Abstract
To understand how chromosome shapes are determined by actions of condensins and cohesin, we devised a series of protocols in which their levels are precisely changed in Xenopus egg extracts. When the relative ratio of condensin I to II is forced to be smaller, embryonic chromosomes become shorter and thicker, being reminiscent of somatic chromosomes. Further depletion of condensin II unveils its contribution to axial shortening of chromosomes. Cohesin helps juxtapose sister chromatid arms by collaborating with condensin I and counteracting condensin II. Thus, chromosome shaping is achieved by an exquisite balance among condensin I and II and cohesin.
Keywords: Xenopus egg extracts, chromosome condensation, cohesin, condensins, sister chromatid cohesion
Upon entry into mitosis, a mass of chromatin distributed within the interphase nucleus is converted into a discrete set of rod-shaped structures. This process, often referred to as mitotic chromosome condensation, culminates in the assembly of metaphase chromosomes in which two sister chromatids are juxtaposed with each other along their entire lengths. It is thought that chromosome condensation is a critical preparatory step to ensure faithful segregation of sister chromatids in subsequent anaphase (Swedlow and Hirano 2003). The shape of metaphase chromosomes is highly characteristic, yet varies widely among different organisms (Losada and Hirano 2001). Progressive shortening and thickening of metaphase chromosomes during development have also been reported in Drosophila (Belmont et al. 1987) and Xenopus laevis (Micheli et al. 1993), implicating a link between chromosome shapes and developmentally regulated processes such as transcription and replication programs. While early cytologists and modern cell biologists have been describing the dynamic behavior of metaphase chromosomes for more than a century, it remains largely unknown how they might be shaped at a mechanistic level.
A vital clue to the problem of chromosome shaping came from the discovery of a class of protein complexes, known as condensins, required for chromosome condensation (Hirano and Mitchison 1994; Hirano et al. 1997). Most eukaryotes possess two different condensin complexes, condensin I and II, that contain the same pair of core subunits and unique sets of regulatory subunits (Hirano 2005; Hudson et al. 2009). In somatic tissue culture cells, condensin I and II associate with chromosomes in a sequential manner, become enriched on chromatid axes, and have distinct contributions to metaphase chromosome architecture (Ono et al. 2003, 2004; Hirota et al. 2004; Gerlich et al. 2006a). Cohesin is another protein complex that plays a pivotal role in sister chromatid cohesion (Onn et al. 2008; Peters et al. 2008; Nasmyth and Haering 2009). Most, if not all, cohesin dissociates from chromosomes in mitotic prophase, leaving its small fraction at centromeres that contributes to centromeric cohesion until anaphase onset (Losada et al. 1998; Waizenegger et al. 2000). Although several lines of recent evidence implicate a rather cryptic role of residual cohesin along arms in metaphase (Gimenez-Abian et al. 2004; Nakajima et al. 2007; Shintomi and Hirano 2009), its precise function remains to be established.
To directly address the question of how chromosomal proteins might contribute to determining chromosome shapes, one must employ an experimental system that meets the following two requirements: (1) Cell cycle progression can be controlled precisely and as desired, and (2) the level or activity of target proteins can be manipulated quantitatively with high precision. Among numerous experimental systems currently available, perhaps the sole system satisfying these rigid requirements is a cell-free system derived from Xenopus eggs (Lohka and Masui 1983; Murray 1991). In the current study, we took advantage of this system to establish a series of sophisticated protocols, and addressed the question of how an intricate balance among condensin I, II, and cohesin might determine the shape of metaphase chromosomes.
