Abstract
There is compelling evidence that treponemes are involved in the etiology of several chronic diseases, including chronic periodontitis as well as other forms of periodontal disease. There are interesting parallels with other chronic diseases caused by treponemes that may indicate similar virulence characteristics. Chronic periodontitis is a polymicrobial disease, and recent animal studies indicate that co-infection of Treponema denticola with other periodontal pathogens can enhance alveolar bone resorption. The bacterium has a suite of molecular determinants that could enable it to cause tissue damage and subvert the host immune response. In addition to this, it has several non-classic virulence determinants that enable it to interact with other pathogenic bacteria and the host in ways that are likely to promote disease progression. Recent advances, especially in molecular-based methodologies, have greatly improved our knowledge of this bacterium and its role in disease.
Keywords: chronic periodontitis, polymicrobial biofilms, immunomodulation, outer sheath vesicles
The Treponemes and Disease
Oral treponemes, along with over 600 other bacterial species, exist as part of a polymicrobial biofilm accreted to the tooth surface in the gingival crevice (Kolenbrander et al., 2002; Ellen and Galimanas, 2005; Dewhirst et al., 2010). Treponemes play a role in the etiology of several chronic diseases of humans including syphilis and yaws (Treponema pallidum), periodontal diseases including chronic periodontitis and acute necrotizing ulcerative gingivitis (Treponema denticola, Treponema lecithinolyticum, Treponema socranskii, and others), and endodontic infections and some acute dental abscesses (Sela, 2001; Foschi et al., 2006; Holt and Ebersole, 2006; Robertson and Smith, 2009). In addition, treponemes have been implicated in the development of chronic diseases of domestic animals, including periodontal diseases of dogs (Nordhoff et al., 2008b), bovine digital dermatitis of dairy cattle (Choi et al., 1997; Klitgaard et al., 2008; Nordhoff et al., 2008a), and contagious ovine digital dermatitis (Dhawi et al., 2005; Sayers et al., 2009).
Treponemes are members of the Spirochaetes phylum, a clade now believed to be distinct from both Gram-positive and Gram-negative bacteria, that is believed to have undergone extensive horizontal gene transfer with Archae and possibly with eukaryotic organisms (Ibba et al., 1997; Brown et al., 1998, 2001; Bond and Francklyn, 2000; Wolf et al., 2001; Henz et al., 2005; Paster and Dewhirst, 2006). It has been proposed that all treponemes descended from a common spirochetal ancestor and, as a taxon, have evolved a range of unique characteristics, including virulence determinants, metabolic pathways, solute transport systems, and surface-binding proteins due to their evolutionary trajectory (Paster and Dewhirst, 2000; Seshadri et al., 2004). However, this hypothesis remains to be comprehensively tested. The sequencing of the complete genome of T. pallidum in 1998, T. denticola ATCC 35405 in 2004, and the recent release of the T. lecithinolyticum OMZ684T and Treponema vincentii ATCC 35580 sequences have already proven invaluable to treponeme research and will continue to illuminate the evolution and virulence characteristics of these species, especially as other genome sequences become available (Fraser et al., 1998; Seshadri et al., 2004; Dewhirst et al., 2010; http://www.jcvi.org/, www.homd.org/). Currently, genomic comparisons with other treponemes and spirochetes are facilitating a more targeted selection of potential T. denticola virulence factors for detailed investigation.
The numbers of both cultivated and uncultivated Treponema phylotypes reported in recent years have rapidly increased, mainly due to the mass of data obtained from efforts to sequence genes encoding 16S rRNA (Choi et al., 1994; Wyss et al., 2004; Demirkan et al., 2006; Molbak et al., 2006; Nordhoff et al., 2008a; Pringle et al., 2008, 2009; Evans et al., 2009; Sayers et al., 2009; Yano et al., 2009). There are currently 49 species of oral Treponema listed on the Human Oral Microbiome Database (Dewhirst et al., 2010), the best-characterized being T. denticola, Treponema amylovorum, T. lecithinolyticum, Treponema maltophilum, Treponema medium, Treponema parvum, Treponema pectinovorum, T. socranskii, and T. vincentii (www.homd.org/).
Within the oral cavity, treponemes are most often associated with diseases of the periodontium. However, these treponemes are members of the normal oral microbiota of healthy individuals, albeit in very low numbers, and even those associated with disease cannot be considered frank pathogens. In this article, we will concentrate on the virulence characteristics of oral treponemes, and particularly T. denticola, in relation to chronic periodontitis. When considering the virulence characteristics of T. denticola, it is imperative to understand that it is part of a pathogenic bacterial consortium, and its interactions with other bacterial species are important for disease pathology. Therefore, the relationship between T. denticola and other bacteria involved in disease progression will also be addressed.
