Abstract
Transcriptional regulation of developmentally controlled genes is at the heart of differentiation and organogenesis. In this study, we performed global genomic analyses in murine embryonic stem (ES) cells and in human cells in response to activation signals. We identified an essential role for the ELL (eleven–nineteen lysine-rich leukemia gene)/P-TEFb (positive transcription elongation factor)-containing super elongation complex (SEC) in the regulation of gene expression, including several genes bearing paused RNA polymerase II (Pol II). Paused Pol II has been proposed to be associated with loci that respond rapidly to environmental stimuli. However, our studies in ES cells also identified a requirement for SEC at genes without paused Pol II, which also respond dynamically to differentiation signals. Our findings suggest that SEC is a major class of active P-TEFb-containing complexes required for transcriptional activation in response to environmental cues such as differentiation signals.
Keywords: transcription elongation, immediate early genes, ELL2, AFF4
Transcriptional regulation by RNA polymerase II (Pol II) is a multifaceted process requiring the concerted action of a large number of factors for the steadfast synthesis of full-length messenger RNA (mRNA) (Workman and Kingston 1998; Shilatifard et al. 2003; Sims et al. 2004; Bres et al. 2008; Boettiger and Levine 2009). Transcription by Pol II is divided into four stages: initiation, promoter clearance, elongation, and termination. The initiation stage of transcription requires nucleosomal remodeling around the enhancer and promoter regions followed by the recognition of the promoter elements by the basal transcription machinery and Pol II. Once the basal factors and Pol II are recruited to the promoter elements, the catalysis of the first phosphodiester bond marks the initiation of transcription (Shilatifard 1998; Sims et al. 2004). For many years, it was considered that transcription initiation was the rate-limiting step to the transcription process as a whole. However, a large number of studies demonstrated that the elongation stage of transcription regulated by a number of factors is essential for productive transcription (Shilatifard et al. 2003; Sims et al. 2004; Levine 2011). In support of a vital role for the elongation stage of transcription in development, it has been demonstrated that the perturbation of this stage of transcription or the factors involved in this process results in the pathogenesis of human diseases, including cancer (Shilatifard et al. 2003; Mohan et al. 2010).
In addition to the control of the productive elongation stage of transcription by Pol II elongation factors, many developmentally regulated genes in mammalian cells are marked by stalled or paused Pol II at their proximal-promoter regions (Muse et al. 2007; Zeitlinger et al. 2007; Core et al. 2008; Boettiger and Levine 2009; Rahl et al. 2010; Levine 2011). Such polymerases have already been initiated and are awaiting proper developmental signals to enter the processive stage of transcription elongation (Rougvie and Lis 1988). Some studies have suggested that marking such developmentally regulated genes by paused Pol II could enhance their ability to be induced rapidly in a robust manner (Nechaev and Adelman 2008). Other studies, however, have proposed that the presence of paused Pol II at developmentally regulated genes allows for a synchronous induction of the same set of genes in distinct cell populations at the appropriate stage of development (Boettiger and Levine 2009).
Multiple factors have been identified to achieve proper promoter clearance and the processive elongation stage of transcription during development. These factors include Elongin A, DSIF (DRB sensitivity-inducing factor), NELF (negative elongation factor), P-TEFb (positive transcription elongation factor), and ELL (eleven–nineteen lysine-rich leukemia gene) (Jones and Peterlin 1994; Shilatifard et al. 2003; Sims et al. 2004; Peterlin and Price 2006; Bres et al. 2008; Levine 2011). Both Elongin A and DSIF are capable of increasing the catalytic rate of the productive transcription by Pol II; however, in addition to its role in this process, DSIF also works with NELF to regulate Pol II arrest (Yamaguchi et al. 1999; Shilatifard et al. 2003; Cheng and Price 2007). Such arrested Pol IIs are released by the Cdk9 kinase activity of P-TEFb, which phosphorylates the C-terminal domain (CTD) of Pol II and many of the other transcription factors, signaling the release of the stalled Pol II into productive transcription (Jones and Peterlin 1994; Fuda et al. 2009). ELL was purified based on its catalytic properties to increase the Vmax rate of transcription elongation by Pol II (Shilatifard et al. 1996; Shilatifard 1998). Translocations of ELL involving the mixed-lineage leukemia gene MLL are associated with the pathogenesis of childhood leukemia and misregulation of developmental genes (Thirman et al. 1994). In addition to ELL, a large number of genes with very little sequence or obvious functional similarities are found in translocations with MLL in leukemia (Mohan et al. 2010).
In support of the hypothesis that the elongation stage of transcription by Pol II has an essential role in development and cancer pathogenesis, ELL and several other MLL translocation partners were biochemically isolated as part of the super elongation complex (SEC) that contains P-TEFb (Lin et al. 2010). SEC has also been shown to play a role in regulating the elongation stage of transcription controlled by HIV-1 Tat (He et al. 2010; Sobhian et al. 2010). These studies suggest that MLL translocations function by regulating the elongation stage of transcription on developmentally regulated genes, such as the HOX loci, through the association of MLL chimeras with P-TEFb within the ELL-containing SEC. This association of SEC with MLL through chromosomal translocations can result in the premature release of paused Pol II at developmental loci (Mohan et al. 2010; Smith et al. 2011).
P-TEFb participates in a variety of complexes, both active and inactive (Bres et al. 2008; He and Zhou 2011). Both Brd4 and Myc-containing P-TEFb complexes have been considered to be major regulators of transcription elongation (Zhou and Yik 2006; Zippo et al. 2009; Donner et al. 2010; Rahl et al. 2010). To investigate to what degree SEC functions genome-wide in transcription elongation control, we performed chromatin immunoprecipitation (ChIP) and sequencing (ChIP-seq) studies in both mouse embryonic stem (ES) cells in response to retinoic acid (RA) induction and human HCT-116 cells in response to serum stimulation. Our studies in mouse ES cells identified gene targets for SEC, many of which are developmental regulators with paused Pol II that were rapidly induced to high, but relatively uniform, levels. Our studies in human HCT-116 cells found that SEC is also a major regulator of immediate early genes induced by growth factors.