Results and Discussion
Condensin I and II differently contribute to the assembly of duplicated chromosomes in Xenopus egg extracts
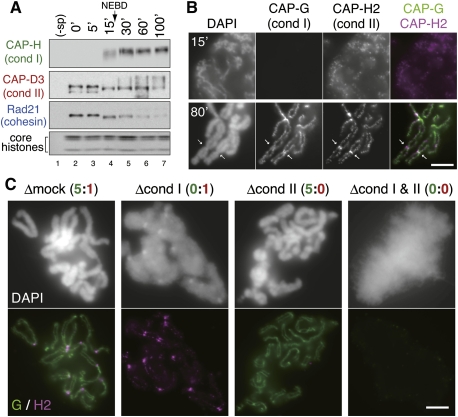
To dissect spatiotemporal regulation of condensins and cohesin, we exploited a cell-free system derived from Xenopus eggs that recapitulate a series of cell cycle-regulated chromosomal events. It should be noted that this study is the first to describe the behaviors of condensin I and II in the cell-free system using duplicated DNA substrates (hereafter referred to as the duplicated chromosome assembly assay). In this assay, sperm chromatin was first duplicated by incubating with an interphase extract for 100 min, and converted into metaphase chromosomes by adding a nondegradable form of Xenopus cyclin B into the extract. At various time points after cyclin B addition, chromatin was isolated and analyzed by immunoblotting and immunofluorescence with antibodies for condensin I- and II-specific subunits, and cohesin subunits (see Supplemental Fig. S1A for the subunit composition of these complexes). Association of condensin I with chromosomes became prominent after nuclear envelope breakdown (NEBD), which occurred ∼20 min after cyclin addition (Fig. 1A; Supplemental Fig. S1B). On the other hand, condensin II already bound to chromatin during interphase (at 0 min), and stayed on chromosomes after mitotic entry when additional modifications were apparently introduced, as judged by its electrophoretic mobility shift (Fig. 1A). Concomitantly with bulk dissociation of cohesin from chromatin, duplicated chromatids gradually became visible and culminated in the assembly of metaphase chromosomes (at 80 min) in which two rod-shaped sister chromatids were juxtaposed to each other. While both condensin I and II formed axis-like structures within each chromatid, the signals of condensin I were more continuous and robust than those of condensin II (Fig. 1B). A subfraction of condensin II was also detectable as discrete signals at the centromere/kinetochore regions (Fig. 1B, arrows). Thus, this in vitro assembly assay can faithfully recapitulate the unique behaviors of the two condensin complexes observed in vivo (Maeshima and Laemmli 2003; Ono et al. 2003, 2004; Hirota et al. 2004; Kireeva et al. 2004; Gerlich et al. 2006a). It is most likely that condensin II continuously contributes to chromosome assembly from prophase through metaphase, whereas condensin I participates in this process after NEBD.
Figure 1.
Differential contributions of condensin I and II to duplicated chromosome assembly in Xenopus egg extracts. (A) Sperm chromatin was preincubated in an interphase extract for 100 min, and cyclin B was then added at time 0 to convert the cell cycle state of the extract to mitosis. Chromatin was isolated at various time points and analyzed by immunoblotting with the antibodies indicated. The lower part of the gel was stained with Coomassie brilliant blue to determine the level of core histones. A mock sample without sperm chromatin was included as a negative control (−sp). (B) Prophase nuclei (15 min) and metaphase chromosomes (80 min) were fixed, isolated, and subjected to immunofluorescence labeling with anti-CAP-G (condensin I) and anti-CAP-H2 (condensin II). DNA was counterstained with DAPI. The arrows indicate the centromere/kinetochore regions. Bar, 5 μm. (C) Sperm chromatin was duplicated in an interphase extract depleted of condensin I (Δcond I), condensin II (Δcond II), or both of them (Δcond I & II). Cyclin B was added to trigger entry into mitosis, and the mixtures were incubated for another 80 min. Then chromatin was isolated, fixed, and labeled as described in A. Chromosomes assembled in an extract that had been treated with nonimmune rabbit IgG (Δmock) were also analyzed as a control. Bar, 5 μm.
