Abstract
Autophagy is a conserved cellular process for bulk degradation of intracellular protein and organelles in lysosomes. In contrast to elegant studies of beclin1 using mouse models and cultured cells demonstrating a tumor suppression function for autophagy, knockout of other essential autophagy proteins such as ATG5, ATG7, or FIP200 (FAK family-interacting protein of 200 kDa) in various tissues did not lead to malignant tumor development in vivo. Here, we report that inhibition of autophagy by FIP200 ablation suppresses mammary tumor initiation and progression in a mouse model of breast cancer driven by the PyMT oncogene. Deletion of FIP200 resulted in multiple autophagy defects including accumulation of ubiquitinated protein aggregates and p62/SQSTM1, deficient LC3 conversion, and increased number of mitochondria with abnormal morphology in tumor cells. FIP200 deletion did not affect apoptosis of mammary tumor cells or Ras-transformed mouse embryonic fibroblasts (MEFs), but significantly reduced their proliferation in both systems. We also observed a reduced glycolysis and cyclin D1 expression in FIP200-null mammary tumor cells and transformed MEFs. In addition, gene profiling studies revealed significantly elevated expression of interferon (IFN)-responsive genes in the early tumors of FIP200 conditional knockout mice, which was accompanied by increased infiltration of effector T cells in the tumor microenvironment triggered by an increased production of chemokines including CXCL10 in FIP200-null tumor cells. Together, these data provide strong evidence for a protumorigenesis role of autophagy in oncogene-induced tumors in vivo and suggest FIP200 as a potential target for cancer therapy.
Keywords: autophagy, FIP200, conditional knockout, mouse models, breast cancer
Autophagy is an evolutionarily conserved cellular process for bulk degradation of regions of the cytoplasm through sequestration within double-membrane-bound vesicles called autophagosomes and delivery to lysosomes (Mizushima 2007; Simonsen and Tooze 2009; Yang and Klionsky 2010). It is induced by starvation and other stresses to clear the damaged proteins and organelles and provide cellular energy and building blocks for biosynthesis by enabling the recycle of the degraded cellular components. Thus, autophagy is crucial to maintain cellular nutrient and energy homeostasis, and dysfunctions in autophagy have been associated with a variety of human diseases including cancer (Levine and Kroemer 2008; Mizushima et al. 2008; Virgin and Levine 2009; Kroemer et al. 2010; Rabinowitz and White 2010).
Deficient autophagy was first linked to tumorigenesis by the observation of very frequent monoallelical deletion of the mammalian autophagy gene beclin1 in sporadic human breast cancers and ovarian cancers (Aita et al. 1999) and the demonstration that haploinsufficiency in beclin1 promoted spontaneous malignancies including lung and liver cancers and lymphomas in mouse models (Liang et al. 1999; Qu et al. 2003; Yue et al. 2003). Recent studies using cancer cell lines provided significant mechanistic insights into the increased tumorigenesis upon inhibition of autophagy (Mathew et al. 2007; White and DiPaola 2009). It was found that in apoptosis-defective tumor cells, inhibition of autophagy caused by heterozygous loss of beclin1 or homozygous deletion of Atg5 induced accumulation of p62, damaged mitochondria, and reactive oxygen species (ROS), and increased DNA damage, leading to tumor promotion (Karantza-Wadsworth et al. 2007; Mathew et al. 2007, 2009). Moreover, autophagy inhibition has also been shown to reduce oncogene-induced senescence to increase tumorigenesis (Young et al. 2009).
Contrary to the better characterized tumor-suppressive roles of autophagy, other studies suggested that the prosurvival function of autophagy under stress conditions (e.g., nutrient deprivation, hypoxia, and therapeutic stress) could promote tumor growth and progression (Dalby et al. 2010; Liang and Jung 2010; Roy and Debnath 2010; Tschan and Simon 2010). Pharmacological or genetic inhibition of autophagy has been shown to sensitize tumor cells to the cytotoxic effects of chemotherapy and ionizing irradiation to enhance cancer treatments (Degenhardt et al. 2006; Abedin et al. 2007; Amaravadi et al. 2007; Gonzalez-Polo et al. 2007; Nishikawa et al. 2010). Interestingly, recent studies also suggested that autophagy may facilitate proliferation and survival of oncogenic Ras-transformed cells by promoting cancer cell metabolism, as inhibition of autophagy results in deficient aerobic glycolysis and depletion of tricarboxylic acid cycle metabolites and cellular energy level in these cells (Guo et al. 2011; Lock et al. 2011). Despite these studies using cancer cell lines and immuno-compromised nude mice, the protumorigenesis function of autophagy has not been evaluated directly by using loss-of-function approaches directed at essential autophagy genes in oncogene-driven mouse models of breast or other cancers in vivo.
FIP200 (FAK family-interacting protein of 200 kDa) encodes an evolutionarily conserved protein characterized by a large coiled-coil region containing a leucine zipper motif, which was initially identified based on its interaction with FAK and Pyk2 (Ueda et al. 2000; Abbi et al. 2002; Chano et al. 2002a). It has been shown to regulate a number of intracellular signaling pathways through interactions with other proteins such as TSC1, p53, and PIASy (Gan and Guan 2008; Martin et al. 2008). Several recent studies have also suggested an important function for FIP200 in the regulation of autophagy in mammalian cells. FIP200 was identified as a component of the ULK1–ATG13–FIP200–ATG101 complex and shown to be essential for autophagosome formation (Hara et al. 2008; Ganley et al. 2009; Hara and Mizushima 2009; Hosokawa et al. 2009; Jung et al. 2009; Behrends et al. 2010). Moreover, our recent studies showed that mouse tissue-specific FIP200 deletion in neurons and hematopoietic stem cells (Liang et al. 2010; Liu et al. 2010) caused defective phenotypes overlapping with those observed in mutant mice with deletion of other autophagy genes (e.g., Atg5 and Atg7) in these tissues (Hara et al. 2006; Komatsu et al. 2006, 2007; Mortensen et al. 2010). In this study, we created and analyzed FIP200 conditional knockout in the MMTV-PyMT mouse model of human breast cancer to study the potential protumorigenesis functions of autophagy in vivo. Our studies showed that FIP200 deletion in mammary epithelial cells (MaECs) reduced tumor initiation and progression by both impairing tumor cell proliferation and inducing increased immune surveillance. These results provide strong support for a positive role of autophagy in oncogene-driven tumor models in vivo and also identify FIP200 as a potential therapeutic target based on the recently proposed “autophagy addiction” model in certain cancers (Guo et al. 2011).
Results
Conditional deletion of FIP200 in MaECs suppresses breast cancer initiation, progression, and metastasis
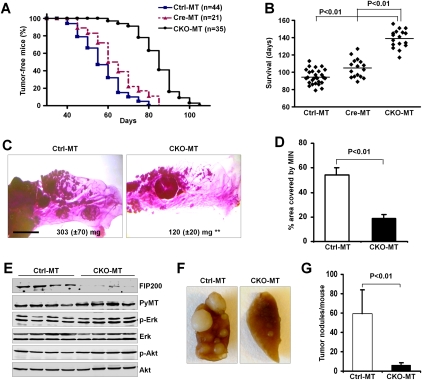
To investigate the potential protumorigenesis functions of autophagy in vivo, we introduced conditional knockout (CKO) of the essential autophagy protein FIP200 into the MMTV-PyMT mouse model of breast cancer. MMTV-PyMT is a well-characterized model that undergoes multistep progression to the rapid appearance of multifocal mammary adenocarcinomas following PyMT-mediated activation of Ras, Src, and PI3K (Guy et al. 1992; Lin et al. 2003). FIP200 CKO mice (Wei et al. 2009) were crossed with MMTV-PyMT mice to generate three cohorts of female mice with the genotypes of FIP200f/f;MMTV-Cre;MMTV-PyMT (designated CKO-MT mice), FIP200f/+;MMTV-Cre;MMTV-PyMT (designated Cre-MT mice; with deletion of FIP200 in one allele in MaECs), and FIP200f/f;MMTV-PyMT (designated Ctrl-MT mice; equivalent to MMTV-PyMT mice as the floxed FIP200 allele does not affect the function of FIP200 or other genes in the absence of Cre). Mammary tumor development in these mice was monitored by physical palpation as described previously (M Luo et al. 2009). As shown in Figure 1A, Ctrl-MT and Cre-MT mice developed palpable mammary tumors rapidly, with mean tumor onset ages (T50) of 56 and 62 d, respectively. Surprisingly, CKO-MT mice showed a significantly increased tumor-free interval with a T50 of 85 d. Moreover, the decreased tumor burden also prolonged the survival time for CKO-MT females as compared with Ctrl-MT and Cre-MT mice (Fig. 1B). Consistent with the decreased tumorigenesis in CKO-MT mice, whole-mount analyses of mammary glands at 10 wk of age revealed a significant reduction of PyMT-induced mammary intraepithelial neoplasia (MIN) lesions in CKO-MT mice compared with Ctrl-MT mice (Fig. 1C). Quantification of multiple samples indicated >60% decrease in the epithelial surface occupied by the hyperplastic nodules for CKO-MT mice compared with that for Ctrl-MT mice (Fig. 1D). Consistent with these results, we found an ∼2.5-fold reduction in the average mass of the mammary glands of CKO-MT mice compared with that of Ctrl-MT mice (Fig. 1C, insets).
Figure 1.
