Abstract
Small cell lung cancer (SCLC) is an aggressive cancer often diagnosed after it has metastasized. Despite the need to better understand this disease, SCLC remains poorly characterized at the molecular and genomic levels. Using a genetically engineered mouse model of SCLC driven by conditional deletion of Trp53 and Rb1 in the lung, we identified several frequent, high-magnitude focal DNA copy number alterations in SCLC. We uncovered amplification of a novel, oncogenic transcription factor, Nuclear factor I/B (Nfib), in the mouse SCLC model and in human SCLC. Functional studies indicate that NFIB regulates cell viability and proliferation during transformation.
Keywords: small cell lung cancer, mouse model, Nuclear factor I/B
Small cell lung cancer (SCLC) is a highly lethal form of cancer that comprises ∼20% of all lung cancer cases (Wistuba et al. 2001; Meuwissen and Berns 2005). Unfortunately, SCLC is frequently diagnosed only after metastatic spread of the disease, and at present only 5% of patients survive beyond 5 years after diagnosis (Worden and Kalemkerian 2000; Cooper and Spiro 2006). Some insight has been gained as to the underlying mechanisms of this aggressive disease, including the identification of loss-of-function mutations in the tumor suppressor genes Trp53 (Yokota et al. 1987; Takahashi et al. 1989, 1991) and RB1 (Harbour et al. 1988; Yokota et al. 1988), which are observed in 75% and 90% of SCLC cases, respectively (Wistuba et al. 2001; Meuwissen and Berns 2005). In addition, MYC family members (C-MYC, L-MYC, and N-MYC) are frequently amplified in SCLC (Nau et al. 1985; Meuwissen and Berns 2005). However, very little is known about other functionally relevant alterations in SCLC, and a more complete understanding of the disease is required to allow the development of new targeted treatments.
Whole-genome profiling has been used to gain information about copy number alterations, point mutations, and translocations in tumors (Campbell et al. 2008; Ley et al. 2008; Mardis et al. 2009). One recent examination of 33 primary SCLC tumors and 13 SCLC cell lines identified MYC family amplifications in 82% of tumors and 62% of cell lines (Voortman et al. 2010). Another study identified 22,000 point mutations in a SCLC cell line, the majority of which were G–T transversions, a hallmark of carcinogens present in tobacco smoke (Toyooka et al. 2003; Lewis and Parry 2004; Pleasance et al. 2010). In other cancer types, comparative studies using mouse models have aided in narrowing lists of candidate genes (Kim et al. 2006; Zender et al. 2006, 2008). Thus, we analyzed the genomic alterations that occur during tumor progression in a mouse model of SCLC to identify oncogenes in this cancer type.
Results and Discussion
Genetically engineered mouse model of metastatic SCLC
Berns and colleagues (Jonkers et al. 2001; Vooijs et al. 2002; Meuwissen et al. 2003; Sage et al. 2003) have developed a mouse model of SCLC (mSCLC) that involves the inactivation of the Trp53 and Rb1 tumor suppressor genes using conditional (“floxed”) alleles in p53fl/fl;Rbfl/fl mice (Supplemental Fig. S1). Inhalation of adenovirus containing Cre recombinase results in infection of lung epithelial cells that develop into tumors resembling human SCLC histopathologically (Supplemental Fig. S1; Meuwissen et al. 2003; DuPage et al. 2009). These mice have a median survival time of 350 d, during which the tumors become malignant and metastatic (Supplemental Fig. S1; Meuwissen et al. 2003). Similar to human SCLC, the mSCLC metastasized to the thoracic lymph nodes, liver, adrenal glands, and bone (Supplemental Fig. S1; Meuwissen et al. 2003). Thus, this model provided a platform with which to identify genetic alterations that occur during tumor progression.