Together, these findings suggest that the presence of paused Pol II at promoter-proximal regions and recruitment of SEC upon activation may represent a major cellular mechanism for rapid and uniform induction of gene expression upon exposure to key developmental signals. Intriguingly, our global genomic studies in ES cells also identified a requirement for SEC at Cyp26a1, a gene that does not bear paused Pol II at its promoter-proximal region, yet responds dynamically to RA in an even more rapid manner than other genes that have paused Pol II at their promoter-proximal regions. Our findings suggest that the recruitment of SEC allows genes to respond in a rapid and dynamic manner to developmental signals in different cell types in mammals, and that SEC is involved in transcriptional induction that is both dependent on and independent of the presence of paused Pol II.
Results
The SEC is enriched on highly transcribed genes in ES cells
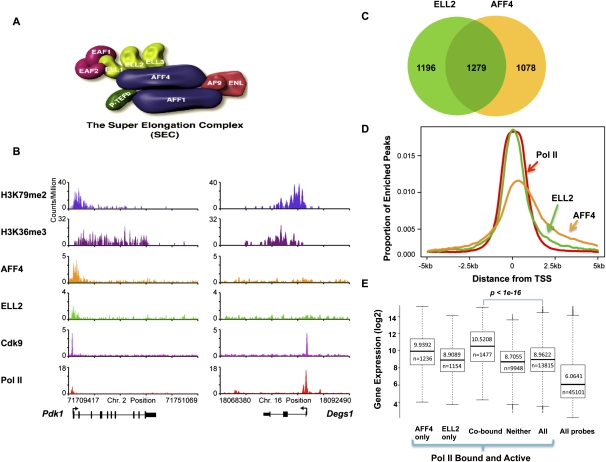
We previously identified the SEC as a transcription elongation complex containing the known elongation factors ELL1–3, EAF1–2, P-TEFb, and several other translocation partners of MLL found in leukemia (Fig. 1A), including AFF1, AFF4, AF9, and ENL (Lin et al. 2010). SEC was previously shown to be required for a normal cellular function, the induction of the HSP70 gene upon stress, and the misregulation of transcription of the HOXA9 and HOXA10 genes by MLL chimeras (Lin et al. 2010). SEC was later found to be required for Tat-mediated HIV transactivation (He et al. 2010; Sobhian et al. 2010). Our previous studies demonstrated a close relationship between AFF4 and ELL2; AFF4 was central for the formation of SEC, and the RNAi-mediated knockdown of AFF4 led to the destabilization of the ELL2 protein (He et al. 2010; Lin et al. 2010). To investigate a possible role of SEC in the control of developmental genes poised for activation in early development, we developed antibodies to SEC components (Supplemental Fig. S1; Lin et al. 2010) and performed a genome-wide occupancy analysis of the SEC components in mouse ES cells using ChIP-seq of AFF4, ELL2, Cdk9, and RNA Pol II. These SEC components co-occupy many of the same genes, including highly expressed housekeeping genes such as the histone genes (Supplemental Fig. S2); however, SEC is only found at a subset of highly transcribed genes (Fig. 1B,C,E). SEC components are enriched at the transcription start site (TSS) regions of these genes and within the gene body similar to the Pol II distribution (Fig. 1D; Supplemental Fig. S2). The co-occupancy of the AFF4 and ELL2 components of SEC correlates with a high level of expression of genes in mouse ES cells (Fig. 1E) suggesting that SEC is frequently associated with highly transcribed regions.
Figure 1.
Global occupancy of the SEC subunits in mouse ES cells. (A) Schematic representation of the SEC. The SEC is a P-TEFb-containing complex that contains various combinations of four types of proteins: ELL1–3; EAF1–2; AFF1 and AFF4; and AF9 and ENL. P-TEFb itself consists of Cdk9 and CycT1/2 and is best characterized as an RNA Pol II CTD kinase. (B) Genome-wide analysis of SEC components AFF4, ELL2, and Cdk9 by ChIP-seq in ES cells found SEC enriched at a subset of actively transcribed genes. Shown are two genes with high levels of expression in ES cells. (Left panel) The Pdk1 gene is occupied by the SEC subunits ELL2 and AFF4. (Right panel) The Degs1 gene does not have significant levels of the SEC components AFF4 and ELL2. H3K36 trimethylation (H3K36me3) and H3K79 dimethylation (H3K79me2) data from Marson et al. (2008) are shown as markers of actively transcribed genes. (C) Venn diagram analysis of AFF4- and ELL2-occupied genes. Around 50% of AFF4-enriched genes are also occupied by ELL2, demonstrating that in mouse ES cells, these two proteins share a similar global occupancy. (D) Histogram of the genome-wide occupancy of AFF4, ELL2, and Pol II. The canonical TSS of each gene in the genome was used to measure the distance to the nearest bound region, which is plotted if falling within 5 kb of the TSS. This analysis shows that SEC components are enriched over the TSS, similar to Pol II occupancy. (E) AFF4 and ELL2 co-occupy highly transcribed genes. The dark lines in the box plots and the number above the line indicate the median level of expression for the gene subset indicated. The number below the line indicates the number of Affymetrix probe sets that correspond to the gene subset. Probe sets for ELL2 and AFF4 cobound genes show significantly higher expression compared with all Pol II-bound and active genes (P < 1 × 10−16 by Wilcoxon two-sample rank sum test). The gene subset containing neither AFF4 nor ELL2 also shows some highly expressed genes. Genes were called active if they were determined present on the array by the MAS5 algorithm.
Paused Pol II and recruitment of SEC at the developmentally regulated Hox clusters in ES cells
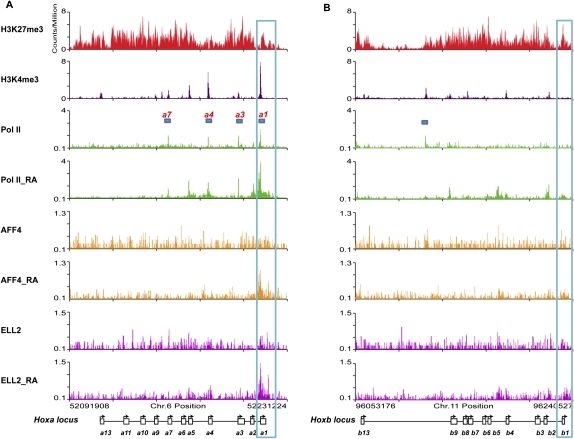
The SEC was discovered based on the purification of several of the MLL chimeras that are commonly found in MLL-rearranged leukemias (Lin et al. 2010). It is not currently known why SEC components are so frequently found to be translocation partners with MLL. One possibility is that the genes misregulated by MLL chimeras, such as the HOX genes, are normal SEC targets. In these leukemias, MLL target genes become misregulated when SEC is recruited inappropriately and prematurely activates transcription by releasing paused Pol II (Lin et al. 2010; Mohan et al. 2010; Smith et al. 2011). Many developmentally regulated genes in flies and mammals have paused Pol II at the TSS before their activation during development (Muse et al. 2007; Zeitlinger et al. 2007). In mammalian stem cells, these genes are characterized by a bivalent mark of both H3K4 and H3K27 trimethylation on the same gene (Bernstein et al. 2006). Looking within the Hox clusters in ES cells, we found bivalent marks co-occurring with Pol II at the TSS at four of the Hoxa cluster genes (Hoxa1, Hoxa3, Hoxa4, and Hoxa7), but not at the promoters of the Hoxb genes (Fig. 2).