To evaluate how condensin I and II might contribute to the assembly of metaphase chromosomes in this assay, we performed immunodepletion experiments (Fig. 1C; see also Supplemental Fig. S1C). When both condensin I and II were depleted from an extract, no individual chromosomes were discernible and only fuzzy masses of chromatin were observed (Δcond I and II). An extract depleted of condensin I alone produced fuzzy, yet slightly ordered, masses of chromatin, in which condensin II were concentrated on axial structures as well as at the centromere/kinetochore regions (Δcond I). In striking contrast, individual chromosomes with a reasonably good morphology were assembled in a condensin II-depleted extract (Δcond II). Thus, unlike in HeLa cells (Ono et al. 2003), the functional contribution of condensin II was much smaller than that of condensin I in this cell-free system. Our previous study had estimated the relative ratio of condensin I to II to be ∼5:1 in Xenopus egg extracts and ∼1:1 in nuclear extracts from HeLa cells (Ono et al. 2003), providing a reasonable explanation for the differential contribution of condensin I and II to chromosome condensation between Xenopus egg extracts and HeLa cells.
The relative ratio of condensin I to II is a critical determinant of chromosome shapes
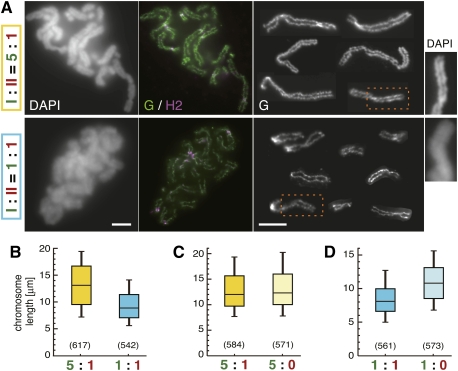
It is known that metaphase chromosomes in early embryonic cells tend to be longer and thinner compared with those in somatic cells (Belmont et al. 1987; Micheli et al. 1993), and the chromosomes assembled in Xenopus egg extracts recapitulate the embryonic properties (Fig. 1B,C). One attractive possibility would be that the relative ratio of condensin I to II contributes to such structural differentiation between embryonic and somatic chromosomes. We therefore sought to test what would happen if the ratio of condensin I to II in the extracts (5:1) were converted to 1:1. It should be emphasized that such precise manipulation of protein levels is virtually impossible in any other experimental systems. To achieve this in our cell-free assay, one volume of a mock-depleted extract was mixed with four volumes of a condensin I-depleted extract to reconstitute an extract containing the equal level of condensin I and II (Supplemental Fig. S2A,B). Remarkably, chromosomes assembled in the 1:1 extract indeed displayed a characteristic appearance reminiscent of somatic chromosomes, being shorter than chromosomes assembled in the mock-depleted extract (5:1) (Fig. 2A). The chromosomes assembled in the 5:1 and 1:1 extracts had lengths of 13.3 ± 4.9 μm (average ± standard deviation) and 9.3 ± 3.3 μm, respectively (Fig. 2B). The width of the chromosomes was apparently greater in the 1:1 extract than in the 5:1 extract (Fig. 2A, right panels). One could argue, however, that the 1:1 extract produced the short and thick chromosomes simply because the absolute level of condensin I present in the extract was reduced (not because the relative ratio of condensin I to II was altered). To exclude this possibility, we prepared another extract in which the absolute levels of both condensin I and II were reduced to 20% of the original level while their relative ratio was kept at 5:1 (referred to as a 5:1* extract). Chromosomes assembled in the 5:1* extract were similar to those assembled in the original 5:1 extract (Supplemental Fig. S2A–C). Immunoblotting analyses of chromosome-bound fractions showed that indistinguishable amounts of condensin I and II were targeted to chromosomes in the 5:1* and 5:1 extracts, while a reduced amount of condensin I was loaded onto chromosomes in the 1:1 extract (Supplemental Fig. S2D). Thus, the relative ratio of the two condensin complexes targeted to chromosomes is mirrored by their ratio present in the manipulated extracts. Taken all together, our results clearly demonstrate that the relative ratio of condensin I to II is one of the critical determinants in shaping metaphase chromosomes.
Figure 2.