Identification of Nuclear factor I/B (Nfib) amplifications
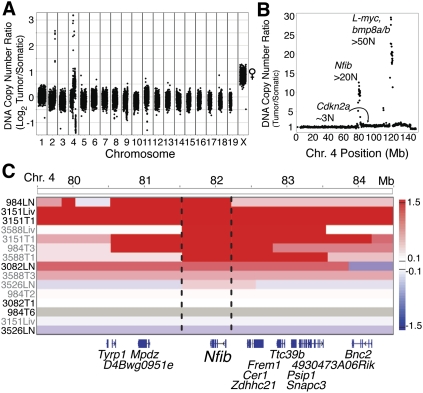
To determine the genetic alterations that occur in mSCLC, primary tumors and metastases were dissected and used for histology, DNA and RNA isolation, and the derivation of cell lines (Supplemental Fig. S1). Each tumor was verified histopathologically to be SCLC, and tumor purity was assessed by PCR for the recombined Trp53 and Rb1 alleles (Supplemental Fig. S1; data not shown). The analysis of DNA copy number alterations in murine tumor models has previously aided in the identification of functionally important genes in several human cancers (Kim et al. 2006; Zender et al. 2006, 2008). Thus, we analyzed mSCLC tumors and metastases using next-generation sequencing-based DNA copy number analysis (Chiang et al. 2009). These data show that while the majority of the genome was surprisingly unaltered, several high-level focal amplifications and deletions were observed in tumor specimens (Fig. 1A; Supplemental Figs. S2–S4; Supplemental Table 1). In particular, we identified two recurrent focal amplifications centered around 82 Mb and 122 Mb on mouse chromosome 4 and a heterozygous deletion spanning from ∼148.5 Mb to the end of chromosome 4 (Fig. 1B). Although one of the focal amplifications on chromosome 4 contained a known proto-oncogene involved in SCLC, L-myc (Mycl1) (Nau et al. 1985), the other focal amplification contained no genes previously implicated in this disease (Fig. 1C; Supplemental Fig. S3). To identify the relevant targets within the amplified region, the amplification breakpoints were mapped using statistical change point analysis of the normalized copy number ratios (Chiang et al. 2009). Nuclear factor I/B (Nfib) was the only gene within this region amplified in each of the samples (Fig. 1C). Furthermore, Nfib is located at the apex of the amplified peak in tumors and tumor-derived cell lines (Supplemental Fig. S2). Thus, Nfib represents a newly identified amplified gene in SCLC.
Figure 1.
Nfib is amplified in mSCLC tumors. (A) Log2 ratio of tumor to somatic DNA copy number across the whole genome of a mSCLC lymph node metastasis cell line. The X chromosome has a copy number ratio of 2 due to the male reference genome, while the sample was derived from a female mouse. (B) DNA copy number ratio of chromosome 4 of the same sample as in A. Interestingly, the region between the two focal amplifications is near diploid and contains the tumor suppressor gene Cdkn2a. Despite the well-known role of Cdkn2a in regulating p53 and Rb, which are already deleted in tumors, the fact that the copy number of this gene is kept low is consistent with this locus regulating other Rb family members (Schaffer et al. 2010). Copy number data are plotted as the tumor to somatic copy number ratio. (C) Integrated genome viewer (IGV) plot of the DNA copy number of position 79.5–84.5 Mb on chromosome 4. The bar indicates the log2 copy number ratio of tumor to somatic reference sample. The dotted line indicates the boundaries of the minimally conserved region. (T) Primary lung tumor; (Liv) liver metastasis; (LN) lymph node metastasis; (gray labels) tumor samples; (black labels) cell lines.
Nfib is a CCAAT-box-binding transcription factor that regulates the expression of lung differentiation genes (Santoro et al. 1988; Steele-Perkins et al. 2005). Nfib knockout mice have lung hypoproliferation and differentiation defects, in addition to brain defects, and die shortly after birth (Gründer et al. 2002). The chromosomal region containing Nfib has been reported to be frequently amplified in a mouse model of prostate cancer (Zhou et al. 2006) and in patients with triple-negative breast cancer (Han et al. 2008). Based on the identification of Nfib as an amplified gene in SCLC and its potential importance in other prevalent tumor types, we chose to examine Nfib further.
