Abstract
In this review, the authors survey the large number of antibacterial and antiviral proteins present in human saliva. Of interest, most of these antibacterial proteins display antiviral activity, typically against specific viral pathogens. The review focuses on one protein that interacts with both bacteria and viruses—gp340, originally referred to as salivary agglutinin. In the oral cavity, soluble gp340 binds to and aggregates a variety of bacteria, and this is thought to increase bacterial clearance from the mouth. However, when bound to the tooth surface, gp340 promotes bacterial adherence. In the oral cavity, most gp340 is found soluble in saliva and can function as a specific inhibitor of infectivity of HIV-1 and influenza A. In contrast, in the female reproductive track, most gp340 is bound to the cell surface, where it can promote HIV-1 infection.
Keywords: HIV, AIDS, viral, antiviral, innate immune system
Like many other mucosal surfaces, the oral cavity is exposed to a variety of infectious agents and toxic compounds. To provide a variety of biological functions (communication, nutrient intake, respiratory activity, etc), it needs to be an open system, yet this requires an equally diverse group of activities/biomolecules to insure its continued viability. Thus, copious salivary flow is important for mastication, taste, the maintenance of hard tissue structure, and the prevention of a range of toxic and infectious assaults. In this article, we focus on one aspect of the role of saliva—namely, protection from infection, specifically via the innate immune system. This system is an important first-line defense against bacterial and viral infection. In addition, mucosal surfaces are typically coated by a protective layer of mucins. Although salivary flow plays a major role in the continuous cleansing of the oral cavity (Dawes, 2008), we focus here on proteins in the oral cavity that demonstrate both antibacterial and antiviral activity.
To obtain an overview of antimicrobial proteins present in saliva, we carried out a PubMed search of the literature (http://www.ncbi.nlm.nih.gov/pubmed/). The Table lists the major proteins found in saliva and documented to demonstrate antibacterial activity. Though not exhaustive, this list contains proteins shown to be either bacteriostatic or bacteriocidal against multiple bacterial species. Of interest, most of these proteins have been reported to be antiviral for at least one virus. As noted on the table, the only entities reported having antibacterial but not antiviral activity are the histatins and calprotectin.
Table.
Salivary Antibacterial and Antiviral Activities
| Salivary Constituent | Antibacterial | Antiviral |
|---|---|---|
| Cathelcidin (LL-37) | ✓ | ✓ |
| Lactoferrin | ✓ | ✓ |
| Lysozyme | ✓ | ✓ |
| Mucins | ✓ | ✓ |
| Peroxidase | ✓ | ✓ |
| Salivary agglutinin (gp340, DMBT1) | ✓ | ✓ |
| sIgA | ✓ | ✓ |
| SLPI | ✓ | ✓ |
| α,β Defensins | ✓ | ✓ |
| Calprotectin (calgranulin) | ✓ | |
| Histatins | ✓ |
Despite the range of antiviral activities found in saliva, many reports still identify viruses in saliva, often infectious—including HSV, HIV, VZV, EBV, HPV, hepatitis A, hepatitis C, Ebola, Norwalk virus, HHV 6 and 8, measles, rabies, adenoviruses, and prions. Although infectious virus in the presence of potent antiviral activity appears counterintuitive, there are potential explanations (as suggested in the Discussion section).
Our studies have focused on a salivary protein initially identified as possessing antibacterial activity but subsequently found to have potent inhibitory activity against HIV-1. Significantly, this activity is manifest against only HIV-1 and influenza A (Wu et al., 2003; White et al., 2005a), with little or no activity against HSV, adenovirus, HIV-2, or SIV. This protein was referred to as salivary agglutinin and is now more commonly called gp340 or DMBT-1 (Wu et al., 2004). Figure 1 outlines the timeline of these discoveries. From 1986 to 1988, a series of publications—first by Fultz (1986) and then Fox et al. (1988, 1989)—reported that incubation of HIV-1 in human or chimpanzee saliva resulted in a loss of infectivity. Many groups subsequently began to report a variety of salivary proteins demonstrating anti-HIV activity (Archibald and Cole, 1990; Bergey et al., 1993a, 1993b; Malamud et al., 1993; McNeely et al., 1995). In 1997, we reported that the effect of saliva on HIV inhibition was directly on the virus and specific for HIV-1 (Malamud et al., 1997), and in 1998 we demonstrated that salivary agglutinin could inhibit HIV infectivity by binding to gp120 on the surface of the virus (Nagashunmugam et al., 1998). Subsequent cloning of gp340 (Holmskov et al., 1999) and proteomic analysis by Prakobphol et al. (2000) identified salivary agglutinin as a previously cloned protein, DMBT-1 or gp340.
