Abstract
Stem cells derived from the dental pulp of extracted human third molars (DPSCs) have the potential to differentiate into odontoblasts, osteoblasts, adipocytes, and neural cells when provided with the appropriate conditions. To advance the use of DPSCs for dentin regeneration, it is important to replicate the permissive signals that drive terminal events in odontoblast differentiation during tooth development. Such a strategy is likely to restore a dentin matrix that more resembles the tubular nature of primary dentin. Due to the limitations of culture conditions, the use of ex vivo gene therapy to drive the terminal differentiation of mineralizing cells holds considerable promise. In these studies, we asked whether the forced expression of TWIST1 in DPSCs could alter the potential of these cells to differentiate into odontoblast-like cells. Since the partnership between Runx2 and Twist1 proteins is known to control the onset of osteoblast terminal differentiation, we hypothesized that these genes act to control lineage determination of DPSCs. For the first time, our results showed that Twist1 overexpression in DPSCs enhanced the expression of DSPP, a gene that marks odontoblast terminal differentiation. Furthermore, co-transfection assays showed that Twist1 stimulates Dspp promoter activity by antagonizing Runx2 function in 293FT cells. Analysis of our in vitro data, taken together, suggests that lineage specification of DPSCs can be modulated through ex vivo gene modifications.
Keywords: TWIST1, RUNX2, dental stem cells, gene transfer, odontoblast, tooth development
When compared with the engineering of an entire biologic tooth, the targeted regeneration of individual tissues, such as dentin and bone, offers a more practical alternative that can be applied toward early treatment interventions during tooth decay and periodontal disease (Cordeiro et al., 2008). While significant progress has been made in the field of bone regeneration (Kimelman et al., 2007), stem-cell-mediated bioengineering strategies for dentin regeneration present several challenges. For example, it has been difficult to recapitulate the developmental mechanisms that drive the terminal differentiation of odontoblasts, a specialized group of cells that form dentin extracellular matrix in a highly vectorial and unidirectional manner. During primary dentin formation, odontoblasts retreat from the mineralization front, leaving behind cellular processes that remain trapped in dentinal tubules (D’Souza and Qin, 2010). Clearly, regenerating a functional dentin matrix that more closely resembles the tubular matrix of primary dentin will better impart properties of permeability and vitality to the restored tooth.
Advances made in the field of bone regeneration contribute useful information that can be extrapolated to studies on dentin regeneration (Cancedda et al., 2007; Kimelman et al., 2007). The use of allogeneic bone matrix showed inconsistent osteogenic activity, while therapy with recombinant bone morphogenetic proteins (rBMPs) resulted in variables like the lack of a sustained delivery and a short half-life that are truly difficult to modulate (Franceschi et al., 2004). Hence, ex vivo gene therapy, in which cells are made to express the growth factor or morphogen of choice, has helped overcome the deficiencies of protein therapy by allowing for the sustained delivery of inductive molecules to specific target sites (Lieberman et al., 1999; Cheng et al., 2001). As a next step in advancing the use of stem cells for bone regeneration, it was recognized that culture conditions cannot provide all the permissive signals needed to trigger terminal events in differentiation. Hence, approaches of transducing marrow stromal cells with Runx2, a transcription factor that is critical for osteoblast differentiation, resulted in substantially more bone and enhanced healing of a critical-sized cranial defect in mice (Zhao et al., 2007). Advances in skeletal biology research revealed another layer of complexity when studies revealed that Runx2’s role in osteoblast differentiation was controlled by its protein partner, Twist1. Relief of the functional antagonism between Runx2 and Twist1 causes the onset of osteoblast differentiation, as marked by the appearance of genes that mark a commitment to mineralization (Bialek et al., 2004). The precise relationship of Runx2 and Twist1 during odontoblast differentiation is not known. Such an understanding is important for translation to applications for dentin-specific tissue regeneration.
