ABSTRACT
Carbonate chimneys at the Lost City hydrothermal field are coated in biofilms dominated by a single phylotype of archaea known as Lost City Methanosarcinales. In this study, we have detected surprising physiological complexity in single-species biofilms, which is typically indicative of multispecies biofilm communities. Multiple cell morphologies were visible within the biofilms by transmission electron microscopy, and some cells contained intracellular membranes that may facilitate methane oxidation. Both methane production and oxidation were detected at 70 to 80°C and pH 9 to 10 in samples containing the single-species biofilms. Both processes were stimulated by the presence of hydrogen (H2), indicating that methane production and oxidation are part of a syntrophic interaction. Metagenomic data included a sequence encoding AMP-forming acetyl coenzyme A synthetase, indicating that acetate may play a role in the methane-cycling syntrophy. A wide range of nitrogen fixation genes were also identified, many of which were likely acquired via lateral gene transfer (LGT). Our results indicate that cells within these single-species biofilms may have differentiated into multiple physiological roles to form multicellular communities linked by metabolic interactions and LGT. Communities similar to these Lost City biofilms are likely to have existed early in the evolution of life, and we discuss how the multicellular characteristics of ancient hydrogen-fueled biofilm communities could have stimulated ecological diversification, as well as unity of biochemistry, during the earliest stages of cellular evolution.
IMPORTANCE
Our previous work at the Lost City hydrothermal field has shown that its carbonate chimneys host microbial biofilms dominated by a single uncultivated “species” of archaea. In this paper, we integrate evidence from these previous studies with new data on the metabolic activity and cellular morphology of these archaeal biofilms. We conclude that the archaeal biofilm must contain cells that are physiologically and possibly genetically differentiated with respect to each other. These results are especially interesting considering the possibility that the first cells originated and evolved in hydrothermal systems similar to Lost City.
Introduction
The set of geochemical reactions known as serpentinization presents unique opportunities and challenges for biological communities, but little is known about the adaptations required of organisms that inhabit serpentinization-dominated habitats. Serpentinization is exothermic anywhere water reacts with the mineral olivine, and it results in warm, alkaline fluids and abiotic synthesis of hydrogen (H2), methane, and larger hydrocarbons (1, 2). It typically occurs in ultramafic oceanic crust and is also expected to occur on other planetary bodies where liquid water and olivine are present (3). Alkaline hydrothermal systems driven by serpentinization were probably widespread on the seafloor of the ancient Archean ocean, and H2 and methane generated by serpentinization may have supported the earliest biological communities (4–7). A modern example of such an ecosystem is the Lost City hydrothermal field, where microbial biofilm communities attached to the carbonate chimneys are fueled by ≤90°C, pH 9 to 11, serpentinization-derived fluids rich in H2 and methane (1). Therefore, the Lost City ecosystem provides an opportunity to directly investigate geochemical and biological processes thought to be important in the early evolution of life.
The densely populated (up to 109 cells/g carbonate) mucilaginous biofilms within Lost City carbonate chimneys appear to be dominated by a single “species” of archaea. In the hottest and highest-pH zones of actively venting chimneys, nearly 100% of the archaeal sequences in 16S rRNA gene clone libraries and tag pyrosequencing data sets belong to a single phylotype referred to as Lost City Methanosarcinales (LCMS) (8, 9). Furthermore, a FISH (fluorescent in situ hybridization) probe specific to the LCMS phylotype comprises up to 32.5% of the cells in the interior of actively venting chimneys. A FISH probe universal to bacteria detected only 4.2% of the cells in the same sample, so the LCMS phylotype represents ~80% of the cells detectable by FISH (8). The bacteria in these samples are dominated by taxa expected to be mesophilic and aerobic or microaerophilic and therefore would not occupy the same hot, anoxic zones as the members of the order Methanosarcinales (9, 10). Such extreme dominance by a single species is remarkable, especially considering the longevity of the Lost City hydrothermal field. Radiometric dating of Lost City chimneys indicates that hydrothermal activity has been ongoing for at least 100,000 years and possibly up to 1 million years (11, 12). Although we cannot know whether the LCMS phylotype has been present throughout that history, it has been found to dominate actively venting chimneys at least ~100 years old (9). In general, ecosystem stability is thought to require high biological diversity (13). If this model applies to the Lost City ecosystem, then two possible explanations for the extremely low diversity of the Lost City archaeal biofilms are apparent. One possibility is that the low-diversity biofilms are inherently unstable but do not experience environmental perturbations or do not experience competition. A potential example of this scenario is a deep mine ecosystem apparently consisting of a single bacterial species (14). The extreme energy limitation of this habitat, along with its isolation, may contribute to ecological simplicity/stability by reducing the number of available competitors. This explanation is consistent with the long-lived nature (compared to other hydrothermal systems) and physiological constraints of Lost City chimneys, but significant changes in chimney fluid chemistry, temperature, and mineralogy do occur over time (15). Therefore, the chimney biofilms are not completely protected from environmental perturbations, but the unusual stability and extreme conditions of their habitat may contribute to their low diversity.
A second explanation (that is not mutually exclusive with respect to the first) is that the archaeal biofilms harbor more ecologically relevant diversity than is apparent in molecular surveys of 16S rRNA genes. It is now commonly accepted that organisms with nearly identical 16S rRNA genes may differ greatly in genomic content and physiological characteristics. For example, Vibrio splendidus isolates with >99% 16S rRNA gene sequence similarity can have widely different genome sizes and contain extensive heterogeneity in protein-coding genes (16). Hunt et al. (17) have suggested that these isolates represent recent or ongoing sympatric speciation events resulting from fine partitioning of resources. Niche partitioning among closely related species is particularly well documented for cyanobacteria. Several studies have delineated multiple cyanobacterial “ecotypes” that inhabit distinct regions within oceanic water columns (18, 19) or within hot spring microbial mats (20). Although the 16S rRNA genes of cyanobacterial ecotypes may be highly similar, the genomic contents can vary widely (21–23). In another study, metagenomic sequencing revealed two coexisting strains of “Cenarchaeum symbiosum” (an archaeal symbiont of marine sponges) with >99% 16S rRNA gene sequence similarity (24). The authors conclude that the two strains may occupy different niches within the host sponge or else participate in a metabolic interdependence that maintains their coexistence.
Recent evidence suggests that the LCMS phylotype also comprises multiple closely related, coexisting strains. Tag pyrosequencing of the V6 hypervariable region of the 16S rRNA gene of natural LCMS biofilm populations revealed hundreds of very rare sequences that differed by 1 to 2 bp from the most common sequence, possibly representing a dormant pool of variants (9). Clone library sequencing of the intergenic transcribed spacer (ITS) region between the 16S and 23S rRNA genes of LCMS biofilms revealed multiple ITS genotypes associated with the same V6 genotype (25). The relative abundance of its genotypes differed among samples, suggesting a possible ecological signature.