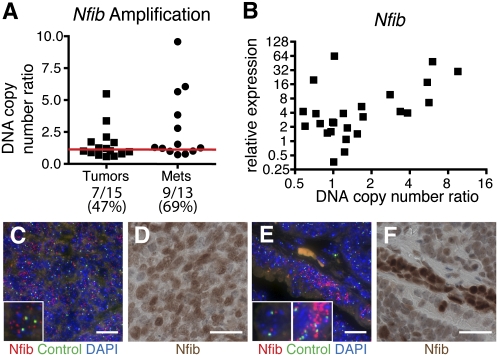
The DNA copy number of Nfib and the expression of Nfib mRNA was determined using real-time PCR in a panel of 28 mSCLC-derived cell lines (Fig. 2A,B). Out of 28 cell lines, 16 had Nfib and six had L-myc amplifications (Fig. 2A; Supplemental Fig. S5). Notably, four mSCLC cell lines had amplified both Nfib and L-myc (Supplemental Fig. S5). mSCLC cell lines with increased Nfib copy number also expressed high levels of Nfib (Fig. 2B). Interestingly, two cell lines with normal Nfib copy number expressed high levels of Nfib mRNA, suggesting that mechanisms other than genomic alteration may increase Nfib levels in SCLC (Fig. 2B).
Figure 2.
Nfib is expressed in mSCLC. (A) Nfib is amplified in tumor- and metastasis-derived cell lines. A DNA copy number ratio >1.2 was considered to be amplified, as determined by real-time PCR on genomic DNA. (B) Cell lines with increased copy number of the Nfib locus also have increased expression of Nfib. (C) FISH analysis on a lymph node metastasis confirming known amplification of Nfib from copy number analysis. The inset shows a cell with amplified Nfib. (D) Matching Nfib immunohistochemistry (IHC) on the same lymph node metastasis as in C, which has high Nfib expression. (E) FISH analysis of a lung tumor illustrating amplification of Nfib in a subset of tumor cells. The left inset illustrates a cell with normal Nfib copy number, and the right inset is a cell with amplified Nfib. (F) Matching Nfib IHC on the same lung tumor as in E, demonstrating increased Nfib expression in the region with Nfib amplification. Bar: C–F, 20 μm. (C,E) (Red) Nfib probe; (green) control chromosome 4qA1 probe; (blue) DAPI.
To confirm Nfib amplification in mouse tumors, we performed fluorescence in situ hybridization (FISH). FISH confirmed the amplification of Nfib in the lymph node metastasis analyzed in Figure 1 (Fig. 2C,D). Interestingly, in a primary tumor, we found that Nfib amplification clearly correlated with a region of increased Nfib expression (Fig. 2E,F). In a normal lung, Nfib protein was localized appropriately to the nucleus of alveolar type II cells (Steele-Perkins et al. 2005) and was also nuclear in mSCLC. Additionally, Nfib protein was detected in a subset of lung neuroendocrine cells (Supplemental Fig. S6). Consistent with data from human tumor samples (Bhattacharjee et al. 2001), Nfib was not detected in lung adenomas that occasionally arise in this mouse model (Supplemental Fig. S7). Furthermore, we observed that both lymph node and liver metastases very frequently expressed high levels of Nfib (Supplemental Fig. S7). These data confirm the amplification and increased expression of Nfib in mSCLC tumors and local and distant metastases.