The availability of tooth-derived stem cell populations has opened up new and exciting avenues for the use of cell-mediated approaches for dentin regeneration (Gronthos et al., 2000; Zhang et al., 2006; Sloan and Smith, 2007; Cordeiro et al., 2008). The first pulp-derived stem cell population was isolated from impacted adult wisdom teeth and was termed “DPSC” (dental pulp stem cells) (Gronthos et al., 2000). Three years later, stem cells were harvested from human exfoliated deciduous teeth (SHED) (Miura et al., 2003). Both cell lines are clonogenic, form adherent cell clusters, and are capable of multilineage differentiation (Gronthos et al., 2000; Miura et al., 2003). Interestingly, SHED and DPSC can be maintained after cryopreservation (Zhang et al., 2006) and hence provide a valuable and reliable tool for tissue engineering. SHED and DPSC express mineralization markers and form mineralization nodules after culture in the presence of inductive media containing ascorbic acid, dexamethasone, and inorganic phosphate (Gronthos et al., 2000; Miura et al., 2003; Gay et al., 2007; Wei et al., 2007; Yang et al., 2007). After transplantation in vivo, increased population doublings were observed for SHED that appeared more immature and highly proliferative when compared with DPSC (Gronthos et al., 2000; Miura et al., 2003). Our previous work on SHED and DPSC in peptide amphiphile hydrogels confirmed these observations and revealed further differences. SHED maintain their high proliferation rates and a spindle-shaped, fibroblast-like morphology. They produce large amounts of collagen and thus form coherent soft tissues during a four-week culture period. DPSC growth is significantly slower in the three-dimensional environment. Up-regulation of marker genes of differentiated osteoblasts/odontoblasts and a higher potential for mineral deposition indicate that DPSC can be driven further toward terminal differentiation (Galler et al., 2008). Whether the modulation of gene expression in odontoblast progenitors mimics odontoblast differentiation during tooth development has yet to be determined. These studies were specifically designed to assess the roles of TWIST1 in driving the potential of DPSCs to differentiate into odontoblasts by lentiviral methods of gene transfer.
Materials & Methods
Plasmid Construction
The human TWIST1 cDNA (Open Biosystem, Rockford, IL, USA) was subcloned into the EcoRI and HincII sites of the pWPI lentiviral vector (Addgene, Cambridge, MA, USA). The empty pWPI vector, encoding enhanced green fluorescent protein (EGFP), was used as the control. Two TWIST1 silencing vectors, expressing different short hairpin RNA (shRNAmir) sequences targeting different segments of human TWIST1 mRNA, were obtained from Open Biosystem. The silencing effects of each shRNA on the target gene were determined by Western blot analysis of target protein expression following infection of DPSC cells with shRNA-expressing lentiviruses (see below). The shRNA sequence yielding the highest suppression level against the target gene was used for following studies. The empty pGIPZ silencing vector, expressing Turbo green fluorescent protein (tGFP), from Open Biosystem, was used as control.
To generate the pDspp-luc construct, we released a 5.7-kb promoter fragment of Dspp gene from the pBS II SK+ vector (provided by Dr. Ashok Kulkarni, NIH/NIDCR) by XbaI and NruI digestion and subcloned it into the NheI and HindIII sites of the pGL3-basic vector (Promega, Madison, WI, USA). The pDmp1-luc construct, containing a 9.6-kb promoter region, exon 1 and intron 1 of the Dmp1 gene, was prepared as described earlier (Lu et al., 2005). The human RUNX2 cDNA, encoding FLAG-RUNX2, was obtained from Dr. Brendan Lee.
Culture of DPSCs
DPSCs isolated from adult human third molars were kindly provided by Dr. Songtao Shi at the University of Southern California. DPSCs were maintained in growth medium, containing alpha modification of Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with 15% fetal calf serum (Hyclone, Logan, UT, USA), 100 µg/mL L-ascorbic acid 2-phosphate (Sigma, St. Louis, MO, USA), 2 mmol/L glutamine (Invitrogen), 100 IU/mL penicillin, and 100 µg/mL streptomycin (Invitrogen). For differentiation assays, DPSCs were grown to confluence in growth medium, then in differentiation medium containing normal growth medium supplemented with 100 µg/mL L-ascorbic acid 2-phosphate, 2 mM β-glycerophosphate, and 10 nM dexamethasone for 2 or 4 wks.