In this paper, we present additional evidence for microdiversity within the LCMS biofilm population in the form of syntrophic physiological activity, cell morphological differentiation, and functional gene diversity. In particular, we focus on the ability of LCMS biofilms to generate or oxidize methane. LCMS has resisted laboratory cultivation because of the difficulty in replicating its in situ environmental conditions and/or because its growth rate is too low to be detected by typical laboratory experiments. Consequently, the physiology of LCMS is unknown. All known members of the order Methanosarcinales either generate or oxidize methane; therefore, LCMS cells are presumed to utilize either H2 or methane, both of which are present at high concentrations (1 to 14 mmol/kg and 1 to 2 mmol/kg, respectively) in Lost City hydrothermal fluids. Carbon dioxide (CO2), however, is virtually absent from Lost City fluids (2), so if methanogenesis occurs in active chimneys, it is either severely carbon limited (26) or else driven by a substrate other than CO2.
In large part because of the unusual combination of high H2, high methane, and low CO2 concentrations, previous studies have provided apparently conflicting data regarding whether LCMS cells are generating or oxidizing methane. Isotopic analyses of methane from Lost City fluids indicate a negligible biogenic contribution (2, 27), suggesting that LCMS biofilms are unlikely to be methanogenic. However, isotopic ratios of archaeal lipids extracted from Lost City carbonate chimneys are inconsistent with LCMS being methanotrophic (26). Therefore, whether the LCMS phylotype represents methanogenic or methanotrophic organisms is unresolved. We present evidence for the ability of LCMS biofilms to mediate both methanogenesis and methanotrophy and describe observations that are consistent with cellular differentiation within the biofilm.
RESULTS AND DISCUSSION
Metabolic diversity: methane production and oxidation.
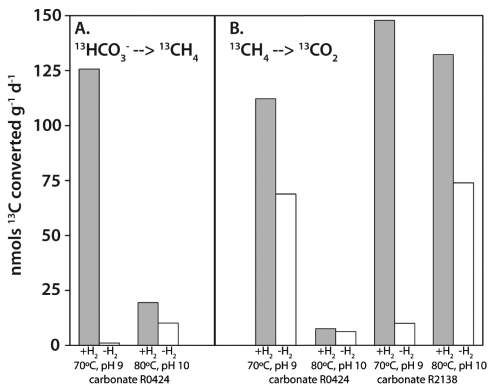
To test the capability of LCMS to mediate methane production and oxidation, we incubated at high temperature ~3-g subsamples of carbonate (aragonite and calcite) chimneys in anaerobic 50-ml serum bottles (containing high-pH artificial seawater medium including sulfate) with either 13C-labeled sodium bicarbonate (NaH13CO3, which exchanges with CO2) or 13C-labeled methane (13CH4). Incubations were stopped after 8 days, and the bottles were analyzed for the 13C contents of CO2 and methane by gas chromatography-mass spectrometry. The results (Fig. 1) show that in the absence of oxygen and at 70 to 80°C and pH 9 to 10, both methanogenesis (NaH13CO3 conversion to 13CH4) and methanotrophy (13CH4 conversion to 13CO2) proceeded at similar rates (5 to 150 nmol g−1 day−1). These rates are in the same range as those reported for anaerobic oxidation of methane (AOM) in flowthrough bioreactors (9 to 138 nmol g−1 day−1) (28) at 5°C but much lower than rates measured for bottle incubations at 9°C of methanotrophic microbial mats from the Black Sea (7,000 to 20,000 nmol g−1 day−1) (29). In situ rates at Lost City chimneys, however, could be much greater than those measured in this experiment due to the increased solubility of H2 and methane at water depths of 700 to 900 m and because of the high fluid flux from the carbonate chimneys. Although analyses of biomarkers (30) and laboratory incubations (31) of marine sediments have indicated that AOM is possible at high temperatures, our results represent the first experimental evidence that AOM occurs in high-temperature hydrothermal chimneys.
FIG 1 .
The rates (in nmol·C·day−1 g−1 of carbonate chimney material) of methane (CH4) production (A) and oxidation (B) are both stimulated by the addition of hydrogen (H2) gas to the incubation vessels. The H2 stimulation effect was observed in two different carbonate samples under two sets of temperature and pH conditions simulating natural mixing of hydrothermal fluid and seawater.
The biochemical pathway of AOM remains unknown (32), but genomic and biochemical analyses suggest that AOM utilizes many of the same enzymes required for methanogenesis but in the opposite direction (33). Accordingly, we hypothesized that high concentrations of H2 would increase the rate of methanogenesis (4H2 + CO2 → CH4 + 2H2O) and inhibit AOM in Lost City chimney biofilm samples. Intriguingly, our results (Fig. 1) show that both methanogenesis and AOM were stimulated by the addition of H2, indicating that the two processes are not in competition. It has been shown that methane is reoxidized at trace levels during methanogenesis (34) such that the rate of methane oxidation is correlated with the rate of methane production (35). In such studies, however, methane oxidation rates are >200-fold lower than methane production rates, in contrast to our experiment, in which production and oxidation were roughly equal (Fig. 1). Furthermore, Fig. 1 shows that methane oxidation does not require the addition of H2 and is therefore not dependent on H2-fueled methanogenesis from CO2 occurring in the same bottle. This result is consistent with the chemical composition of Lost City fluids, which are nearly devoid of CO2 but rich in methane, and suggests that methane is the root carbon source fueling the biofilm community. Alternatively, it is possible that the methane oxidation in our experiment is a small fraction of the total methane production from a substrate other than CO2, such as formate or acetate. The stimulation of methane oxidation by H2 as measured in our experiment, however, is not easily explained by this scenario because no known pathway involves the production of methane via oxidation of H2 by formate or acetate. Regardless of the methanogenic substrate, it is clear that both methanogenesis and AOM occurred during the experiment and both reactions were stimulated by H2.
Thus, it appears that Lost City chimney biofilms are capable of consuming both H2 and methane, the two most abundant reactive species in Lost City fluids. Our previous work has shown that the LCMS phylotype comprises >80% of the cells detectable by FISH (8) and nearly 100% of the archaeal 16S rRNA gene sequences (9) in Lost City chimneys. It is possible that representatives of the ANME-1 group (anaerobic methane oxidizer group 1) of anaerobic methanotrophic archaea were present in our experiment, but ANME-1 sequences are infinitesimally rare (0.0001% of the 16S rRNA gene pyrotags) (9) in hot, actively venting chimneys such as the one used in this experiment. Furthermore, there is no evidence for the presence or activity of ANME-1 organisms in any high-temperature environment.
Bacteria are also unlikely to have played a role in this experiment. Sequences corresponding to potential sulfate-reducing bacteria related to the genus Desulfotomaculum have been detected at low levels in Lost City chimneys, but there is no evidence linking these organisms to AOM (10). The most abundant bacterial sequences at this chimney derive from the Thiomicrospira group of microaerophilic, sulfur-oxidizing bacteria (9, 10, 36). These organisms are not expected to be active under the conditions of this experiment, nor are they likely to be involved in methane cycling.
Therefore, both methanogenesis and AOM were most likely mediated by a single organism or a syntrophic assemblage of organisms belonging to the LCMS phylotype. Other studies have reported production of methane associated with AOM in marine sediments, suggesting that the associated archaea (all of which are phylogenetically affiliated with or closely related to Methanosarcinales) are capable of both methane production and oxidation (reference 37 and references therein). This interpretation is consistent with micrometer-scale diffusion-reaction models of these habitats, which indicate that archaea which are expected to be methanotrophs may be net producers of methane when H2 and methane are both available (38, 39). Our experiments provide the first evidence for the stimulation of both methane production and oxidation by H2 in a biofilm community dominated by a single phylotype.