Figure 2.
The Hoxa1 promoter is preloaded with Pol II and recruits SEC after RA treatment in ES cells. (A) Bivalent marks, paused Pol II, and SEC recruitment to the Hoxa cluster. In ES cells, the whole Hoxa cluster is highly enriched for H3K27me3, and also contains H3K4me3 at the promoters of a subset of genes, including Hoxa1, Hoxa3, Hoxa4, and Hoxa7. These regions are preloaded with Pol II (bars indicate regions that have both a bivalent mark and Pol II). (B) Bivalent marks and paused Pol II are both largely absent from the Hoxb genes, which do not recruit SEC after 6 h of RA treatment. While H3K27me3 marks the whole cluster of Hoxb genes, only Hoxb4, Hoxb7, and Hoxb9 contain H3K4me3 at their promoters and can be considered bivalent. There is no significant Pol II detected on the promoters of the Hoxb genes in ES cells. The bar marks a peak of significant Pol II that does not correspond to a known gene feature. Before RA treatment, there is no detectable AFF4 and ELL2 signal on the Hoxa or Hoxb cluster genes. Both AFF4 and ELL2 are recruited to the Hoxa1, but not the Hoxb1, gene promoter after exposure to RA for 6 h. Blue boxes highlight the Hoxa1 and Hoxb1 genes. Expanded views of the Hoxa1 and Hoxb1 regions are shown in Supplemental Figure S4.
The regulation of gene transcription at the level of paused Pol II and its controlled release have been best studied at the heat-shock genes such as HSP70, as well as in the control of HIV transcription, and both processes require SEC (He et al. 2010; Lin et al. 2010; Sobhian et al. 2010). Genes with paused Pol II, such as HSP70, are transcriptionally engaged with pausing 30–40 nucleotides downstream from the TSS (Core et al. 2008; Nechaev et al. 2010). These genes contain basal transcriptional machinery at their promoters and have a form of Pol II phosphorylated on Ser5, but not Ser2, of the CTD, and the Pol II is associated with DSIF/NELF (Nechaev and Adelman 2011). By all of these criteria, Hoxa1, but not Hoxb1, is occupied and engaged by paused Pol II (Supplemental Fig. S3).
This led us to ask whether SEC was differentially recruited to Hoxa1 and not Hoxb1 upon induction by RA treatment. We performed AFF4, ELL2, and Pol II ChIP-seq after 6 h of RA treatment of mouse ES cells. Pol II was recruited to both Hoxa1 and Hoxb1 promoters after exposure to RA for 6 h (Fig. 2; Supplemental Fig. S4). Interestingly, AFF4 and ELL2 were only recruited to Hoxa1, and not Hoxb1, by 6 h of RA treatment (Fig. 2; Supplemental Fig. S4). However, we cannot rule out the possibility that SEC was not detected at Hoxb1 due to lower levels of Pol II and a concomitant decrease in SEC that falls below our detection level. Our genome-wide analyses suggest that our ability to detect SEC occupancy on a gene is not strictly dependent on levels of Pol II or transcription levels (Fig. 1B,E).
SEC is required for the rapid induction of Hoxa1
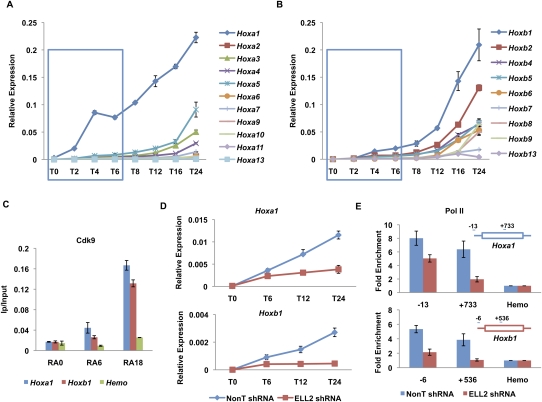
Promoter-proximal paused Pol II has been proposed to allow for a more rapid induction of genes upon differentiation cues (Nechaev and Adelman 2008). Therefore, we assayed the induction kinetics of Hoxa and Hoxb cluster genes by RT-qPCR after RA treatment from 2–24 h (Fig. 3A,B). We found that Hoxa1 and Hoxb1 were the first genes rapidly induced within their respective clusters, followed more slowly by other members of the clusters, in general agreement with the collinearity of expression that occurs during normal embryonic development (Duboule and Dolle 1989; Graham et al. 1989; McGinnis and Krumlauf 1992). The Hoxa1 and Hoxb1 paralogs functionally synergize in regulating the hindbrain pattern formation and cranial nerve patterning (Gavalas et al. 2001). During normal mouse development, Hoxa1 is the first Hox gene expressed in neural tissue directly induced by RA through a 3′ RA response element (RARE). It is closely followed by RA-mediated induction of Hoxb1 through a similar 3′ RARE. Hoxa1 also participates in the proper activation of Hoxb1 by binding to Hoxb1's 5′ autoregulatory element (ARE), and Hoxb1 further stimulates transcription of its own gene (Popperl et al. 1995; Studer et al. 1998). Indeed, when looking within the first 6-h window of RA treatment of mouse ES cells, we observe that Hoxa1 is induced more rapidly than Hoxb1, mirroring their normal kinetics of induction in neural development (Fig. 3A,B, blue boxes). The more rapid induction of the Hoxa1 locus compared with Hoxb1 could result from the presence of paused Pol II before RA treatment.
Figure 3.