Conditional deletion of FIP200 in MaEC suppresses breast cancer initiation, progression, and metastasis. (A) Kaplan-Meier analysis of mammary tumor development in the Ctrl-MT (n = 44), Cre-MT (n = 21), and CKO-MT (n = 35) mice. CKO-MT versus Ctrl-MT or Cre-MT: P < 0.01 by the log-rank test. (B) Scatter plots showing days of survival for indicated mouse strains before they succumb to their tumor burden. Horizontal line represents the mean. (C) Representative mammary gland whole mounts from 10-wk-old Ctrl-MT (left) and CKO-MT mice (right). The average mass of inguinal mammary glands from Ctrl-MT (left, n = 6) and CKO-MT (right, n = 7) mice are also shown. Bar, 5 mm. (**) P < 0.01. (D) Quantification of the area occupied by hyperplastic lesions expressed as a percentage of the total mammary gland surface. (E) Lysates were prepared from four tumors in four different Ctrl-MT (left four lanes) and CKO-MT (right four lanes) mice and analyzed by immunoblotting using antibodies against various proteins as indicated. (F,G) Lungs were harvested from Ctrl-MT (n = 17) and CKO-MT (n = 12) mice at 7 wk after initial detection of palpable primary tumors. (F) Representative images from these mice. (G) The mean ± SD of the number of metastatic nodules on the surfaces of lungs per mouse.
To ensure that the reduced tumorigenesis in the CKO-MT mice was not due to changes in the expression of PyMT oncoprotein, lysates were prepared from multiple primary tumors and analyzed by Western blotting. Figure 1E shows the abolished expression of FIP200 in the tumors from CKO-MT mice but not those from Ctrl-MT mice, as expected. Comparable levels of PyMT oncoprotein were detected in tumors from both mice. Furthermore, similar activation as measured by their phosphorylation status of PyMT downstream targets Akt and Erk was observed for tumors from CKO-MT and Ctrl-MT mice. Lastly, immunofluorescent staining of tumor sections also showed expression of PyMT oncoprotein at similar levels in CKO-MT and Ctrl-MT mice (data not shown). These results suggest that the reduced tumorigenesis in CKO-MT mice was not due to changes in the expression of PyMT oncoprotein.
We also examined the effect of FIP200 inactivation on metastasis of mammary tumors developed in CKO-MT mice. At 7 wk after the initial detection of palpable primary tumors for both mice, CKO-MT and Ctrl-MT mice were sacrificed and examined for metastatic nodules on the lung surface. As shown in Figure 1F, significantly fewer nodules were found for CKO-MT mice compared with Ctrl-MT mice. Quantification of multiple samples indicated a 10-fold reduction in metastasis for the CKO-MT mice (Fig. 1G). At dissection time, the size of primary tumors was generally smaller in CKO-MT compared with that in Ctrl-MT mice; therefore, we could not exclude that the reduced primary tumor burden in CKO-MT mice may contribute to the decreased metastasis in CKO-MT mice. Together, these results suggest that suppression of autophagy by FIP200 inactivation significantly inhibited mammary tumor initiation, progression, and metastasis, providing support for a protumorigenesis role of autophagy in vivo.
Ablation of FIP200 leads to autophagic defects and decreased proliferation in mammary tumor cells
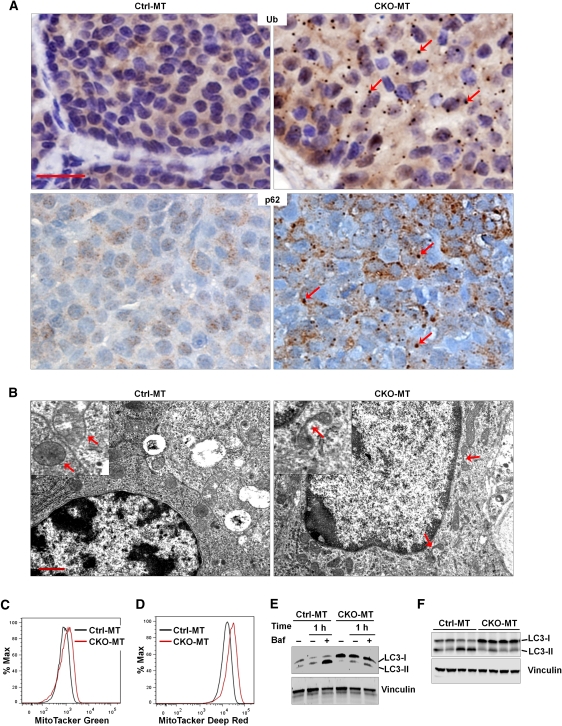
To investigate the cellular and molecular mechanisms of the decreased mammary tumorigenesis in CKO-MT mice, we first evaluated autophagy deficiency in FIP200-null tumors as well as isolated tumor cells. Consistent with its crucial role in autophagy induction (Hara et al. 2008; Ganley et al. 2009; Hara and Mizushima 2009; Hosokawa et al. 2009; Jung et al. 2009) and our previous findings in neurons (Liang et al. 2010), large ubiquitin-positive aggregates and p62-positive aggregates, which are both associated with defective autophagy (Hara et al. 2006; Komatsu et al. 2006; Liang et al. 2010), were found in mammary tumors of CKO-MT mice but not those in Ctrl-MT mice (Fig. 2A). Further analysis of mammary tumors by transmission electron microscopy revealed deformed and branched mitochondria in FIP200-null tumor cells in contrast to the smooth and extended morphology of mitochondria in control tumor cells (Fig. 2B, arrows). Similar to findings in other autophagy-deficient cells by inactivation of various autophagy genes (Komatsu et al. 2005; Nakai et al. 2007; Ebato et al. 2008), we also detected an increased number of mitochondria per nucleus in FIP200-null mammary tumor cells (25 ± 3) compared with that in Ctrl-MT mice (14 ± 2) (Fig. 2B). We next examined the number and respiratory status of mitochondria in isolated tumor cells by staining with the MitoTracker Green (for total) and MitoTracker Deep Red (for live or respiring), respectively, followed by flow cytometry. We found an increased accumulation of both total and healthy mitochondria mass in FIP200-null tumor cells compared with control cells (Fig. 2C,D). Lastly, a reduced LC3 conversion was observed in FIP200-null mammary tumor cells under starvation condition in the presence of bafilomycin A1, an inhibitor for the vacuolar H+ ATPase that can block autophagosome–lysosome fusion and thus LC3-II degradation (Fig. 2E; Yamamoto et al. 1998). The defective autophagosome formation was also found in CKO-MT tumors in vivo, as measured by accumulation of LC3-I (i.e., reduced conversion of LC3-I to LC3-II) in lysates prepared from tumors of CKO-MT mice compared with those from Ctrl-MT mice (Fig. 2F). Together, these results demonstrate that FIP200 deletion results in autophagy defects in mammary tumor cells.
Figure 2.
Ablation of FIP200 leads to autophagic defects in mammary tumor cells. (A) Sections from mammary tumors of Ctrl-MT and CKO-MT mice were analyzed by immunohistochemistry using anti-ubiquitin (top panels) or anti-p62. Note the ubiquitin- and p62-positive aggregates in tumor cells from CKO-MT mice (red arrows, right panels), but not Ctrl-MT mice (left panels). Bars, 25 μm. (B) Mammary tumor sections of Ctrl-MT and CKO-MT mice were examined by transmission electron microscopy. Note the abnormal morphology of mitochondria in CKO-MT tumor cells (red arrows, right panel). Bar, 1 μm. (C,D) Mammary tumor cells from CKO-MT or Ctrl-MT mice were analyzed for total mitochondrial mass by MitoTracker Green staining (C) or for respiring mitochondria mass by MitoTracker Deep Red staining (D). (E) Mammary tumor cells from CKO-MT or Ctrl-MT mice were incubated in the complete or starvation medium (HBSS) for 1 h with or without 100 nM bafilomycin A1 (Baf), as indicated. The cell lysates were then prepared and analyzed by immunoblotting using anti-LC3 antibody. (F) Lysates were prepared from four tumors in four different Ctrl-MT (left four lanes) and CKO-MT (right four lanes) mice and analyzed by immunoblotting using antibodies against various proteins as indicated.
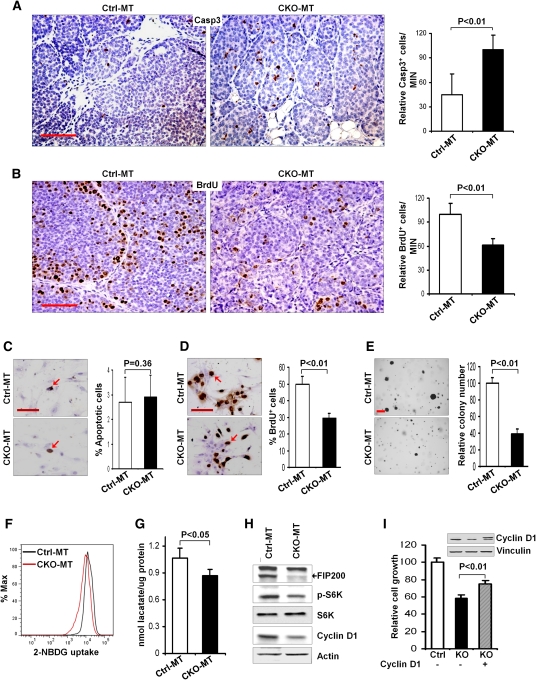
Given the previous findings of the prosurvival function of autophagy (Degenhardt et al. 2006; Hara et al. 2006; Komatsu et al. 2006), it is possible that defective autophagy upon FIP200 ablation increases apoptosis of tumor cells to inhibit tumorigenesis in CKO-MT mice. Consistent with previous findings in conditional deletion of FIP200 in neurons (Liang et al. 2010), examination of MIN lesions by cleaved caspase-3 staining showed an increased apoptosis in CKO-MT tumors compared with Ctrl-MT tumors (Fig. 3A). Analysis by BrdU incorporation assays also indicated a reduction of tumor cell proliferation in CKO-MT mice (Fig. 3B). Surprisingly, however, we did not detect significant difference in apoptosis between mammary tumor cells isolated from primary tumors of CKO-MT and Ctrl-MT mice, as determined by TUNEL analysis (Fig. 3C), suggesting that the increased apoptosis in vivo is likely due to multiple unique microenvironmental stresses (e.g., increased immune surveillance; see below) not present in ex vivo tumor cell cultures. In contrast, measurement of cell cycle progression by BrdU incorporation assay in primary tumor cell culture showed a significant decrease in proliferation of mammary tumor cells from CKO-MT mice compared with those from Ctrl-MT mice (Fig. 3D). CKO-MT tumor cells also showed a significant decrease in anchorage-independent growth in soft agar compared with Ctrl-MT tumor cells (Fig. 3E). Interestingly, we also observed a decreased glucose uptake and intracellular lactate production in CKO-MT tumor cells (Fig. 3F,G), which is consistent with a recent study showing that inhibition of autophagy reduced proliferation as well as glycolysis in Ras-transformed cells (Lock et al. 2011). Lastly, similar to the observation in CKO-MT mice, although they appear grossly normal with functional mammary glands for nursing (Wei et al. 2009), CKO mice also exhibited a reduced proliferation in terminal end buds (TEBs) of developing mammary glands as measured by BrdU incorporation and consequently retarded mammary ductal outgrowth compared with Ctrl mice (Supplemental Fig. S1), supporting a role of FIP200 in promoting cell proliferation.