Metabolic diversity: formate and acetate.
CO2 is scarce (0.1 to 26 µmol/kg) in Lost City fluids (2), and the low methanogenesis rates shown in Fig. 1 could reflect substrate limitation at high pH. Indeed, isotopic measurements of lipids extracted from Lost City chimneys could indicate in situ carbon limitation if the chimney biofilms are dependent on CO2 utilization (26). Recently it has been reported that formate and acetate are present in relatively high concentrations (36 to 158 µmol/kg and 1 to 35 µmol/kg, respectively) in Lost City fluids (40); therefore, these species may be more suitable as substrates for methanogenesis than is CO2.
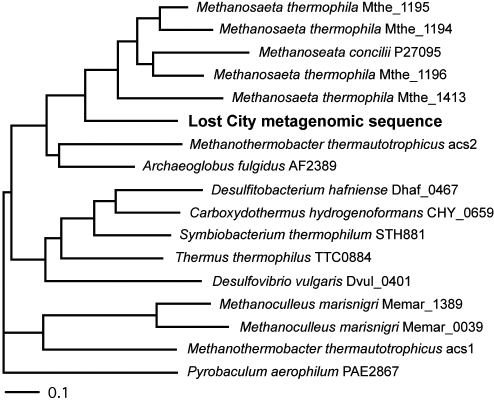
During a preliminary analysis of metagenomic data (41) from the same Lost City carbonate chimney sample with which the methane production and oxidation experiments were conducted, we identified an open reading frame encoding AMP-forming acetyl coenzyme A (CoA) synthetase (ACS). The Lost City ACS sequence has high overall sequence similarity to that from the Methanosaeta genus of aceticlastic methanogens (Fig. 2) and shares several conserved residues diagnostic of a strong preference for acetate as a substrate (42) (see Fig. S1 in the supplemental material). Therefore, it is possible that at least some of the cells within the LCMS biofilm are capable of utilizing acetate as a carbon source and/or a methanogenic substrate. Further work is necessary to determine whether the consumption of acetate is also stimulated by the addition of H2 and whether acetate may serve as a shuttle between methanogenic and methanotrophic cells (43). In addition to acetate, formate may also be utilized as a methanogenic substrate. Although no members of the order Methanosarcinales have been shown to utilize formate, the genome of Methanosarcina barkeri contains a complete formate dehydrogenase operon (44). We attempted, but failed, to amplify with degenerate primers the gene encoding the alpha subunit of formate dehydrogenase from Lost City chimney samples. Rapid cycling of methane with formate and/or acetate within the LCMS biofilm may account for the apparent discrepancies between isotopic ratios of methane and organic lipids described above, but further work is required to fully characterize the putative processes.
FIG 2 .
Phylogenetic tree illustrating the relationship between AMP-forming ACS of aceticlastic methanogens and the metagenomic sequence from a Lost City carbonate chimney. The Lost City partial sequence includes 276 amino acids 65% identical and 79% similar to Methanosaeta thermophila Mthe_1413. Quartet puzzling support values are shown at nodes. Note that other aceticlastic methanogens (e.g., Methanosarcina acetivorans) utilize a different pathway featuring the enzyme acetate kinase rather than AMP-forming ACS (75).
Morphological diversity.
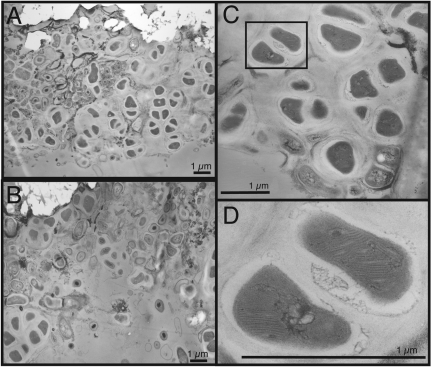
Additional evidence for the ability of the LCMS biofilm to oxidize methane is shown by transmission electron micrographs (TEMs) of a carbonate chimney in thin section (Fig. 3). Many of the cells in the TEMs contain stacks of what appear to be intracellular membranes (Fig. 3B). These structures bear some resemblance to the putative intracellular membranes observed in cylinder-shaped ANME-1 cells by Reitner et al. (45), who noted the similarity to membrane stacks utilized by aerobic methanotrophic bacteria. The cells in our TEMs that contain putative intracellular membranes are abundant and exhibit the characteristic sarcinal morphology of the Methanosarcinales (46, 47), contrasting sharply with the expected morphology of either bacterial or ANME-1 cells. Furthermore, previous work has shown that 80% of the cells visible by microscopy via FISH belong to the LCMS phylotype (8). Therefore, it seems plausible that the LCMS phylotype includes both morphologies and that some LCMS cells oxidize methane with the aid of intracellular membranes. Future studies should test this hypothesis with single-cell imaging and sorting techniques (48, 49).
FIG 3 .
TEMs of carbonate chimney thin sections. (A and B) A cell type with sarcinal morphology is prevalent, but multiple cell types are present and closely associated with each other. Furthermore, a viscous matrix appears to surround each cell cluster and may aid in the attachment of cells to the carbonate minerals (bright white areas). (C) At least three lightly stained cells surround a densely stained cell near the bottom of the panel. The outlined area corresponds to the area shown in panel D, which shows that the sarcinal cells contain intracellular membrane stacks putatively involved in methane oxidation.
Other remarkable features of the TEMs include (i) the close association between most of the cells and the carbonate minerals, (ii) the lightly stained matrix that encloses cell clusters, and (iii) the range of cell morphologies in a sample dominated by the LCMS phylotype. Densely stained cells that are enclosed by the lightly stained matrix exhibit the characteristic sarcinal morphology, appear to have recently divided, and commonly contain stacks of intracellular membranes. The more lightly stained, slightly smaller cells, in contrast, are enclosed by a grainier, darker matrix and are rarely in sarcinal packets. Some of the latter cells contain a central densely stained body, while others show more diffuse staining.
We have previously reported low but significant levels of diversity within the ITS region of LCMS sequences (25). Diversity in the ITS region has been linked to important ecological and physiological variations within populations previously considered to be one species (17–20), so it would be very interesting to know whether the morphological diversity observed in our TEMs is correlated with genetic diversity. Alternatively, cell differentiation may arise within genetically homogenous populations; indeed, the diversity of cell morphologies in Fig. 3 is similar to that of multicellular aggregates seen in pure cultures of Methanosarcina mazei. Such aggregates commonly contain multiple cell types, some of which contain cytoplasmic granules similar to the densely stained bodies in Fig. 3, and some have groups of intracellular tubules that may form membranes (47). A closely related species, Methanosarcina acetivorans, also forms cell clusters containing dense cytoplasmic granules and intracellular vesicles (46). This species can mediate trace levels of AOM during methanogenesis (34), so it is possible that the diversity of cell morphologies in Methanosarcina spp. and in the LCMS biofilm is related to the ability to mediate simultaneous methanogenesis and AOM.
Functional gene diversity.