Lentivirus Packaging
293FT cells (Invitrogen) were maintained in DMEM supplemented with 10% fetal bovine serum (Hyclone), 2 mmol/L glutamine (Invitrogen), 100 IU/mL penicillin, and 100 µg/mL streptomycin (Invitrogen). To prepare lentivirus for DPSC cell infection, we co-transfected 293FT cells with lentiviral vector, packaging vector psPAX2, and envelope vector pMD2.G, using Fugene6 (Roche, Indianapolis, IN, USA), as per the manufacturer’s instructions. The supernatant was collected 48 hrs after transfection and filtered with 0.45-µm filters for removal of cell debris.
Lentiviral Transduction and Cell-sorting
For lentiviral transduction, sub-confluent DPSCs were incubated with lentiviral supernatant in the presence of 10 µg/mL of polybrene (Sigma) for 48 hrs, with lentiviral supernatant changed once. DPSCs stably expressing green fluorescent protein (GFP) were selected by fluorescent-activated cell-sorting (FACS), with the use of a Vantage SE Diva cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA). Sorted DPSCs were expanded for two passages for further studies. Cells overexpressing or silencing TWIST1 were confirmed by Western blot analysis.
Western Blot Analysis and von Kossa Staining
Blots were first immunolabeled with the following primary antibodies, anti-TWIST1 monoclonal antibody (Abcam, 1:50), anti-alkaline phosphatase monoclonal antibody (Abcam, Cambridge, MA, USA; 1:100), anti-osteocalcin monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:1000), rabbit anti-dentin matrix protein 1 polyclonal antibody (gift from Larry W. Fisher, 1:500), anti-osteopontin monoclonal antibody (Santa Cruz Biotechnology, 1:1000), rabbit anti-dentin sialoprotein polyclonal antibody (gift from Larry W. Fisher, 1:250), or anti-β-actin monoclonal antibody (Sigma, 1:20,000). They were then incubated with horseradish-peroxidase-conjugated goat anti-mouse or anti-rabbit secondary antibody (Santa Cruz Biotechnology). The blot was finally detected with ECL reagents (GE Healthcare Bio-Sciences, Piscataway, NJ, USA).
For von Kossa staining, DPSC cells were fixed with 4% paraformaldehyde-0.1M phosphate buffer, and stained with 10% silver nitrate for von Kossa staining.
Promoter-luciferase Assays
We used promoter-luciferase assays to determine the effects of RUNX2 and TWIST1 interactions on Dmp1 and Dspp promoter activities in 293FT cells. The cells were transiently transfected with pDmp1-Luc or pDspp reporter construct and empty vector or vectors expressing FLAG-RUNX2 or TWIST1 or both, with Fugene 6 as described above. pRL-TK was co-transfected as an internal control for normalizing transfection efficiency. Firefly and Renilla luciferase activities were analyzed 48 hrs after transfection with the use of a dual luciferase assay system (Promega) following the manufacturer’s instructions. Each assay was performed in triplicate and repeated at least three times.
Results
TWIST1 Enhances the Odontoblast-like Differentiation of DPSCs
We first assessed whether the transduction of dental stem cells with transcription factors like TWIST1 would alter their differentiation into osteoblasts and odontoblasts. DPSCs that were stably transduced with lentivirus expressing TWIST1 or their silencing RNA probe or control vector were carefully selected by cell-sorting. Western blot analysis showed that the overexpression of TWIST1 in DPSCs stably transduced with TWIST1 lentiviral vectors (Fig. 1A). The TWIST1 silencing lentiviral vectors, ShT1 and ShT2, reduced the levels of TWIST1 protein 50% and 90%, respectively (Fig. 1B). Therefore, ShT2 were selected for subsequent TWIST1 silencing experiments.
Figure 1.