SEC is required for the rapid induction of the Hoxa1 gene. (A,B) RT-qPCR analysis of Hoxa and Hoxb cluster genes upon RA treatment. ES cells were treated with RA for different time points as indicated. Total RNAs were extracted from these cells and then subjected to RT-qPCR analysis using an Applied Biosystems' custom TaqMan array card. Hoxa1 was the first Hox gene to be induced by RA. Compared with Hoxa1, the induction of Hoxb1 was much slower within the first 6 h of RA treatment. The blue boxes indicate the first three RA induction time points. (C) Cdk9 is recruited to both the Hoxa1 and Hoxb1 gene promoters. Cdk9 ChIP was performed to measure its enrichment on Hoxa1 and Hoxb1 after RA treatment. A hemoglobin gene, Hba (Hemo), serves as a nontranscribed control gene. (D) ELL2 RNAi inhibits the induction of Hoxa1 and Hoxb1 by RA. shRNA targeting ELL2 or nontargeting shRNA (NonT) was introduced by lentiviral infection for 3 d before RA treatment. (E) Knockdown of ELL2 reduces Pol II occupancy at Hoxa1 and Hoxb1 after 6 h of RA treatment. Pol II occupancy was assayed by ChIP at the start site of transcription and in the ORF of Hoxa1 and Hoxb1 in RA-induced cells. Pol II is reduced in the ORF of both Hoxa1 and Hoxb1, and Hoxb1 also shows dramatically reduced levels of Pol II at its promoter after ELL2 RNAi. The Hoxa1 promoter, but not the Hoxb1 promoter, has prebound Pol II before RA treatment (see Fig. 2; Supplemental Fig. S4). Error bars represent the standard deviation.
Since SEC was only detected at the Hoxa1 promoter, and not at the Hoxb1 promoter, we asked whether the Pol II CTD kinase Cdk9 was also differentially localized to these two genes at early induction time points. Direct comparisons of Cdk9 at Hoxa1 and Hoxb1 show that Cdk9 is recruited to both genes as early as 6 h and has increased occupancy at 18 h (Fig. 3C). When ES cells are induced with RA for various time points, in the presence or absence of the Cdk9 inhibitor flavopiridol (Chao and Price 2001), the induction of both Hoxa1 and Hoxb1 are diminished (Supplemental Fig. S5). This indicates that Cdk9 is required for the activation of both genes, even though the kinetics of their induction differ. This indicates that the recruitment of P-TEFb within SEC, specifically to Hoxa1, functions in its rapid induction. In support of this statement, ELL2 RNAi also reduces the induction of Hoxa1 (Fig. 3D; Supplemental Fig. S5). ELL2 is also required for the induction of Hoxb1; however, this observation could be explained by the requirement of the Hoxa1 protein for the full induction of Hoxb1 (Studer et al. 1998). Accordingly, in the absence of ELL2 (ELL2 RNAi), we also observe the loss of Pol II in the body of the Hoxa1 gene with no significant change or slight reduction in occupancy of Pol II at the Hoxa1 promoter (Fig. 3E, top panel). Furthermore, since Hoxb1 expression requires Hoxa1 activity and lacks prior paused Pol II in ES cells, in the absence of ELL2, we detect a loss in Pol II occupancy both at the promoter and in the body of the Hoxb1 locus (Fig. 3E, bottom panel). Therefore, Hoxa1 is likely to be a direct target of SEC, and Hoxb1 is likely to be an indirect target of SEC. In summary, given the fact that Hoxa1, and not Hoxb1, possesses paused Pol II and recruits SEC upon a differentiation signal, we hypothesized that the recruitment of SEC to genes bearing paused Pol II is associated with rapid induction.
SEC is required for the induction of other rapidly induced genes in ES cells bearing paused Pol II
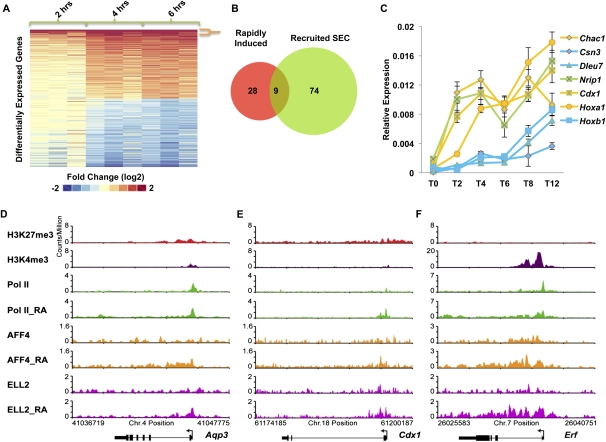
Using genome-wide approaches, we asked whether there were other genes that were regulated similarly to Hoxa1. We performed gene expression analyses of ES cells treated for 2–6 h with RA using Affymetrix expression arrays with probes representing ∼30,000 genes (Fig. 4A). Sorting the gene expression data by fold expression over time showed that only a small number of genes demonstrated rapid and sustained induction over this time frame in a manner similar to Hoxa1 (Fig. 4A,B). We found that 37 genes were rapidly induced at least twofold at 2, 4, and 6 h post-induction (Fig. 4A,B). Among these genes was Hoxb1, which our RT-qPCR data had shown was not as rapidly induced as Hoxa1 (Fig. 3A,B). We therefore performed RT-qPCR analyses of other genes from the top of this list to confirm their patterns of induction (Fig. 4C). These RT-qPCR studies demonstrated that two of the genes identified by microarray, Dleu7 and Csn3 (Fig. 4C, blue), behaved similarly to Hoxb1 and were not as rapidly induced as Hoxa1, while two others showed the kinetics of rapid induction similar to Hoxa1 (Fig. 4C, yellow).
Figure 4.
SEC regulates the rapid induction of RA signaling. (A, left panel) Microarray analyses of RA induction of ES cells as a function of time (2, 4, and 6 h) in biological triplicate. Differentially expressed probes (twofold or more) at the sixth hour post-induction compared with no induction are shown. Thirty-seven genes were induced twofold or more at each of the 2-, 4-, and 6-h time points (demarcated by the orange bracket). (B) Of the 37 induced genes, nine of them recruited SEC (ELL2 and AFF4). Newly recruited SEC genes are cobound at 6 h post-induction and are not cobound before induction. (C) RT-qPCR analysis of some of the induced genes identified from the microarray analysis. ES cells were treated with RA for the indicated time points, 0 h (T0), 2 h (T2), 4 h (T4), 6 h (T6), 8 h (T8), and 12 h (T12). Genes that recruit SEC are shown in yellow and genes that do not recruit SEC are shown in blue. Nrip1, which does not recruit SEC but is rapidly induced, is shown in green. Error bars represent the standard deviation. (D–F) Examples of ChIP-seq data showing SEC recruitment to RA-induced genes. Shown are Aqp3, Cdx1, and Erf, three of the nine genes from B.
