Abstract
Background
Classical swine fever virus (CSFV) is the member of the genus Pestivirus under the family Flaviviridae. The 5' untranslated region (UTR) of CSFV contains the IRES, which is a highly structured element that recruits the translation machinery. The 3' UTR is usually the recognition site of the viral replicase to initiate minus-strand RNA synthesis. Adenosine-uridine rich elements (ARE) are instability determinants present in the 3' UTR of short-lived mRNAs. However, the presence of AREs in the 3' UTR of CSFV conserved in all known strains has never been reported. This study inspects a possible role of the ARE in the 3' UTR of CSFV.
Results
Using RNA pull-down and LC/MS/MS assays, this study identified at least 32 possible host factors derived from the cytoplasmic extracts of PK-15 cells that bind to the CSFV 3' UTR, one of which is HuR. HuR is known to bind the AREs and protect the mRNA from degradation. Using recombinant GST-HuR, this study demonstrates that HuR binds to the ARE present in the 3' UTR of CSFV in vitro and that the binding ability is conserved in strains irrespective of virulence.
Conclusions
This study identified one of the CSFV 3' UTR binding proteins HuR is specifically binding to in the ARE region.
Background
Classical swine fever virus (CSFV), along with Bovine viral diarrhea virus (BVDV) and Border disease virus (BDV), is a member of the genus Pestivirus under the family Flaviviridae. CSFV infects pigs and causes high economic losses especially in Europe and Southeast Asia. CSFV infection causes lymphopenia and thrombocytopenia, thereby evading and compromising the immune system [1]. CSFV is an enveloped icosahedral virus containing a positive sense RNA genome that is devoid of a 5' cap or a 3' poly(A) tail. The 5' untranslated region (UTR) comprises an internal ribosomal entry site (IRES) that drives the translation of a single open reading frame (ORF; Figure 1A) and encoding a polyprotein approximately 4,000 amino acids long, which is co- and post-translationally processed by viral and host proteases into individual structural and non-structural proteins [2]. Due to the limitation of its genome size and the number and functions of encoded proteins, the virus exploits the cellular machinery and components to complete its lifecycle. Cellular host factors could either aid in or interfere with the various stages of the virus lifecycle participating in diverse functions, such as translation, replication, countering host defense, and so on [3-6]. Host factors could either bind to the viral proteins or to the viral RNA.
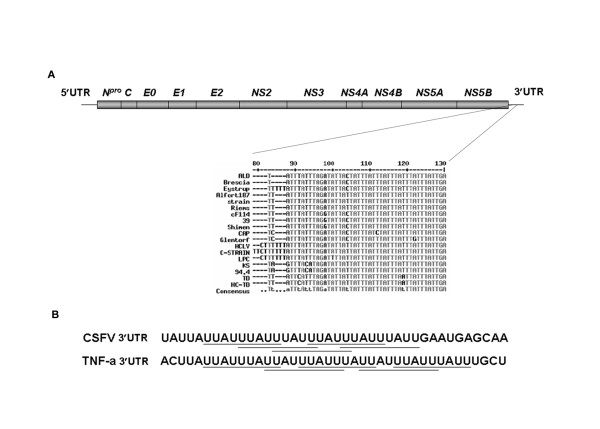
Figure 1.
The illustration of the sequence of adenosine-uridine rich elements present in the 3'UTR of CSFV. (A) Genome organization of CSFV. The conserved AREs present in the 3' UTR of various CSFV strains are shown. (B) CSFV ARE compared with a type II ARE present in the 3' UTR of tumor necrosis factor alpha (TNF-α) mRNA. The overlapping ARE nonamers are underlined.