NFIB amplifications in human SCLC
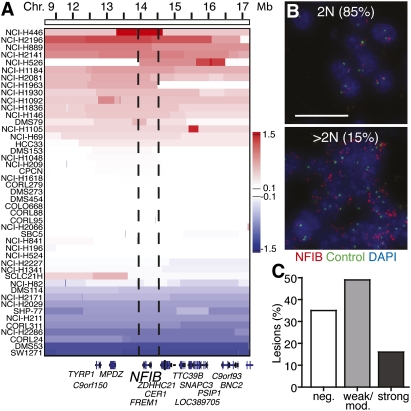
We next examined whether NFIB is amplified and/or expressed in human SCLC. Copy number analysis revealed a broad region of amplification on chromosome 9p23 encompassing 210 genes. GISTIC analysis identified an ∼200-kb minimal region of amplification containing only one gene, NFIB (Beroukhim et al. 2007). In total, 16 of 46 human SCLC cell lines had NFIB copy number gains (Fig. 3A). Interestingly, 11 of the cell lines with NFIB amplification also had L-MYC amplification, and 15 out of the 16 cell lines with amplification of NFIB displayed additional amplification of one of the MYC family members (Supplemental Fig. S5; data not shown). Increased NFIB copy number was confirmed by real-time PCR (Supplemental Fig. S8). Additionally, NFIB amplification was detected by FISH in 15% of primary human tumor samples (Fig. 3B). We next addressed whether NFIB protein was expressed in human SCLC tumor samples by performing immunohistochemistry (IHC) on a tissue microarray containing 68 distinct human SCLC samples. High-level NFIB protein expression was noted in 16% of samples, and the protein was detectable in 65% of tumors (Fig. 3C; Supplemental Fig. S9). Collectively, these data from both the mouse model and human patients suggest a potentially oncogenic role for NFIB in SCLC.
Figure 3.
NFIB is amplified and expressed in human SCLC. (A) IGV plot of 46 human SCLC cell lines from position 11.1 to 17.2 Mb on human chromosome 9. The dotted line indicates the boundaries of the minimally conserved region. The bar indicates the log2 copy number ratio of tumor to somatic reference sample. (B) NFIB was amplified in human tissue samples, as detected by NFIB FISH. The top panel is a representative sample with normal NFIB copy number. The bottom panel is a representative sample with amplified NFIB. (Red) NFIB probe; (green) control chromosome 9 9q12 probe; (blue) DAPI. Bar, 10 μm. (C) Quantification of NFIB expression in human SCLC tissue samples as detected by IHC for NFIB.
NFIB controls apoptosis and viability in human SCLC
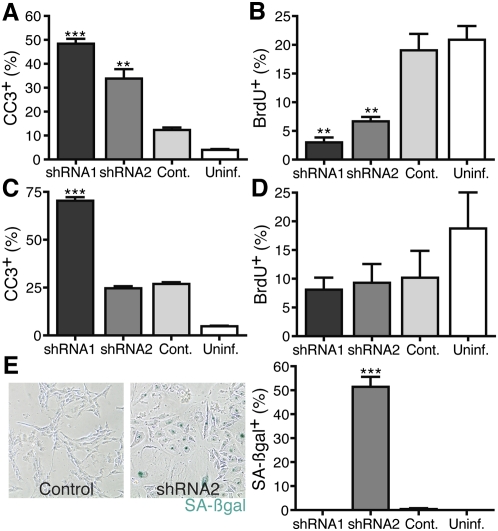
To determine which cellular processes are regulated by NFIB, we used RNAi to inhibit NFIB in several human SCLC cell lines. In one adherent cell line (NCI-H446), which has high-level NFIB amplification and expresses very high levels of NFIB, RNAi-mediated NFIB knockdown caused a dramatic increase in apoptosis and a corresponding decrease in proliferation (Fig. 4A,B; Supplemental Fig. S10). In a second SCLC cell line (NCI-H196), which lacked NFIB amplification, NFIB knockdown led to different outcomes depending on the shRNA used, with one inducing apoptosis and the other cellular senescence (Fig. 4C–E; Supplemental Fig. S10). In one final human SCLC cell line (NCI-H82), NFIB knockdown reduced proliferation (Supplemental Fig. S10). Collectively, these data suggest that NFIB expression is integral to human SCLC cell line viability and/or continued proliferation, likely depending on the NFIB levels or the cellular context of each individual tumor.
Figure 4.