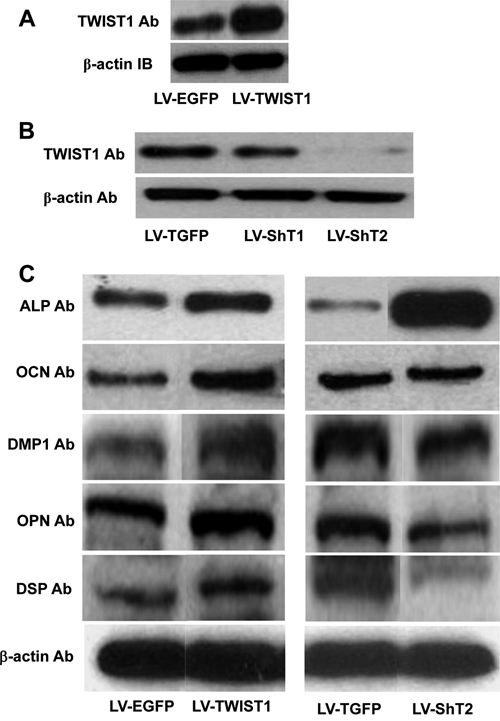
TWIST1 modulates the odontoblast-like differentiation of human DPSCs. (A) Human DPSCs were stably transduced with lentiviruses expressing TWIST1 (LV-TWIST1) or empty lentiviral vectors expressing only enhanced green fluorescent protein (LV-EGFP). Western blot analysis showed the overexpression of Twist1 in DPSCs transduced with LV-TWIST1 compared with LV-EGFP. (B) Human DPSCs were transduced with lentiviruses expressing 2 short hairpin RNA targeting human TWIST1 mRNA (LV-ShT1 or LV-ShT2, respectively) or empty lentiviral vectors expressing only Turbo green fluorescent protein (LV-TGFP). Western blot analysis showed that LV-ShT1 and LV-ShT2 knocked down TWIST1 expression by 50% and 90%, respectively. (C) Western blot analysis of total proteins extracted from DPSCs stably transduced with TWIST1 overexpressing (LV-TWIST1, left panel) or silencing (LV-ShT2, right panel) lentiviral vectors or their control vectors. DPSCs stably transduced with the indicated lentiviral vectors were allowed to grow for 2 wks in the differentiation medium. Total proteins were extracted and analyzed with antibodies for ALP, OCN, DMP1, OPN, and DSP, with β-actin as a loading control. TWIST1 overexpression enhanced the expression of OCN, DMP1, and OPN as well as DSP, but had no apparent effect on ALP expression. Consistently, TWIST1 silencing dramatically enhanced the expression of ALP, but inhibited DMP1, OPN, and DSP, and had no effect on OCN expression.
Next, we analyzed the effects of Twist1 overexpression and silencing on the differentiation potential of DPSCs, including differentiation markers and mineral deposition. The differentiation markers analyzed for both osteoblasts and odontoblasts include an early-differentiation marker, alkaline phosphatase (ALP), and 4 late-differentiation markers, osteocalcin (OCN), dentin matrix protein 1 (DMP1), osteopontin (OPN), and dentin sialophosphoprotein (DSPP). Western blot analysis showed that TWIST1 overexpression enhanced the expression of OCN, DMP1, and OPN as well as DSP, but had no apparent effect on ALP expression (Fig. 1C, left panel). Consistently, TWIST1 silencing dramatically enhanced the expression of ALP, but had differential effects on the late-differentiation markers, inhibiting DMP1, OPN, and DSP expression while exerting no effect on OCN expression (Fig. 1C, right panel). Consistent with the Western blot data, von Kossa staining showed that TWIST1 overexpression enhanced the mineral deposition of DPSCs, whereas its silencing appeared to inhibit mineralization (Fig. 2A). DSPP is an odontoblast-specific differentiation marker, which is processed into N-terminal fragment, DSP, and C-terminal fragment, DPP after synthesis. Analysis of these data thus suggests that TWIST1 enhances the mineralizing capacity of DPSCs, most likely through promoting the expression of DSPP, a late-differentiation maker of odontoblast differentiation.
Figure 2.