Polymicrobial Nature of Chronic Periodontitis
Chronic periodontitis is a polymicrobial disease that results from the overgrowth of a limited number of bacterial species that are normal members of the oral microbiota. It is widely accepted that T. denticola, Porphyromonas gingivalis, and Tannerella forsythia form a bacterial consortium, often referred to as the ‘Red Complex’, that is strongly associated with the clinical progression of chronic periodontitis (Lamont and Jenkinson, 1998; Socransky et al., 1998; Holt and Ebersole, 2005). The levels of P. gingivalis and T. denticola in subgingival plaque have been shown to allow for prediction of periodontal breakdown in a longitudinal clinical trial (Byrne et al., 2009). The unifying features of the Red Complex bacteria are their extracellular proteolytic activity, their complex anaerobic fermentations of amino acids, production of toxic metabolites, and outer membrane (or sheath) vesicles. Of the three species, only the treponeme is motile and able to respond chemotactically to environmental stimuli. The in vivo interactions of these species are still poorly characterized, but some studies have indicated that P. gingivalis may be needed for T. denticola colonization and presence in subgingival plaque (Simonson et al., 1992; Sela, 2001). A recent study of the bacterial composition of subgingival plaque in individuals with chronic periodontitis (Byrne et al., 2009) showed that P. gingivalis and T. denticola and T. forsythia were routinely found together in subgingival plaque (Fig. 1). Interestingly, P. gingivalis or T. denticola were rarely found in subgingival plaque without T. forsythia (Fig. 1). Mineoka et al. (2008) have recently shown a similar relationship between T. forsythia and P. gingivalis. T. forsythia has also been found to be more prevalent than P. gingivalis in subgingival plaque (Haffajee et al., 2006; Colombo et al., 2009). This may suggest that T. forsythia colonizes plaque before P. gingivalis and T. denticola. Recently, Zijnge et al. (2010) used fluorescent in situ hybridization to show that Tannerella sp. were located in the intermediate layer of subgingival plaque, whereas P. gingivalis. tended to be found in micro-colonies in the top layer, and Treponemes were found outside the top layer. These results are consistent with the proposal that T. forsythia may be a necessary precursor species for colonization by T. denticola and P. gingivalis. The interactions within the Red Complex species and with other newly identified species and the timing of colonization and proliferation clearly need further study.
Figure 1.
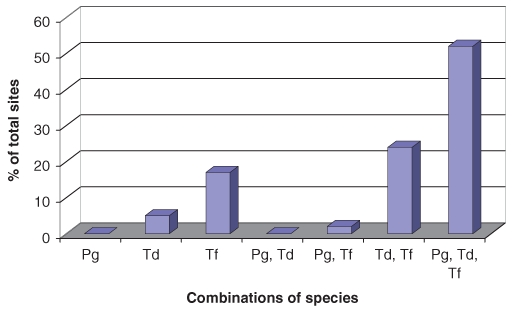
Combinations in which P. gingivalis, T. denticola, and T. forsythia were found in subgingival plaque that had quantifiable levels of at least one of these species, by real-time PCR, in 41 sites of 37 individuals previously diagnosed with chronic periodontitis. The majority of sites were colonized by all three species, and P. gingivalis was rarely detected without the presence of T. denticola and T. forsythia The individuals had completed initial treatment and been on a maintenance program for a minimum of six months in the Specialist Periodontics Department of the Royal Melbourne Dental Hospital, Australia (Byrne et al., 2009).