The 5' UTR of CSFV contains the IRES, which is a highly structured element that recruits the translation machinery. The 3' UTR is usually the recognition site of the viral replicase to initiate minus-strand RNA synthesis [7,8]. The 3' UTR is also known to influence translation, either by interacting with the 5' UTR or by other means [9]. Adenosine-uridine rich elements (ARE) containing mRNA are considered highly unstable, and the translation of which is necessary only for a short period, as in the case of transcription factors, cytokines, and oncogenes [10], and in certain RNA transcripts of DNA viruses [11]. Apart from mRNAs, AREs are also found in non-coding RNAs of viruses [12-14]. Cellular proteins, such as Tristetraprolin (TTP), ARE/poly(U)-binding/degradation factor 1 (AUF 1), Hu antigen R (HuR), and T-cell internal antigen-1 (TIA-1), bind to cellular mRNAs that contain AREs. Upon binding, the ARE binding proteins (AREBPs) mostly target the mRNA to the degradation pathway, for instance, for TTP and AUF1 [15,16], or protect the mRNA from degradation, for example, with HuR [17], or suppress translation, as is the case with HuR and TIA [18,19].
AU-rich sequences have previously been reported to be present in the 3' UTR of all known strains [20,21]. This study found that this AU-rich sequence that CSFV harbors at its 3' UTR is indeed a typical AU-rich element classified as type II ARE, which contains at least two AUUUA pentamer motifs in a U-rich region [10]. This ARE is conserved in all strains of CSFV. Using CSFV 3' UTR as bait, we isolated the interacting host factors and identified several of them employing LC/MS/MS. HuR is one of the proteins identified. This study found via EMSA that HuR binds to the 3' UTR of CSFV RNA in vitro. In addition, we also found that HuR binding ability is independent of strain virulence.
Methods
Constructs
Plasmids templates for T7 in vitro transcription containing the 3'UTR of CSFV strains ALD, LPC, 93KS, and 95TD are derived from the partial cDNA clone of each strain. In brief, the primers used in the reaction are the 5' primer CSFV/T7+12,058 (5'TAATACGACTCACTATAGGGTATGAGCGCGGGTAACCCGGGATCTGGA3'), and the 3' primers ALD3'12328 (5'GGGCCGTTAGGAAATTACCTTAGTC3') and LPC3'12269 (5'GGGCCGTTAGAAATTACCTTA3') for ALD and LPC, respectively. For 93KS and 95TD clones, the 5' primers are CSFV/T7+93KS (5'TAATACGACTCACTATAGGGCACATGAGTGCGGGTAG3') and CSFV/T7+95TD (5'TAATACGACTCACTATAGGGTGTGAGAACGGCCGGCCC3') with the common 3' primer used for ALD strain, ALD3'12328. The T7 promoter sequence is underlined. PCR was performed using the kinased primers and pfu DNA polymerase. The insert was ligated into SmaI digested and dephosphorylated pUC18 vector; and the positive clones were verified by sequencing. These clones were designated as pT7-ALD3'UTR, pT7-LPC3'UTR, pT7-93KS3'UTR and pT7-95TD 3'UTR.
RNA labeling
A typical 10 μl in vitro transcription reaction for RNA body-labeling contains 2 μl 5X T7 buffer [200 mM Tris-HCl pH-8.0, 40 mM MgCl2, 10 mM spermidine-(HCl)3 and 125 mM NaCl], 1 μl of 100 mM Dithiotheratol, 20 units T7 RNA polymerase, 1 μg linearized template, 0.25 μl containing 25 mM each of ATP, CTP, and GTP, 0.25 μl containing 250 μM UTP, 3.5 μl of [α-32P]UTP containing a total of 11.65 pmole (35 μCi) and 20 units HPRI (Human placental RNAse inhibitor). The reaction was scaled up proportionally as required. Plasmids pT7-ALD3'UTR, pT7-LPC3'UTR, pT7-93KS 3'UTR and pT7-95TD 3'UTR DNA clones were linearized using SmaI for run-off transcription RNA labeling. The transcription reaction was performed at 37°C for 2 hrs and the samples were run on a 5% polyacrylamide gel. The appropriate bands were cut and the RNA was eluted from the gel. The 32P body-labeled RNA was checked for quality and then quantified.
