing by the number of bacteria present prior to introduction of acid. At least three independent experiments were used to generate the results. A nonlinear, one-phase decay curve was fit to each escape curve, and statistical significance was assessed using an F test. For quantification of H. pylori swimming behavior, we used single-particle tracking with Metamorph software (Molecular Device) and analyzed with an Excel macro as described by Astling et al. (61). A reversal is defined in our analysis as a ≥110° turn. Statistical differences between the number of reversals per minute of the WT, ∆ChePep, ∆CheY, ∆CheY ∆ChePep, ∆CheW, and ∆ChePep ∆CheW strains were evaluated by a two-tailed Student’s t test.
Animal infections.
All animal experiments were performed in accordance with NIH guidelines and with approval from the Institutional Animal Care and Use Committee of Stanford University. Female 6- to 8-week-old C57BL/6J mice were purchased from Jackson Laboratories (Bar Harbor, ME). Animals were orally infected with 108 CFU of H. pylori grown in broth. For competition infections, mice were infected with an equal mixture of WT and ΔChePep cells, totaling 108 CFU. For sequential infections, mice were infected with the ΔChePep mutant for 2 weeks and then infected with the WT for 4 weeks (total, 6-week infection). Animals were sacrificed at 2 or 6 weeks postinfection. For competition and sequential infections, the entire glandular stomach was weighed and mechanically homogenized in Brucella broth for CFU counts. For single infections, the glandular stomach was dissected into two pieces—one for CFU determination and another for confocal and electron microscopy.
Epifluorescence and confocal microscopy.
For subcellular localization of ChePep, H. pylori cells were grown overnight on blood plates, resuspended in phosphate-buffered saline (PBS) and adhered to poly-l-lysine-coated coverslips. The bacteria were fixed and permeabilized as previously described (32) and stained with chicken anti-H. pylori antibodies (19), rabbit anti-ChePep antibodies, and mouse anti-FLAG antibodies (Sigma). Alexa Fluor-conjugated antibodies of appropriate fluorescence and species reactivity (Molecular Probes) were used for secondary detection. Bacteria were imaged with an Orca100 CCD camera coupled to an Olympus BX60 microscope. For animal infections, tissue samples were processed for confocal immunofluorescence as previously described (32), except that tissue was embedded in agar and 100-µm-wide sections were cut on a vibratome (Leica). Tissue sections were stained with rabbit anti-H. pylori antibodies and appropriate secondary antibodies. DAPI (4′,6-diamidino-2-phenylindole) and Alexa Fluor 594-phalloidin (Molecular Probes) were used for visualization of the nuclei and actin cytoskeleton. Samples were imaged with a Zeiss LSM 700 confocal microscope, and z-stacks were reconstructed into 3D using Volocity software (Improvision). For quantification of bacterial colonization of the gastric glands, we analyzed antral tissue of infected mice by volumetric image analysis as previously described (17).
DIC and phase video microscopy of H. pylori reversals and swimming direction.
H. pylori cells were grown to mid-log phase in broth as described above. One microliter of the culture was placed onto an 8-mm multiwell slide (Cel-Line Associates) and sealed with a coverslip. Reversal behavior of WT and ∆ChePep cells was recorded by DIC video microscopy using a Hamamatsu high-resolution ORCA-285 digital camera and Nikon TE2000E microscope. The bacterial position in each frame of the movie was traced using ImageJ and the plugin MtrackJ. Forward movement was arbitrarily assigned the color green, while reverse movement was assigned red. Tracking of bacterial forward movement (flagella pushing) and backwards movement (flagella pulling) was recorded with a Zeiss Axiovert-35 inverted microscope equipped with 100× phase-contrast optics, and a heated stage. A Hammamatsu C2400 video CCD camera recorded via an Argus-20 image processor onto Quicktime at 30 frames per s. To determine the percentage of time bacteria spent swimming forward or in reverse, the position of the flagella was first marked. Then, the number of frames was counted for which the bacteria moved forward or backwards over approximately 5 to 60 s. The percentage of time spent moving forward or backward was calculated by dividing the number of frames for which the bacteria moved forward or backward by the total number of frames.
Immunoblotting.