Determination of Virulence - Animal Models of Disease
Virulence is defined as the capacity of a pathogen, usually a micro-organism, to cause disease. For the virulence factors of T. denticola to be determined, the role of this bacterium in disease must be clearly defined, and in recent times, animal models have been developed to examine this role. The animal model of disease allows key elements of the disease process to be investigated under the complex conditions found in vivo; however, the initiation of “disease” in animal models is, by necessity, induced and, of course, differs from natural pathogenesis in humans (Arnett and Viney, 2007). Animal models of chronic inflammatory diseases are problematic (Radbruch and Isaacs, 2009), especially for those diseases such as chronic periodontitis that are caused by an overgrowth of members of the normal microbiota. Despite these limitations, animal models of disease have proven invaluable for determining the pathogenicity of bacteria and providing insight into their potential virulence factors. Murine subcutaneous abscess models of disease have been used to investigate the virulence of T. denticola and other oral treponemes and the pathogenicity of polymicrobial infections (Kesavalu et al., 1997; Gemmell et al., 2002). As a subcutaneous monoinfection T. denticola, T. pectinovorum, or T. vincentii produced localized abscesses (Kesavalu et al., 1997, 1998). In comparison, at similar inoculum sizes, P. gingivalis produced more severe, spreading ulcerative lesions remote from the site of infection (Kesavalu et al., 1998; O’Brien-Simpson et al., 2000). At high doses of P. gingivalis in the subcutaneous abscess model, addition of T. denticola had no effect on the formation and size of the spreading lesion; however, at low doses of P. gingivalis, T. denticola co-infection significantly enhanced tissue damage (Kesavalu et al., 1998). Analysis of these data indicates not only that the presence of P. gingivalis was needed for invasion and extensive tissue damage, but also that T. denticola greatly facilitated this process.
Although broadly predictive of disease, the relevance of these abscess models to chronic periodontitis has been questioned, and more appropriate models have been developed. In recent years, murine alveolar (periodontal) bone loss models have been adapted to test the virulence of T. denticola and polymicrobial infections. These studies reported that intra-oral inoculations with T. denticola resulted in colonization of the oral cavity of the animal, induction of a specific immune response, and significant alveolar bone loss (Kesavalu et al., 2007; SF Lee et al., 2009). Furthermore, a polymicrobial intra-oral inoculation of P. gingivalis, T. denticola, and T. forsythia at a 1:1:1 cell ratio with the same total number of bacterial cells caused significantly higher levels of bone resorption than those caused by mono-inoculations (Kesavalu et al., 2007). In a recent study using a murine model of periodontal disease (O’Brien-Simpson et al., 2005; Pathirana et al., 2007), intra-oral inoculation with a 1:1 ratio of T. denticola:P. gingivalis cells caused the same level of alveolar bone loss as intra-oral inoculation with a 40-fold greater number of P. gingivalis cells inoculated alone (unpublished data). Analysis of these data together suggests a significant role of T. denticola in the tissue damage associated with chronic periodontitis, and demonstrates the importance of a polymicrobial infection for the initiation and progression of disease. The continuing development of these models will allow for the testing of T. denticola mutants lacking specific gene products, which will greatly improve our knowledge of T. denticola virulence factors.
Interbacterial binding and localization of T. Denticola with other species
T. denticola exists as part of a complex, structured multispecies biofilm (subgingival plaque) in the relatively protected region of the gingival crevice or periodontal pocket, which is largely protected from the shear forces associated with salivary flow and the effects of mastication. T. denticola tends to inhabit the deeper periodontal pockets and is not an early colonizer of subgingival plaque (Kolenbrander et al., 2002). Treponemes, including T. denticola, have been reported to be located on the surface of the dense subgingival bacterial biofilms, at the interface of the biofilms and the gingival epithelium (Kigure et al., 1995; Ellen and Galimanas, 2005; Zijnge et al., 2010).
Interspecies bacterial binding plays a vital role in biofilm development and in the co-location of bacterial species. T. denticola binds weakly to fusobacteria in an interaction mediated by the carbohydrate moiety of the T. denticola major sheath protein (Msp, see below) and the galactose-binding lectin receptor of the fusobacteria (Kolenbrander et al., 1995; Rosen et al., 2008; Kaplan et al., 2009). T. denticola also binds to the commensal, early-colonizing Streptococcus crista (Yao et al., 1996). These interactions may be important for T. denticola to initially colonize sites and persist during health. T. denticola has been shown to bind specifically to both P. gingivalis and T. forsythia with similar avidity, which may explain the close association of the species (Fig. 2a), and these interactions are mediated to some extent by T. denticola dentilisin or Msp (see below). However, co-aggregation of the bacteria was not reduced in T. denticola dentilisin- or Msp-deficient mutants, and pre-incubation of T. denticola with P. gingivalis fimbriae did not prevent its ability to co-aggregate with P. gingivalis (Yao et al., 1996; Ishihara et al., 1998; Hashimoto et al., 2003b; Rosen et al., 2008). This indicates that this interaction is multimodal, involving different proteins and possibly carbohydrate moieties. This close association of species will enhance the repertoire of proteolytic and other enzymes available to each species.
Figure 2.