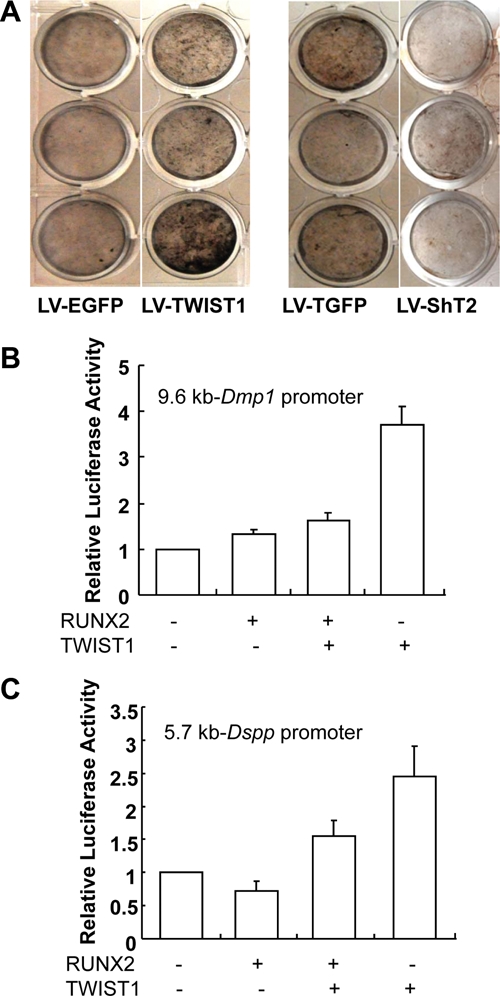
TWIST1 regulates mineralization of DPSCs through antagonizing RUNX2 function. (A) Von Kossa staining. DPSCs stably transduced with the indicated lentiviral vectors were cultured for 4 wks in the differentiation medium. Mineralization was determined by von Kossa staining. TWIST1 overexpressing enhanced the mineral deposition of DPSCs (LV-TWIST1, left panel), whereas its silencing inhibited the mineral deposition (LV-ShT2, right panel). (B) pDmp1-Luc reporter construct was transiently co-transfected into 293FT cells with expression vectors for RUNX2 or TWIST1 alone or both. While RUNX2 stimulated Dmp1 promoter activity, it inhibited the stimulatory effects of TWIST1 on Dmp1 promoter activity. Data are mean ± the standard error of mean (SEM), n = 3. (C) pDspp-Luc reporter construct was transiently co-transfected into 293FT cells with expression vectors for RUNX2 or TWIST1 alone or both. RUNX2 inhibited Dspp promoter activity, and further inhibited the stimulatory effects of TWIST1 on Dspp promoter activity. Data are mean ± SEM, n = 3.
TWIST1 Stimulates Dmp1 and Dspp Promoter Activities
Previous studies have shown that Twist1 inhibits the transactivation functions of Runx2 by binding to its DNA-binding domain. It is the release of this functional antagonism between Twist1 and Runx2 that triggers the onset of osteoblast differentiation (Bialek et al., 2004). To determine the effects of TWIST1 and RUNX2 protein interaction on odontoblast differentiation, we studied the functional consequences of their interactions on the promoter activities of 2 odontoblast differentiation markers, Dmp1 and Dspp. pDspp-Luc or pDmp1-Luc were transiently co-transfected into 293FT cells with expression vectors for FLAG-RUNX2 or TWIST1 alone or both. Although co-transfection of RUNX2 expression vector increased Dmp1 promoter activity by 33%, TWIST1 expression vector resulted in a 3.7-fold increase in Dmp1 promoter activity (Fig. 2B). More interestingly, co-transfection of RUNX2 expression vector decreased Dspp promoter activity by 28%, in contrast to a 2.5-fold increase in Dspp promoter activity with TWIST1 (Fig. 2C). Further, co-transfection of both RUNX2 and TWIST1 expression vectors resulted in a 63% decrease in Dmp1 promoter activity and a 55% decrease in Dspp promoter activity, compared with TWIST1 expression vector alone (Figs. 2B, 2C). Analysis of these data suggests that TWIST1 activates Dmp1 and Dspp promoter activities, and antagonizes the effects of Runx2 function on these promoters.
Discussion
In this study, we have demonstrated that the differentiation capacity of human-tooth-derived pulpal stem cells, DPSCs, could be altered by overexpressing or silencing TWIST1 levels with lentiviral vectors. We showed that DPSCs could be stably transduced with lentivirus vectors that either increased or silenced expression levels of TWIST1. Analyses of these lentivirus-transduced DPSCs revealed that TWIST1 has the potential to enhance the odontoblastic potential of DPSCs in a manner that will benefit the regeneration of dentin.