Figure 3.
Inactivation of FIP200 decreases proliferation of mammary tumor cells. (A,B) The apoptosis and proliferation of tumor cells in MIN lesions of Ctrl-MT and CKO-MT mice in vivo were measured by immunohistochemistry using anti-cleaved caspase 3 (A) and BrdU incorporation assays (B), respectively. Bars, 50 μm. Data on the right side graphs are mean ± SD (n = 4 for each sample) of relative numbers of cleaved caspase 3+ or BrdU+ cells in MIN lesions. (C–E) Mammary tumor cells isolated from Ctrl-MT and CKO-MT mice were measured for apoptosis using TUNEL assay (C), proliferation by BrdU incorporation assay (D), and anchorage-independent growth in soft agar (E). Data on the right side graphs are mean ± SD. Bars: C,D, 50 μm; E, 300 μm. (F,G) CKO-MT and Ctrl-MT tumor cells were analyzed for the level of glucose uptake (2-NBDG uptake) following 6 h of incubation (F) or for intracellular level of lactate in growth medium (G), as described in the Materials and Methods. (H) Lysates were prepared from mammary tumor cells isolated from Ctrl-MT and CKO-MT mice and analyzed by immunoblotting using antibodies against various proteins as indicated. (I) Tumor cells from FIP200f/f;MMTV-PyMT mice were infected by recombinant adenoviruses encoding Cre (Ad-Cre) or a control insert (Ad-lacZ) to produce FIP200 KO and Ctrl tumor cells, respectively. These tumor cells were then infected with recombinant retroviruses encoding HA-tagged cyclin D1 (cyclin D1) or control viruses (Mock) to prepare stable pools of the infected cells. After 6 d of incubation in growth medium, the cells were counted, and the mean ± SD of relative growth is shown. The inset shows expression of HA-tagged ectopic cyclin D1 in KO cells (right lane) but not vector-infected Ctrl cells (left lane) or KO cells (middle line).
Previous studies suggested that FIP200 can bind to the TSC1–TSC2 complex and inhibit its activity, leading to the activation of mTOR and its target S6K, and that FIP200 down-regulation causes compromised mTOR activation (Gan et al. 2005). To study mTOR and other signaling pathways in FIP200-null mammary tumor cells, lysates were prepared from the isolated tumor cells and analyzed by Western blotting (Fig. 3E). Similar to direct analysis of tumor lysates (see Fig. 1E), FIP200 expression was abolished in the tumor cells from CKO-MT mice. Consistent with previous results in other cells (Gan et al. 2005, 2006), decreased phosphorylation of S6K was found in CKO-MT tumor cells compared with Ctrl-MT tumor cells. However, direct analysis of tumor lysates only detected a slight, but statistically insignificant, decrease in phosphorylation of S6K in tumors from CKO-MT mice compared with those from Ctrl-MT mice (Supplemental Fig. S2). No apparent differences in the activation status of Erk or Akt were detected between tumor cells from CKO-MT and Ctrl-MT mice (data not shown). Lastly, consistent with the reduced proliferation, the expression level of cyclin D1 was decreased in the tumor cells from CKO-MT mice compared with those from Ctrl-MT mice. Moreover, ectopic expression of cyclin D1 in FIP200-null tumor cells partially rescued the growth of CKO-MT tumor cells (Fig. 3I), suggesting that reduced cyclin D1 expression upon FIP200 deletion could contribute to the intrinsic defective cell growth of FIP200-null tumor cells. Together, these data suggested that defective autophagy upon FIP200 ablation could lead to reduced cell proliferation in mammary tumor cells, contributing to the suppression of tumorigenesis in CKO-MT mice.
Deletion of FIP200 reduces proliferation of Ras-transformed primary mouse embryonic fibroblasts (MEFs)
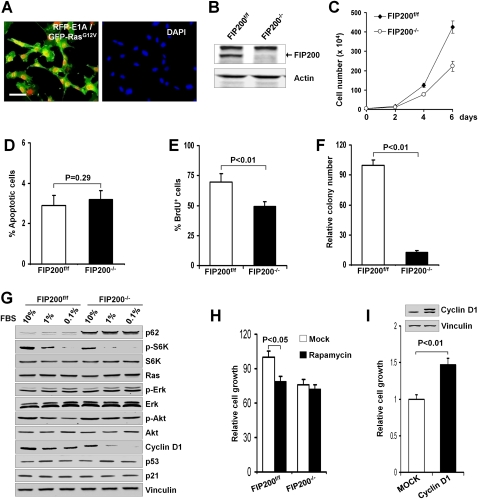
While consistent with the reduced tumorigenesis of CKO-MT mice, the reduced proliferation of FIP200-null mammary tumor cells appears to be in apparent conflict with our previous results showing inhibition of cell cycle progression by overexpression of FIP200 in breast cancer cell lines, although the effect of FIP200 inactivation in these cell lines was not examined directly (Melkoumian et al. 2005). To address whether endogenous FIP200 may also play a positive role in oncogenesis in other cell types, we investigated the effect of the genetic ablation of FIP200 on transformation of MEFs by oncogenic Ras. Primary MEFs isolated from embryos of floxed FIP200 mice (i.e., FIP200f/f MEFs) (Gan et al. 2006) were immortalized by infection with a retrovirus encoding E1A, followed by infection with a retrovirus encoding the oncogenic Ha-RasV12, as described previously (Deng et al. 2005; Sun et al. 2007). Coexpression of E1A and oncogenic Ras in the transformed FIP200f/f MEFs is shown in Figure 4A. These cells were then infected by recombinant adenoviruses encoding Cre recombinase (Ad-Cre) or LacZ (Ad-LacZ) as a control to generate E1A/RasV12-transformed MEFs with FIP200−/− and FIP200f/f alleles (designated FIP200 KO and control cells, respectively). As shown in Figure 4B, efficient deletion of the FIP200 gene and removal of endogenous FIP200 protein was obtained by Ad-Cre infection compared with that infected by Ad-LacZ.
Figure 4.
Analysis of FIP200 deletion in Ras-transformed primary MEFs. (A) Primary MEFs from FIP200 floxed mice were infected with retroviral vectors encoding RFP-E1A and GFP-Ha-RasV12. After staining with DAPI, the cells were viewed under a fluorescent microscope for the expression of E1A and RasV12 (left) and nuclei (right). Bar, 50 μm. (B–F) RFP-E1A/GFP-RasV12-transformed primary MEFs from FIP200 floxed mice were infected with recombinant adenoviruses encoding Cre (Ad-Cre) or a control insert (Ad-lacZ) to produce FIP200 KO (FIP200−/−) and control (FIP200f/f) cells, respectively. (B) Lysates from the cells were analyzed by immunoblotting using antibodies against FIP200 or actin as indicated. The cells were measured for multiplication (C), apoptosis using TUNEL assay (D), proliferation by BrdU incorporation assay (E), and anchorage-independent growth in soft agar (F). Data in C–F are mean ± SD. (G) FIP200 KO (FIP200−/−) and control (FIP200f/f) cells were grown in medium containing the indicated concentration of serum for 24 h. Cell lysates were then prepared and analyzed by immunoblotting using antibodies against various proteins as indicated. (H) FIP200 KO (FIP200−/−) and control (FIP200f/f) cells were incubated in growth medium in the presence or absence (Mock) of rapamycin (100 nM), as indicated. After 24 h, the cells were counted, and the mean ± SD of relative growth is shown. (I) FIP200 KO cells were infected by recombinant retroviruses encoding HA-tagged cyclin D1 (cyclin D1) or control viruses (Mock) to prepare stable pools of the infected cells. After 3 d of incubation in growth medium, the cells were counted, and the mean ± SD of relative growth is shown. (Inset) Expression of ectopic cyclin D1 in cyclin D1 cells (right lane) but not Mock cells (left lane).
The effect of FIP200 inactivation in the transformed MEFs was first assessed by examining multiplication of the cells in vitro. Figure 4C shows an ∼50% decrease in the number of FIP200 KO cells compared with control cells over a period of 6 d of culture under normal conditions. We next investigated the effects of FIP200 inactivation on the proliferation and apoptosis of the transformed MEFs to determine their possible contributions to the reduced growth of FIP200 KO cells in culture. Analysis of cell apoptosis by TUNEL assays showed little difference between FIP200 KO and control cells (Fig. 4D). Figure 4E shows a decreased cell cycle progression of FIP200 KO cells in comparison with control cells, as measured by BrdU incorporation. In addition, analysis of anchorage-independent growth in soft agar showed a significant reduction in the number of colonies (Fig. 4F) as well as their average size (data not shown) from FIP200 KO cells compared with control cells. Similar to FIP200-null mammary tumor cells, we found a decreased glucose uptake and intracellular lactate production in FIP200 KO cells compared with control cells (Supplemental Fig. S3A,B). Also consistent with previous results (Lock et al. 2011), FIP200 KO cells were insensitive to reduced glucose availability, whereas it decreased proliferation of control cells (Supplemental Fig. S3C). Together, these results suggested that abrogation of endogenous FIP200 expression also inhibited cell proliferation in Ras-transformed primary MEFs.