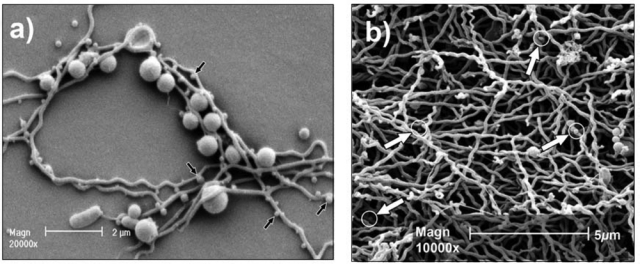
Electron micrographic images of T. denticola co-cultured with P. gingivalis and as a monospecies biofilm. (a) Electron micrograph of P. gingivalis and T. denticola growing in continuous co-culture, showing the intimate association of the bacteria and the putative vesicles on the surface of the treponeme (arrows). The bacteria were co-cultured under conditions similar to those described previously for the culture of T. denticola in Oral Bacterial Growth Medium (Orth et al., 2010) and P. gingivalis (Ang et al., 2008). T. denticola was initially inoculated into the chemostat, and once cell density had stabilized, P. gingivalis was added to the continuous culture. (b) Electron micrograph of T. denticola biofilm formed on a glass rod submerged in continuous culture after 14 days’ incubation as described previously (Mitchell et al., 2010). Outer sheath vesicles are indicated by arrows. Morphological variations of T denticola cells in the form of “spherical bodies” can also be seen in this micrograph.
Leucine-Rich Repeat Proteins
A T. denticola leucine-rich repeat protein (LrrA) has recently been shown to play a role in binding to T. forsythia, but not to P. gingivalis or F. nucleatum, and to mediate binding to epithelial cells and to promote swarming (Ikegami et al., 2004; Rosen et al., 2008). Interestingly, Lrr proteins have been shown to be important for epithelial cell invasion and biofilm formation by P. gingivalis. They are also important for epithelial cell invasion and virulence in a mouse alveolar bone loss model by T. forsythia (Sharma et al., 1998, 2005; Capestany et al., 2006; Inagaki et al., 2006; Dashper et al., 2009). In both of these species, the leucine-rich repeat proteins are members of the CTD family of proteins that are secreted and attached to the surface by novel mechanisms (Seers et al., 2006; Veith et al., 2009). There are two characterized Lrr proteins in P. gingivalis. In the T. forsythia genome, one Lrr protein is characterized (BspA) and another 5 predicted. Six Lrr proteins are predicted in the T. denticola genome. Together, this suggests that this family of proteins plays an important role in the virulence of these species, and that these Lrr proteins warrant further attention.
Metabolic Cooperativity
Close association can minimize the dilution of metabolites and signaling molecules exchanged between interacting species, thereby facilitating efficient metabolic communication (Grenier, 1992). It is likely that some of the bacterial species to which oral treponemes bind have some form of metabolic interaction with them. Consistent with this concept, it has been shown that T. denticola and P. gingivalis display symbiosis in protein degradation, nutrient utilization, and growth promotion (Grenier, 1992; Nilius et al., 1993; Kigure et al., 1995; Hollmann and Van der Hoeven, 1999; Grenier and Mayrand, 2001; Yoneda et al., 2005).
When grown together in continuous culture, P. gingivalis and T. denticola bound to each other (Fig. 2a) and developed a stable cell ratio of 6:1, respectively, as determined by real-time PCR (unpublished data). This indicates that they do not directly compete under these specific growth conditions, since if this were the case, one species would eventually displace the other.