We have presented evidence that the LCMS biofilm has adapted to utilize the most abundant electron donor (H2) and two of the most abundant sources of carbon (methane and acetate) in its environment. Our evidence suggests that this biofilm community has also adapted to utilize the most abundant source of nitrogen in Lost City fluids—dissolved nitrogen gas (N2). Although N2 has not yet been measured in Lost City fluids, the N2 concentrations at the Rainbow and Logatchev hydrothermal fields (1.8 mM and 3.0 mM, respectively), which are also hosted on serpentinized peridotites on the Mid-Atlantic Ridge, are 3- to 5-fold greater than those in ambient seawater (50). Fixed nitrogen, in contrast, is relatively scarce in end-member Lost City fluids (nitrate and ammonium concentrations are <4 µM [D. Butterfield, personal communication]). Although micromolar quantities of fixed nitrogen may be enough to sustain the densely populated LCMS biofilms, N2 fixation is likely to occur because biofilms are typically nitrogen limited due to their ability to quickly remove fixed nitrogen from their surroundings (51, 52). Furthermore, the highly reducing conditions within Lost City chimneys may be much more favorable for N2 fixation than those in surface environments, where N2 fixation is often inhibited by oxygen.
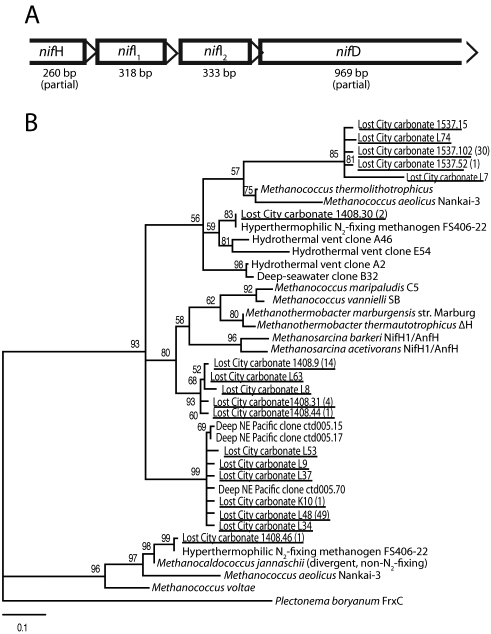
These observations led us to investigate LCMS biofilms for the presence of genes encoding the nitrogenase enzyme complex, which is required for biological N2 fixation. We identified a partial nitrogenase operon containing four open reading frames in preliminary metagenomic data from the same carbonate chimney sample utilized for the incubation experiments described above (Fig. 4A). The 5′ end consists of a partial open reading frame with 75% amino acid sequence identity to the 3′ end of the product of nifH from Methanothermobacter thermautotrophicus, a thermophilic, N2-fixing methanogen (53). The other open reading frames contain nifI1, nifI2, and nifD (Fig. 4A) in the same order as in M. thermautotrophicus, and the amino acid sequences they encode are 54 to 60% identitical to those encoded by the corresponding genes in that genome. Although nifH sequences have been detected in organisms unable to fix N2, our identification of an apparent nitrogenase operon containing at least four genes, including nifH, with high sequence similarity to a known thermophilic N2 fixer strongly supports the existence of N2 fixers in high-temperature Lost City chimneys.
FIG 4 .
(A) A partial nitrogenase operon recovered from metagenomic sequencing of a carbonate chimney including the 3′ end of nifH (258 bp), nifI1 (106 bp), nifI2 (111 bp), and the 5′ end of nifD (323 bp). The sequence assembly contig (GenBank accession no. ACQI01004781) is included in the metagenomic data set described in references 36 and 41. (B) Diversity of NifH sequences in clone libraries constructed from actively venting carbonate chimneys. NifH sequences from this study are underlined and followed by the number of additional NifH sequences with ≥97% amino acid identity in parentheses. Although the partial metagenomic nifH sequence in panel A does not overlap the region included in the nifH clone libraries, it most likely corresponds to clone 1408.9 in panel B. Quartet puzzling support values are shown at nodes. The tree is outgroup rooted with Plectonema boryanum FrxC, a dinitrogenase reductase-like protein involved in the light-independent reduction of protochlorophyllide.
We further explored the phylogenetic diversity of nitrogenase genes in actively venting Lost City chimneys by targeted amplification and sequencing of nifH. A total of 181 nifH clones were sequenced from the four samples shown in Table 1. These 181 NifH sequences fell into 31 clusters based on >97% amino acid identity. Eighteen clusters (representing 121 sequences and forming at least 5 distinct clades) are most closely related to methanogenic NifH and are shown in Fig. 4B. This represents a surprisingly wide range of NifH diversity, given that these samples were dominated by a single archaeal 16S rRNA gene phylotype and that no other methanogens have been detected at Lost City. The Lost City NifH sequences represented by clone 1408.30 (Fig. 4B) are identical to that of NifH expressed at 92°C by the N2-fixing methanogen isolated from deep-sea hydrothermal fluid on the Juan de Fuca Ridge in the northeastern Pacific (54). These sequences were detected only in the hottest Lost City carbonate chimney sampled, which vented fluids reaching 81°C at the time of sampling. Interestingly, this sequence is phylogenetically distinct from the NifH sequence encoded by the partial nitrogenase operon (Fig. 4A), indicating that there are at least two different nifH genes in Lost City chimneys with high sequence similarity to known thermophilic, methanogenic N2 fixers. The phylogenetic affiliations of the many other NifH sequences in Fig. 4B are less clear, and further work is necessary to determine whether they encode functional nitrogenases.
TABLE 1 .
Lost City carbonate chimney samples that were used to create nifH clone libraries
| Sample | Marker | Description | Temp (°C) | No. of unique NifH sequences/total no. sequenced | Dominant archaeal 16S phylotypea |
|---|---|---|---|---|---|
| LC1149 | 2 | Active flange covered in biofilm | 55 | 20/40 | LCMS |
| 3864-1537 | 2 | Active flange covered in biofilm | 53.5 | 28/50 | LCMS |
| LC1022 | 3 | Active chimney covered in biofilm | 75 | 26/52 | LCMS |
| 3881-1408 | 3 | Active chimney covered in biofilm | 81 | 21/39 | LCMS |
Regardless of their expression and function, it is clear that Lost City chimneys contain a remarkable diversity of nifH sequences, given that the most likely host for these genes is the LCMS phylotype. One possible explanation is that LCMS cells have variable genomes, with each cell containing different nifH genes. Members of the order Methanosarcinales are known to have acquired a significant fraction of their genomes, including nitrogenase genes, via lateral gene transfer (LGT) (55), so it may be that many of these nifH genes were acquired via lateral transfer from other organisms. We have previously reported an extremely high abundance and diversity of transposase sequences in Lost City metagenomic data (41), which is consistent with LGT influencing the genomic content of LCMS biofilms. Nitrogen fixation and LGT may be commonly associated with AOM, as two recent studies have also recovered a wide diversity of nifH genes from N2-fixing anaerobic methanotrophic archaea in marine sediments (48, 49).
Differentiation and syntrophy within a single-species biofilm.