H. pylori cells were harvested from blood plates and lysed with 2× SDS sample buffer. Lysates were boiled and then separated on an 8% to 16% gradient or 14% SDS-PAGE gels. After transfer onto polyvinylidene difluoride (PVDF) or nitrocellulose membranes, H. pylori ChePep, 3×FLAG-tagged C. mediatlanticus ChePep, and 3×FLAG-tagged C. jejuni ChePep were detected by blotting with rabbit anti-ChePep and mouse anti-3×FLAG followed by goat anti-mouse Alexa Fluor 660 and goat anti-rabbit Alexa Fluor 800. The blot was then scanned with a Licor-Odyssey scanner.
Supplemental methods.
For methods of antibody production, sequence analysis, mass spectrometry, and scanning and transmission electron microscopy, see Text S1 in the supplemental material.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download Text S1, DOC file, 0.053 MB.
Identification and sequence analysis of ChePep. (A) Coomassie-stained gel of the polypeptide immunoprecipitated (IP) from H. pylori lysates using anti-Par-3 antibody. (B) Immunoblot of lysates from E. coli expressing recombinant ChePep using the anti-Par-3 antibody confirms that ChePep contains an epitope recognized by this antibody. (C) Phylogenetic tree of ChePep homologs from across the Epsilonproteobacteria. (D) Alignment of H. pylori CheY with the amino terminus of ChePep from 16 species across the class of Epsilonproteobacteria. The first arrowhead and box point to the phosphoryl group accepting aspartic acid 53 of CheY (Jimenez-Pearson MA, et al., Microbiology 151:3299–3311, 2005). The second arrowhead and box show the conserved KPFLPT motif in ChePep, which contains the critical Lys 109 (Lukat GS, et al., J. Biol. Chem. 266:8348–8354, 1991). Nonconserved residues are indicated with a value of zero and the color blue, while completely conserved residues are indicated with an asterisk and are colored red. Download Figure S1, TIF file, 2.186 MB.
ChePep localizes to the flagellar pole, and ∆ChePep mutants are slightly impaired in swarming but have normal flagella and cell growth. (A) Fluorescence intensity of ChePep at the new pole from Fig. 1A plotted on the y axis and the corresponding bacterial length plotted on the x axis. A linear regression curve was fit to the data. (B) Immunofluorescence images of wild-type (WT) and ChePep mutant (∆ChePep) H. pylori cells stained with antibodies to H. pylori (red) and recombinant ChePep (green). The same anti-ChePep antibody was used to determine ChePep expression in immunoblots of bacterial lysates from the WT and ∆ChePep mutant. (C) Growth curves of WT H. pylori and ∆ChePep in Brucella broth with 10% fetal bovine serum. (D) Photograph of 3-day-old swarm plate with the WT and ∆ChePep and ∆FliF (aflagellate, nonmotile) mutants. The diameters of swarming halos were measured for each strain and are expressed as percentages of WT swarming (n = 4 for each strain). (E) Scanning (left) and negative-stained transmission (right) electron micrographs comparing the morphologies and locations of flagella from WT and ∆ChePep strains. Both bacteria have tufts of two to six sheathed flagella at one pole. The tips of the sheathed flagella are shown by negative staining on the right and contain a typical end bulb. Both WT and ∆ChePep strains have an average of approximately 4 flagella per bacteria (n = 14). P values are from the two-tailed Student t test. NS, no statistical difference. The scale bar is 1 µm in panel B. Download Figure S2, TIF file, 2.729 MB.
ChePep confers an advantage to in vivo colonization in multiple strains of H. pylori and is required for persistent infection. (A) Colony-forming units (CFU) of H. pylori in the stomachs of mice colonized with either the SS2000 WT or ∆ChePep mutant strain for 2 weeks. Each marker represents an individual mouse. (B) CFU counts from mice coinfected with both the SS2000 WT and ∆ChePep mutant in a 1:1 ratio for 2 weeks. The dashed red line indicates the limit of detection. P < 0.0001. (C) Mice were infected with either WT SS1 (n = 4), or ∆ChePep SS1 (n = 10). After 2 weeks, two ∆ChePep strain-infected mice were harvested to document that they were colonized, and five others were superinfected with the WT. After 4 additional weeks, all mice were harvested, and bacterial CFU were determined. Download Figure S3, TIF file, 0.613 MB.
A stable pH gradient is formed by a point source of HCl. A microinjection needle (Eppendorf Femtotip II) with 0.5-µm inner diameter was filled with 1 N HCl, and constant pressure was adjusted to flow at 0.8 pmol/min. In this figure, the needle is imaged by phase-contrast microscopy while inserted into 3 ml of water containing Lysosensor dye. Fluorescence imaging of the same field followed for 30 s with 2-s exposure times. The color image is a pseudocolor rendering of the fluorescence panel. Markings show 10-µm rings up to a 60-µm radius. The bottom graph plots the fluorescence intensity as a function of distance from the needle tip. The different plots show the gradient is stable over time by measuring the fluorescence intensity at different times after inserting the needle (at 30 s and at 1, 2, 3, 4, and 5 min). Download Figure S4, TIF file, 2.743 MB.