NFIB knockdown induces death and reduces proliferation in human SCLC. (A) NFIB knockdown increases the percentage of cleaved caspase 3 (CC3)-positive cells in the adherent cell line NCI-H446 by FACS compared with control (Cont.) infected cells. (B) Quantification of BrdU incorporation by FACS following NFIB knockdown in the same cell line as in A. (C) NFIB shRNA1 induces apoptosis, as observed in an increase in the percentage of CC3-positive cells in the adherent cell line NCI-H196 by FACS. (D) Reduced BrdU incorporation as detected by FACS following NFIB knockdown in the same cell line as in C. (E) Representative pictures of senescence-associated β-galactosidase staining (SA-βgal) in an adherent cell line, NCI-H196, following control (left) or NFIB (right) knockdown with shRNA2. Quantification of the SA-βgal staining is shown in the right panel. (**) P-value <0.005; (***) P-value <0.0005, compared with control.
Nfib is an oncogene in mSCLC
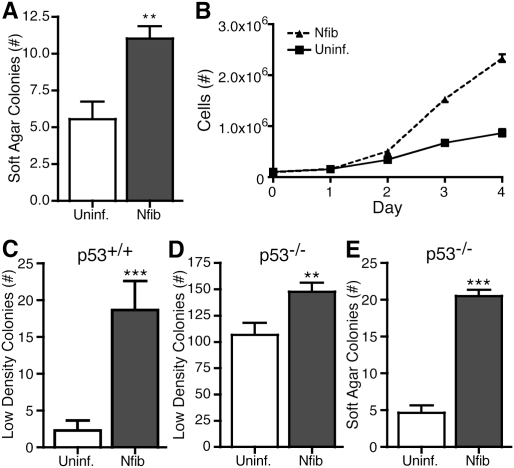
We next examined whether Nfib plays an oncogenic role in the cellular transformation of murine small cell lung tumors. For initial functional studies, we used mSCLC cell lines that expressed low levels of endogenous Nfib, in which we then stably expressed Nfib (Supplemental Fig. S11). Ectopic expression of Nfib in two independent mSCLC cell lines increased the number and size of anchorage-independent colonies compared with uninfected cells (Fig. 5B; Supplemental Fig. S11). Furthermore, cells overexpressing Nfib proliferated more quickly under standard culture conditions (Fig. 5; Supplemental Fig. S11). To assess which pathways were altered by the overexpression of Nfib, we performed gene expression arrays and a gene set enrichment analysis (GSEA). Within curated gene sets, a number of cancer-related gene sets correlated with overexpression of Nfib (Supplemental Fig. S12). These data support our hypothesis that Nfib has oncogenic properties.
Figure 5.
Nfib is a novel oncogene in murine SCLC. (A) Quantification of the number of soft agar colonies in the uninfected or stably expressing Nfib cell line. The values represent the mean ± standard deviation of triplicate plates. (B) Growth curve of a primary tumor cell line, either uninfected or stably expressing Nfib. (C) Wild-type MEFs either uninfected or infected with Nfib-expressing viruses were plated at a low density and assayed for colony formation. Colonies were visualized using crystal violet and quantified. Values represent the mean ± standard error of the mean (SEM) of the total number of colonies in triplicate plates of both experiments using two different MEF preparations. (D) p53−/− MEFs either uninfected or infected with Nfib-expressing viruses were plated at a low density and assayed for colony formation. Colonies were visualized using crystal violet and quantified. Values represent the mean ± SEM of the total number of colonies in triplicate plates of both experiments using two different MEF preparations. (E) p53−/− MEFs either uninfected or infected with Nfib-expressing viruses were plated in soft agar and assayed for anchorage-independent colony formation. Colonies were visualized using crystal violet and quantified. Values represent the mean ± SEM of the number of colonies in nine camera views of triplicate plates in experiments using two different MEF preparations. (**) P-value <0.005; (***) P-value <0.0005.