Electrophoretic mobility shift assay (EMSA)
Binding was performed in a 10 μl reaction containing 1 μl of 10X binding buffer, 100 ng of yeast total RNAs, about 1 picomole of radio-labeled probe and 1 μg of protein (either GST or GST-HuR, in the case of BSA 2 μg was used ) and 20 units HPRI. For competition assay equal mass (about 5 ng) of cold competitor RNA was used. For super shift assay, 1 μg of anti-HuR antibody was included in the reaction. The reaction was incubated for 10 min at room temperature and then run on a 5% native polyacrylamide gel. The gel was exposed to a phosphorimage plate for a few minutes and then scanned using PhosphorImager Fujifilm BAS 2500.
PK-15 cytoplasmic extract preparation
PK-15 (Porcine Kidney-15) cells were grown at 37°C in Eagle's minimal essential medium (EMEM) with 5% newborn calf serum (NCS) and 100 U/ml penicillin and 100 μg/ml streptomycin. Sub-cellular fractions were prepared as described [22]. Briefly, PK-15 cells grown in twelve 15 cm2 dishes were washed with PBS, harvested and then washed in 5 volumes of hypotonic buffer and finally suspended in 3 volumes of hypotonic buffer and placed on ice for 10 min. Cells were lysed using a type B pestle Dounce homogenizer by using 10 to 20 strokes on ice. The lysate was centrifuged at 3300 g for 15 min at 4°C and the supernatant was saved as the cytoplasmic extract and its protein content estimated. The pellet contains nuclei.
RNA-agarose beads cross-linking and protein recovery
In order to be cross-linked, 25 μg of RNA was first oxidized in a 50 μl reaction containing 100 mM sodium acetate pH 5.0 and 5 mM sodium meta-periodate which was incubated for 1.5 hrs at RT in a shaker shielded from light. About 80 μl of agarose beads (Adipic acid dihydrazide Agarose; Sigma) were washed 4 times with and then re-suspended in 60 μl of 100 mM sodium acetate pH 5.0. Oxidized RNA was added to the agarose beads and incubated for 2.5 hr at RT. Then the RNA-agarose beads were washed thrice with 1 ml of 2 M NaCl then washed thrice in buffer D (20 mM HEPES-KOH pH 7.6, 100 mM KCl, 200 mM EDTA, 500 mM DTT, and 5% v/v glycerol).
About 60, 90, and 120 μg of PK-15 cytoplasmic extracts along with 10, 15, and 20 μg yeast total RNAs, respectively, were incubated for 5 min at RT, then added to RNA-agarose beads and placed in a shaker for 25 min at RT (HPRI was added to all reactions at 2 U/μl reaction ratio). The beads were spun down and washed four times in 1 ml of buffer D containing 125 mM NaCl. Then the beads were suspended in 10 μl 4× SDS-PAGE loading buffer and boiled for 5 min and loaded on to a 12% SDS-polyacrylamide gel and run for 65 min at 200V. The gel was stained with silver SNAP and bands of the indicated sizes were cut and sent for mass spectroscopy.
Plasmids and Purification of GST-HuR Proteins
For constructing the GST-HuR expression vectors, the coding region of HuR was amplified by PCR from human A431 cells cDNA using primers 5'TCTAATGGTTATGAAGACCACATG3' and 5'TTATTTGTGGGACTTGTTGG3', and cloned into pGEX-6P-1 vector (Amersham Bioscience). GST-HuR was prepared using E. coli BL21/DE3 cells transformed with pGEX-HuR. GST fusion protein was purified on glutathione-Sepharose beads as recommended by the manufacturer, and the concentration determined by comparison with a bovine serum albumin (BSA) curve in SDS-PAGE stained with Coomassie brilliant blue.