All of the evidence that has been presented in this paper and published previously (1, 9, 10, 25) is consistent with the model illustrated in Fig. 5, in which the LCMS phylotype includes genetically heterogenous populations of cells that are differentiated into multiple cell types engaging in metabolic cooperation and LGT. Metabolic and genetic interactions among organisms within biofilm communities are well documented. For thorough reviews, see references 56 to 58. Because individual cells benefit in numerous ways from their membership in a diverse community, a successful biofilm requires at least some level of cooperation or, at the very least, noncompetition among the members of the community. Although selection still acts on competing lineages, the ability to participate in the community is also a selectable trait (58). Therefore, biofilm communities can be considered to be inherently social and multicellular. The observations reported in this paper highlight the potential for multicellular characteristics in biofilms even when they are dominated by a single 16S rRNA gene phylotype (e.g., LCMS). Our results are consistent with the provocative study by Boles et al. (59), who showed that during biofilm growth, a clonal population of Pseudomonas aeruginosa rapidly generates specialized subpopulations that have increased fitness.
FIG 5 .
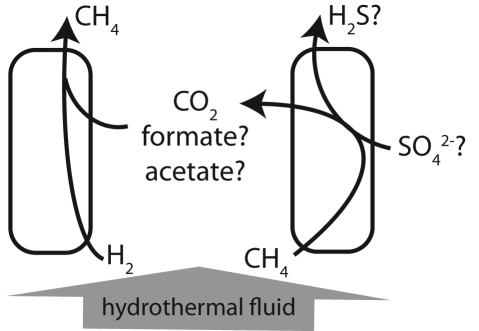
Hypothetical syntrophic reactions between methanogenic and methanotrophic cell types that are consistent with experimental data. The observation that both the production and oxidation of methane (CH4) are stimulated by H2 is explained by the dependence of methane oxidation on the uptake of its waste product by a nearby H2-utilizing cell. CO2, formate, and acetate are potential transfer molecules. The terminal electron acceptor of the overall reaction is unknown, but sulfate (SO42−) is a likely candidate.
In the case of the LCMS biofilms, differentiation and syntrophy likely evolved as adaptations to maximize the metabolic potential of Lost City fluids, which are among the most H2- and methane-rich fluids ever measured. To our knowledge, no organisms capable of utilizing both H2 and methane have been characterized; indeed, the simultaneous consumption of H2 and methane does not seem thermodynamically sensible for a single cell. This process is feasible, however, for a population that is differentiated into H2-utilizing and methane-utilizing cell types that consume each other’s waste products (Fig. 5). Any other explanation of the data would involve novel and seemingly implausible metabolic pathways. This putative differentiation might involve only a few specialized cells, akin to heterocyst formation in cyanobacteria, or all cells may differentiate into one of the possible types according to environmental stimuli. The exact reactions and metabolites involved in our proposed syntrophic assemblage have yet to be determined, but initial metagenomic data indicate that acetate may play an important role as a carbon source, methanogenic substrate, or metabolite shuttle between cell types (Fig. 2). The terminal electron acceptor is also unknown, but the most obvious candidate is sulfate, which is surprisingly abundant in end-member Lost City fluids (1 to 4 mM) and increases with seawater mixing (1).
LGT, as evidenced in the LCMS biofilms by the high diversity of nitrogenase (Fig. 4) and transposase (41) genes, may be directly related to the multicellular nature of the LCMS biofilm community. In addition to increasing the genetic diversity of the community by introducing novel DNA from unrelated organisms, a moderate rate of LGT within a biofilm community is also predicted to stabilize the coexistence of multiple phenotypes (60). Thus, LGT provides another mechanism, in addition to metabolic interactions, whereby genetic variants can evolve into codependent populations.
Biofilms and the unity of biochemistry.
There is no reason to suppose that the multicellular nature of biofilms should be considered a novelty or a recently derived trait of a few lineages. Although traditional microbiological research has focused on clonal cultures in nutrient-rich broth, the predominant mode of growth for most microorganisms in natural environments is as surface-attached, multispecies biofilms (61). It is therefore plausible that the earliest biological communities utilized the biofilm mode of growth (56) and that evolutionary dynamics on the ancient Earth were strongly shaped by physiological interactions and LGT events within biofilms.
In particular, we propose that these processes acting within ancient biofilm communities would have stimulated the diversification of organisms that shared biochemical pathways. One of the most fundamental observations of biology is the unity of biochemistry (62); i.e., all forms of life on Earth use the same biochemicals and relatively minor variations of the same biochemical pathways. The unity of biochemistry strongly argues that all modern organisms have a common ancestor. The nature of the most recent common ancestor is widely debated, but it would be naive to assume that it consisted of only a single cell living in isolation. On the contrary, it seems plausible that it inhabited a biofilm-like community in which cells interacted with each other by exchanging nutrients, metabolites, and genes. Differentiation of certain populations to fill niches within the community would have led to the diversification of life from a common ancestral pool. It has been previously proposed that promiscuous LGT within an ancestral “multiphenotypical population” (63) of “precells” (64) or “progenotes” (65) could explain the vast diversity observed in extant organisms within the unity of biochemistry. We suggest that the LCMS biofilms within Lost City carbonate chimneys, where a single ancestral population appears to have differentiated into metabolically linked cells undergoing frequent LGT, are reminiscent of this hypothetical ancestral community and provide an intriguing model for the study of potentially ancient evolutionary processes.
Conclusions
It is unclear whether the functional types identified in this study correlate with the genotypic microdiversity reported previously (25). It is possible that the methanogenic and methanotrophic cell types within the LCMS phylotype are not phylogenetically distinct; i.e., they may not represent ecotypes, as defined by Cohan (66). Differentiation into multiple cell types seems to imply a kind of niche partitioning, similar to that reported for vibrioplankton populations (16, 17), but it may be that genetically identical LCMS cells are capable of filling any of multiple niches, as necessitated by local conditions within the biofilm. The same LCMS phylotype dominates chimneys differing in age by at least 100 years (9), so the LCMS community pangenome may encode the necessary adaptations to fill the many niches that arise during chimney development. The available data are still limited, but if further work corroborates this model, then the LCMS community could be considered one evolutionary unit comprising multiple ecological units, making it a “poster child” for a pluralistic view of microbial species (67, 68).
It has long been known that clonal bacterial populations can form multiple cell types (57); the results of this study implicate syntrophic metabolic interactions and LGT as potentially important aspects of cellular differentiation within biofilms. The Lost City biofilms are also excellent models for studying the diversification of life from a common ancestor, and they inhabit environmental conditions that were probably widespread on the ancient Earth (4, 5). Frequent LGT during the early stages of evolution (Fig. 4) (41) would have favored the utilization of identical biochemical building blocks, leading to a unity of biochemistry. Syntrophic metabolic interactions among closely related organisms (Fig. 1 and 3) would have increased the overall efficiency and fitness of the community while promoting diversification into multiple ecological niches. In short, the ecological dynamics operating in Lost City biofilms today can help explain how the tremendous physiological diversity of life could have evolved from a community of organisms sharing a unity of biochemistry.
MATERIALS AND METHODS
Methane production/oxidation experiments.