Motility tracings of WT and ∆ChePep and ChePep* (∆ChePep complemented in trans) mutant H. pylori swimming behavior. Phase-contrast video microscopy movies in real time (30 frames per s) of WT versus various mutant bacteria were digitized, and individual bacteria were manually tracked using the MTrackJ plugin for ImageJ software. Images were inverted to show bacteria in white with their individual motility tracings in various colors. The movie is best viewed in loop mode. Download Movie S1, MOV file, 1.127 MB.
The ∆ChePep cell swims with frequent reversals of direction rather than end-over-end tumbles. WT and ∆ChePep cell swimming was recorded by high-resolution digitally enhanced DIC microscopy, and the cells’ trajectory was traced. At the end of this movie, an inset image appears showing detailed reversal events for the ∆ChePep cell. This video was used to generate Fig. 3C. Download Movie S2, MOV file, 8.739 MB.
∆ChePep cells can sustain prolonged backward runs, with the flagella “pulling” the bacteria. WT and ∆ChePep H. pylori cells were recorded using high-magnification phase-contrast video microscopy at 30 frames per s. The locations of their respective flagella are initially indicated with arrows. Download Movie S3, MOV file, 8.375 MB.
H. pylori swimming response to a point source of acid. The right frame shows a log-phase culture of WT H. pylori imaged by video phase-contrast microscopy for 5 s before and 12 s after introduction of a micropipette as a point source of acid. The left frame shows a culture of ∆CheW H. pylori (defective in chemotaxis) under identical conditions. The WT changes in behavior and redistributes away from the needle, while the ∆CheW mutant does not. The movie can be appreciated in real time, at 30 frames per s. Download Movie S4, MOV file, 6.339 MB.
Movies from motility tracings of Fig. 5E. H. pylori chemotactic response to acid. Phase-contrast video microscopy movies, similar to Movie S4, were digitized, and individual bacteria were manually tracked using the MTrackJ plugin for ImageJ software. The upper panels show the motility paths for 2 s prior to the introduction of the acid point source. These 2-s tracts repeat over the duration of the movie. The bottom panels show 10-s tracings of the motility paths of the bacteria during exposure to the pH gradient. The position of the needle is also shown. Tracings disappear from the movie when the bacteria escape the field of view. Download Movie S5, MOV file, 5.506 MB.
ACKNOWLEDGMENTS
We thank Constantino Vetriani for providing C. mediatlanticus genomic DNA and Victor DiRita for providing C. jejuni. We also thank Stan Falkow, Lucy Tompkins, Denise Monack, and Mickey Pentecost for helpful discussions. We are grateful to Brooke Lane, Lee Shaughnessy, Shumin Tan, and Elizabeth Joyce for comments on the manuscript. We thank Julie Theriot for providing Metamorph tracking software and Nafisa Ghori for assistance with TEM. We are thankful to Stefano Censini for help with the initial experiments.
M.R.H. and J.Y.L. were partially supported by NIH Ruth L. Kirschtein National Research Service Award 5 T32 GM07276. The project described was supported by Award no. R01 CA92229, R01 AI38459, and AI050000 from the National Cancer Institute and the National Institute of Allergy and Infectious Diseases.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute and the National Institute of Allergy and Infectious Diseases.
Footnotes
Citation Howitt MR, et al. 2011. ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio 2(4):e00098-11. doi:10.1128/mBio.00098-11.