Fig. 1.
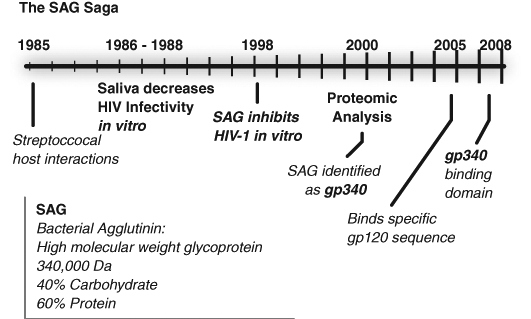
Timeline of salivary agglutinin (SAG; i.e., gp340) antibacterial and anti-HIV-1 activity.
gp340 Overview
To briefly summarize the published data on gp340, we reviewed the literature in the field. Salivary agglutinin, gp340, and the tumor suppressor DMBT1 (deleted in malignant brain tumors) are encoded by the dmbt1 gene. Proteins in the scavenger receptor cysteine-rich (SRCR) superfamily are either secreted or membrane-bound, demonstrating a variety of biologic functions (Resnick et al., 1994; Kang and Reid, 2003; Sarrias et al., 2004; Ligtenberg et al., 2007a, 2007b). Members of this family have at least one conserved domain, with either 6 or 8 cysteines forming characteristic disulfide bonds (S-S). The sequences of SRCR domains consist of about 110 amino acids that are conserved in evolution from sponges to humans. In sum, gp340 contains 14 SRCR domains, with the first 13 being highly conserved and the 14th somewhat less so (Holmskov et al., 1999). In general, the biological functions of the SRCR family of proteins consist of either tumor suppressor activity or innate immune defense properties. Salivary gp340 is able to aggregate a range of oral bacteria (Golub et al., 1985) and demonstrate antiviral activity against HIV-1 (Wu et al., 2003) as well as influenza A (White et al., 2005a, 2005b).
Identification of a Bioactive Peptide Within N-Terminal SRCR
In a series of elegant studies, a research team in Amsterdam, Netherlands, identified a bioactive peptide within a consensus SRCR sequence that binds to streptococcal receptors and mediates bacterial aggregation (Bikker et al., 2002). Using alanine scanning, they were able to identify a 5–amino acid motif required within this sequence for antibacterial activity (Bikker et al., 2004).
Molecular Modeling Studies
Our group examined the reported antibacterial sequence in SRCR, and we used an available crystal structure for a single domain from the Mac-2 binding proteins to create a model for the active domain identified by Bikker et al. (2004) (see Fig. 2). Of interest, all the key amino acids identified by Bikker et al. occur on one face of the structure in this predicted model.
Fig. 2.
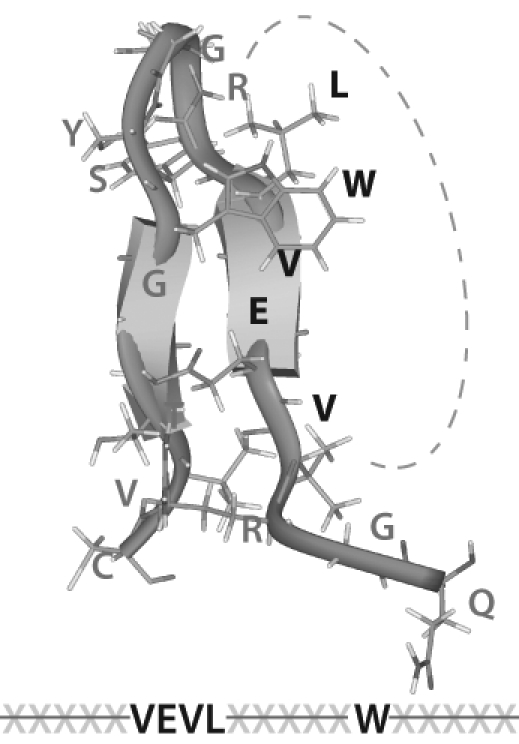
Tertiary structure model for the active bacterial-binding motif in a scavenger receptor cysteine-rich (SRCR) domain that is proposed as being responsible for anti-HIV activity.