The relative ratio of condensin I to II determines the shape of duplicated chromosomes in Xenopus egg extracts. (A) Duplicated chromosomes were assembled in a control extract (I:II = 5:1) or a reconstituted extract (I:II = 1:1) (for details, see Supplemental Fig. S2A,B), and processed for immunofluorescence. The third panels on each row are galleries of CAP-G-labeled images of individualized chromosomes. Magnified DAPI images of the selected regions of chromosomes (indicated by the broken boxes) are shown at the right. Bars, 5 μm. (B) The lengths of chromosomes assembled in A are shown in the box plot. The middle line of each box is the median. The top and bottom lines are the third and first quartiles, and the whiskers indicate the 90th and 10th percentiles. The numbers of chromosomes measured are shown in parentheses (P = 1.2 × 10−51, t-test). (C,D) The lengths of chromosomes assembled in different extracts are compared pairwise in the box plots. A statistically significant difference was seen only between the right pair (P = 0.22, left; P = 2.2 × 10−31, right). The distribution of chromosome lengths in each condition examined in B–D is also shown in histograms (Supplemental Fig. S6).
We then tested whether depletion of condensin II from extracts containing different levels of condensin I might affect the length of chromosomes assembled (Supplemental Fig. S3). As expected from the images shown in Figure 1C, little if any difference in the chromosome length was detectable when condensin II was depleted (the 5:0 extract, 13.1 ± 4.6 μm) from the standard extract (the 5:1 extract, 13.3 ± 4.7 μm) (Fig. 2C). In contrast, when condensin II was depleted from the 1:1 extract in which short chromosomes were assembled (8.6 ± 2.9 μm), the resulting 1:0 extract produced chromosomes whose length became closer to that of control chromosomes (11.0 ± 3.4 μm) (Fig. 2D). These observations suggest that condensin II primarily contributes to axial shortening of chromosomes, and that this action of condensin II is rather cryptic when the action of condensin I is predominant under the standard condition.
Functional interplay between cohesin and condensins in chromosome shaping
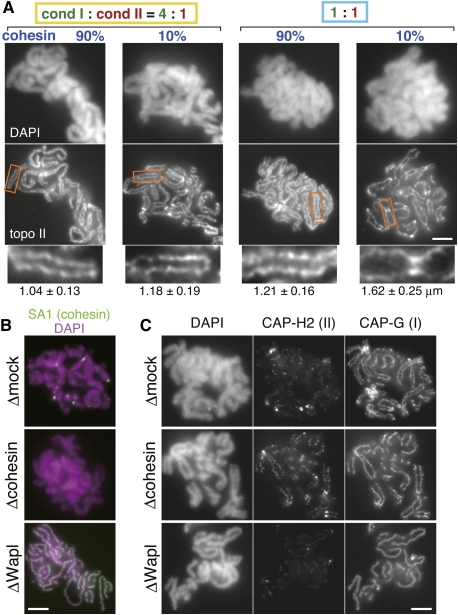
Despite some descriptive observations implicating cohesin in metaphase chromosome structures (Gimenez-Abian et al. 2004; Nakajima et al. 2007; Shintomi and Hirano 2009), no systematic approach had been taken to address this problem. In particular, we wished to test how cohesin's action might potentially be coordinated with the action of condensins. To this end, we established a protocol in which the levels of cohesin and condensin I were partially reduced individually or in combinations (Supplemental Fig. S4A). To clearly visualize the chromosome axes regardless of the level of condensins targeted, an antibody against topoisomerase II was used for immunofluorescence analysis in this experiment (Fig. 3A). We found that, under the condition where condensin I existed dominantly over condensin II (4:1), reducing the cohesin level from 90% to 10% increased the distance between sister chromatids only modestly (from 1.04 ± 0.13 μm to 1.18 ± 0.19 μm). A similarly modest increase in the distance between sisters was also observed when the relative ratio of condensin I to II was forced to be 1:1 with a near-original level of cohesin (1.21 ± 0.16 μm). In striking contrast, when the cohesin level was reduced to 10% in an extract with the 1:1 ratio of condensin I to II, the distance between chromatid arms (but not between sister centromeres) was drastically increased (1.62 ± 0.25 μm), resulting in the formation of highly characteristic, X-shaped chromosomes. Admittedly speculative, we interpret these observations as follows. Parallel