To further examine the potential mechanisms of reduced cell proliferation upon FIP200 deletion in Ras-transformed MEFs, we analyzed the activation status of signaling pathways downstream from Ras as well as those known to be affected by FIP200. Lysates were prepared from FIP200 KO and control cells grown in media containing decreasing concentrations of serum and then used for Western blotting with various antibodies (Fig. 4G). As expected, p62 showed increased accumulation in FIP200 KO cells compared with control cells, consistent with defective autophagy after FIP200 deletion (Hara et al. 2008; Ganley et al. 2009; Hara and Mizushima 2009; Hosokawa et al. 2009; Jung et al. 2009). Similar to the observation in mammary tumor cells (see Fig. 3E) and in other cells previously (Gan et al. 2005), phosphorylation of S6K was significantly decreased in FIP200 KO cells, suggesting reduced mTOR signaling upon deletion of FIP200 in these cells. Consistent with the idea that reduced mTOR signaling could contribute to the decreased proliferation of FIP200 KO cells, further inhibition of mTOR by rapamycin did not affect proliferation of these cells, although it readily inhibited the control cells as expected (Fig. 4H). As expected, deletion of FIP200 did not affect the expression level of Ras itself. Moreover, neither Erk nor Akt signaling pathways were affected in FIP200 KO cells compared with control cells, although a slight increase of Akt phosphorylation was found in FIP200 KO cells at 0.1% serum. These data suggest that deletion of FIP200 did not reduce cell proliferation through these major Ras downstream pathways. The increased phosphorylation of Akt in FIP200 KO cells in low serum concentrations might be caused by feedback mechanisms of the deregulated mTOR pathway (Huang and Manning 2009). Although our previous studies suggested that FIP200 can interact with p53 and stabilize p53 protein when overexpressed in breast cancer cells (Melkoumian et al. 2005), we did not observe differences in the expression of p53 or its target p21 between FIP200 KO and control cells. However, we found that, similar to mammary tumor cells, the level of cyclin D1 expression was markedly reduced in FIP200 KO cells under different serum concentrations, which is consistent with their decreased proliferation (see Fig. 4D). Moreover, expression of ectopic cyclin D1 rescued the proliferation defect in FIP200 KO cells (Fig. 4I). Together, these studies suggest that, in agreement with mammary tumor cells, FIP200 deletion could lead to reduced proliferation of transformed primary MEFs.
Increased expression of immune responsive genes in mammary tumors of CKO-MT mice
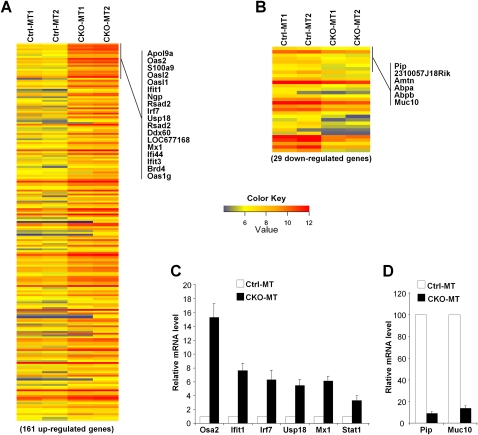
To further explore potential cellular and molecular mechanisms by which deletion of FIP200 suppressed mammary tumorigenesis, we examined the gene expression profile of mammary tumor tissues by microarray analysis. Mammary tumors at early stages from CKO-MT or Ctrl-MT mice were isolated and pooled for mRNA preparation and reverse transcription to generate probes for the Affymetrix gene chip mouse genome 430 2.0 array. Two independent samples were generated for CKO-MT mice (each from a pool of four mice) and analyzed against two samples from Ctrl-MT mice (each from a pool of four mice). The microarray analyses identified 161 up-regulated genes (by 209 probe sets) and 29 down-regulated genes (by 31 probe sets) with at least twofold changes in tumors from CKO-MT mice compared with those from Ctrl-MT mice (Fig. 5A,B; Supplemental Tables 1, 2). Further evaluation of the data using Gene Ontology (Ashburner et al. 2000) indicated that the most prominent feature of the genome-wide alterations of CKO-MT tumors is the increased expression of genes encoding proteins involved in response to IFN stimulation and other immune responses (Supplemental Fig. S4). The up-regulated genes linked to IFNs include the transcription factors Irf7, Stat1, and Stat2; the GTPases Mx1 and Mx2; the 2′,5′-oligoadenylate synthetases Oas2, Oasl2, Oasl1, and Oas3; and the IFN-induced proteins with tetratricopeptide repeats Ifit1–Ifit3. We next performed quantitative RT–PCR (qRT–PCR) analyses to verify these results for a subset of genes (both up-regulated and down-regulated) using the same set of mRNAs. We found that the relative expression levels of a wide range of IFN-responsive genes, including Stat1, Mx1, Usp18, lrf7, Ifit1, and Osa2, are up-regulated from threefold to 15-fold in CKO-MT tumors in relative to Ctrl-MT tumor samples (Fig. 5C). Consistent with microarray data, we found that the expression levels of two down-regulated genes, Pip and Muc10, are severely decreased in CKO-MT tumors compared with Ctrl-MT tumor samples (Fig. 5D). Considering that up-regulated IFN signaling plays an important role in anti-tumor immune responses (Dunn et al. 2006; Swann and Smyth 2007), these results suggested that FIP200 deletion might trigger enhanced anti-tumor immune surveillance and contribute to the suppression of mammary tumorigenesis and progression in these mice.
Figure 5.
Global changes in gene expression profile of mammary tumors from CKO-MT mice. (A,B) Heat map of genes showing differential up-regulation (A) or down-regulation (B) of at least twofold in tumors from CKO-MT mice compared with those from Crtl-MT mice. Representative up-regulated and down-regulated genes are marked on the right. (C,D) A select group of up-regulated genes were validated using qRT–PCR. The mean ± SD (n = 3) of relative levels (normalized to Ctrl-MT tumors) from three independent experiments is shown.
Increased immune cell infiltration in mammary tumors of CKO-MT mice
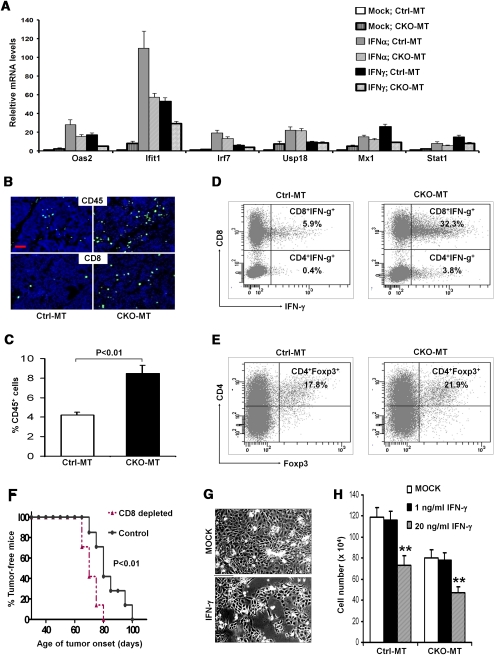
Our microarray analyses revealed that many target genes of type I (IFN-α/β) and type II (IFN-γ) IFN were highly elevated in mammary tumors from CKO-MT mice. The increased expression of these genes could be due to an increased tumor cell response to IFNs and/or an increased production of IFNs in the tumor microenvironment of these mice. To examine the former possibility that FIP200 may be involved in the IFN-responsive pathway per se, mammary tumor cells were isolated from CKO-MT and Ctrl-MT mice and examined for their responses to IFN stimulation in culture. Upon stimulation with IFN-α or IFN-γ (Fig. 6A), expression of several genes including Oas2, Ifit1, Irf7, Usp18, Mx1, and Stat1 did not display a larger increase, but some even decreased response; e.g., Ifit1, in tumor cells from CKO-MT mice compared with those from Ctrl-MT mice. Therefore, these results suggested that the increased expression of IFN-responsive genes is not caused by increased sensitivity of tumor cells to IFN stimulation in CKO-MT mice.
Figure 6.
Analysis of immune responsive genes and infiltration of effector T cells in mammary tumors of CKO-MT mice. (A) Mammary tumor cells were isolated from Ctrl-MT and CKO-MT mice. They were cultured in vitro and then treated with 10 ng/mL IFN-α or IFN-γ, or media mock as a control, as indicated. The expression levels of several IFN-responsible genes were analyzed by qRT–PCR. The mean ± SD of relative levels (normalized to mock Ctrl-MT tumor cells) is shown. (B) Cryosections of mammary tumors from Ctrl-MT (left) and CKO-MT (right) mice were immunostained with antibody against CD45 (top) or CD8 (bottom) to detect infiltrated leukocytes (shown by green FITC fluorescence). Nuclei were counterstained with DAPI (blue). Bar, 100 μm. (C) Freshly isolated immune cells from mammary tumors in Ctrl-MT and CKO-MT mice were analyzed by flow cytometry for CD45+ leukocytes. (D,E) Freshly isolated immune cells from mammary tumors in Ctrl-MT and CKO-MT mice were analyzed for subpopulations of T cells by flow cytometry. CD3 gated T cells were measured for the fraction of CD8+IFN-γ+ and CD4+IFN-γ+ cells (D) or CD4+Foxp3+ regulatory T cells (E). (F) Kaplan-Meier analysis of mammary tumor initiation in CD8+ cells depleted of CKO-MT (n = 7) and IgG control-treated CKO-MT (n = 7) mice. P < 0.01 by the log-rank test. (G,H) Mammary tumor cells from Ctrl-MT or CKO-MT mice were incubated in growth medium in the absence (Mock) or presence of IFN-γ (1 ng/mL or 20 ng/mL), as indicated. (G) Representative image of CKO-MT tumor cells incubated in Mock or 20 ng/mL IFN-γ after 3 d. Bar, 25 μm. (H) The cells were counted after 6 d of incubation, and the mean ± SD of the number of cells (5 × 104 at day 0) is shown. (**) P < 0.01.