REFERENCES
- 1. Campbell BJ, Engel AS, Porter ML, Takai K. 2006. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4:458–468 [DOI] [PubMed] [Google Scholar]
- 2. Blaser MJ. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176(Suppl. 2):S103–S105 [DOI] [PubMed] [Google Scholar]
- 3. Beswick EJ, Suarez G, Reyes VE. 2006. H pylori and host interactions that influence pathogenesis. World J. Gastroenterol. 12:5599–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schreiber S, et al. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. U. S. A. 101:5024–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Larsen SH, Reader RW, Kort EN, Tso WW, Adler J. 1974. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature 249:74–77 [DOI] [PubMed] [Google Scholar]
- 6. Kuo SC, Koshland DE., Jr 1987. Roles of cheY and cheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J. Bacteriol. 169:1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024–1037 [DOI] [PubMed] [Google Scholar]
- 8. Nakagawa S, et al. 2007. Deep-sea vent epsilon-proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. U. S. A. 104:12146–12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campbell BJ, et al. 2009. Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 5:e1000362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3:ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68:301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marchant J, Wren B, Ketley J. 2002. Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 10:155–159 [DOI] [PubMed] [Google Scholar]
- 13. Terry K, Go AC, Ottemann KM. 2006. Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol. Microbiol. 61:871–882 [DOI] [PubMed] [Google Scholar]
- 14. Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM. 2010. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol. 18:494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uemura N, et al. 2001. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345:784–789 [DOI] [PubMed] [Google Scholar]
- 16. Romero-Gallo J, et al. 2008. Effect of Helicobacter pylori eradication on gastric carcinogenesis. Lab. Invest. 88:328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan S, Tompkins LS, Amieva MR. 2009. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 5:e1000407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hazell SL, Lee A, Brady L, Hennessy W. 1986. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J. Infect. Dis. 153:658–663 [DOI] [PubMed] [Google Scholar]
- 19. Amieva MR, et al. 2003. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300:1430–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. West AH, Stock AM. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369–376 [DOI] [PubMed] [Google Scholar]
- 21. Krell T, et al. 2010. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 64:539–559 [DOI] [PubMed] [Google Scholar]
- 22. Maddock JR, Shapiro L. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717–1723 [DOI] [PubMed] [Google Scholar]
- 23. Sourjik V, Berg HC. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740–751 [DOI] [PubMed] [Google Scholar]
- 24. Wadhams GH, Martin AC, Warren AV, Armitage JP. 2005. Requirements for chemotaxis protein localization in Rhodobacter sphaeroides. Mol. Microbiol. 58:895–902 [DOI] [PubMed] [Google Scholar]
- 25. Shapiro L, McAdams HH, Losick R. 2009. Why and how bacteria localize proteins. Science 326:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thompson LJ, et al. 2004. Chronic Helicobacter pylori infection with Sydney strain 1 and a newly identified mouse-adapted strain (Sydney strain 2000) in C57BL/6 and BALB/c mice. Infect. Immun. 72:4668–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williams SM, et al. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 75:3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Terry K, Williams SM, Connolly L, Ottemann KM. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun. 73:803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee A, et al. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386–1397 [DOI] [PubMed] [Google Scholar]
- 30. Correa P, Houghton J. 2007. Carcinogenesis of Helicobacter pylori. Gastroenterology 133:659–672 [DOI] [PubMed] [Google Scholar]
- 31. Shi Y, et al. 2010. Helicobacter pylori-induced Th17 responses modulate Th1 cell responses, benefit bacterial growth, and contribute to pathology in mice. J. Immunol. 184:5121–5129 [DOI] [PubMed] [Google Scholar]
- 32. Pentecost M, Otto G, Theriot JA, Amieva MR. 2006. Listeria monocytogenes invades the epithelial junctions at sites of cell extrusion. PLoS Pathog. 2:e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pentecost M, et al. 2010. Listeria monocytogenes internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog. 6:e1000900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Macnab R, Koshland DE., Jr 1974. Bacterial motility and chemotaxis: light-induced tumbling response and visualization of individual flagella. J. Mol. Biol. 84:399–406 [DOI] [PubMed] [Google Scholar]
- 35. Parkinson JS. 1978. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J. Bacteriol. 135:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lowenthal AC, et al. 2009. A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology 155:1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yonekawa H, Hayashi H, Parkinson JS. 1983. Requirement of the cheB function for sensory adaptation in Escherichia coli. J. Bacteriol. 156:1228–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou J, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 95:6436–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Welch M, Oosawa K, Aizawa S, Eisenbach M. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dyer CM, et al. 2009. A molecular mechanism of bacterial flagellar motor switching. J. Mol. Biol. 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lowenthal AC, et al. 2009. Functional analysis of the Helicobacter pylori flagellar switch proteins. J. Bacteriol. 191:7147–7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Croxen MA, Sisson G, Melano R, Hoffman PS. 2006. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 188:2656–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Voordeckers JW, Starovoytov V, Vetriani C. 2005. Caminibacter mediatlanticus sp. nov., a thermophilic, chemolithoautotrophic, nitrate-ammonifying bacterium isolated from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. Int. J. Syst. Evol. Microbiol. 55:773–779 [DOI] [PubMed] [Google Scholar]