juxtaposition of sister chromatid arms within a metaphase chromosome is sustained by two mechanistically distinct factors: One is cohesin-mediated cohesion between sister chromatid arms, and the other is condensin I-dependent structural rigidity of each chromatid (e.g., Almagro et al. 2004). Our current results clearly demonstrate that reducing either one of the two factors to 10%–20% affects the juxtaposition of sister arms only modestly, whereas reducing both of them has a synergistic effect, leading to hyperdissociation of sister chromatid arms. It should be noted that similar X-shaped chromosomes are often observed in somatic cells arrested at mitosis for a long time, during which cohesin is lost progressively from chromosome arms (Gimenez-Abian et al. 2004; Nakajima et al. 2007). Although previous studies in budding yeast implicated possible contribution of cohesin to “condensation” (Guacci et al. 1997; Lavoie et al. 2002) and of condensin to “cohesion” (Lam et al. 2006), the inability to visualize individual chromosomes in yeast made it difficult to provide lucid explanations for the functional cross-talk between the two processes. Our current study helps clarify this tangled issue, demonstrating that cohesin and condensin I collaborate to shape and maintain metaphase chromosomes, yet through distinct mechanisms.
Figure 3.
Cohesin collaborates with condensins to shape chromosomes. (A) Chromosomes were assembled in extracts containing various levels of cohesin and condensin I (for details, see Supplemental Fig. S4A), and subjected to immunofluorescence analysis with anti-topoisomerase II (topo II). Blowup images of selected regions (indicated by the rectangles) are shown in the bottom row. Bar, 5 μm. The average distance between sister chromatids, together with standard deviation, is provided below each image (n = 22, 28, 21, 20, from left to right). (B) Chromosomes were assembled in a control extract (Δmock), an extract depleted of cohesin (Δcohesin), or Wapl (ΔWapl). To visualize cohesin left on these chromosomes, immunofluorescence was performed with an antibody against its SA1 subunit. Bar, 5 μm. (C) Chromosomes were assembled as described in B, and labeled with anti-CAP-G and anti-CAP-H2 antibodies. Bar, 5 μm.
We next investigated whether more drastic manipulation of cohesin levels left on metaphase chromosomes might affect the behavior of condensins and, if so, how. To this end, we prepared duplicated chromosomes with no cohesin in a cohesin-depleted extract and those with an increased level of cohesin in a Wapl-depleted extract (Fig. 3B; Losada et al. 1998; Shintomi and Hirano 2009). It was noticed that the axial distribution of condensin II became much more prominent in cohesin-free chromosomes (Δcohesin) compared with control ones (Δmock). Conversely, condensin II was hardly detectable in poorly resolved chromosomes with an unusually high level of cohesin (ΔWapl) (Fig. 3C). On the other hand, the signal intensity of condensin I was indistinguishable among the three conditions examined, confirming our previous data that condensin I loading occurs independently of cohesin release in this cell-free system (Losada et al. 1998, 2002). The current results show instead that accumulation of condensin II on chromosome axes is counterbalanced by the amount of cohesin left on metaphase chromosomes (Supplemental Fig. S4B). Notably, cohesin is released from chromatin exactly when condensin II initiates chromosome condensation within the prophase nucleus in somatic cells (Waizenegger et al. 2000; Hirota et al. 2004; Ono et al. 2004; Gerlich et al. 2006a,b). Thus, the balancing actions between cohesin and condensin II observed here would represent a “tug-of-war” between forces holding and resolving sister chromatids.