We then investigated the latter possibility that more IFN-producing immune cells may infiltrate tumor tissues of CKO-MT mice. We first examined the presence of CD45+ leukocytes in the tumors of CKO-MT and Ctrl-MT mice. The levels of leukocytes were significantly increased in the tumor tissues of CKO-MT mice over those of Ctrl-MT mice (Fig. 6B, top panels). Given the crucial function of cytotoxic T cells in controlling tumor growth, we next compared the infiltration of CD8+ T cells in the tumors of these mice. While only a few CD8+ T cells were detected in the tumors from Ctrl-MT mice, the tumors from CKO-MT mice were infiltrated with significantly higher amounts of CD8+ T cells (Fig. 6B, bottom panels). To further investigate the functional subsets of tumor-infiltrating immune cells, single cell suspensions were prepared from mammary tumors (Araki et al. 2009) and analyzed by a flow cytometry analyzer. Consistent with data by immunofluorescent staining, there was a significantly higher percentage of CD45+ cells in the tumors from CKO-MT mice compared with those from Ctrl-MT mice (Fig. 6C). We further observed significantly increased fractions of both IFN-γ+CD8+ and IFN-γ+CD4+ T cells (Th1) in CKO-MT samples compared with Ctrl-MT samples (Fig. 6D). It is known that regulatory T cells (Tregs) contribute to inactivation of antigen-presenting cells and suppression of effector cell functions including IFN-γ-producing Th1 cells, CD8+ cytotoxic T lymphocytes (CTLs), and NK cells (Zou 2006). Th17 cells, a new subset of CD4+ T cells expressing IL-17, might also regulate tumor development (Dong 2008; Zou and Restifo 2010). We did not observe a significant difference in the infiltration of these T-cell subsets (Fig. 6E; data not shown) in CKO-MT and Ctrl-MT samples. Together, these results suggested that the increased infiltration of IFN-γ-producing CD8+ and CD4+ T cells may be responsible for the up-regulation of IFN-γ-responsive genes observed by microarray analyses in the mammary tumors of CKO-MT mice.
The host immune responses and immune surveillance have been widely recognized as a critical barrier to the development and progression of various human cancers including breast cancer (Dunn et al. 2004; Koebel et al. 2007; Finn 2008). While several types of immune cells participate in cancer immune surveillance, the CD8+ effector T cells play a critical role in the elimination of tumors (Dunn et al. 2004; Zou 2006). To examine whether the increased infiltrating CD8+ T cells indeed contribute to suppressed mammary tumor initiation and progression in CKO-MT mice, we depleted CD8+ T cells using specific antibodies in these mice (Supplemental Fig. S5). As shown in Figure 6F, CKO-MT mice in the IgG isotype control-treated group developed mammary tumors with a similar kinetics as the untreated mice (see Fig. 1A). In contrast, CD8+ T-cell-depleted CKO-MT mice showed an accelerated mammary tumor initiation. Moreover, we found that IFN-γ inhibited growth of mammary tumor cells from CKO-MT mice by about a similar degree as those from Ctrl-MT mice in vitro (Fig. 6G,H). Together, these results suggest that the increased infiltration of IFN-γ-producing CD8+ T cells plays an important role in the suppression of mammary tumorigenesis in CKO-MT mice.
FIP200 deletion leads to increased chemokine production in mammary tumor cells
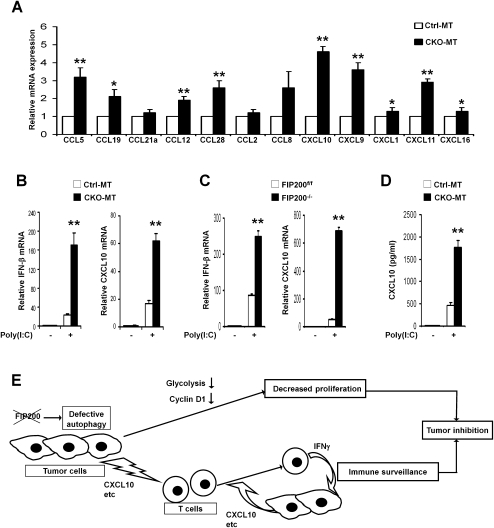
Chemokines play crucial roles in the recruitment of T cells into the tumor microenvironment (Khan et al. 2000; Frigerio et al. 2002; Zou 2005). Our microarray data revealed that CXCL9, CXCL10, and CXCL11, which can attract activated T cells (Farber 1997; Yu et al. 2004; Kryczek et al. 2009), as well as a number of other chemokines are up-regulated in CKO-MT tumors compared with Ctrl-MT tumors (Supplemental Table 3). We confirmed with qRT–PCR that the levels of CXCL9, CXCL10, and CXCL11 were elevated by threefold to fivefold in the purified primary tumor cells from CKO-MT mice compared with those from Ctrl-MT mice (Fig. 7A). Consistent with these results, we found enhanced migration of tumor-associated CD8+ T cells in response to the extracts of tumor tissues from CKO-MT mice compared with those from Ctrl-MT mice as well as an increased amount of CXCL10 in the extracts from CKO-MT mice (Supplemental Fig. S6). These results suggest that the enhanced immune surveillance in CKO-MT mice may be mediated by effector T cells and Th-1-type chemokines produced by FIP200-null tumor cells.
Figure 7.
Increased chemokine production in FIP200-null tumor cells and a working model. (A) Mammary tumor cells were isolated from Ctrl-MT or CKO-MT mice. RNA was isolated from the purified cells and subjected to analysis by qRT–PCR to detect the expression of various chemokines as indicated. The mean ± SD of relative levels (normalized to Ctrl-MT tumors) from three independent experiments is shown. (*) P < 0.05; (**) P < 0.01. (B,C) Mammary tumor cells from Ctrl-MT or CKO-MT mice (B) and FIP200 KO (FIP200−/−) or control (FIP200f/f) cells (C) were transfected with 10 μg/mL poly(I:C) or mock using Lipofectamine. RNA was isolated at 8 h after stimulation and subjected to qRT–PCR analysis for the relative level of IFN-β (left) and CXCL10 (right). (**) P < 0.01. (D) Mammary tumor cells from Ctrl-MT or CKO-MT mice were stimulated by poly(I:C) for 8 h as described in B. The amount of CXCL10 in the medium was then measured by ELISA using a kit (R & D Systems). (**) P < 0.01. (E) A working model summarizing the potential mechanisms of suppression of mammary tumorigenesis and progression in CKO-MT mice. Inactivation of FIP200 results in defective autophagy in tumor cells that may trigger multiple events leading to the inhibition by mammary tumorigenesis through both reduced cell proliferation and increased immune surveillance in the host.
Recent studies suggested that autophagy deficiency could sensitize cellular responses via the RIG-I-like receptor (RLR) signaling to induce type I IFN production (Jounai et al. 2007; Tal et al. 2009; Levine et al. 2011). Moreover, defective autophagy has also been shown to synergize with environmental factors including viral infections in inflammatory diseases in an IFN-γ-dependent manner (Cadwell et al. 2008, 2010; Saitoh et al. 2008). As inflammatory chemokines like CXCL10 are known targets of RLR signaling (Borden et al. 2007), it is possible that suppression of autophagy by FIP200 deletion may lead to enhanced production of CXCL10 through an increased sensitivity to environmental stimuli such as viral infection or dsRNAs released by dead tumor cells (Zitvogel et al. 2010). To investigate such a possibility, mammary tumor cells from CKO-MT and Ctrl-MT mice were treated with poly(I:C), a synthetic dsRNA. Consistent with previous studies in ATG5-null cells (Jounai et al. 2007; Tal et al. 2009), a significantly increased expression of IFN-β is detected in CKO-MT tumor cells compared with Ctrl-MT tumor cells upon poly(I:C) stimulation (Fig. 7B, left). As expected, an increased response in the IFN target gene CXCL10 was also found in CKO-MT tumor cells (Fig. 7B, right). A similar sensitized response was found in FIP200 KO MEFs in comparison with control cells (Fig. 7C). Lastly, ELISA analysis verified the corresponding increase in protein expression and secretion of CXCL10 by CKO-MT tumor cells (Fig. 7D). Together, these results suggest that FIP200 deletion resulted in increased chemokine production and secretion by tumor cells, which could lead to the increased infiltration of CD8+ effector T cells contributing to the suppression of mammary tumorigenesis in CKO-MT mice.
Discussion
Consistent with a well-established tumor suppression function of autophagy based on studies using tumor cell lines and mouse models (Liang et al. 1999; Qu et al. 2003; Yue et al. 2003; Mathew et al. 2009), earlier work showed that deletion of the FIP200 gene was found in a fraction of primary mammary tumor samples (Chano et al. 2002b) and that overexpression of FIP200 inhibited cell cycle progression in several breast cancer cell lines (Melkoumian et al. 2005). Despite these early data implicating a role of FIP200 in suppressing breast cancer, our recent analyses using mouse models indicated that deletion of FIP200 did not lead to spontaneous development of breast cancer or augment lymphomagenesis upon inactivation of p53 (Wei et al. 2009), casting doubts on its putative tumor suppressor function. Here, we present data showing that suppression of autophagy by FIP200 ablation in MaECs suppressed mammary tumor initiation and progression driven by the PyMT oncoprotein in a well-characterized mouse model of human breast cancer. Complementing a number of recent studies using cancer cells and xenograft models (Guo et al. 2011; Lock et al. 2011), our results showing a positive role for FIP200 using mouse models of human breast cancer provide the first direct demonstration of a protumorigenesis role of autophagy in oncogene-induced tumorigenesis and progression in animals with an intact immune system.