Lentivirus-mediated TWIST1 overexpression stimulated expression of all the late-mineralization markers—OCN, DMP1, and OPN—that are common for both osteoblasts and odontoblasts. In addition, TWIST1 overexpression also enhanced the expression of DSPP, a tooth-specific marker. We thus conclude that such transduced DPSCs assumed a phenotype more resembling odontoblasts than osteoblasts. Consistently, lentivirus-mediated Twist1 silencing reduced DSPP protein expression. Furthermore, analysis of data from co-transfection assays confirmed that TWIST1 directly activates the Dspp promoter, and that this activity is significantly reduced in the presence of RUNX2. Similar effects of TWIST1 activation of Dmp1 were observed in the presence and absence of RUNX2. These observations can be interpreted to suggest that the partnership between RUNX2 and TWIST1 is important for the diversification of odontoblasts from osteoblasts, and that TWIST1 may be responsible for driving terminal differentiation of odontoblasts through its activation of Dspp expression.
These data closely align with information derived from developmental studies. During tooth development, Runx2 and Twist1 share overlapping yet unique profiles of expression in dental mesenchyme. Both genes are co-expressed during tooth initiation and morphogenesis (bud, cap, and early bell stages). Interestingly, at the onset of odontoblast differentiation, Runx2 is markedly down-regulated (D’Souza et al., 1999), while Twist1 expression persists in the cells that lie at the interface with dental epithelium and are destined to become odontoblasts (Bourgeois et al., 1998). This is in sharp contrast to expression profiles of Runx2 and Twist1 within the osteogenic zones that are contiguous with dental mesenchyme. Runx2 expression persists in osteoblasts that differentiate and form osteoid at the time that Twist1 expression is markedly down-regulated. These differential profiles of expression of Runx2 and Twist1 during odontoblast and osteoblast differentiation suggest that further lineage determination of mineralizing cells is the result of differences in signaling pathways that control gene expression in odontogenic vs. osteogenic zones. Our earlier studies also showed that the up-regulation of the odontoblast differentiation markers, type I collagen and Dspp, occurred only after Runx2 expression decreased, and was closely correlated to the persistence of Twist1 expression in the pre-odontoblast layer (D’Souza et al., 1999; Galler et al., 2008). Furthermore, previous studies have demonstrated that continuous Runx2 expression inhibits the terminal differentiation of odontoblasts, resulting in their transdifferentiation into osteoblasts, as marked by the loss of Dspp gene expression (Miyazaki et al., 2008). The observation that Twist1 overexpression did not affect ALP levels indicates that genes that mark the earliest commitment to mineralization are not involved. These expression profiles suggest that Runx2 and Twist1 may partner in tooth development at stages that precede cell differentiation and matrix synthesis. However, it is Twist1 that is likely needed for the terminal events that drive odontoblast differentiation and the subsequent deposition of primary dentin matrix.
The present in vitro studies, although limited in approach, will provide a framework for future studies that should test the ability of Twist1 expressing cells to form dentin matrix both in vitro and in vivo. Clearly, ex vivo gene therapy methods may not offer the most practical alternative for enhancing the targeted differentiation of dental stem cells in tissue-engineering strategies for dentin regeneration. However, these studies provide the impetus for future experiments that will identify effector molecules of Twist1 that could be used therapeutically.
Footnotes
This research was supported by NIDCR/NIH Grants R01 DE013368 and U24 DE16472 to RDS.
The authors declare no conflicts of interest.