Additional evidence that balancing actions of condensin I and II are important for chromosome assembly
To gain additional insight into functional interactions between condensin I and II, we set up a different type of assay that induces chromosome assembly from an unduplicated substrate (the single-chromatid assembly assay). In this experimental setup, sperm chromatin was directly incubated with metaphase-arrested egg extracts to produce single chromatids, independently of DNA replication or cohesion. This simple assay for chromosome assembly, which magnifies its individualization process (Swedlow and Hirano 2003), had been used effectively for the original identification of condensin I (Hirano and Mitchison 1994; Hirano et al. 1997). In this assay, both condensin I and II started to associate with chromatin in a very short time window upon incubation (i.e., within 5 min), as judged by immunoblotting analysis (Supplemental Fig. S5A). Consistently, immunofluorescence analysis showed that condensin I and II localized to the axis of each chromatid with distinct patterns at both early and late time points. After an 80-min incubation, condensed chromatids became individualized so that they were readily distinguishable from each other (Supplemental Fig. S5B).
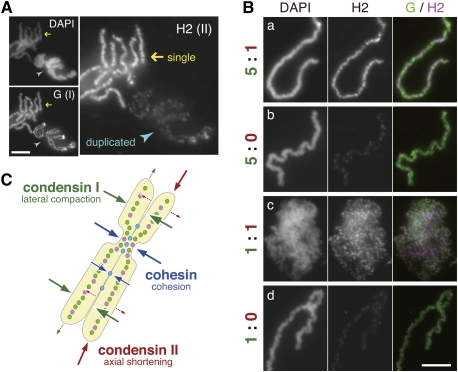
During the course of this study, we noticed unexpectedly that a far larger amount of condensin II was loaded on the single chromatids than the duplicated chromosomes, whereas the signals of condensin I on the two types of chromosomes were comparable (Fig. 4A). Although the absence of cohesion alone would not fully explain the apparent hyperloading of condensin II observed in the single-chromatid assembly assay (and the reason is currently unknown), we realized that this property might give us an opportunity to address balancing actions of condensin I and II in a different context. With this background information in our hand, the same ratio manipulation was applied to the single-chromatid assembly assay. To our surprise, we found that no discrete chromatid structures were assembled in the 1:1 extract; instead, masses of entangled chromatin fibers were observed, although both condensins bound to them (Fig. 4B, row c). This characteristic morphology was distinct from that produced in extract depleted of either condensin I, II, or both (Fig. 4B, row b; Supplemental Fig. S5C; Ono et al. 2003). Interestingly, however, when condensin II was further depleted from the 1:1 extract to produce a 1:0 extract, the defective phenotype was partially rescued and individual chromatids readily became discernible (Fig. 4B, row d). Our interpretation of these observations is as follows: Due to the inherent nature associated with the single-chromatid assembly assay (Fig. 4A), the functional balance between the two condensins in the 1:1 extract was shifted heavily toward condensin II. As a consequence, the action of condensin I was seemingly compromised, leading to the failure in chromatid individualization under this condition. When condensin II is removed from the extract, the balance was restored toward condensin I, thereby partially rescuing the defective phenotype. Thus, these results suggest that balancing actions of condensin I and II are critically important for the chromosome individualization process, which is best addressed in this single-chromatid assembly assay.
Figure 4.
Balancing actions of two condensins are also important for single-chromatid assembly. (A) Single chromatids (yellow arrows) and duplicated chromosomes (blue arrowheads) were assembled separately, fixed on a single slide, and subjected to immunofluorescence. Bar, 5 μm. (B) Sperm chromatin was incubated with metaphase-arrested extracts containing condensin I and II at the indicated ratios. After an 80-min incubation, chromatids were analyzed by immunofluorescence. Bar, 5 μm. (C) A model of how balancing actions of condensins and cohesin determine the shape of chromosomes is depicted in the cartoon.