- 44. Tomb JF, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed] [Google Scholar]
- 45. Alm RA, et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176–180 [DOI] [PubMed] [Google Scholar]
- 46. Lertsethtakarn P, Ottemann KM. 2010. A remote CheZ orthologue retains phosphatase function. Mol. Microbiol. 77:225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Backert S, et al. 2005. Subproteomes of soluble and structure-bound Helicobacter pylori proteins analyzed by two-dimensional gel electrophoresis and mass spectrometry. Proteomics 5:1331–1345 [DOI] [PubMed] [Google Scholar]
- 48. Kiraga J, et al. 2007. The relationships between the isoelectric point and: length of proteins, taxonomy and ecology of organisms. BMC Genomics 8:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uversky VN. 2010. The mysterious unfoldome: structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010:568068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. 2004. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337:635–645 [DOI] [PubMed] [Google Scholar]
- 51. Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605–614 [DOI] [PubMed] [Google Scholar]
- 52. Ottemann KM, Lowenthal AC. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70:1984–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McGee DJ, et al. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73:1820–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qiao XT, et al. 2007. Prospective identification of a multilineage progenitor in murine stomach epithelium. Gastroenterology 133:1989–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hendrixson DR, DiRita VJ. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471–484 [DOI] [PubMed] [Google Scholar]
- 56. Hartley-Tassell LE, et al. 2010. Identification and characterization of the aspartate chemosensory receptor of Campylobacter jejuni. Mol. Microbiol. 75:710–730 [DOI] [PubMed] [Google Scholar]
- 57. Yao R, Burr DH, Guerry P. 1997. CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol. 23:1021–1031 [DOI] [PubMed] [Google Scholar]
- 58. Baar C, et al. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. U. S. A. 100:11690–11695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Amieva MR, Salama NR, Tompkins LS, Falkow S. 2002. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell. Microbiol. 4:677–690 [DOI] [PubMed] [Google Scholar]
- 60. Chalker AF, et al. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Astling DP, Lee JY, Zusman DR. 2006. Differential effects of chemoreceptor methylation-domain mutations on swarming and development in the social bacterium Myxococcus xanthus. Mol. Microbiol. 59:45–55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download Text S1, DOC file, 0.053 MB.
Identification and sequence analysis of ChePep. (A) Coomassie-stained gel of the polypeptide immunoprecipitated (IP) from H. pylori lysates using anti-Par-3 antibody. (B) Immunoblot of lysates from E. coli expressing recombinant ChePep using the anti-Par-3 antibody confirms that ChePep contains an epitope recognized by this antibody. (C) Phylogenetic tree of ChePep homologs from across the Epsilonproteobacteria. (D) Alignment of H. pylori CheY with the amino terminus of ChePep from 16 species across the class of Epsilonproteobacteria. The first arrowhead and box point to the phosphoryl group accepting aspartic acid 53 of CheY (Jimenez-Pearson MA, et al., Microbiology 151:3299–3311, 2005). The second arrowhead and box show the conserved KPFLPT motif in ChePep, which contains the critical Lys 109 (Lukat GS, et al., J. Biol. Chem. 266:8348–8354, 1991). Nonconserved residues are indicated with a value of zero and the color blue, while completely conserved residues are indicated with an asterisk and are colored red. Download Figure S1, TIF file, 2.186 MB.
ChePep localizes to the flagellar pole, and ∆ChePep mutants are slightly impaired in swarming but have normal flagella and cell growth. (A) Fluorescence intensity of ChePep at the new pole from Fig. 1A plotted on the y axis and the corresponding bacterial length plotted on the x axis. A linear regression curve was fit to the data. (B) Immunofluorescence images of wild-type (WT) and ChePep mutant (∆ChePep) H. pylori cells stained with antibodies to H. pylori (red) and recombinant ChePep (green). The same anti-ChePep antibody was used to determine ChePep expression in immunoblots of bacterial lysates from the WT and ∆ChePep mutant. (C) Growth curves of WT H. pylori and ∆ChePep in Brucella broth with 10% fetal bovine serum. (D) Photograph of 3-day-old swarm plate with the WT and ∆ChePep and ∆FliF (aflagellate, nonmotile) mutants. The diameters of swarming halos were measured for each strain and are expressed as percentages of WT swarming (n = 4 for each strain). (E) Scanning (left) and negative-stained transmission (right) electron micrographs comparing the morphologies and locations of flagella from WT and ∆ChePep strains. Both bacteria have tufts of two to six sheathed flagella at one pole. The tips of the sheathed flagella are shown by negative staining on the right and contain a typical end bulb. Both WT and ∆ChePep strains have an average of approximately 4 flagella per bacteria (n = 14). P values are from the two-tailed Student t test. NS, no statistical difference. The scale bar is 1 µm in panel B. Download Figure S2, TIF file, 2.729 MB.