Our analysis of FIP200-null mammary tumor cells from the MMTV-PyMT mouse model as well as Ras-transformed primary MEFs provides several lines of evidence that deficient cell proliferation contributes to the suppressed tumor formation and progression upon deletion of FIP200. Primary tumor cells isolated from CKO-MT mice, in comparison with those from Ctrl-MT mice, exhibited significantly reduced proliferation without apparent difference in apoptosis in vitro. Consistent with these data, decreased cell cycle progression as measured by BrdU incorporation was also found in early primary tumors in CKO-MT mice. Moreover, reduced proliferation was found in TEBs of developing mammary glands with a correspondingly retarded mammary ductal outgrowth in CKO mice compared with Ctrl mice. Lastly, deletion of FIP200 in E1A/Ras-transformed primary MEF cells also reduced their proliferation. While these data suggest a role of FIP200 in promoting tumor cell proliferation that at least partially accounts for the suppressed tumorigenesis in CKO-MT mice, it should be noted that our previous studies showed an inhibition of cell cycle progression by overexpression of FIP200 in breast cancer cell lines and fibroblasts (Abbi et al. 2002; Melkoumian et al. 2005). This earlier study suggesting a negative role of FIP200 in cell proliferation appears to be in apparent conflict with data presented in the present study, although the effect of FIP200 inactivation in cultured cell lines was not previously examined directly. It is possible that FIP200 may function differently in its regulation of cell proliferation depending on cellular context. In this regard, we also found that FIP200 deletion led to decreased cell proliferation in primary keratinocytes (Wei et al. 2009). A more intriguing possibility to explain the apparent discrepancy, however, is that FIP200 plays a key role in cell cycle regulation, and its level of expression needs to be tightly controlled; thus, either protein ablation or overexpression could inhibit cell proliferation, perhaps through influencing different signaling pathways. Despite these remaining issues to be resolved in future studies, data presented here strongly indicate that intrinsic defects in tumor cell proliferation upon deletion of FIP200 contribute at least in part to suppressed tumorigenesis in vivo.
Although the mechanisms by which inactivation of FIP200 led to reduced tumor cell proliferation are still not well understood at present, defects in several cellular and molecular processes could be involved. Consistent with recent findings of FIP200 as an essential component of mammalian autophagy (Hara et al. 2008; Ganley et al. 2009; Hara and Mizushima 2009; Hosokawa et al. 2009; Jung et al. 2009), deletion of FIP200 in MaECs resulted in increased accumulation of large ubiquitin-positive aggregates and p62-positive aggregates as well as reduced LC3 conversion. Interestingly, recent studies showed that inhibition of autophagy in tumor cell lines driven by oncogenic Ras decreased their proliferation and compromised glycolytic capacity in these cells (Lock et al. 2011). Consistent with their findings, we also found impaired glucose uptake and intracellular lactate production as well as decreased sensitivity to reduced glucose availability in FIP200-null primary MEFs transformed by Ras. Therefore, deficient glycolysis after deletion of FIP200 could contribute to the decreased proliferation of mammary tumor cells in CKO-MT mice. We also found a decreased expression of cyclin D1 upon FIP200 deletion in both mammary tumor cells and transformed MEFs. Moreover, ectopic expression of cyclin D1 rescued the cell proliferation defect in these cells. Therefore, reduced glycolysis and cyclin D1 expression could be responsible for the intrinsic defects in proliferation of FIP200-null tumor cells in CKO-MT mice.
In addition to intrinsic mechanisms of tumor cells such as cell proliferation, tumor microenvironments and tumor–host cell interactions also play important roles in tumor initiation and progression of breast and other cancers in vivo (Hanahan and Weinberg 2000, 2011; Wiseman and Werb 2002; J Luo et al. 2009). In particular, the host immune defense mechanisms play a crucial role in tumor immune surveillance and removal of cancerous cells (Dunn et al. 2004; Koebel et al. 2007; Finn 2008). De Palma et al. (2008) showed recently that PyMT-driven tumor growth and metastasis were effectively inhibited by tumor-targeted interferon-α delivery through up-regulation of interferon target genes such as Irf7. Interestingly, our microarray analysis of mammary tumors revealed significantly increased expression of interferon target genes including Irf7 in tumors isolated from CKO-MT mice compared with those from Ctrl-MT mice. Moreover, further analysis showed that this is not caused by an increased response of tumor cells to interferons upon FIP200 deletion, but rather is due to the increased infiltration of CD8+IFN-γ+ and CD4+IFN-γ+ T cells in the tumor microenvironment of CKO-MT mice. While several types of immune cells participate in cancer immune surveillance, the CD8+ effector T cells play a critical role in the elimination of tumors (Dunn et al. 2004; Zou 2006). Indeed, impaired interferon signaling is a very common immune defect (Critchley-Thorne et al. 2009), and tumor-infiltrating effector T cells are associated with improved prognoses in multiple human cancers (Zhang et al. 2003; Sato et al. 2005; Galon et al. 2006; Kryczek et al. 2009). Recent studies using mouse models demonstrate that developing tumors are indeed recognized and destroyed by the intact immune systems through anti-tumor immune surveillance mechanisms (Shankaran et al. 2001; Dunn et al. 2002; Bui and Schreiber 2007). It is also well established that IFN-γ produced by both CD8+ and CD4+ cells (i.e., infiltrated CD8+IFN-γ+ and CD4+IFN-γ+ T cells in the tumor microenvironment) mediates anti-tumor immunity (Dunn et al. 2004; Finn 2008). Consistent with previous findings of inhibition of cell growth by IFN-γ in various cells (Gooch et al. 2000; Platanias 2005), we also observed that IFN-γ could inhibit growth of mammary tumor cells driven by PyMT. Moreover, we showed that depletion of CD8+ T cells restored the more rapid mammary tumorigenesis in CKO-MT mice. Thus, our results suggest that an increased anti-tumor immunity might also contribute to the decreased tumorigenesis in CKO-MT mice.
The mechanisms by which FIP200 inactivation in mammary tumor cells led to the increased immune surveillance of the host is not well understood at present. Interestingly, several chemokines including CXCL9 and CXCL10 were found to be increased in the tumor cells of CKO-MT mice, suggesting that increased production and/or secretion of these chemokines in FIP200-null tumor cells could initiate the increased immune surveillance. Moreover, we detected a significantly increased production of IFN-β as well as CXCL10 upon stimulation of the RLR signaling in FIP200-null mammary tumor cells compared with control cells. dsRNAs may be released by dead cells and activate RLR signaling pathway (Zitvogel et al. 2010). Interestingly, we found increased apoptosis in MIN lesions of CKO-MT mice. This suggests that dead cells may trigger IFN production by the RLR signaling pathway in CKO-MT mice. These results are consistent with several recent studies showing sensitized cellular responses in autophagy gene deletion or knockdown cells with increased production of IFNs and their target genes in viral infection and inflammatory diseases (Jounai et al. 2007; Cadwell et al. 2008; Saitoh et al. 2008; Tal et al. 2009; Virgin and Levine 2009). Although further studies will be required to clarify the mechanism, our results suggest the intriguing possibility that autophagy deficiency upon FIP200 ablation may lead to increased production of chemokines like CXCL10 for the increased recruitment of effector T cells and immune responses in CKO-MT mice.
In summary, our studies identify a protumorigenesis function of an essential autophagy protein FIP200 in mouse models of human breast cancer in vivo and show that ablation of FIP200 in mammary tumor cells inhibits tumor initiation and progression through both intrinsic and extrinsic mechanisms, as summarized in a working model (Fig. 7E). These results provide novel insights into the modulation of breast cancer development and progression by autophagy pathway and proteins. They also suggest that inactivation of FIP200 may serve as a potent new strategy for human anticancer therapy on multiple levels for tumor inhibition by not only inducing cell cycle arrest but also enhancing immune surveillance through augmenting the effectiveness of CD8+ T cells. This “kill two birds with one stone” strategy is urgently needed in treating patients with cancer.
Materials and methods
Mouse strains and tumor monitoring
Floxed FIP200, FIP200 CKO mice, and MMTV-PyMT transgenic mice have been described previously (Gan et al. 2006; M Luo et al. 2009; Wei et al. 2009). Mouse genotyping for floxed FIP200, Cre, and PyMT alleles was performed using genomic DNA prepared from tails, as described previously (M Luo et al. 2009; Wei et al. 2009). Floxed FIP200 and FIP200 CKO mice were kept in 129 and C57BL/6 mixed background and used for mammary gland development studies. For tumorigenesis studies, FIP200 CKO mice were backcrossed into FVB/n strain for more than five generations, and then intercrossed with MMTV-PyMT mice (FVB/n) to generate breeding cohorts. After weaning, female mice were examined twice a week for the development of mammary tumors by palpation. Mice were sacrificed when tumor burden became excessive. In some experiments, mice were injected intraperitoneally with 100 μg of anti-CD8 antibody (clone 2.43) or control IgG every 5 d after weaning to monitor the effect of CD8+ cell depletion, as described previously (Loeser et al. 2007). The antibody was produced by University of Michigan Hybridoma Core facility. For metastasis studies, mouse lungs were harvested for analyses at 7 wk after palpable mammary tumors were found. Lung metastasis burden was determined by counting the number of visible surface tumor nodules on the lungs. All animals were maintained under pathogen-free conditions and handled according to local, state, and federal regulations, and all Materials and Methods were carried out according to the guidelines of Institutional Animal Care and Use Committee at the University of Michigan.
Whole-mount analysis, immunohistochemical and immunofluorescent staining, and transmission electron microscopy
For whole-mount analysis, the number 4 inguinal mammary glands were dissected at the indicated ages. They were fixed in Carnoy's solution, stained in carmine alum, and then viewed under a Leica dissecting microscope (Leica Microsystems GmbH) and photographed, as described previously (M Luo et al. 2009).
For immunohistochemical analysis, tumor samples were fixed in formalin and embedded in paraffin. They were sectioned at 4-μm thickness, deparaffinized, rehydrated, and subjected to antigen retrieval procedures, as described previously (M Luo et al. 2009). Immunostainings were performed using VECTASTAIN ABC peroxidase/DAB staining kits (Vector Laboratories) with antibodies against ubiquitin (DAKO), p62 (Enzo Life Sciences), or cleaved caspase-3 (Cell Signaling).
For immunofluorescent staining, mammary tumors were embedded into OCT compound (Tissue-Tek) and frozen on dry ice. Sections were prepared at 8-μm thickness and staining with antibodies against CD45 or CD8 (both from eBioscience) followed by FITC conjugated secondary antibodies. Cell nuclei were counterstained by 4′,6′-diamino-2-phenylindole (DAPI).