Conclusions and perspective
While the unique shape of metaphase chromosomes had been observed and described by cytogeneticists and cell biologists for more than a century, addressing the molecular basis of chromosome shaping became possible only recently after the discovery of condensins (Hirano and Mitchison 1994; Hirano et al. 1997; Ono et al. 2003) and cohesin (Guacci et al. 1997; Michaelis et al. 1997; Losada et al. 1998). The current study represents the first systematic attempt to address this question by precisely manipulating the level of these essential components for chromosome morphogenesis. From a mechanistic point of view, the assembly of a metaphase chromosome would involve at least three different processes: axial shortening, lateral compaction of each chromatid, and cohesion between sister chromatids (Losada and Hirano 2001; Hirano 2005). Given the fixed volume of a chromatin fiber whose density within a chromosome has been shown to be constant from prometaphase through metaphase (Mora-Bermudez et al. 2007), axial shortening would create shorter and thicker chromatids, whereas lateral compaction would make chromatids longer and thinner. In fact, it has been demonstrated that chromosomes progressively get shorter and thicker during animal development (Belmont et al. 1987; Micheli et al. 1993). On the basis of the ratio manipulation experiments reported here, we propose that the primary actions of condensin I and II are aimed at lateral compaction and axial shortening, respectively, and that their intricate balance acts as one of the critical determinants in shaping chromatids (Fig. 4C). While this is clearly an oversimplified model, it is consistent with the recent finding that caspase-mediated cleavage of a condensin I subunit results in the formation of short and thick chromosomes in cells arrested at metaphase for a long time (Lai et al. 2011). On the other hand, our results suggest that cohesin contributes to chromosome shaping in two ways. First, release of cohesin itself from chromosome arms facilitates condensin II-mediated axis formation. Second, residual cohesin holds sister chromatid arms together, whose juxtaposition could further be reinforced by condensin I-mediated chromatid rigidity. In the future, it will be of great interest to combine the sophisticated cell-free protocols reported here with other assays such as micromanipulation of chromosomes (Marko 2008). Furthermore, it will be important to examine exactly how expression of condensins and cohesin might be under the control of developmental cues, and how their intricate balance might be established and fine-tuned among different organisms. Such considerations would undoubtedly help broaden our horizon, linking chromosome biology to other neighboring areas such as developmental biology and evolutionary biology.
Materials and methods
Duplicated chromosome and single-chromatid assembly assays
Interphase low-speed supernatants and cytostatic factor-arrested high-speed supernatants of Xenopus eggs were prepared as described previously (Hirano et al. 1997; Shintomi and Hirano 2009). Throughout this study, we refer to them as interphase egg extracts and metaphase-arrested egg extracts, respectively. To prepare duplicated chromosomes, sperm chromatin was mixed with interphase egg extracts at a final concentration of 1000 per microliter. After a 100-min incubation at 22°C, the mixture was supplemented with a nondegradable cyclin B (60 nM) to induce mitosis, and incubated for another 80 min. To assemble single chromatids, sperm chromatin was directly incubated with metaphase-arrested egg extracts. Immunodepletion was performed as described previously (Shintomi and Hirano 2009). Detailed protocols for depletion and manipulation are described in the Supplemental Material.
Immunofluorescence
Immunofluorescence was performed as described previously (Shintomi and Hirano 2009) with the following modifications. For double labeling with rabbit antibodies against condensin I- and II-specific subunits, chromosomes recovered on coverslips were first incubated with 2 μg/mL unlabeled anti-CAP-H2, followed by Alexa Fluor 594-labeled anti-rabbit IgG secondary antibody (Invitrogen). The coverslips were soaked in TBS containing 1 mg/mL rabbit IgG to saturate the open IgG-binding site, and then incubated with anti-CAP-G (1 μg/mL) that had been covalently labeled with Alexa Fluor 488 (by using a kit from Invitrogen). After counterstaining with DAPI, the coverslips were mounted on slides with VectaShield mounting medium (Vector Laboratories) and examined with an epifluorescent microscope (BX51, Olympus).
Quantification of chromosome shapes
For quantification of chromosome lengths, chromosomes were randomly selected and the lengths of lines manually traced along CAP-G-positive structures were quantified with the measure function of the ImageJ software (http://rsb.info.nih.gov/ij). An unpaired two-tailed t-test was used for statistical analysis. For quantification of chromosome arm distances, lines were drawn perpendicularly to the path of sister chromatids at a couple of points on each chromosome (with avoiding centromeric regions), and the distance between topo II-peak positions on sister chromatids was measured.