Carbonate chimney samples were collected from the central “Poseidon” edifice of the Lost City hydrothermal field by the deep submergence vehicle (DSV) Hercules during the 2005 Lost City Expedition aboard the research vessel (R/V) Ronald H. Brown. Incubation experiments were conducted shipboard immediately after sample collection according to the following protocol. A portion (~500 g) of carbonate chimney samples H03_072705_R0424 and H02_072705_R2138 from the Poseidon edifice (very similar to sample 3881-1408, Table 1) was crushed and homogenized in a sterile mortar and pestle. Within a plastic anoxic glove bag continually flushed with N2 gas, subsamples (~3 g) of the crushed carbonate were added to 50-ml glass serum bottles containing 35 ml of artificial medium (recipe below) buffered at pH 9 or 10. The bottles were sealed with sterile butyl rubber stoppers and aluminum crimps. To each bottle, either NaHCO3 or NaH13CO3 (Cambridge Isotope Laboratories) was added to a final concentration of 10 mM. The bottles were injected with 10 ml of CH4 or 13CH4 (Cambridge Isotope Laboratories), and half of the bottles were also injected with 20 ml of H2. The bottles were incubated at 70 or 80°C for 7 to 8 days, after which 5 to 7 ml of fluid was withdrawn and injected into gas-tight glass Exetainers (Labco Limited, High Wycombe, Buckinghamshire, United Kingdom) flushed with argon and poisoned with HgCl2.
On land, the Exetainers were injected with 0.2 ml of 50% phosphoric acid to convert all of the dissolved inorganic carbon into CO2, and the 13C/12C ratios of CO2 and CH4 were measured with a Finnegan 253 mass spectrometer (using a PLOT-Q gas chromatography column) at the University of Washington Stable Isotope Laboratory. The mole fractions of 13C in CO2 and CH4 in postincubation Exetainers were compared with the 13C fraction of CO2 in “time zero” Exetainers and the 13C fraction of CH4 from the source container (Scott Specialty Gases) in order to calculate methane production/oxidation rates. Specifically, rates were calculated according to the equation (Npost – Npre)/t, where Npost and Npre are the moles of 13C postincubation and preincubation, respectively, and t is the number of days of incubation. The calculation assumes that all of the dissolved inorganic carbon converted to CO2 by phosphoric acid was biologically available for methanogenesis during incubation. In the methane oxidation experiments, the total 13CO2 signal (Fig. 1) was up to 8% of the added 13CH4 label [(150 nmol g−1 day−1 × 3 g × 8 days)/(44,600 nmol 13CH4 added)], and the purity of the 13CH4 was 99.8% according to the manufacturer’s quality control analysis.
The incubation medium contained the following (in grams per liter): NaCl, 19.6; Na2SO4, 3.3; KCl, 0.5; KBr, 0.05; H3BO3, 0.02; MgCl2 · 6H2O, 8.8; KH2PO4, 0.072; 2-(N-cyclohexylamino)ethanesulfonic acid (CHES buffer), 5; resazurin, 0.001. After autoclaving of the medium described above, the following solutions were combined: 5 parts N/P solution, 5 parts Trace Elements F, 2.5 parts 1% CaCl2 · 2H2O solution, and 1 part Fe-EDTA solution. Twenty milliliters of this mixture was filter sterilized and added to a liter of autoclaved medium after cooling. The N/P solution contained the following (in grams per liter): (NH4)2SO4, 43.0; NaNO3, 60.5; KH2PO4, 3.6. The Trace Elements F and Fe-EDTA solutions were described previously by Mehta and Baross (54).
AMP-forming ACS phylogeny.
A metagenomic sequencing read (GenBank accession no. ACQI01017844) obtained from carbonate chimney sample H03_072705_R0424 encoded 276 amino acids corresponding to amino acid positions 131 to 406 of Methanosaeta thermophila AMP-forming ACS Mthe_1413. Start and stop codons were not present in the sequence. The multiple amino acid sequence alignment (see Fig. S1 in the supplemental material) was constructed with Muscle 3.6 (69), and the phylogeny was calculated with tree-Puzzle 5.2 (70) with default values plus exact parameter estimation and gamma-distributed rate heterogeneity. The multiple-sequence alignment contained complete sequences and was not trimmed down to the partial Lost City sequence in order to maintain previously published phylogenetic relationships (42). Two residues (L385 and G386 of M. thermophila ACS1) that are diagnostic of a strong substrate preference for acetate over other substrates containing acyl groups (42) and four acetate binding pocket residues (71) are all present in the Lost City metagenomic sequence (see Fig. S1).
Transmission electron micrography.
TEM images were obtained from carbonate chimney sample 3867-1228i (a subsample of the interior portion of sample 3867-1228), collected at marker 7 of the Lost City hydrothermal field. The sample showed a mineralogy in the interior zones markedly different (more aragonite, less calcite) from that of the darker, more lithified exterior zones. Fluid temperatures between 28 and 51°C have been measured near marker 7. Because these measurements reflect at least some mixing with seawater, the interior zones of the chimney included in sample 3867-1228i were likely exposed to temperatures of >51°C. Terminal restriction fragment length polymorphism analysis confirmed that subsample 3867-1228i contained a single dominant archaeal fragment corresponding to LCMS, like all of the other high-temperature chimneys at Lost City. Shipboard, carbonate minerals were submerged in 4% paraformaldehyde (diluted in 1× phosphate-buffered saline buffer) overnight at 4°C and then placed in 70% ethanol and stored at 4°C. Carbonate mineral thin sections were prepared and examined by the Department of Pathology Electron Microscopy Resource Center, University of Washington, according to the following procedure. Carbonate minerals were rehydrated through a graded alcohol series and fixed for 1 h at room temperature in 2% paraformaldehyde−2.5% glutaraldehyde−0.1 M sodium cacodylate−3.4 mM calcium chloride. Samples were then washed three times for 10 min each with 0.1 M cacodylate−sucrose buffer, postfixed with 1% OsO4−cacodylate buffer, washed with double-stranded H2O three times for 10 min each, and embedded in 2% agar. Embedded minerals were dehydrated through an ascending ethanol series and propylene oxide, infiltrated with Spurr’s low-viscosity embedding medium, and polymerized at 65°C overnight. Ultrathin sections (80 to 100 nm) were cut and stained with saturated uranyl acetate. The material was examined and photographed with a JEM 1200EXII (JEOL) at 80 kV.
Nitrogenase sequencing.
The carbonate chimney samples from the Lost City hydrothermal field that were used to create the NifH phylogeny are shown in Table 1. The samples were collected in 2000 and 2003 aboard R/V Atlantis during cruises AT03-6 and AT07-34 using DSV Alvin on dives 3,651 and 3,862 through 3,881. Samples were frozen at −80°C until DNA extraction. For all of the carbonate chimney samples, the same DNA extract was used in both 16S rRNA gene analysis and nifH analysis in order to compare the two sets of data confidently. Sequencing and phylogenetic analyses of nifH clones were conducted as described previously (72). Amino acid sequences were aligned (see Fig. S2 in the supplemental material) using ClustalX version 2.0.11 (73), and phylogenetic groups were created using mothur version 1.4.1 (74). The quartet puzzling maximum-likelihood phylogenetic tree was constructed with tree-Puzzle 5.2 (70) using the Whelan and Goldman model of protein evolution. Other NifH sequences recovered in this study not shown in Fig. 4 include a single sequence related to the anaerobic bacterium Desulfomicrobium baculatum and belonging to cluster III of the NifH phylogeny and 59 sequences belonging to cluster IV of the NifH phylogeny that are related to divergent archaeal and bacterial nitrogenases that are not expected to be involved in nitrogen fixation.