We also investigated whether Nfib has oncogenic activity in a heterologous setting by testing whether Nfib could transform wild-type or p53−/− mouse embryonic fibroblasts (MEFs) (Fig. 5C–E; Supplemental Fig. S13). Expressing Nfib significantly increased colony formation in a low-density colony formation assay (Fig. 5C,D; Supplemental Fig. S13). Additionally, while neither control nor Nfib-expressing wild-type MEFs could grow under anchorage-independent conditions, expression of Nfib in p53−/− MEFs dramatically enhanced growth of anchorage-independent colonies (Fig. 5E; Supplemental Fig. S13). Finally, when wild-type MEFs were plated at high density to assay for the ability to overcome contact inhibition of growth, significantly more three-dimensional foci formed when MEFs expressed Nfib (Supplemental Fig. S13).
Interestingly, we observed frequent concurrent amplification and expression of both Nfib and L-myc (Fig. 1B; Supplemental Figs. S3, S5). In some cell culture assays, including the formation of three-dimensional foci of wild-type MEFs in high-density conditions, we observed cooperativity between exogenous L-myc and Nfib, which was significant by the Bliss Independence test (Supplemental Fig. S13). Thus, L-myc and Nfib may act synergistically in some settings.
The use of a genetically engineered mouse model of SCLC enabled us to interrogate the genomic alterations that occur in SCLC. We identified several high-level focal amplifications and deletions and, notably, uncovered amplification of a novel potential proto-oncogene, Nfib. This mouse model provides several advantages over studying this cancer type in humans. First, the tumors are initiated by defined genetic events and the disease progresses in the absence of smoking carcinogen-induced passenger mutations and alterations. Second, a wealth of tumor and metastasis samples can be collected and used for DNA, RNA, and protein analyses as well as histology and cell line derivation. Third, tumors can be collected at different stages, with our data indicating that Nfib is highly expressed in the most advanced stages of mSCLC. Interestingly, the human SCLC cell lines in which we demonstrated that NFIB is critical for tumor maintenance were also derived from patient pleural effusions and a lymph node metastasis. Thus, advanced lesions still appear to critically depend on NFIB for cell viability. Given our success in analyzing copy number alterations in mSCLC, future efforts to catalog point mutations in mSCLC, followed by cross-species analyses, would likely prioritize the daunting number of potentially meaningful mutations being identified in human SCLC (Pleasance et al. 2010). Our study highlights the power of rationally designed mouse models to uncover novel cancer-promoting alterations and uncovered important proto-oncogenic transcription factors in SCLC.
Materials and methods
Mice
p53fl/fl;Rbfl/fl and p53fl/fl;Rbfl/fl;Rosa26LSL-Luciferase/LSL-Luciferase mice have been described (Jonkers et al. 2001; Vooijs et al. 2002; Meuwissen et al. 2003; Sage et al. 2003) (Rosa26LSL-Luciferase/LSL-Luciferase mice are unpublished, from Erica Jackson and Tyler Jacks). They were infected intranasally or intratracheally with 2.5 × 107 or 1 × 108 plaque-forming units (PFU) of adenovirus, as described previously (DuPage et al. 2009). Mouse research was approved by the Committee for Animal Care and conducted in compliance with the Animal Welfare Act Regulations and other federal statutes relating to animals and experiments involving animals, and adheres to the principles set forth in the National Research Council's 1996 Guide for the Care and Use of Laboratory Animals (institutional animal welfare assurance no. A-3125-01).
Isolation of tumor DNA and production of cell lines
Primary tumors from the mouse model were snap-frozen for DNA isolation and stored at −80°C. In addition, part of each tumor was kept for histology, used for cell line preparation, and snap-frozen in RNALater (Ambion). To isolate DNA, tumors were digested in 800 μg/mL proteinase K overnight at 55°C and phenol/chloroform-extracted. To dissociate tumors for cell lines, minced tumors were digested in HBSS (without calcium and magnesium), 1 mg/mL Collagenase IV (Worthington Biochemicals), and 0.025% trypsin-EDTA (Gibco) for 30 min at 37°C. Following dissociation, samples were quenched in PBS with 10% FBS, incubated in 25 μg/mL DNase I (Sigma), centrifuged, filtered, and plated in standard tissue culture medium (DME, 10% FBS, 1% L-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin).