Acknowledgments
We thank J. Gannon and T. Hunt (Cancer Research UK) for Xenopus cyclin cDNAs. We are grateful to members of the Hirano laboratory for critically reading the manuscript. This work was supported by Grant-in-Aid for Specially Promoted Research (20002010 to T.H.). K.S. was a RIKEN Special Post-doctoral Researcher and was supported by Grant-in-Aid for Young Scientists B.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2060311.
References
- Almagro S, Riveline D, Hirano T, Houchmandzadeh B, Dimitrov S 2004. The mitotic chromosome is an assembly of rigid elastic axes organized by structural maintenance of chromosomes (SMC) proteins and surrounded by a soft chromatin envelope. J Biol Chem 279: 5118–5126 [DOI] [PubMed] [Google Scholar]
- Belmont AS, Sedat JW, Agard DA 1987. A three-dimensional approach to mitotic chromosome structure: evidence for a complex hierarchical organization. J Cell Biol 105: 77–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J 2006a. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol 16: 333–344 [DOI] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J 2006b. Live-cell imaging reveals a stable cohesin–chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578 [DOI] [PubMed] [Google Scholar]
- Gimenez-Abian JF, Sumara I, Hirota T, Hauf S, Gerlich D, de la Torre C, Ellenberg J, Peters JM 2004. Regulation of sister chromatid cohesion between chromosome arms. Curr Biol 14: 1187–1193 [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T 2005. Condensins: organizing and segregating the genome. Curr Biol 15: R265–R275 doi: 10.1016/j.cub.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ 1994. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79: 449–458 [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89: 511–521 [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM 2004. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117: 6435–6445 [DOI] [PubMed] [Google Scholar]
- Hudson DF, Marshall KM, Earnshaw WC 2009. Condensin: architect of mitotic chromosomes. Chromosome Res 17: 131–144 [DOI] [PubMed] [Google Scholar]
- Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS 2004. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J Cell Biol 166: 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SK, Wong CH, Lee YP, Li HY 2011. Caspase-3-mediated degradation of condensin Cap-H regulates mitotic cell death. Cell Death Differ 18: 996–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam WW, Peterson EA, Yeung M, Lavoie BD 2006. Condensin is required for chromosome arm cohesion during mitosis. Genes Dev 20: 2973–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie BD, Hogan E, Koshland D 2002. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol 156: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y 1983. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science 220: 719–721 [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano T 2001. Shaping the metaphase chromosome: coordination of cohesion and condensation. Bioessays 23: 924–935 [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T 2002. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev 16: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Laemmli UK 2003. A two-step scaffolding model for mitotic chromosome assembly. Dev Cell 4: 467–480 [DOI] [PubMed] [Google Scholar]
- Marko JF 2008. Micromechanical studies of mitotic chromosomes. Chromosome Res 16: 469–497 [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Micheli G, Luzzatto AR, Carri MT, de Capoa A, Pelliccia F 1993. Chromosome length and DNA loop size during early embryonic development of Xenopus laevis. Chromosoma 102: 478–483 [DOI] [PubMed] [Google Scholar]
- Mora-Bermudez F, Gerlich D, Ellenberg J 2007. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat Cell Biol 9: 822–831 [DOI] [PubMed] [Google Scholar]
- Murray AW 1991. Cell cycle extracts. Methods Cell Biol 36: 581–605 [PubMed] [Google Scholar]
- Nakajima M, Kumada K, Hatakeyama K, Noda T, Peters JM, Hirota T 2007. The complete removal of cohesin from chromosome arms depends on separase. J Cell Sci 120: 4188–4196 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH 2009. Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE 2008. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 24: 105–129 [DOI] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115: 109–121 [DOI] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector DL, Hirano T 2004. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell 15: 3296–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J 2008. The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Shintomi K, Hirano T 2009. Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev 23: 2224–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow JR, Hirano T 2003. The making of the mitotic chromosome: modern insights into classical questions. Mol Cell 11: 557–569 [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103: 399–410 [DOI] [PubMed] [Google Scholar]