For transmission electron microscopy, tumor samples were prepared and analyzed as described previously (Liang et al. 2010).
Isolation of mammary tumor cells
Mammary tumor cells were isolated essentially as described previously (M Luo et al. 2009) with minor modifications. Briefly, the tumors were dissected and then digested for 2–3 h at 37°C in DME:F12 medium containing 0.125% Collagenase III (Worthington) and 0.1% Hyaluronidase (Worthington) plus 5% FBS. Cells were spun down, resuspended in 0.25% trypsin (Invitrogen), and incubated for 10 min at 37°C. They were then spun down again and resuspended in HBSS (Invitrogen) with 2% FBS and 20 U/mL DNase I (Worthington) for an additional 10-min incubation before passing through 40-μm filters to remove debris. The isolated mammary tumor cells were cultured in plates coated with the growth factor-reduced Matrigels (1:20 dilution) in 1:1 DME:F12 medium (Invitrogen) supplemented with 2% FBS, 10 ng/mL EGF (Invitrogen), 10 μg/mL insulin (Sigma), 1 μg/mL hydrocortisone (Sigma), and 100 ng/mL cholera toxin (Sigma).
Preparation of recombinant retroviruses and infection of primary MEFs
A cDNA fragment containing Ha-RasV12 mutation was obtained by PCR using a plasmid containing Ha-RasV12 (generous gift of Drs. Richard Cerione and Jianbin Wang, Cornell University) as a template, and then inserted into pLEGFP-C1 (Clontech) or pLERFP-C1 (derived from pLEGFP-C1 by replacing GFP with RFP) to produce pLEGFP-RasV12 or pLERFP-RasV12. Similarly, a cDNA fragment containing full-length adenovirus 5 E1A was prepared by PCR using a plasmid containing the gene (kind gift of Dr. Scott Lowe, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). It was then cloned into pLERFP-C1 to generate pLERFP-E1A. HA-tagged cyclin D1 plasmid (Addgene plasmid 9050 from Dr. William Hahn, Harvard University) was purchased from Addgene. FIP200 floxed primary MEFs were isolated from embryonic day 13.5 (E13.5) embryos and cultured in DME supplemented with 10% FBS, as described previously (Gan et al. 2006). They were infected by viral supernatants of recombinant retroviruses encoding E1A and RasV12 followed by antibiotic selection, essentially as previously described (Wei et al. 2006), to obtain the transformed primary MEFs. They were then infected with recombinant adenoviruses encoding Cre recombinase or lacZ control (Gene Transfer Vector Core, University of Iowa), as described previously (Shen et al. 2005). In some experiments, FIP200 KO cells were infected by viral supernatants of recombinant retroviruses encoding HA-tagged cyclin D1 followed by puromycin selection.
Western blotting
Protein extracts were prepared from mammary tumors, isolated tumor cells, or transformed primary MEFs by lysis in modified RIPA buffer as described previously (Wei et al. 2009). They were then analyzed by Western blotting analysis as described previously (Wei et al. 2006). Antibodies against the following proteins were used: FIP200 (Ueda et al. 2000), PyMT, S6K, cyclin D1, actin, p53, p21, and GFP (for detection of GFP-RasV12) (all from Santa Cruz Biotechnology); p-Erk (Y204), Erk, p-Akt (S473), Akt and p-S6K (T389), and LC3B (all from Cell signaling); or p62 (Enzo Life Sciences).
Flow cytometric analysis
For analysis of mitochondria content, mammary tumor cells were trypsinized and resuspended. They were then stained with Mitotracker Green (100 nM) and Mitotracker Deep Red 633 (100 nM; Molecular Probes) for 30 min at 37°C. The cells were then washed and resuspended for FACS analysis using a FACStarPLUS (Becton Dickinson) flow cytometer. DAPI staining was used to distinguish live from dead cells.
For analysis of mammary tumor-infiltrating immune cells, the thoracic and inguinal mammary tumors were dissected at 1 wk after appearance of palpable tumors. Single-cell suspensions were then prepared, and immune cells were separated from tumor cells by Ficoll gradient centrifugation (GE Healthcare), as described previously (Kryczek et al. 2007a,b; Araki et al. 2009). They were then subjected to FACS analysis using the following antibodies: anti-CD45, anti-CD3, anti-CD4, anti-CD8, anti-IFN-γ (all from BD pharmingen), or anti-Foxp3 (eBiosciences). For intracellular cytokine stimulation, cells were treated with phorbol myristate acetate (50 ng/mL; Sigma), ionomycin (1 μM; Sigma) for 4 h at 37°C before staining. Cells were first stained extracellularly with specific antibodies, then were fixed and permeabilized with Perm/Fix solution (eBioscience), and finally were stained intracellularly for anti-IFN-γ antibody. FACS analyses were performed on an LSR II sorter (BD Biosciences), and data were analyzed with DIVA software (BD Biosciences).
BrdU incorporation and TUNEL assays
For BrdU incorporation assay in cell culture, cells were deprived of growth factors for 24 h, incubated in the complete medium with 100 μM BrdU for 15 h, and then subjected to anti-BrdU staining using a BrdU staining kit according to the manufacturer's instructions (Invitrogen). For BrdU incorporation in vivo assays, mice were injected intraperitoneally with BrdU (Sigma; 100 μg per gram body weight). After 3 h, mice were sacrificed, and mammary glands were prepared, fixed overnight in formalin, and embedded in paraffin. BrdU uptake was detected using the same BrdU staining kit (Invitrogen). Apoptosis analyses in cell culture were performed using the ApopTag Peroxidase In Situ Apoptosis Detection kit (Chemicon International).
Cell growth assays
To measure multiplication of cells, the same number of cells were plated on cultured dishes and incubated in DMEM containing 10% FBS for various times as indicated. They were then counted to determine growth under the various conditions. In some experiments, rapamycin was included in the culture medium as indicated. In other experiments, the cells were incubated in DMEM lacking glucose (Invitrogen), which was supplemented with 25 mM or 5.5 mM glucose, 10% dialyzed FBS, penicillin, and streptomycin to determine the sensitivity of cells to diminished glucose availability.
To measure anchorage-independent growth, cells (1 × 104) were resuspended in culture medium with 0.3% agarose and plated on the top of 0.5% agarose in culture medium in 35-mm culture dishes. Three weeks after seeding, colonies were stained with p-iodonitrotetrazolium violet (Sigma), and those >50 μm in diameter were counted under a microscope.
Measurements of glucose uptake and lactate levels
Glucose uptake was measured by FACS detection of uptake of 2-[N-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl) amino]-2-deoxyglucose (2-NBDG; Invitrogen) in cultured cells as described previously (Lock et al. 2011). Intracellular lactate level was measured using Lactate Assay Kit II (Biovision).
RNA isolation, microarray gene expression profiling, and qRT–PCR analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For microarray gene expression profiling, two independent mammary tumor samples were prepared each from a pool of four mice for both CKO-MT and Ctrl-MT mice (designated CKO-MT1, CKO-MT2, Ctrl-MT1, and Ctrl-MT2) at 1 wk after the appearance of palpable tumors. Microarray and data analyses were carried out by the University of Michigan Comprehensive Cancer Center Affymetrix and Microarray Core Facility using the Affymetrix mouse 430 2.0 gene chip according to standard protocols. Genes were selected if their signals were up-regulated or down-regulated at least twofold in CKO-MT1 and CKO-MT2 samples compared with Ctrl-MT1 and Ctrl-MT2 samples. For qRT–PCR analyses, equal amounts of RNA were reverse-transcribed by SuperScript III first-strand synthesis system (Invitrogen) with oligo(dT) as a primer, and then the resulting cDNA templates were subjected to qRT–PCR using the SYBR Green PCR Core reagents system (Qiagen). Primer sequences are available upon request.
Statistical analysis
Statistical significance was evaluated by a paired T-test, using P < 0.05 as indicative of statistical significance. Kaplan-Meier tumor-free survival data were compared using the log-rank test.
Acknowledgments
We are grateful to Drs. Jianbin Wang and Rick Cerione of Cornell University and Dr. Scott W. Lowe of Cold Spring Harbor Laboratory for generously providing Ha-RasV12 and E1A constructs, respectively. We are indebted to Linhua Vatan for assistance in FACS analysis; Dr. Ilona Kryczek for discussion of immune cell analyses; and Drs. Ming Luo, Fei Liu, Chenran Wang, and Chun-Chi Liang for discussions and assistance on different methods used in this study. We thank our laboratory colleagues for helpful comments on the manuscript. This research was supported by NIH grants to J.-L.G.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2051011.