Many of the induced genes that recruited SEC had bivalent histone marks and paused Pol II prior to RA induction. Genome browser track files for some examples are shown in Figure 4, D–F. Regardless of whether genes are rapidly or more slowly induced, Cdk9 was recruited and required for their induction (Supplemental Fig. S7). This analysis indicates that several genes that recruit SEC with Cdk9 respond more rapidly and uniformly to developmental signals than genes recruiting Cdk9 without SEC. However, the existence of genes like Nrip1, which is induced with similar kinetics to Hoxa1 (Fig. 4C, green) but does not recruit SEC, suggests that while SEC is a major form of the Cdk9 complexes recruited to genes for rapid gene activation, other pathways to rapid gene activation are also possible.
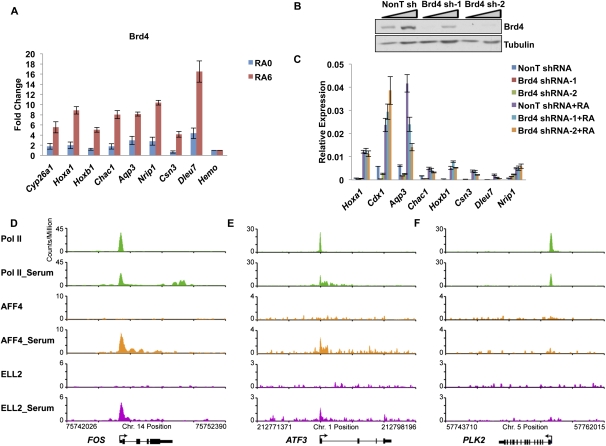
We tested for the presence of another P-TEFb interactor, Brd4, on these genes and demonstrated that although Brd4 is recruited to these loci upon RA induction (Fig. 5A), its reduction by RNAi has very little to do with their activation by RA, except for the Aqp3 gene (Fig. 5B,C). This observation suggests that although Brd4 is also recruited to those SEC target sites, it might not play a major role for their activation (Fig. 5A–C). Perhaps, as in the case of HIV-1 transcriptional regulation, Brd4 has a role in maintaining basal levels of transcription, but not in the activation of these genes (Yang et al. 2005).
Figure 5.
Brd4 is broadly present, but not broadly required, for RA induction of genes. (A) ChIP of Brd4 at RA-6-induced genes. Brd4 levels significantly increase at all RA-6-induced genes tested. The Hba gene serves as a nontranscribed control gene. (B) shRNA-mediated knockdown of Brd4. Two different shRNA constructs targeting Brd4 and a nontargeting shRNA (NonT) were introduced by lentiviral infection for 3 d before RA treatment. Brd4 levels were significantly reduced by Western analysis. Triangles indicate titrations of cell extracts. Tubulin serves as a loading control. (C) Induction of genes with RA is not broadly affected by Brd4 knockdown. Several genes, identified in Figure 4 as rapidly induced, were assayed for expression levels before and after RA treatment. Only Aqp3 showed a significant decrease in its induction. Error bars represent the standard deviation. (D–F) SEC is recruited to the immediate early genes in HCT-116 cells after serum stimulation, genes previously identified as regulated by Brd4-containing P-TEFb complexes. HCT-116 cells were starved for 40 h before the 30-min add-back of serum. (D–F) Genome browser track files of three serum-induced genes, which were previously shown to be rapidly induced after serum stimulation (Donner et al. 2010). SEC is recruited to ATF3 and FOS concomitant with release of paused Pol II into the gene body. PLK2 is shown for comparison as a gene induced by serum that does not recruit SEC.
SEC is also required for the rapid transcriptional induction of many growth-related immediate early genes in human cells
Given the small number of RA-induced genes in the mouse ES system, we sought another system to determine to what degree SEC regulates rapid transcriptional responses to environmental signals. Therefore, we investigated the role of SEC in the induction of genes in response to serum in human cells (Fig. 5D–F; Supplemental Fig. S8). The immediate early genes induced by growth factors are some of the best-characterized genes regulated at the level of the release of paused Pol II (Simone et al. 2001; Kong et al. 2005). We performed ChIP-seq of SEC and Pol II in HCT-116 cells before and after serum stimulation. SEC components are also enriched at the TSS in HCT-116 cells, consistent with their distribution in ES cells (Fig. 1E, Supplemental Fig. S8A). SEC was newly recruited to 55 genes within 30 min of serum stimulation (Supplemental Fig. S8B). Similar to what we observed in ES cells (Fig. 1E), genes bound by AFF4 and ELL2 showed higher levels of expression than those that lacked SEC (Supplemental Fig. S8C,D). Previous gene expression analysis of serum-inducible genes in HCT-116 cells identified 29 genes that were up-regulated twofold or more within 30 min of serum stimulation (Donner et al. 2010), 12 of which recruited both AFF4 and ELL2. We also performed RNA-seq analysis in these cells in the presence and absence of serum stimulation and identified 66 genes, which were induced above twofold, including 26 out of the 29 genes identified by Donner et al. (2010). To more precisely characterize the induction kinetics of these genes, we performed RT-qPCR on 17 serum-induced genes at different times after serum stimulation (Supplemental Fig. S8E). As we had seen with RA induction, serum-responsive genes were induced at varying rates, with SEC recruitment frequently occurring on the most rapidly induced genes (Supplemental Fig. S8D,E). Thus, SEC appears to be one of the major factors in the rapid release of paused Pol II in response to developmental and environmental stimuli.