Results
CSFV 3' UTR harbors AREs
A detailed analysis revealed five overlapping AUUUA pentamers. This pentamer is a typical adenosine and uridine-rich element (ARE), an instability determinant found in the 3' UTR of short-lived cellular mRNAs, which usually encode proteins required only for a short period, such as transcription factors, oncogenes, and cytokines [10]. AREs are most often found as repeats within U-rich regions. In CSFV 3' UTR, this pentamer is found as five repeats within three UUAUUUAUU tandem-overlapping nonamer repeats (Figure 1B) indicative of type II ARE [10,23]. Type II ARE is typical of mRNAs of cytokines such as TNFα. This ARE repeat is present at approximately 105 nt to 130 nt (nucleotides are numbered from the stop codon) of the 3' UTR of all the known strains of CSFV, and is approximately conserved across these strains, as determined by multiple sequence alignment (Figure 1A). Since viruses usually employ strategies to increase RNA synthesis and evade detection and degradation by host defenses, discovering an instability determinant in the genomic RNA of a virus was intriguing. AREs are the target of a set of proteins that bind to ARE containing RNA collectively, called ARE-binding proteins (AREBPs), which degrade or protect ARE containing RNA or repress its translation upon binding [23].
Identification of host factors that bind to CSFV 3' UTR
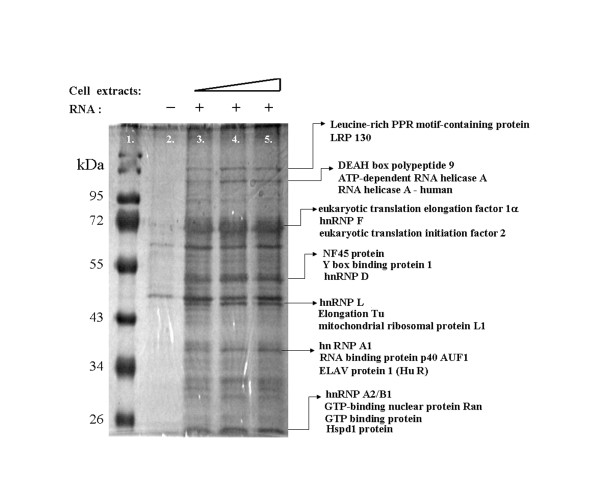
Agarose beads covalently cross-linked to CSFV 3' UTR RNA were used to pull down the proteins binding to the 3' UTR present in cytoplasmic extracts of PK-15 cells [24]. The eluted proteins were resolved on an SDS-gel, and selected bands (Figure 2) were cut and sent for LC/MS/MS. Results revealed approximately 32 proteins listed in Table 1. Three of the proteins identified above hnRNP A1, hnRNP D, and HuR are bona fide AREBPs [23]. This study chose HuR for future experiments due to its function of protecting or at least delaying the onset of ARE RNA degradation [17,25].
Figure 2.
RNA binding protein isolated from RNA pull-down experiment. Silver stained SDS-polyacrylamide gel containing the proteins pulled down from PK-15 cytoplasmic extracts using agarose beads linked to CSFV ALD 3' UTR. Lane 1 is the protein size marker; lane 2 is proteins from only beads without RNA, lane 3-5 shows 60, 90, and 120 μg of PK-15 cytoplasmic extract used in the pull down reaction. Some of the identity of the proteins are shown with arrows to represent the bands identified by LC/MS/MS.
Table 1.