Because we had previously identified the sequence in HIV-1 gp120 involved in binding gp340 (Wu et al., 2004), we carried out a best-fit model of the binding between SRCR and HIV-1 env, using a docking module in the Discovery Studio software suite (Accelyris RDOCK, V 2.0; Accelrys, San Diego, California). Figure 3 presents the predicted interaction of gp340 SRCR with HIV gp120. Although it is not certain that the same peptide sequence of the SRCR binds both a bacterial and a viral target, the molecular modeling studies provide a hypothesis-generating approach that allows one to carry out targeted mutations. These studies are currently ongoing.
Fig. 3.
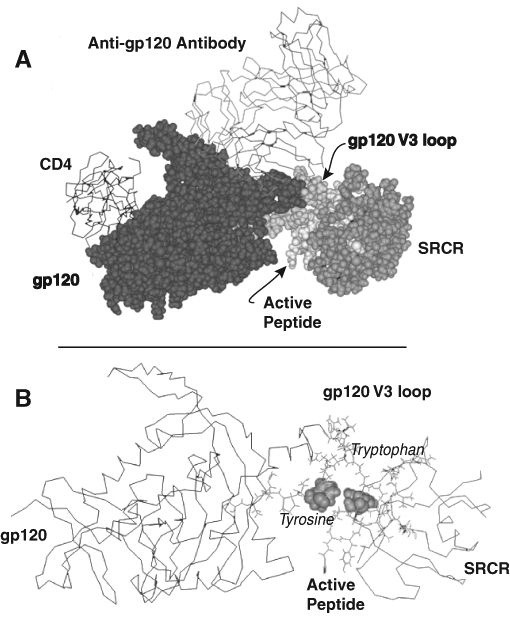
Space filling (A) and wire frame (B) models showing the putative interactions of the active peptides on an scavenger receptor cysteine-rich (SRCR) domain with the V3 loop of gp120. The positions of tryptophan and tyrosine on the intersecting face are indicated.
Parallel patterns?
We recently reported that, in contrast to the oral cavity, the female reproductive track is characterized by low levels of soluble gp340 (Cummins et al., 2006) but has high levels of gp340 on the surface of vaginal and cervical epithelial cells, as detected by immunohistochemistry and FACS analyses (Stoddard et al., 2007; Cannon et al., 2008). When incubated with HIV-1, cells derived from the oral cavity do not bind virus; in contrast, when vaginal or cervical epithelial cells are incubated with HIV-1, they bind virus and are subsequently able to transfer the captured virions to CD4+ T cells (Stoddard et al., 2007). It is thus possible that in the mouth, soluble gp340 aggregates bacteria and inhibits HIV infection whereas bound gp340 increases bacterial adhesion (Rosan et al., 1982a, 1982b) and promotes oral disease by increasing bacterial adherence. Instead, in the female reproductive track, gp340 is primarily on the cell surface and is able to bind HIV; these virions can then remain infectious for > 4 days, thereby potentially promoting HIV infection.
Discussion
In this brief overview, we describe a variety of antibacterial proteins present in saliva, most of which also demonstrate antiviral activity. Despite this range of antimicrobial proteins, the oral cavity remains a site for colonization by multiple bacterial species and numerous viruses. These findings suggest a complex interplay between microbial colonization and clearance in the oral cavity. The antimicrobial proteins may be in solution or bound to hard and soft tissues in the oral cavity. Thus, the levels of the antibacterial proteins and the site within the oral cavity where these proteins are found modulate the balance between bacterial adherence and clearance. If salivary agglutinin (gp340) is in its soluble form in saliva, then it acts primarily as a bacterial aggregator leading to clearance. In contrast, when salivary agglutinin is localized to the tooth surface, it serves to foster bacterial adhesion (Carlen et al., 1998). In addition, this same glycoprotein can be involved in biofilm formation (Demuth et al., 2001; Ahn et al., 2008) and thereby interact with other bacteria to promote either attachment to a surface or clearance from the oral cavity.
In attempting to understand the persistence of infectious viruses in the oral cavity despite the presence of potent antiviral activities, it is important to note that most of the antiviral proteins have relatively limited antiviral potency and a narrow range of activity. As such, gp340 interacts with both bacteria and only 2 viruses: HIV-1 and influenza A. Note that these are both enveloped RNA viruses with a susceptible glycoprotein target on their surfaces—gp120 and hemagglutinin, respectively—which suggests a common mechanism of action of gp340 with these 2 viruses. The concentration of specific salivary proteins differs among individuals such that some individuals have higher levels of specific antibacterial and/or antiviral proteins. These levels could be part of the explanation for the differential infectivity with bacteria such as Streptococcus pyogenes and Streptococcus mutans and viruses such as influenza A and HIV-1, which infect some individuals more efficiently than they do others. It also appears that the oral cavity is susceptible to colonization by a variety of microorganisms and that there is a diversity of antimicrobial activities with unique modes of action. In addition to “backup” in the case of resistance mutations, the existence of multiple active antimicrobial molecules with different modes of bioactivity ensures protection against diverse pathogens.