Dynamic transcriptional induction requiring SEC without the presence of paused Pol II
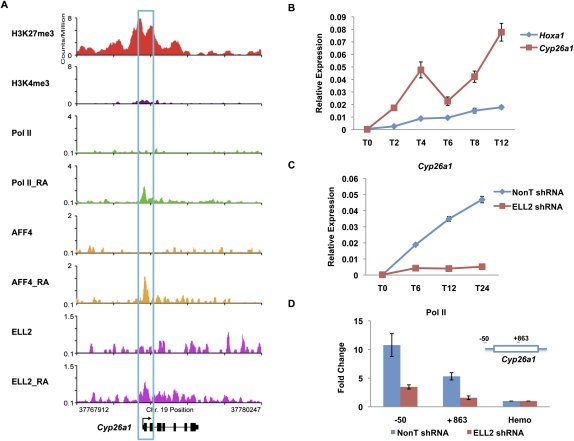
To date, published studies indicate that paused Pol II functions in the rapid and robust induction of many developmentally regulated genes (Rougvie and Lis 1988; Muse et al. 2007; Zeitlinger et al. 2007; Nechaev and Adelman 2008; Boettiger and Levine 2009). However, our genome-wide expression and ChIP-seq data identified one gene that is extremely rapidly induced by RA: the Cyp26a1 gene (Fig. 6). Cyp26a1 encodes a cytochrome P450 that metabolizes RA (Duester 2008). The Cyp26a1 gene bears several RAREs in its promoter and is known to be one of the most rapidly induced genes after exposure to RA (Alexander et al. 2009). Loss of Cyp26a1 is toxic to development in mice, but this toxicity can be rescued by the loss of RA receptor γ (RARγ) (Abu-Abed et al. 2001; Sakai et al. 2001). While the Cyp26a1 gene appears to have high levels of H3K27 trimethylation, it contains very low levels of H3K4 trimethylation compared with Hoxa1 (see Figs. 2A,B, 6A). Also, this gene lacks paused Pol II in the untreated ES cells (Fig. 6A). After RA addition, Pol II and SEC are recruited to Cyp26a1 by 6 h post-induction (Fig. 6A). In mouse ES cells, Cyp26a1 is more rapidly induced when compared with Hoxa1 and Hoxb1 (Figs. 4C, 6B). Knockdown of ELL2 by shRNA treatment causes a reduction in Cyp26a1 activation and also affects the recruitment of Pol II in its promoter and gene body (Fig. 6C,D), while flavopiridol completely eliminates Cyp26a1 induction, indicating that this gene requires Cdk9 for its rapid induction by RA treatment (Supplemental Fig. S9). Furthermore, reduction of the Brd4 level by RNAi did not significantly affect Cyp26a1 induction, suggesting that it is the SEC version of P-TEFb that regulates this gene. The dynamic induction of Cyp26a1 without pre-existing paused Pol II suggests that there are other mechanisms for rapid induction of transcription during early development, which involves SEC.
Figure 6.
The rapid induction of Cyp26a1 does not involve preloaded Pol II. (A) Pol II, H3K4me3, and H3K27me3 occupancy analysis of the Cyp26a1 gene before RA induction. Before RA treatment, the Cyp26a1 promoter is significantly enriched for H3K27me3, with lower levels of H3K4me3. However, there is no detectable Pol II on the promoter. AFF4, ELL2, and Pol II are newly recruited to the Cyp26a1 gene promoter upon RA treatment. (B) RT-qPCR analysis of Cyp26a1 mRNA levels upon RA treatment. ES cells were treated with RA for the indicated time points, 0 h (T0), 2 h (T2), 4 h (T4), 6 h (T6), 8 h (T8), and 12 h (T12). Total RNAs were extracted from these treated cell samples and then subjected to RT-qPCR analysis. (C) ELL2 RNAi inhibits the induction of Cyp26a1 by RA. shRNA targeting ELL2 or a nontargeting shRNA (NonT) was introduced by lentiviral infection for 3 d before RA treatment. (D) Knockdown of ELL2 reduces Pol II occupancy at Cyp26a1 after 24 h RA treatment. The Hba gene serves as a nontranscribed control gene. Error bars represent the standard deviation.
Discussion
Our study reports that genes recruiting SEC in human and mouse cells are capable of rapidly responding in a dynamic manner to developmental cues. Although the presence of paused Pol II has been associated with rapid transcriptional induction, the response to some of the genes we identified in our study does not require the presence of paused Pol II. These findings are supported by the following observations. (1) Hoxa1, but not Hoxb1, contains paused Pol II on its promoter-proximal region and, upon developmental cues, SEC is recruited to Hoxa1, but not Hoxb1. (2) Hoxa1 is induced more rapidly than Hoxb1 after RA induction in ES cells. (3) Genome-wide analyses in ES cells identified a set of rapidly induced genes that contain paused Pol II, some of which also recruit SEC, and transcriptionally respond in a relatively uniform manner to a differentiation signal. (4) SEC is recruited to some of the immediate early genes upon serum stimulation in human cells, suggesting that SEC is one of the regulators of rapidly induced genes in different cellular contexts. (5) One rapidly induced gene, Cyp26a1, does not possess paused Pol II prior to induction, but still requires SEC for its rapid and dynamic activation. Therefore, while rapid induction is a key function of SEC, the function of the paused Pol II state is not simply to mediate rapid induction. It is likely that paused Pol II has some additional role(s) in developmentally controlled gene expression, as genes containing paused Pol II at their promoter-proximal regions transcriptionally respond in a well-regulated and uniform manner upon induction.
Genome-wide occupancy of SEC components
The SEC was identified through the purification of several of the MLL chimeric proteins that mediate leukemogenesis in mixed-lineage leukemias (Lin et al. 2010). SEC contains two classes of elongation factors, ELL (represented by ELL1–3) and P-TEFb, which consists of Cdk9 and CycT1 or CycT2, which were isolated through biochemical approaches for the discovery of transcription elongation factors (Marshall and Price 1995; Shilatifard et al. 1996). The fact that ELL is a translocation partner of MLL and associates with other translocation partners of MLL such as AFF1, AFF4, ENL, and AF9 indicates that SEC function is important for the misregulation of key developmental genes like the Hox loci that are part of the leukemogenic process. Our genome-wide approaches sought to discover the normal developmental role of SEC using mouse ES cells before and after induction of differentiation.
SEC is recruited to genes with paused Pol II for rapid and coordinated transcriptional induction
Many developmentally regulated genes are marked by the presence of bivalent histone marks, the methylation of H3K4 and H3K27, DSIF/NELF, and paused Pol II at the TSS (Bernstein et al. 2006; Stock et al. 2007; Rahl et al. 2010). Since P-TEFb complexes such as the SEC are proposed to release paused Pol II via phosphorylation of the CTD and other general factors within the transcription complex, we asked whether SEC is recruited to these genes after induction of differentiation. We first focused on the Hox loci, because misregulation of Hox transcription is strongly implicated in leukemogenesis by MLL chimeras. The mammalian Hox genes exist in four clusters and are expressed in a collinear manner during development, with the most anterior Hox gene (e.g., Hoxa1 in mammals) being expressed first from its location at the 3′ end of the cluster, and a more posteriorly expressed Hox gene (e.g., Hoxa13) expressed later during development from its location at the 5′ end of the cluster. Although a large number of developmentally regulated genes contain bivalent marks and paused Pol II at their promoters, we found that only a subset of Hox genes followed this pattern. For example, four genes within the Hoxa cluster bear bivalent histone marks and paused Pol II at their promoters, but no Hoxb genes are marked in this manner in the ES cells (Fig. 2). Importantly, after induction of differentiation, Hoxa1 was induced more rapidly than its paralog, Hoxb1 (Fig. 3). Furthermore, the SEC was specifically recruited to Hoxa1, and not Hoxb1, suggesting that SEC releases paused Pol II for rapid induction of transcription during development. This mechanism helps to explain the more rapid induction and regulatory roles of Hoxa1 compared with Hoxb1 in early neural development (Alexander et al. 2009).