ChePep confers an advantage to in vivo colonization in multiple strains of H. pylori and is required for persistent infection. (A) Colony-forming units (CFU) of H. pylori in the stomachs of mice colonized with either the SS2000 WT or ∆ChePep mutant strain for 2 weeks. Each marker represents an individual mouse. (B) CFU counts from mice coinfected with both the SS2000 WT and ∆ChePep mutant in a 1:1 ratio for 2 weeks. The dashed red line indicates the limit of detection. P < 0.0001. (C) Mice were infected with either WT SS1 (n = 4), or ∆ChePep SS1 (n = 10). After 2 weeks, two ∆ChePep strain-infected mice were harvested to document that they were colonized, and five others were superinfected with the WT. After 4 additional weeks, all mice were harvested, and bacterial CFU were determined. Download Figure S3, TIF file, 0.613 MB.
A stable pH gradient is formed by a point source of HCl. A microinjection needle (Eppendorf Femtotip II) with 0.5-µm inner diameter was filled with 1 N HCl, and constant pressure was adjusted to flow at 0.8 pmol/min. In this figure, the needle is imaged by phase-contrast microscopy while inserted into 3 ml of water containing Lysosensor dye. Fluorescence imaging of the same field followed for 30 s with 2-s exposure times. The color image is a pseudocolor rendering of the fluorescence panel. Markings show 10-µm rings up to a 60-µm radius. The bottom graph plots the fluorescence intensity as a function of distance from the needle tip. The different plots show the gradient is stable over time by measuring the fluorescence intensity at different times after inserting the needle (at 30 s and at 1, 2, 3, 4, and 5 min). Download Figure S4, TIF file, 2.743 MB.
Motility tracings of WT and ∆ChePep and ChePep* (∆ChePep complemented in trans) mutant H. pylori swimming behavior. Phase-contrast video microscopy movies in real time (30 frames per s) of WT versus various mutant bacteria were digitized, and individual bacteria were manually tracked using the MTrackJ plugin for ImageJ software. Images were inverted to show bacteria in white with their individual motility tracings in various colors. The movie is best viewed in loop mode. Download Movie S1, MOV file, 1.127 MB.
The ∆ChePep cell swims with frequent reversals of direction rather than end-over-end tumbles. WT and ∆ChePep cell swimming was recorded by high-resolution digitally enhanced DIC microscopy, and the cells’ trajectory was traced. At the end of this movie, an inset image appears showing detailed reversal events for the ∆ChePep cell. This video was used to generate Fig. 3C. Download Movie S2, MOV file, 8.739 MB.
∆ChePep cells can sustain prolonged backward runs, with the flagella “pulling” the bacteria. WT and ∆ChePep H. pylori cells were recorded using high-magnification phase-contrast video microscopy at 30 frames per s. The locations of their respective flagella are initially indicated with arrows. Download Movie S3, MOV file, 8.375 MB.
H. pylori swimming response to a point source of acid. The right frame shows a log-phase culture of WT H. pylori imaged by video phase-contrast microscopy for 5 s before and 12 s after introduction of a micropipette as a point source of acid. The left frame shows a culture of ∆CheW H. pylori (defective in chemotaxis) under identical conditions. The WT changes in behavior and redistributes away from the needle, while the ∆CheW mutant does not. The movie can be appreciated in real time, at 30 frames per s. Download Movie S4, MOV file, 6.339 MB.
Movies from motility tracings of Fig. 5E. H. pylori chemotactic response to acid. Phase-contrast video microscopy movies, similar to Movie S4, were digitized, and individual bacteria were manually tracked using the MTrackJ plugin for ImageJ software. The upper panels show the motility paths for 2 s prior to the introduction of the acid point source. These 2-s tracts repeat over the duration of the movie. The bottom panels show 10-s tracings of the motility paths of the bacteria during exposure to the pH gradient. The position of the needle is also shown. Tracings disappear from the movie when the bacteria escape the field of view. Download Movie S5, MOV file, 5.506 MB.