Footnotes
This research project was supported by the National Institute of Dental and Craniofacial Research (U19 DE018385, DE14825) and a New York State Foundation for Science, Technology and Innovation award to D.M.
References
- Ahn SJ, Wen ZT, Brady LJ, Burne RA. (2008). Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect Immun 76:4259-4268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald DW, Cole GA. (1990). In vitro inhibition of HIV-1 infectivity by human salivas. AIDS Res Hum Retroviruses 6:1425-1432 [DOI] [PubMed] [Google Scholar]
- Bergey EJ, Cho MI, Hammarskjold ML, Rekosh D, Levine MJ, Blumberg BM, et al. (1993a). Aggregation of human immunodeficiency virus type 1 by human salivary secretions. Crit Rev Oral Biol Med 4:467-474 [DOI] [PubMed] [Google Scholar]
- Bergey EJ, Gu M, Collins AR, Bradway SD, Levine MJ. (1993b). Modulation of herpes simplex virus type 1 replication by human salivary secretions. Oral Microbiol Immunol 8:89-93 [DOI] [PubMed] [Google Scholar]
- Bikker FJ, Ligtenberg AJ, End C, Renner M, Blaich S, Lyer S, et al. (2004). Bacteria binding by DMBT1/SAG/gp-340 is confined to the VEVLXXXXW motif in its scavenger receptor cysteine-rich domains. J Biol Chem 279:47699-47703 [DOI] [PubMed] [Google Scholar]
- Bikker FJ, Ligtenberg AJ, Nazmi K, Veerman EC, van’t Hof W, Bolscher JG, et al. (2002). Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J Biol Chem 277:32109-32115 [DOI] [PubMed] [Google Scholar]
- Cannon G, Yi Y, Ni H, Stoddard E, Scales DA, Van Ryk DI, et al. (2008). HIV envelope binding by macrophage-expressed gp340 promotes HIV-1 infection. J Immunol 181:2065-2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlen A, Bratt P, Stenudd C, Olsson J, Stromberg N. (1998). Agglutinin and acidic proline-rich protein receptor patterns may modulate bacterial adherence and colonization on tooth surfaces. J Dent Res 77:81-90 [DOI] [PubMed] [Google Scholar]
- Cummins JE, Christensen L, Lennox JL, Bush TJ, Wu Z, Malamud D, et al. (2006). Mucosal innate immune factors in the female genital tract are associated with vaginal HIV-1 shedding independent of plasma viral load. AIDS Res Hum Retroviruses 22:788-795 [DOI] [PubMed] [Google Scholar]
- Dawes C. (1998). Recent research on calculus. N Z Dent J 94(416):60-62 [PubMed] [Google Scholar]
- Demuth DR, Irvine DC, Costerton JW, Cook GS, Lamont RJ. (2001). Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect Immun 69:5736-5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PC, Wolff A, Yeh CK, Atkinson JC, Baum BJ. (1988). Saliva inhibits HIV-1 infectivity. J Am Dent Assoc 116:635-637 [DOI] [PubMed] [Google Scholar]
- Fox PC, Wolff A, Yeh CK, Atkinson JC, Baum BJ. (1989). Salivary inhibition of HIV-1 infectivity: functional properties and distribution in men, women, and children. J Am Dent Assoc 118:709-711 [DOI] [PubMed] [Google Scholar]
- Fultz PN. (1986). Components of saliva inactivate human immunodeficiency virus. Lancet 2:1215. [DOI] [PubMed] [Google Scholar]
- Golub EE, Cheruka J, Boosz B, Davis C, Malamud D. (1985). A comparison of bacterial aggregation induced by saliva, lysozyme, and zinc. Infect Immun 48:204-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmskov U, Mollenhauer J, Madsen J, Vitved L, Gronlund J, Tornoe I, et al. (1999). Cloning of gp-340, a putative opsonin receptor for lung surfactant protein D. Proc Natl Acad Sci U S A 96:10794-10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Reid KB. (2003). DMBT1, a regulator of mucosal homeostasis through the linking of mucosal defense and regeneration? FEBS Lett 540:21-25 [DOI] [PubMed] [Google Scholar]