Illumina sequencing and data analysis
Libraries for DNA copy number analysis were prepared from 5 μg of DNA using the Illumina Genomic DNA Sample Preparation kit. Single-end 35-nucleotide reads were generated using the Illumina Genome Analyzer IIx. Reads were aligned to the mm9 reference genome with MAQ and filtered for mapping quality >30 (Supplemental Table 2). For each 100-kb genomic window, the number of reads aligning to that window were normalized by the total number of aligned reads in the sample. Copy number ratios were calculated as the number of normalized reads from the tumor sample, divided by the number of normalized reads from the reference 129/SVJ strain. Chromosomal boundaries of copy number changes were identified by change point analysis (Chiang et al. 2009). DNA copy number was visualized using the Broad Institute's Integrated Genome Viewer (http://www.broadinstitute.org/igv).
Human SCLC cell line copy number profiling
Affymetrix SNP 6.0 data were obtained from the Cancer Cell Line Encyclopedia (http://www.broadinstitute.org/ccle). DNA isolation and hybridization arrays were performed as recommended by Affymetrix. Probe normalization, segmentation, and copy number profiles were determined as described previously (The Cancer Genome Atlas Research Network 2008). Significantly recurrent regions of somatic copy number alterations were identified using the latest GISTIC methodology as described (Beroukhim et al. 2007, 2010).
IHC
Anti-Nfib antibody (1:500; Abcam) was used for IHC using standard methods.
Acknowledgments
We acknowledge D. Feldser, E. Meylan, N. Dimitrova, and T. Papagiannakopoulos for advice, and C. Kim-Kiselak for critical reading of the manuscript. We thank Jacks laboratory members and T. Parisi for reagents. We are indebted to the Koch Institute Core Facilities: D. Cook and A. Leshinsky (Biopolymers), E. Vasile (Microscopy), and G. Paradis (Flow Cytometry). We thank M. Luo (BioMicro Center) for microarray support and M. Leversha at Sloan Kettering Cancer Center Cytogenetics core facility for FISH. This work was supported by the Ludwig Center for Molecular Oncology at MIT, the Howard Hughes Medical Institute, and in part by the Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute. T.J. is a Howard Hughes Investigator, the David H. Koch Professor of Biology, and a Daniel K. Ludwig Scholar. M.M.W was a Merck Fellow of the Damon Runyon Cancer Research Foundation and a Genentech Post-doctoral Fellow. D.Y.C. is supported by an Alfred P. Sloan Foundation Research Fellowship, and S.B. is supported by an International Association for the Study of Lung Cancer Fellowship. A.L.D., M.M.W., and T.J. designed the experiments. A.L.D., M.M.W., T.D., and S.S. performed the experiments. D.Y.C. analyzed the mouse copy number analysis, and C.W. provided bioinformatics support. S.B., N.S., J.B., L.G., and M.M. performed the human copy number analysis. E.S. and R.B. provided histopathological analysis, and D.C. provided histological support.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2046711.