LC/MS/MS results of cellular proteins associating with the CSFV 3'UTR
| No | Cellular protein | Mr (kDa) | Coverage* |
|---|---|---|---|
| 1 | Leucine-rich PPR motif-containing protein | 153.2 | 12% |
| 2 | RNA helicase A | 142 | 13% |
| 3 | ATP-dependent RNA helicase A isoform 2 | 141.1 | 16% |
| 4 | Nucleolin | 76.6 | 13% |
| 5 | ATP-dependent RNA helicase DDX3X | 73.2 | 12% |
| 6 | hnRNP Q | 69.6 | 7% |
| 7 | ATP-dependent RNA helicase DDX5X | 69.1 | 6% |
| 8 | RNA binding protein 47 | 64 | 6% |
| 9 | Far upstream element binding protein 3 | 61.6 | 7% |
| 10 | 60 kDa heat shock protein | 60.9 | 6% |
| 11 | hnRNP L | 60.4 | 14% |
| 12 | RNA binding protein FUS | 52.3 | 5% |
| 13 | Eukaryotic translation initiation factor 2 | 51 | 11% |
| 14 | Eukaryotic translation elongation factor 1A (EF1A) | 50 | 19% |
| 15 | hnRNP H | 49.2 | 3% |
| 16 | Lupus La protein | 46.8 | 4% |
| 17 | hnRNP F | 45.6 | 15% |
| 18 | NF45 protein | 44.7 | 29% |
| 19 | DNA binding protein A | 40 | 13% |
| 20 | hnRNP A2/B1 | 37.4 | 27% |
| 21 | Mitochondria ribosomal protein L1 | 36.6 | 10% |
| 22 | ELAV protein 1 (HuR) | 36 | 3% |
| 23 | Y box binding protein 1 | 35.9 | 9% |
| 24 | hnRNP A1 | 34.2 | 15% |
| 25 | hnRNP D | 33.4 | 16% |
| 26 | RNA binding protein p40 AUF1 | 32.6 | 25% |
| 27 | hnRNP A/B | 30.8 | 16% |
| 28 | hnRNP D isoform c | 29.6 | 21% |
| 29 | 40S ribosomal protein S3 | 27.7 | 33% |
| 30 | Hspd1 protein | 26.9 | 11% |
| 31 | GTP-binding nuclear protein Ran | 25.5 | 23% |
| 32 | GTP-binding protein | 24.4 | 31% |
* The coverage is used to indicate the percentage of the database protein sequence covered by matching peptides.
HuR binds to the ARE of CSFV 3' UTR in vitro
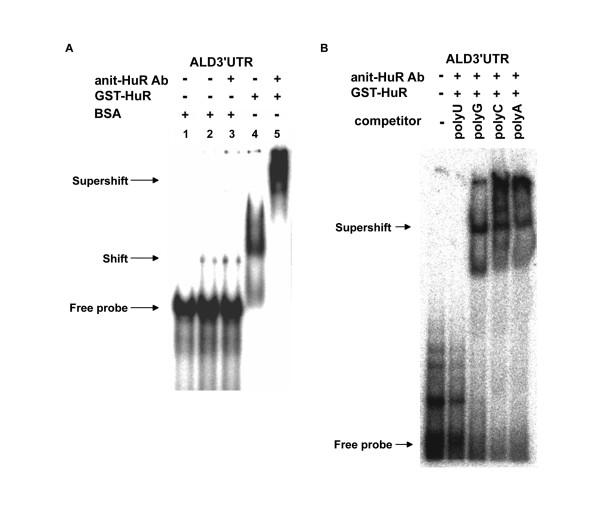
To assess whether CSFV 3' UTR binds to HuR, E. coli-purified GST-HuR was used in EMSA. Radioactive-labeled CSFV ALD 3' UTR RNA probe (about 1 picomole) interacts with GST-HuR, though not with BSA, even at a higher concentration (Figure 3A). To further confirm this, an antibody against HuR was included in EMSA reaction, resulting in a super-shift only in GST-HuR, and not when incubated with BSA (Figure 3A). This confirms that CSFV 3' UTR binds to HuR in vitro. To discover which region of the CSFV 3' UTR binds to HuR, a super-shift competition assay was performed using CSFV 3' UTR as a probe, and equal masses (about 5 ng) of polyA, polyC, polyG, and polyU were used as competitors (Figure 3B). Only polyU was able to out-compete the CSFV 3' UTR (Figure 3B), which is an indirect indication that HuR binds to the ARE of the CSFV 3' UTR, since the ARE is rich in uridines compared to any other region of the CSFV 3' UTR.