References
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, et al. (2004). A twist code determines the onset of osteoblast differentiation. Dev Cell 6:423-435 [DOI] [PubMed] [Google Scholar]
- Bourgeois P, Bolcato-Bellemin AL, Danse JM, Bloch-Zupan A, Yoshiba K, Stoetzel C, et al. (1998). The variable expressivity and incomplete penetrance of the twist-null heterozygous mouse phenotype resemble those of human Saethre-Chotzen syndrome. Hum Mol Genet 7:945-957 [DOI] [PubMed] [Google Scholar]
- Cancedda R, Giannoni P, Mastrogiacomo M. (2007). A tissue engineering approach to bone repair in large animal models and in clinical practice. Biomaterials 28:4240-4250 [DOI] [PubMed] [Google Scholar]
- Cheng SL, Lou J, Wright NM, Lai CF, Avioli LV, Riew KD. (2001). In vitro and in vivo induction of bone formation using a recombinant adenoviral vector carrying the human BMP-2 gene. Calcif Tissue Int 68:87-94 [PubMed] [Google Scholar]
- Cordeiro MM, Dong Z, Kaneko T, Zhang Z, Miyazawa M, Shi S, et al. (2008). Dental pulp tissue engineering with stem cells from exfoliated deciduous teeth. J Endod 34:962-969 [DOI] [PubMed] [Google Scholar]
- D’Souza R, Qin C. (2010). The development of the pulp-dentin complex. In: Seltzer and Bender’s The dental pulp. 2nd ed. Hargreaves K, Goodis H, editors. Chicago: Quintessence International [Google Scholar]
- D’Souza RN, Aberg T, Gaikwad J, Cavender A, Owen M, Karsenty G, et al. (1999). Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development 126:2911-2920 [DOI] [PubMed] [Google Scholar]
- Franceschi RT, Yang S, Rutherford RB, Krebsbach PH, Zhao M, Wang D. (2004). Gene therapy approaches for bone regeneration. Cells Tissues Organs 176:95-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galler KM, Cavender A, Yuwono V, Dong H, Shi S, Schmalz G, et al. (2008). Self-assembling peptide amphiphile nanofibers as a scaffold for dental stem cells. Tissue Eng Part A 14:2051-2058 [DOI] [PubMed] [Google Scholar]
- Gay IC, Chen S, MacDougall M. (2007). Isolation and characterization of multipotent human periodontal ligament stem cells. Orthod Craniofac Res 10:149-160 [DOI] [PubMed] [Google Scholar]
- Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. (2000). Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo . Proc Natl Acad Sci USA 97:13625-13630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman N, Pelled G, Helm GA, Huard J, Schwarz EM, Gazit D. (2007). Review: gene- and stem cell-based therapeutics for bone regeneration and repair. Tissue Eng 13:1135-1150 [DOI] [PubMed] [Google Scholar]
- Lieberman JR, Daluiski A, Stevenson S, Wu L, McAllister P, Lee YP, et al. (1999). The effect of regional gene therapy with bone morphogenetic protein-2-producing bone-marrow cells on the repair of segmental femoral defects in rats. J Bone Joint Surg Am 81:905-917 [DOI] [PubMed] [Google Scholar]
- Lu Y, Zhang S, Xie Y, Pi Y, Feng JQ. (2005). Differential regulation of dentin matrix protein 1 expression during odontogenesis. Cells Tissues Organs 181:241-247 [DOI] [PubMed] [Google Scholar]
- Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, et al. (2003). SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA 100:5807-5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Kanatani N, Rokutanda S, Yoshida C, Toyosawa S, Nakamura R, et al. (2008). Inhibition of the terminal differentiation of odontoblasts and their transdifferentiation into osteoblasts in Runx2 transgenic mice. Arch Histol Cytol 71:131-146 [DOI] [PubMed] [Google Scholar]
- Sloan AJ, Smith AJ. (2007). Stem cells and the dental pulp: potential roles in dentine regeneration and repair. Oral Dis 13:151-157 [DOI] [PubMed] [Google Scholar]
- Wei X, Ling J, Wu L, Liu L, Xiao Y. (2007). Expression of mineralization markers in dental pulp cells. J Endod 33:703-708 [DOI] [PubMed] [Google Scholar]
- Yang X, van den Dolder J, Walboomers XF, Zhang W, Bian Z, Fan M, et al. (2007). The odontogenic potential of STRO-1 sorted rat dental pulp stem cells in vitro. J Tissue Eng Regen Med 1:66-73 [DOI] [PubMed] [Google Scholar]
- Zhang W, Walboomers XF, van Kuppevelt TH, Daamen WF, Bian Z, Jansen JA. (2006). The performance of human dental pulp stem cells on different three-dimensional scaffold materials. Biomaterials 27:5658-5668 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Wang Z, Ge C, Krebsbach P, Franceschi RT. (2007). Healing cranial defects with AdRunx2-transduced marrow stromal cells. J Dent Res 86:1207-1211 [DOI] [PubMed] [Google Scholar]