Nucleotide sequence accession numbers.
The 18 Lost City NifH sequences shown in Fig. 4B have been assigned GenBank accession no. GU474815 to GU474832.
SUPPLEMENTAL MATERIAL
Alignment of ACS sequences used to construct the phylogenetic tree displayed in Fig. 2. The four residues forming the acetate binding pocket (71) are indicated by asterisks, and two residues diagnostic of a strong preference for acetate as a substrate (42) are indicated by the symbol ^. Download Figure S1, DOC file, 0.046 MB.
Alignment of partial dinitrogenase reductase (NifH) sequences that were used to construct the NifH phylogenetic tree in Fig. 4. Sequences from the Lost City and other hydrothermal vent environments are identified by their clone names; all other sequences are identified by their GenBank accession numbers. Download Figure S2, DOC file, 0.029 MB.
ACKNOWLEDGMENTS
We express our appreciation to the crews of R/V Atlantis, DSV Alvin, R/V Ronald H. Brown, and DSV Hercules and the scientific party of the 2003 and 2005 Lost City expeditions. We are deeply grateful to Matt Schrenk for mentoring and many helpful discussions. We also thank Sheryl Bolton, Andrew Rice, Dave Wilbur, Giora Proskurowski, and Clara Fuchsman for technical assistance.
This research was supported by the NASA Astrobiology Institute through the Carnegie Institution for Science (J.A.B.), NSF grant OCE0137206 (D.S.K.), and NOAA Ocean Exploration support (D.S.K.). W.J.B. was supported by an NSF IGERT fellowship awarded to the University of Washington astrobiology program and Washington Sea Grant (NA76RG0119).
Footnotes
Citation Brazelton WJ, Mehta MP, Kelley DS, Baross JA. 2011. Physiological differentiation within a single-species biofilm fueled by serpentinization. mBio 2(4):e00127-11. doi:10.1128/mBio.00127-11.
REFERENCES
- 1. Kelley DS, et al. 2005. A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307:1428–1434 [DOI] [PubMed] [Google Scholar]
- 2. Proskurowski G, et al. 2008. Abiogenic hydrocarbon production at Lost City hydrothermal field. Science 319:604–607 [DOI] [PubMed] [Google Scholar]
- 3. Schulte M, Blake D, Hoehler T, McCollom T. 2006. Serpentinization and its implications for life on the early Earth and Mars. Astrobiology 6:364–376 [DOI] [PubMed] [Google Scholar]
- 4. Martin W, Baross J, Kelley D, Russell MJ. 2008. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 6:805–814 [DOI] [PubMed] [Google Scholar]
- 5. Nisbet EG, Sleep NH. 2001. The habitat and nature of early life. Nature 409:1083–1091 [DOI] [PubMed] [Google Scholar]
- 6. Olson JM. 2006. Photosynthesis in the Archean era. Photosynth. Res. 88:109–117 [DOI] [PubMed] [Google Scholar]
- 7. Sleep NH, Meibom A, Fridriksson T, Coleman RG, Bird DK. 2004. H2-rich fluids from serpentinization: geochemical and biotic implications. Proc. Natl. Acad. Sci. U. S. A. 101:12818–12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schrenk MO, Kelley DS, Bolton SA, Baross JA. 2004. Low archaeal diversity linked to subseafloor geochemical processes at the Lost City Hydrothermal Field, Mid-Atlantic Ridge. Environ. Microbiol. 6:1086–1095 [DOI] [PubMed] [Google Scholar]
- 9. Brazelton WJ, et al. 2010. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1000-year time scales in hydrothermal chimneys. Proc. Natl. Acad. Sci. U. S. A. 107:1612–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. 2006. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl. Environ. Microbiol. 72:6257–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Früh-Green GL, et al. 2003. 30,000 years of hydrothermal activity at the Lost City vent field. Science 301:495–498 [DOI] [PubMed] [Google Scholar]
- 12. Ludwig KA, Shen C-C, Kelley DS, Cheng H, Edwards RL. 2011. U–Th systematics and 230Th ages of carbonate chimneys at the Lost City Hydrothermal Field. Geochim. Cosmochim. Acta 75:1869–1888 [Google Scholar]
- 13. Tilman D, Wedin D, Knops J. 1996. Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature 379:718–720 [Google Scholar]
- 14. Chivian D, et al. 2009. Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322:275–278 [DOI] [PubMed] [Google Scholar]
- 15. Ludwig KA, Kelley DS, Butterfield DA, Nelson BK, Fruh-Green G. 2006. Formation and evolution of carbonate chimneys at the Lost City Hydrothermal Field. Geochim. Cosmochim. Acta 70:3625–3645 [Google Scholar]
- 16. Thompson JR, et al. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311–1313 [DOI] [PubMed] [Google Scholar]
- 17. Hunt DE, et al. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085 [DOI] [PubMed] [Google Scholar]
- 18. Johnson ZI, et al. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737–1740 [DOI] [PubMed] [Google Scholar]
- 19. West NJ, et al. 2001. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology 147:1731–1744 [DOI] [PubMed] [Google Scholar]
- 20. Ferris MJ, Kühl M, Wieland A, Ward DM. 2003. Cyanobacterial ecotypes in different optical microenvironments of a 68 degrees C hot spring mat community revealed by 16S-23S rRNA internal transcribed spacer region variation. Appl. Environ. Microbiol. 69:2893–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rocap G, et al. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042–1047 [DOI] [PubMed] [Google Scholar]
- 22. Dufresne A, et al. 2008. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9:R90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kettler GC, et al. 2007. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 3:e231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schleper C, et al. 1998. Genomic analysis reveals chromosomal variation in natural populations of the uncultured psychrophilic archaeon Cenarchaeum symbiosum. J. Bacteriol. 180:5003–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brazelton WJ, Sogin ML, Baross JA. 2010. Multiple scales of diversification within natural populations of archaea in hydrothermal chimney biofilms. Environ. Microbiol. Rep. 2:236–242 [DOI] [PubMed] [Google Scholar]
- 26. Bradley AS, Hayes JM, Summons RE. 2009. Extraordinary C-13 enrichment of diether lipids at the Lost City Hydrothermal Field indicates a carbon-limited ecosystem. Geochim. Cosmochim. Acta 73:102–118 [Google Scholar]
- 27. Proskurowski G, Lilley MD, Kelley DS, Olson EJ. 2006. Low temperature volatile production at the Lost City Hydrothermal Field, evidence from a hydrogen stable isotope geothermometer. Chem. Geol. 229:331–343 [Google Scholar]
- 28. Girguis PR, Orphan VJ, Hallam SJ, DeLong EF. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69:5472–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Treude T, et al. 2007. Consumption of methane and CO2 by methanotrophic microbial mats from gas seeps of the anoxic Black Sea. Appl. Environ. Microbiol. 73:2271–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schouten S, Wakeham SG, Hopmans EC, Sinninghe Damsté JS. 2003. Biogeochemical evidence that thermophilic archaea mediate the anaerobic oxidation of methane. Appl. Environ. Microbiol. 69:1680–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kallmeyer J, Boetius A. 2004. Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl. Environ. Microbiol. 70:1231–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thauer RK, Shima S. 2008. Methane as fuel for anaerobic microorganisms. Ann. N. Y. Acad. Sci. 1125:158–170 [DOI] [PubMed] [Google Scholar]