Figure 3.
The specific interaction between HuR and the 3' UTR of CSFV RNA. GST-HuR binds to CSFV ALD 3' UTR. An autoradiogram of electrophoretic mobility shift assay (EMSA) runs on a 5% non-denaturing PAGE. (A) Free probe (32P labeled CSFV ALD3'UTR) without any proteins is loaded in lane 1, 1 μg BSA in lanes 2 and 3, and GST-HuR in lanes 4 and 5 are used in an EMSA. The extra1 μg of anti-HuR antibody is included in lanes 3 and 5. (B) EMSA with unlabeled homo-polyribonucleotides. Free probe without protein components is loaded in lane 1, about 1 μg each of GST-HuR and anti-HuR antibody are loaded with equal mass cold homo-polyribonucleotides, polyU in lane 2, polyG in lane 3, polyC in lane 4, and polyA in lane 5. Arrows indicate the positions of the free, shifted, and super-shifted radiolabeled probes.
ARE conserved across all strains of CSFV retains in vitro HuR binding ability
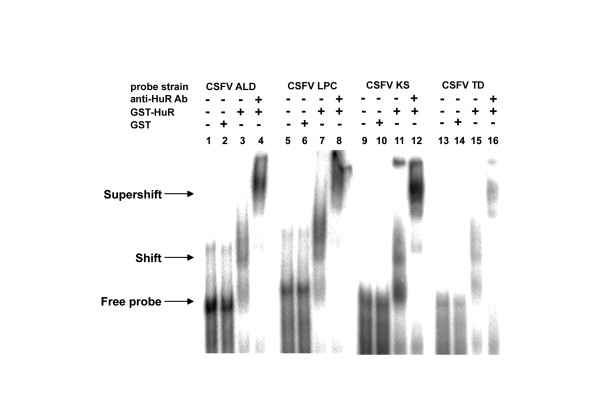
CSFV comprises numerous strains of varied virulence and infectivity that cause diseases of varied severity, ranging from acute to sub-acute and chronic symptoms. They all possess the ARE in their 3' UTR, irrespective of virulence. Even the avirulent vaccine strain LPC contains this sequence in addition to the polypyrimidine stretch that precedes the ARE. This study sought to discover if HuR could bind to the 3' UTR of CSFV strains irrespective of virulence, and chose four strains, ALD (a highly virulent strain), 93KS and 96TD (both moderately virulent Taiwanese isolates), and LPC (an avirulent vaccine strain).32P-labeled 3' UTR of the aforementioned strains were used in EMSA containing purified GST or purified GST-HuR (Figure 4). The anti-HuR antibody was then included in the EMSA super-shift reaction containing GST-HuR. Results show a shift in mobility in reactions containing purified GST-HuR compared to the free probe or the negative control that contains purified GST only (Figure 4). In reactions that contain the anti-HuR antibody in addition to GST-HuR, a clear super-shift is observed, indicating that HuR binds to the ARE of CSFV 3' UTR, irrespective of its nature of infection.
Figure 4.
The interaction of HuR with the 3' UTR conserved in CSFV strains irrespective of virulence. The 3' UTR of CSFV strains of varied virulence bind to GST-HuR in vitro. Autoradiogram of EMSA of the 32P labeled RNA of the 3' UTR belonging to four different CSFV strains ALD, LPC, 93KS, and 96TD run on a 5% non-denaturing polyacrylamide gel. Free probe derived from each strain is loaded in lanes 1, 5, 9, and 13. The purified GST only as the control is loaded in lanes 2, 6, 10, and 14; and GST-HuR as the binding partner (lanes 3, 7, 11, and 15) and anti-HuR antibody are included in the super-shift reactions (lanes 4, 8, 12, and 16) indicated above the lanes. Arrows indicate the positions of the free, shifted, and super-shifted radiolabeled probes.