References
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. 2007. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci 104: 20007–20012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. 2001. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci 98: 13790–13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang DY, Getz G, Jaffe DB, O'Kelly MJT, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES 2009. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods 6: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Spiro SG 2006. Small cell lung cancer: treatment review. Respirology 11: 241–248 [DOI] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T 2009. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4: 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründer A, Ebel TT, Mallo M, Schwarzkopf G, Shimizu T, Sippel AE, Schrewe H 2002. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mech Dev 112: 69–77 [DOI] [PubMed] [Google Scholar]
- Han W, Jung E-M, Cho J, Lee JW, Hwang K-T, Yang S-J, Kang JJ, Bae J-Y, Jeon YK, Park I-A, et al. 2008. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer 47: 490–499 [DOI] [PubMed] [Google Scholar]
- Harbour JW, Lai SL, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ 1988. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science 241: 353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29: 418–425 [DOI] [PubMed] [Google Scholar]
- Kim M, Gans JD, Nogueira C, Wang A, Paik J-H, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al. 2006. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell 125: 1269–1281 [DOI] [PubMed] [Google Scholar]
- Lewis PD, Parry JM 2004. In silico p53 mutation hotspots in lung cancer. Carcinogenesis 25: 1099–1107 [DOI] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. 2008. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R, Berns A 2005. Mouse models for human lung cancer. Genes Dev 19: 643–664 [DOI] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A 2003. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 4: 181–189 [DOI] [PubMed] [Google Scholar]
- Nau MM, Brooks BJ, Battey J, Sausville E, Gazdar AF, Kirsch IR, McBride OW, Bertness V, Hollis GF, Minna JD 1985. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature 318: 69–73 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin M-L, Beare D, Lau KW, Greenman C, et al. 2010. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424: 223–228 [DOI] [PubMed] [Google Scholar]
- Santoro C, Mermod N, Andrews PC, Tjian R 1988. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature 334: 218–224 [DOI] [PubMed] [Google Scholar]
- Schaffer BE, Park K-S, Yiu G, Conklin JF, Lin C, Burkhart DL, Karnezis AN, Sweet-Cordero EA, Sage J 2010. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res 70: 3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM 2005. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol 25: 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, Levitt M, Pass H, Gazdar AF, Minna JD 1989. p53: a frequent target for genetic abnormalities in lung cancer. Science 246: 491–494 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Takahashi T, Suzuki H, Hida T, Sekido Y, Ariyoshi Y, Ueda R 1991. The p53 gene is very frequently mutated in small-cell lung cancer with a distinct nucleotide substitution pattern. Oncogene 6: 1775–1778 [PubMed] [Google Scholar]
- Toyooka S, Tsuda T, Gazdar AF 2003. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat 21: 229–239 [DOI] [PubMed] [Google Scholar]
- Vooijs M, te Riele H, van der Valk M, Berns A 2002. Tumor formation in mice with somatic inactivation of the retinoblastoma gene in interphotoreceptor retinol binding protein-expressing cells. Oncogene 21: 4635–4645 [DOI] [PubMed] [Google Scholar]
- Voortman J, Lee JH, Killian JK, Suuriniemi M, Wang Y, Lucchi M, Smith WI Jr, Meltzer P, Giaccone G 2010. Array comparative genomic hybridization-based characterization of genetic alterations in pulmonary neuroendocrine tumors. Proc Natl Acad Sci 107: 13040–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistuba II, Gazdar AF, Minna JD 2001. Molecular genetics of small cell lung carcinoma. Semin Oncol 28: 3–13 [PubMed] [Google Scholar]
- Worden FP, Kalemkerian GP 2000. Therapeutic advances in small cell lung cancer. Expert Opin Investig Drugs 9: 565–579 [DOI] [PubMed] [Google Scholar]
- Yokota J, Wada M, Shimosato Y, Terada M, Sugimura T 1987. Loss of heterozygosity on chromosomes 3, 13, and 17 in small-cell carcinoma and on chromosome 3 in adenocarcinoma of the lung. Proc Natl Acad Sci 84: 9252–9256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota J, Akiyama T, Fung Y-KT, Benedict WF, Namba Y, Hanaoka M, Wada M, Terasaki T, Shimosato Y, Sugimura T, et al. 1988. Altered expression of the retinoblastoma (RB) gene in small-cell carcinoma of the lung. Oncogene 3: 471–475 [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan S-T, Luk JM, Wigler M, Hannon GJ, et al. 2006. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125: 1253–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. 2008. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, Nikitin AY 2006. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res 66: 7889–7898 [DOI] [PubMed] [Google Scholar]