References
- Abbi S, Ueda H, Zheng C, Cooper LA, Zhao J, Christopher R, Guan JL 2002. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol Biol Cell 13: 3178–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A 2007. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ 14: 500–510 [DOI] [PubMed] [Google Scholar]
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B 1999. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 59: 59–65 [DOI] [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB 2007. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 117: 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Wang Z, Zang C, Wood WH III, Schones D, Cui K, Roh TY, Lhotsky B, Wersto RP, Peng W, et al. 2009. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30: 912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW 2010. Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov 6: 975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui JD, Schreiber RD 2007. Cancer immunosurveillance, immunoediting and inflammation: independent or interdependent processes? Curr Opin Immunol 19: 203–208 [DOI] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW 2010. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141: 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chano T, Ikegawa S, Kontani K, Okabe H, Baldini N, Saeki Y 2002a. Identification of RB1CC1, a novel human gene that can induce RB1 in various human cells. Oncogene 21: 1295–1298 [DOI] [PubMed] [Google Scholar]
- Chano T, Kontani K, Teramoto K, Okabe H, Ikegawa S 2002b. Truncating mutations of RB1CC1 in human breast cancer. Nat Genet 31: 285–288 [DOI] [PubMed] [Google Scholar]
- Critchley-Thorne RJ, Simons DL, Yan N, Miyahira AK, Dirbas FM, Johnson DL, Swetter SM, Carlson RW, Fisher GA, Koong A, et al. 2009. Impaired interferon signaling is a common immune defect in human cancer. Proc Natl Acad Sci 106: 9010–9015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B 2010. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy 6: 322–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Li Y, Tedesco D, Liao R, Fuhrmann G, Sun P 2005. The ability of E1A to rescue ras-induced premature senescence and confer transformation relies on inactivation of both p300/CBP and Rb family proteins. Cancer Res 65: 8298–8307 [DOI] [PubMed] [Google Scholar]
- De Palma M, Mazzieri R, Politi LS, Pucci F, Zonari E, Sitia G, Mazzoleni S, Moi D, Venneri MA, Indraccolo S, et al. 2008. Tumor-targeted interferon-α delivery by Tie2-expressing monocytes inhibits tumor growth and metastasis. Cancer Cell 14: 299–311 [DOI] [PubMed] [Google Scholar]
- Dong C 2008. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol 8: 337–348 [DOI] [PubMed] [Google Scholar]
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD 2002. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 3: 991–998 [DOI] [PubMed] [Google Scholar]
- Dunn GP, Old LJ, Schreiber RD 2004. The immunobiology of cancer immunosurveillance and immunoediting. Immunity 21: 137–148 [DOI] [PubMed] [Google Scholar]
- Dunn GP, Koebel CM, Schreiber RD 2006. Interferons, immunity and cancer immunoediting. Nat Rev Immunol 6: 836–848 [DOI] [PubMed] [Google Scholar]
- Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, et al. 2008. Autophagy is important in islet homeostasis and compensatory increase of β cell mass in response to high-fat diet. Cell Metab 8: 325–332 [DOI] [PubMed] [Google Scholar]
- Farber JM 1997. Mig and IP-10: CXC chemokines that target lymphocytes. J Leukoc Biol 61: 246–257 [PubMed] [Google Scholar]
- Finn OJ 2008. Cancer immunology. N Engl J Med 358: 2704–2715 [DOI] [PubMed] [Google Scholar]
- Frigerio S, Junt T, Lu B, Gerard C, Zumsteg U, Hollander GA, Piali L 2002. β Cells are responsible for CXCR3-mediated T-cell infiltration in insulitis. Nat Med 8: 1414–1420 [DOI] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. 2006. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313: 1960–1964 [DOI] [PubMed] [Google Scholar]
- Gan B, Guan JL 2008. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal 20: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Melkoumian ZK, Wu X, Guan KL, Guan JL 2005. Identification of FIP200 interaction with the TSC1–TSC2 complex and its role in regulation of cell size control. J Cell Biol 170: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL 2006. Role of FIP200 in cardiac and liver development and its regulation of TNFα and TSC–mTOR signaling pathways. J Cell Biol 175: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284: 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Polo RA, Niso-Santano M, Ortiz-Ortiz MA, Gomez-Martin A, Moran JM, Garcia-Rubio L, Francisco-Morcillo J, Zaragoza C, Soler G, Fuentes JM 2007. Inhibition of paraquat-induced autophagy accelerates the apoptotic cell death in neuroblastoma SH-SY5Y cells. Toxicol Sci 97: 448–458 [DOI] [PubMed] [Google Scholar]
- Gooch JL, Herrera RE, Yee D 2000. The role of p21 in interferon γ-mediated growth inhibition of human breast cancer cells. Cell Growth Differ 11: 335–342 [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. 2011. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25: 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12: 954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hara T, Mizushima N 2009. Role of ULK–FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy 5: 85–87 [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. 2009. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD 2009. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans 37: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, et al. 2007. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci 104: 14050–14055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH 2009. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20: 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E 2007. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 21: 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IA, MacLean JA, Lee FS, Casciotti L, DeHaan E, Schwartzman JD, Luster AD 2000. IP-10 is critical for effector T cell trafficking and host survival in Toxoplasma gondii infection. Immunity 12: 483–494 [DOI] [PubMed] [Google Scholar]
- Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD 2007. Adaptive immunity maintains occult cancer in an equilibrium state. Nature 450: 903–907 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B 2010. Autophagy and the integrated stress response. Mol Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Wei S, Vatan L, Escara-Wilke J, Szeliga W, Keller ET, Zou W 2007a. Cutting edge: opposite effects of IL-1 and IL-2 on the regulation of IL-17+ T cell pool IL-1 subverts IL-2-mediated suppression. J Immunol 179: 1423–1426 [DOI] [PubMed] [Google Scholar]
- Kryczek I, Wei S, Zou L, Altuwaijri S, Szeliga W, Kolls J, Chang A, Zou W 2007b. Cutting edge: Th17 and regulatory T cell dynamics and the regulation by IL-2 in the tumor microenvironment. J Immunol 178: 6730–6733 [DOI] [PubMed] [Google Scholar]
- Kryczek I, Banerjee M, Cheng P, Vatan L, Szeliga W, Wei S, Huang E, Finlayson E, Simeone D, Welling TH, et al. 2009. Phenotype, distribution, generation, functional and clinical relevance of Th17 cells in the human tumor environments. Blood 114: 1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW 2011. Autophagy in immunity and inflammation. Nature 469: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Jung JU 2010. Autophagy genes as tumor suppressors. Curr Opin Cell Biol 22: 226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676 [DOI] [PubMed] [Google Scholar]
- Liang CC, Wang C, Peng X, Gan B, Guan JL 2010. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem 285: 3499–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW 2003. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 163: 2113–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, Guan JL 2010. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood 116: 4806–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J 2011. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell 22: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeser S, Loser K, Bijker MS, Rangachari M, van der Burg SH, Wada T, Beissert S, Melief CJ, Penninger JM 2007. Spontaneous tumor rejection by cbl-b-deficient CD8+ T cells. J Exp Med 204: 879–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ 2009. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Fan H, Nagy T, Wei H, Wang C, Liu S, Wicha MS, Guan JL 2009. Mammary epithelial-specific ablation of the focal adhesion kinase suppresses mammary tumorigenesis by affecting mammary cancer stem/progenitor cells. Cancer Res 69: 466–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Schwamborn K, Urlaub H, Gan B, Guan JL, Dejean A 2008. Spatial interplay between PIASy and FIP200 in the regulation of signal transduction and transcriptional activity. Mol Cell Biol 28: 2771–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E 2007. Role of autophagy in cancer. Nat Rev Cancer 7: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkoumian ZK, Peng X, Gan B, Wu X, Guan JL 2005. Mechanism of cell cycle regulation by FIP200 in human breast cancer cells. Cancer Res 65: 6676–6684 [DOI] [PubMed] [Google Scholar]
- Mizushima N 2007. Autophagy: process and function. Genes Dev 21: 2861–2873 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, Simon AK 2010. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci 107: 832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, et al. 2007. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 13: 619–624 [DOI] [PubMed] [Google Scholar]
- Nishikawa T, Tsuno NH, Okaji Y, Shuno Y, Sasaki K, Hongo K, Sunami E, Kitayama J, Takahashi K, Nagawa H 2010. Inhibition of autophagy potentiates sulforaphane-induced apoptosis in human colon cancer cells. Ann Surg Oncol 17: 592–602 [DOI] [PubMed] [Google Scholar]
- Platanias LC 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5: 375–386 [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112: 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E 2010. Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Debnath J 2010. Autophagy and tumorigenesis. Semin Immunopathol 32: 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456: 264–268 [DOI] [PubMed] [Google Scholar]
- Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, Jungbluth AA, Frosina D, Gnjatic S, Ambrosone C, et al. 2005. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci 102: 18538–18543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD 2001. IFNγ and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410: 1107–1111 [DOI] [PubMed] [Google Scholar]
- Shen TL, Park AY, Alcaraz A, Peng X, Jang I, Koni P, Flavell RA, Gu H, Guan JL 2005. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J Cell Biol 169: 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 186: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, et al. 2007. PRAK is essential for ras-induced senescence and tumor suppression. Cell 128: 295–308 [DOI] [PubMed] [Google Scholar]
- Swann JB, Smyth MJ 2007. Immune surveillance of tumors. J Clin Invest 117: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A 2009. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci 106: 2770–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschan MP, Simon HU 2010. The role of autophagy in anticancer therapy: promises and uncertainties. J Intern Med 268: 410–418 [DOI] [PubMed] [Google Scholar]
- Ueda H, Abbi S, Zheng C, Guan JL 2000. Suppression of Pyk2 kinase and cellular activities by FIP200. J Cell Biol 149: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Levine B 2009. Autophagy genes in immunity. Nat Immunol 10: 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wang X, Gan B, Urvalek AM, Melkoumian ZK, Guan JL, Zhao J 2006. Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J Biol Chem 281: 16664–16671 [DOI] [PubMed] [Google Scholar]
- Wei H, Gan B, Wu X, Guan JL 2009. Inactivation of FIP200 leads to inflammatory skin disorder, but not tumorigenesis, in conditional knock-out mouse models. J Biol Chem 284: 6004–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, DiPaola RS 2009. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res 15: 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman BS, Werb Z 2002. Stromal effects on mammary gland development and breast cancer. Science 296: 1046–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y 1998. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 23: 33–42 [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ 2010. Eaten alive: a history of macroautophagy. Nat Cell Biol 12: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM, et al. 2009. Autophagy mediates the mitotic senescence transition. Genes Dev 23: 798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Lee Y, Liu W, Chin RK, Wang J, Wang Y, Schietinger A, Philip M, Schreiber H, Fu YX 2004. Priming of naive T cells inside tumors leads to eradication of established tumors. Nat Immunol 5: 141–149 [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci 100: 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN, et al. 2003. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med 348: 203–213 [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Kepp O, Kroemer G 2010. Decoding cell death signals in inflammation and immunity. Cell 140: 798–804 [DOI] [PubMed] [Google Scholar]
- Zou W 2005. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer 5: 263–274 [DOI] [PubMed] [Google Scholar]
- Zou W 2006. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol 6: 295–307 [DOI] [PubMed] [Google Scholar]
- Zou W, Restifo NP 2010. T(H)17 cells in tumour immunity and immunotherapy. Nat Rev Immunol 10: 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]