Discussion
We have demonstrated that CSFV harbors bona fide AREs in the 3' UTR of its genomic RNA and that it is conserved in all analyzed strains. To the best of our knowledge, this is the first report showing that a viral genomic RNA of a positive-sense RNA animal virus harbors ARE motifs in its 3' UTR. The presence of an instability determinant such as the ARE in the genomic RNA of positive-sense RNA virus is seemingly suicidal. The presence of ARE in cellular mRNAs or certain RNAs of specific DNA viruses is understandable, though its presence in the sole RNA of CSFV appears harmful to its existence. However, since CSFV is extremely efficient in infectivity and multiplication and has a broad tissue tropism, the presence of the ARE hints that the virus could use the ARE interacting machinery of the host to its advantage.
Using RNA agarose beads, we isolated host factors from PK-15 cytoplasmic extracts that interact with the CSFV 3' UTR, and identified many of them using LC/MS/MS. Data attained via LC/MS/MS indicate more than one AREBP; we chose to focus on HuR first because it has been reported to protect ARE containing mRNAs from degradation. The coverage of HuR is only 3%, which could be because HuR is a nuclear/cytoplasmic protein, though it is predominantly nuclear. This study has shown that purified recombinant GST-HuR binds to CSFV 3' UTR in vitro (Figures 3 and 4).
Though HuR is predominantly nuclear, various types of stress, such as UV exposure, chemical stress, and heat shock, cause HuR to relocalize to the cytoplasm [26-29]. While CSFV genomic RNA is restricted to the cytoplasm and HuR is predominantly nuclear, viral infection is a type of stress and, as the name implies, the typical symptom of CSFV infection is high fever, which is a heat shock stress. In addition, reports show that HuR shuttles between the nucleus and the cytoplasm at various stages of the cell cycle. HuR is predominantly cytoplasmic during S-phase to M-phase transition [30], whereas in T cells, HuR is predominantly cytoplasmic during the early G1 phase [26]. The facts stated above hint at a possibility that when HuR relocalizes to the cytoplasm during such conditions, it might play a role in the CSFV lifecycle. The binding of HuR to CSFV 3' UTR and the repression of translation [19,31] may be the trigger that switches the process of translation to RNA replication.
The advent of low molecular weight inhibitors of HuR, which have demonstrated its drugability, could open exciting possibilities into the functional and mechanistic studies of HuR, and also in the therapeutic potential of combating viral infections [32]. The role of HuR in the CSFV lifecycle, and the interaction of other AREBPs with the CSFV 3' UTR ARE and their functions, is the scope of our future study.
Conclusions
In conclusion, this study used the RNA pull-down and proteomics techniques to identify several host proteins interacting with the 3' UTR of CSFV RNA. One of the CSFV 3' UTR binding proteins, HuR specifically binds to the ARE region in the 3' UTR.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MN prepared the HuR protein and did the EMSA analysis. MYC did the RNA pull down experiments. SWH prepare the labeled transcripts. CCH provided critical review of the manuscript. JTT and CHT were project supervisors, participated in the discussion of all experiments of the project and preparation of the manuscript.
All authors read and approved the manuscript
Contributor Information
Muthukumar Nadar, Email: muthukumarmsa@yahoo.com.
Meng-Yu Chan, Email: g9141011@mail.nchu.edu.tw.
Shi-Wei Huang, Email: g9241019@mail.nchu.edu.tw.
Chin-Cheng Huang, Email: cchuang@mail.nvri.gov.tw.
Joseph T Tseng, Email: tctseng@mail.ncku.edu.tw.
Ching-Hsiu Tsai, Email: chtsai1@dragon.nchu.edu.tw.