In order to extend these findings beyond the Hox loci, we undertook genome-wide analyses of expression and SEC occupancy before and after induction of differentiation with RA. We found additional examples of rapidly induced genes bearing paused Pol II at their promoter-proximal region that also recruited SEC, and many of these were among the most rapidly induced (Fig. 4). These findings were shown to be more general by studying the recruitment of SEC to the immediate early genes in HCT-116 cells after serum induction. The immediate early genes, including the FOS, JUN, and EGR families, are well characterized as genes containing paused Pol II and exhibiting rapid induction kinetics (Galbraith and Espinosa 2011). As with RA-induced genes, SEC was recruited to many of the most rapidly induced genes after serum stimulation (Fig. 5).
Dynamic and rapid transcriptional induction does not always require paused Pol II
Most of the rapidly induced genes contained paused Pol II in the undifferentiated ES cells. However, we were able to identify Cyp26a1 as a gene that was even more rapidly induced without having prior Pol II occupancy, however, in an SEC-dependent manner. By all molecular characteristics, Cyp26a1 in the undifferentiated condition appears to be relatively repressed in ES cells, as it bears H3K27 trimethylation, only modest levels of H3K4 methylation, and no detectable paused Pol II on its promoter when compared with other paused Pol II-regulated genes, such as Hoxa1 (cf. Figs. 2 and 6).
Cyp26a1 encodes a cytochrome, P450, that metabolizes RA and is essential for development. One function of Cyp26a1 is to restrict the response to RA to the appropriate regions of the embryo. Not only is Cyp26a1 extremely rapidly induced compared with other early RA response genes, but it is induced to much higher levels than the other very early RA-induced genes containing paused Pol II (Figs. 4, 6, 7).
The function of paused Pol II in modulating transcription of developmentally regulated genes
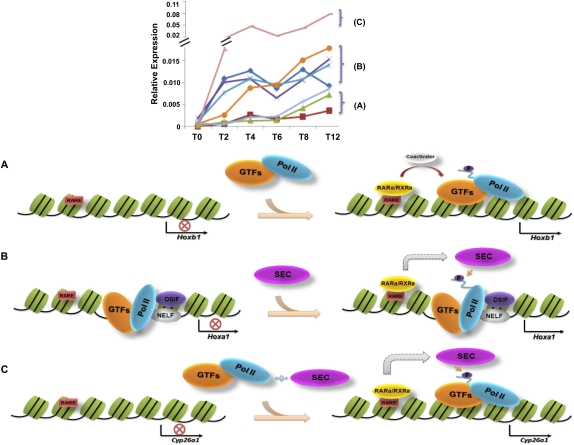
Our genome-wide analyses of RA-induced gene transcription and SEC recruitment identified three classes of genes, two of which require SEC for induction (Fig. 7). One class, which includes Hoxb1, lacks paused Pol II and does not recruit SEC upon induction (Fig. 7A). A second class, which includes Hoxa1, contains paused Pol II, recruits SEC, and is induced more rapidly than the first class (Fig. 7B). A third class, exemplified by Cyp26a1, recruits SEC, is induced just as rapidly as the second class, but to a greater extent than Hoxa1, yet lacks paused Pol II at its promoter-proximal region before induction, and requires SEC (Fig. 7C).
Figure 7.
Diverse mechanisms for rapid activation of genes during development. The top panel shows that rapidly activated genes can be further subdivided into distinct categories, A–C. (A) The Hoxb1 gene newly recruits Pol II and GTFs in a classical gene activation mechanism, where RAR/RXR binds in the presence of RA and, with the help of coactivators, recruits GTFs and Pol II. (B) Paused Pol II, with DSIF/NELF, is present at the TSS of developmentally regulated genes, such as Hoxa1. In the presence of RA, RAR/RXR recruits SEC to stimulate transcription elongation through phosphorylation of the DSIF/NELF and the Pol II CTD. (C) Cyp26a1, a developmentally regulated gene that lacks paused Pol II, is induced by RA in a SEC-dependent manner. All of the same factors are present after RA treatment as seen at Hoxa1, but Cyp26a1 is induced to higher levels, suggesting that paused Pol II may serve to help regulate activation to equivalent levels.
The HSP70, FOS, JUN, and EGR families of genes are well studied and rapidly induced, and contain paused Pol II in the unstimulated state, leading to the paradigm that rapid induction is the primary function of paused Pol II (Nechaev and Adelman 2008; Donner et al. 2010). However, paused Pol II is not present on Cyp26a1 before its rapid induction to high levels of transcription, which suggests that paused Pol II is not a prerequisite for rapid induction, but rather facilitates coordinated and controlled induction. Studies in Drosophila have shown that developmentally regulated genes that have paused Pol II are activated in a synchronous manner, while developmentally regulated genes that lack paused Pol II have a more stochastic pattern of induction during development (Boettiger and Levine 2009; Levine 2011). Having preloaded Pol II and general transcription factors (GTFs) reduces the number of steps for productive transcription, and thus could result in a more equivalent and uniform way to induce gene expression. Genes such as Cyp26a1, while being required for proper development and being induced rapidly to high levels, may not need to be as precisely regulated at the earliest time points of induction.
Overall, our studies demonstrate that SEC is involved in many of the rapid and dynamic inductions of gene expression responses to developmental and environmental cues. P-TEFb was identified >15 years ago (Marshall and Price 1995) and was soon shown to be required for HIV transactivation (Mancebo et al. 1997; Zhu et al. 1997; Wei et al. 1998). Although the majority of P-TEFb is in the inactive Hexim1 complex, P-TEFb has also been shown to associate with a variety of factors that could help recruit it to chromatin in an active form (Bres et al. 2008; He and Zhou 2011). In this study, we show that the recently discovered SEC version of P-TEFb is a major regulator of rapidly induced genes in development. However, our genome-wide analyses indicate that not all rapidly activated genes require the SEC components investigated in this study. How the different P-TEFb complexes are recruited to regulate distinct sets of genes will be an important area of future investigations.