- 33. Caldwell SL, et al. 2008. Anaerobic oxidation of methane: mechanisms, bioenergetics, and the ecology of associated microorganisms. Environ. Sci. Technol. 42:6791–6799 [DOI] [PubMed] [Google Scholar]
- 34. Moran JJ, House CH, Freeman KH, Ferry JG. 2005. Trace methane oxidation studied in several Euryarchaeota under diverse conditions. Archaea 1:303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zehnder AJ, Brock TD. 1979. Methane formation and methane oxidation by methanogenic bacteria. J. Bacteriol. 137:420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brazelton WJ, Baross JA. 2010. Metagenomic comparison of two Thiomicrospira lineages inhabiting contrasting deep-sea hydrothermal environments. PLoS One 5:e13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. House CH, et al. 2009. Extensive carbon isotopic heterogeneity among methane seep microbiota. Environ. Microbiol. 11:2207–2215 [DOI] [PubMed] [Google Scholar]
- 38. Orcutt B, Meile C. 2008. Constraints on mechanisms and rates of anaerobic oxidation of methane by microbial consortia: process-based modeling of ANME-2 archaea and sulfate reducing bacteria interactions. Biogeosciences 5:1587–1599 [Google Scholar]
- 39. Alperin MJ, Hoehler TM. 2009. Anaerobic methane oxidation by archaea/sulfate-reducing bacteria aggregates: 1. Thermodynamic and physical constraints. Am. J. Sci. 309:869–957 [Google Scholar]
- 40. Lang SQ, Butterfield DA, Schulte M, Kelley DS, Lilley MD. 2010. Elevated concentrations of formate, acetate and dissolved organic carbon found at the Lost City hydrothermal field. Geochim. Cosmochim. Acta 74:941–952 [Google Scholar]
- 41. Brazelton WJ, Baross JA. 2009. Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J. 3:1420–1424 [DOI] [PubMed] [Google Scholar]
- 42. Ingram-Smith C, Smith KS. 2007. AMP-forming acetyl-CoA synthetases in Archaea show unexpected diversity in substrate utilization. Archaea 2:95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Valentine DL, Reeburgh WS. 2000. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2:477–484 [DOI] [PubMed] [Google Scholar]
- 44. Maeder DL, et al. 2006. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 188:7922–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reitner J, et al. 2005. Concretionary methane-seep carbonates and associated microbial communities in Black Sea sediments. Palaeogeogr. Palaeoclimatol. Palaeoecol. 227:18–30 [Google Scholar]
- 46. Sowers KR, Baron SF, Ferry JG. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robinson RW. 1986. Life cycles in the methanogenic archaebacterium Methanosarcina mazei. Appl. Environ. Microbiol. 52:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pernthaler A, et al. 2008. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc. Natl. Acad. Sci. U.S.A. 105:7052–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dekas AE, Poretsky RS, Orphan VJ. 2009. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326:422–426 [DOI] [PubMed] [Google Scholar]
- 50. Charlou JL, Donval JP, Fouquet Y, Jean-Baptiste P, Holm N. 2002. Geochemistry of high H2 and CH4 vent fluids issuing from ultramafic rocks at the Rainbow hydrothermal field (36°14°N, MAR). Chem. Geol. 191:345–359 [Google Scholar]
- 51. Jones K, Bradshaw SB. 1997. Synergism in biofilm formation between Salmonella enteritidis and a nitrogen-fixing strain of Klebsiella pneumoniae. J. Appl. Microbiol. 82:663–668 [DOI] [PubMed] [Google Scholar]
- 52. Pynaert K, et al. 2003. Characterization of an autotrophic nitrogen-removing biofilm from a highly loaded lab-scale rotating biological contactor. Appl. Environ. Microbiol. 69:3626–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith DR, et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 179:7135–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mehta MP, Baross JA. 2006. Nitrogen fixation at 92 degrees C by a hydrothermal vent archaeon. Science 314:1783–1786 [DOI] [PubMed] [Google Scholar]
- 55. Deppenmeier U, et al. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453–461 [PubMed] [Google Scholar]
- 56. Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187–209 [DOI] [PubMed] [Google Scholar]
- 57. Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81–104 [DOI] [PubMed] [Google Scholar]
- 58. West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4:597–607 [DOI] [PubMed] [Google Scholar]
- 59. Boles BR, Thoendel M, Singh PK. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 101:16630–16635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chia N, Woese CR, Goldenfeld N. 2008. A collective mechanism for phase variation in biofilms. Proc. Natl. Acad. Sci. U. S. A. 105:14597–14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Davey ME, O'toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thauer R. 1997. Biodiversity and unity in biochemistry. Antonie Van Leeuwenhoek 71:21–32 [DOI] [PubMed] [Google Scholar]
- 63. Kandler O. 1994. The early diversification of life, p 152–160. In Bengtson S (ed), Early life on Earth. Nobel symposium no. 84.Columbia University Press, New York, NY [Google Scholar]
- 64. Baross JA, Hoffman SE. 1985. Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Orig. Life Evol. Biosph. 15:327–345 [Google Scholar]
- 65. Woese C. 1998. The universal ancestor. Proc. Natl. Acad. Sci. U. S. A. 95:6854–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cohan FM. 2006. Towards a conceptual and operational union of bacterial systematics, ecology, and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1985–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bapteste E, et al. 2009. Prokaryotic evolution and the tree of life are two different things. Biol. Direct. 4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Doolittle WF, Zhaxybayeva O. 2009. On the origin of prokaryotic species. Genome Res. 19:744–756 [DOI] [PubMed] [Google Scholar]
- 69. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schmidt HA, Strimmer K, Vingron M, von Haeseler A. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504 [DOI] [PubMed] [Google Scholar]
- 71. Ingram-Smith C, Woods BI, Smith KS. 2006. Characterization of the acyl substrate binding pocket of acetyl-CoA synthetase. Biochemistry 45:11482–11490 [DOI] [PubMed] [Google Scholar]
- 72. Mehta MP, Butterfield DA, Baross JA. 2003. Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca ridge. Appl. Environ. Microbiol. 69:960–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 74. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Smith KS, Ingram-Smith C. 2007. Methanosaeta, the forgotten methanogen? Trends Microbiol. 15:150–155 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of ACS sequences used to construct the phylogenetic tree displayed in Fig. 2. The four residues forming the acetate binding pocket (71) are indicated by asterisks, and two residues diagnostic of a strong preference for acetate as a substrate (42) are indicated by the symbol ^. Download Figure S1, DOC file, 0.046 MB.
Alignment of partial dinitrogenase reductase (NifH) sequences that were used to construct the NifH phylogenetic tree in Fig. 4. Sequences from the Lost City and other hydrothermal vent environments are identified by their clone names; all other sequences are identified by their GenBank accession numbers. Download Figure S2, DOC file, 0.029 MB.