Acknowledgements
This work was supported by a grant from National Science Council Project NSC 96-2311-B-005-004-MY3.
References
- Sanchez-Cordon PJ, Nunez A, Salguero FJ, Pedrera M, Fernandez de Marco M, Gomez-Villamandos JC. Lymphocyte apoptosis and thrombocytopenia in spleen during classical swine fever: role of macrophages and cytokines. Vet Pathol. 2005;42:477–488. doi: 10.1354/vp.42-4-477. [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000;6:1791–1807. doi: 10.1017/S1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Sarnow P. Hijacking the translation apparatus by RNA viruses. J Cell Biol. 2002;158:395–399. doi: 10.1083/jcb.200205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- Lin JW, Ding MP, Hsu YH, Tsai CH. Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucleic Acids Res. 2007;35:424–432. doi: 10.1093/nar/gkl1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JH, Peng CW, Hsu YH, Tsai CH. The synthesis of minus-strand RNA of bamboo mosaic potexvirus initiates from multiple sites within the poly(A) tail. J Virol. 2002;76:6114–6120. doi: 10.1128/JVI.76.12.6114-6120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Chen J, Wang Y, Zhen Y, Lu W, Li B. Sequence, necessary for initiating RNA synthesis, in the 3'-noncoding region of the classical swine fever virus genome. Mol Biol. 2004;38:343–351. [PubMed] [Google Scholar]
- Ito T, Tahara SM, Lai MM. The 3'-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/S0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Tan W, Schwartz S. The Rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3' untranslated region. J Virol. 1995;69:2932–2945. doi: 10.1128/jvi.69.5.2932-2945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HL, Mischo HE, Steitz JA. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Mol Cell Biol. 2004;24:4522–4533. doi: 10.1128/MCB.24.10.4522-4533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseni F, Garcin D, Nishio M, Kedersha N, Anderson P, Kolakofsky D. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 2002;21:5141–5150. doi: 10.1093/emboj/cdf513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. Cell proteins TIA-1 and TIAR interact with the 3' stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J Virol. 2002;76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaria CT, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CH, Hung LY, Hsu C, Le SY, Lee PT, Liao WL, Lin YT, Chang WC, Tseng JT. RNA-binding protein HuR interacts with thrombomodulin 5'untranslated region and represses internal ribosome entry site-mediated translation under IL-1 beta treatment. Mol Biol Cell. 2008;19:3812–3822. doi: 10.1091/mbc.E07-09-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcek S, Belak S. Organization and diversity of the 3'-noncoding region of classical swine fever virus genome. Virus Genes. 1997;15:181–186. doi: 10.1023/A:1007971110065. [DOI] [PubMed] [Google Scholar]
- Vilcek S, Paton D, Lowings P, Bjorklund H, Nettleton P, Belak S. Genetic analysis of pestiviruses at the 3' end of the genome. Virus Genes. 1999;18:107–114. doi: 10.1023/A:1008000231604. [DOI] [PubMed] [Google Scholar]
- Abmayr SM, Yao T, Parmely T, Workman JL. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr Protoc Mol Biol. 2006;Chapter 12:Unit 12 11. doi: 10.1002/0471142727.mb1201s75. [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Pettiford SM, Jacobs BL. Nucleic acid affinity chromatography: preparation and characterization of double-stranded RNA agarose. Protein Expr Purif. 1995;6:25–32. doi: 10.1006/prep.1995.1004. [DOI] [PubMed] [Google Scholar]
- Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J. The 3' untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol Cell Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Sengupta TK, Spicer EK. PMA induces stabilization of oncostatin M mRNA in human lymphoma U937 cells. Biochem J. 2008;410:177–186. doi: 10.1042/BJ20070311. [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle. 2008;7:3124–3126. doi: 10.4161/cc.7.20.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, Ottl J, Oberer L, Guenat C, Moss S. et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol. 2007;3:508–515. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]