Materials and methods
ES cell culture
Mouse ES cells (KH2 and V6.5) were cultured under mouse ES complete medium on irradiated mouse embryonic fibroblasts (MEFs). For the shRNA knockdown analysis, viral supernatants were collected and concentrated after 48–72 h of packaging in 293T cells. ES cells were infected with concentrated lentiviral particles with polybrene at the concentration of 4 μg/mL. After 3 d of infection, the ES cells were treated with RA for 6 h and grown one passage off feeders before harvesting for RT-qPCR analysis. For the flavopiridol (Sigma catalog no. F3680) treatment, ES cells were cultured in feeder-free medium and plated at a density of 4 × 105 cells per well in a six-well plate. For the ChIP-seq analysis, cells were grown under feeder-free medium ESGRO (Millipore).
HCT-116 cell culture, serum stimulation, and RNAi treatment
HCT-116 cells were grown in McCoy's 5A medium supplemented with 10% FBS. For serum stimulation, cells were first starved by washing cells twice in PBS, then culturing for 40 h in McCoy's 5A without serum. Cells were then either left untreated or treated with serum for 30 min before harvesting.
Antibodies
Anti-RNA Pol II antibodies were purchased from Covance (8WG16) and from Santa Cruz Biotechnologies (N-20), and H3K4 trimethylation (H3K4me3), AFF4, and ELL2 antibodies were generated in our laboratory as described previously (Lin et al. 2010). A fragment of human CDK9 (amino acids 204–372) was expressed as His tag fusion protein in pET-16b, purified on NTA-agarose according to Qiagen's protocol, and sent to Pocono Rabbit Farm and Laboratory for immunization into rabbits.
ChIP and gene expression analysis
ChIP was performed according to previously described protocols (Wang et al. 2009). RNA for microarray analysis was isolated from ES cells grown on feeder cells. After induction, cells were washed in PBS, trypsinized, and replated onto a fresh plate for 30 min. Unbound cells were harvested and total RNA was extracted with Trizol. For RT-qPCR, RNA was isolated with the RNeasy (Qiagen) kit, treated with DNase I, and repurified with RNeasy. Hox expression assays were purchased from Applied Biosystems in a custom TaqMan array card. Five reference controls were experimentally validated using geNorm. Gapdh and Tbp were found to be stably expressed and were used for normalization. Relative expression levels were determined using the comparative cycle threshold method. ChIP-seq and other data analyses can be found in the Supplemental Material. ChIP-seq and expression data have been deposited at the Gene Expression Omnibus (GEO) under the accession number GSE30268.
Acknowledgments
We thank Tari Parmely and the Stowers Institute Tissue Culture Facility for assistance with mouse ES cells and other cell culture needs, and the Molecular Biology Facility for Illumina sequencing and help with qPCR analysis. We are also grateful to Laura Shilatifard for editorial assistance. We thank Joaquin Espinosa for providing HCT-116 cells. This work was performed to fulfill, in part, requirements for C.L.'s and B.D.K.'s PhD thesis research as students registered with the Open University. R.K. and A.S. are supported in part by funds provided by the Stowers Institute for Medical Research. This study was also supported by funds provided by the Alex's Lemonade Stand Foundation and funds from the National Institute of Health (R01CA150265) to A.S.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2059211.
References
- Abu-Abed S, Dolle P, Metzger D, Beckett B, Chambon P, Petkovich M 2001. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev 15: 226–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander T, Nolte C, Krumlauf R 2009. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol 25: 431–456 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Levine M 2009. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325: 471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bres V, Yoh SM, Jones KA 2008. The multi-tasking P-TEFb complex. Curr Opin Cell Biol 20: 334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao SH, Price DH 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276: 31793–31799 [DOI] [PubMed] [Google Scholar]
- Cheng B, Price DH 2007. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem 282: 21901–21912 [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol 17: 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D, Dolle P 1989. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J 8: 1497–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G 2008. Retinoic acid synthesis and signaling during early organogenesis. Cell 134: 921–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Espinosa JM 2011. Lessons on transcriptional control from the serum response network. Curr Opin Genet Dev 21: 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavalas A, Trainor P, Ariza-McNaughton L, Krumlauf R 2001. Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development 128: 3017–3027 [DOI] [PubMed] [Google Scholar]
- Graham A, Papalopulu N, Krumlauf R 1989. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 57: 367–378 [DOI] [PubMed] [Google Scholar]
- He N, Zhou Q 2011. New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC). J Neuroimmune Pharmacol 6: 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38: 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Peterlin BM 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem 63: 717–743 [DOI] [PubMed] [Google Scholar]
- Kong SE, Banks CA, Shilatifard A, Conaway JW, Conaway RC 2005. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc Natl Acad Sci 102: 10094–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M 2011. Paused RNA polymerase II as a developmental checkpoint. Cell 145: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37: 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev 11: 2633–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 270: 12335–12338 [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R 1992. Homeobox genes and axial patterning. Cell 68: 283–302 [DOI] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A 2010. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer 10: 721–728 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K 2008. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle 7: 1539–1544 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K 2011. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 1809: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R 1995. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 81: 1031–1042 [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [DOI] [PubMed] [Google Scholar]
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H 2001. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev 15: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 1998. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J 12: 1437–1446 [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW 1996. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271: 1873–1876 [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW 2003. The RNA polymerase II elongation complex. Annu Rev Biochem 72: 693–715 [DOI] [PubMed] [Google Scholar]
- Simone F, Polak PE, Kaberlein JJ, Luo RT, Levitan DA, Thirman MJ 2001. EAF1, a novel ELL-associated factor that is delocalized by expression of the MLL–ELL fusion protein. Blood 98: 201–209 [DOI] [PubMed] [Google Scholar]
- Sims RJ 3rd, Belotserkovskaya R, Reinberg D 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 25: 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 38: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R 1998. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 125: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD 1994. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci 91: 12110–12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29: 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92: 451–462 [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67: 545–579 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97: 41–51 [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19: 535–545 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Yik JH 2006. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev 70: 646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev 11: 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138: 1122–1136 [DOI] [PubMed] [Google Scholar]