- Ligtenberg AJ, de Soet JJ, Veerman EC, Amerongen AV. (2007a). Oral diseases: from detection to diagnostics. Ann N Y Acad Sci 1098:200-203 [DOI] [PubMed] [Google Scholar]
- Ligtenberg AJ, Veerman EC, Nieuw Amerongen AV, Mollenhauer J. (2007b). Salivary agglutinin/glycoprotein-340/DMBT1: a single molecule with variable composition and with different functions in infection, inflammation and cancer. Biol Chem 388:1275-1289 [DOI] [PubMed] [Google Scholar]
- Malamud D, Davis C, Berthold P, Roth E, Friedman H. (1993). Human submandibular saliva aggregates HIV. AIDS Res Hum Retroviruses 9:633-637 [DOI] [PubMed] [Google Scholar]
- Malamud D, Nagashunmugam T, Davis C, Kennedy S, Abrams WR, Kream R, et al. (1997). Inhibition of HIV infectivity by human saliva. Oral Dis 3 Suppl 1:(S58-63). [DOI] [PubMed] [Google Scholar]
- McNeely TB, Dealy M, Dripps DJ, Orenstein JM, Eisenberg SP, Wahl SM. (1995). Secretory leukocyte protease inhibitor: a human saliva protein exhibiting anti-human immunodeficiency virus 1 activity in vitro. J Clin Invest 96:456-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashunmugam T, Malamud D, Davis C, Abrams WR, Friedman HM. (1998). Human submandibular saliva inhibits human immunodeficiency virus type 1 infection by displacing envelope glycoprotein gp120 from the virus. J Infect Dis 178(6):1635-1641 [DOI] [PubMed] [Google Scholar]
- Prakobphol A, Xu F, Hoang VM, Larsson T, Bergstrom J, Johansson I, et al. (2000). Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J Biol Chem 275:39860-39866 [DOI] [PubMed] [Google Scholar]
- Resnick D, Pearson A, Krieger M. (1994). The SRCR superfamily: a family reminiscent of the Ig superfamily. Trends Biochem Sci 19:5-8 [DOI] [PubMed] [Google Scholar]
- Rosan B, Appelbaum B, Golub E, Malamud D, Mandel ID. (1982a). Enhanced saliva-mediated bacterial aggregation and decreased bacterial adhesion in caries-resistant versus caries-susceptible individuals. Infect Immun 38:1056-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosan B, Malamud D, Appelbaum B, Golub E. (1982b). Characteristic differences between saliva-dependent aggregation and adhesion of streptococci. Infect Immun 35:86-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrias MR, Gronlund J, Padilla O, Madsen J, Holmskov U, Lozano F. (2004). The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit Rev Immunol 24:1-37 [DOI] [PubMed] [Google Scholar]
- Stoddard E, Cannon G, Ni H, Kariko K, Capodici J, Malamud D, et al. (2007). gp340 expressed on human genital epithelia binds HIV-1 envelope protein and facilitates viral transmission. J Immunol 179:3126-3132 [DOI] [PubMed] [Google Scholar]
- White MR, Crouch E, van Eijk M, Hartshorn M, Pemberton L, Tornoe I, et al. (2005a). Cooperative anti-influenza activities of respiratory innate immune proteins and neuraminidase inhibitor. Am J Physiol Lung Cell Mol Physiol 288:L831-L840 [DOI] [PubMed] [Google Scholar]
- White MR, Crouch E, Vesona J, Tacken PJ, Batenburg JJ, Leth-Larsen R, et al. (2005b). Respiratory innate immune proteins differentially modulate the neutrophil respiratory burst response to influenza A virus. Am J Physiol Lung Cell Mol Physiol 289:L606-L616 [DOI] [PubMed] [Google Scholar]
- Wu Z, Golub E, Abrams WR, Malamud D. (2004). gp340 (SAG) binds to the V3 sequence of gp120 important for chemokine receptor interaction. AIDS Res Hum Retroviruses 20:600-607 [DOI] [PubMed] [Google Scholar]
- Wu Z, Van Ryk D, Davis C, Abrams WR, Chaiken I, Magnani J, et al. (2003). Salivary agglutinin inhibits HIV type 1 infectivity through interaction with viral glycoprotein 120. AIDS Res Hum Retroviruses 19:201-209 [DOI] [PubMed] [Google Scholar]