Metabolic End-Products
Relatively little is known regarding the metabolism of T. denticola, and many of its metabolic pathways are likely to be novel (Rother et al., 2001; Seshadri et al., 2004; Chu et al., 2008). It primarily utilizes serine, alanine, cysteine, and glycine when grown in vitro and generates fermentation products including acetate, lactate, succinate, formate, pyruvate, ethanol, carbon dioxide, hydrogen sulfide (H2S), and ammonia (Hespell and Canale-Parola, 1971). These metabolites can accumulate in high concentrations and could influence the composition of bacterial species within the polymicrobial biofilm as well as affecting host tissue (Carlsson, 1997; Kuramitsu et al., 2007). These compounds can penetrate periodontal tissue and disrupt host cell activity and the host immune response (Tonetti et al., 1987; Bartold et al., 1991; Eftimiadi et al., 1993; Kurita-Ochiai et al., 1995; Carlsson, 1997; Niederman et al., 1997; Kuramitsu et al., 2007). The short-chain fatty-acid fermentation products have been suggested to play a role in disease progression (Niederman et al., 1997). Volatile sulfur compounds such as methyl mercaptan and H2S, highly toxic metabolites produced by T. denticola and other oral bacteria, are thought to play an important role in periodontal disease (Yoshimura et al., 2000). Methyl mercaptan is derived from methionine by the action of methionine-α-deamino-γ-mercaptomethane lyase (METase), which has been shown to be present in both T. denticola and P. gingivalis, but is absent in mammals (Yoshimura et al., 2000; Fukamachi et al., 2005). Methyl mercaptan reduces protein synthesis by human gingival fibroblasts and inhibits cell migration in periodontal ligament cells (Johnson et al., 1992; Lancero et al., 1996). Mice infected with a P. gingivalis mutant lacking METase had a significantly higher survival rate relative to the wild-type strain W83 (Yoshimura et al., 2000). T. denticola METase has a much higher affinity for methionine than P. gingivalis METase, indicating that, at low concentrations, T. denticola may be the major producer of methyl mercaptan (Fukamachi et al., 2005).
Hydrogen sulfide is a major metabolic end-product of the fermentation of cysteine by some anaerobic bacteria. H2S is cytotoxic for a variety of host cells, including gingival fibroblasts and epithelial cells (Beauchamp et al., 1984; Reiffenstein et al., 1992; Chu et al., 1999; Yoshimura et al., 2000). T. denticola is the only known oral bacterium to contain the three-step pathway required to produce H2S, from the abundant host tripeptide, glutathione (Chu et al., 2008). This pathway contains cysteinylglycinase, γ-glutamyltransferase, and the novel enzyme, cystalysin (Makinen and Makinen, 1997; Chu et al., 2002). H2S production via cystalysin has been shown to be the mechanism by which T. denticola disrupts erythrocyte membranes, thereby explaining its original description as a hemolysin (Chu et al., 1995). H2S also has both pro- and anti-inflammatory effects (Kimura, 2009), and a recent study by Chen et al. showed that H2S production by P. gingivalis stimulated production of the pro-inflammatory cytokine interleukin-8 (IL-8) by gingival epithelial cells (Chen et al., 2010). The stimulation of both pro- and anti-inflammatory mediators may serve to dysregulate the host’s defense.
Biofilms
Survival and virulence of oral treponemes are dependent on their ability to form biofilms, grow in this milieu, interact with the other species in the biofilm, and, presumably, escape from the biofilm, depending on environmental conditions. Direct microscopy of freshly prepared samples of periodontal pocket plaque most often reveals planktonic spirochetes of diverse size swimming among a rich variety of other highly motile bacterial species. The surface of the adjacent biofilm usually has a concentrated sessile population of the same morphotypes (Ellen and Galimanas, 2005; Zijnge et al., 2010).
When co-cultured, P. gingivalis and T. denticola form significantly more biofilm compared with monoculture, with the resultant polymicrobial biofilms adhering more tightly to the substratum than the monospecific biofilms (Yamada et al., 2005). Molecules involved in polymicrobial biofilm formation have been examined in a panel of T. denticola mutants. A T. denticola CfpA-deficient mutant, which lacks cytoplasmic filament, was unable to form a mixed biofilm with P. gingivalis, while an immotile FlgE mutant and an Msp-deficient mutant displayed reduced biofilm-forming ability with P. gingivalis (Yamada et al., 2005).
Obtaining a model system for biofilm growth of pure cultures of T. denticola has been reported as problematic (Vesey and Kuramitsu, 2004), and, as a result, little work has been conducted on T. denticola biofilms. To determine the effects of biofilm growth on T. denticola, investigators have recently developed a continuous-culture system that allowed for the cultivation of biofilm and planktonic cells simultaneously in the one vessel. This system was then used specifically to determine the effects of biofilm growth relative to the planktonic state. In the biofilm, 126 T. denticola genes were differentially expressed, with a fold change of 1.5 or greater (Mitchell et al., 2010). In biofilm cells, there was an up-regulation of genes encoding several putative virulence factors, including cystalysin and the outer membrane dentilisin protease lipoprotein complex, toxin-antitoxin systems, and a family of putative transposases (Mitchell et al., 2010).
Toxin-Antitoxin Systems
Toxin-antitoxin (TA) systems consist of both a toxin, which inhibits essential cell components, and an antitoxin, which counteracts the toxin. The antitoxin component is generally labile, requiring constant synthesis to remain active in the cell, whereas the toxin component is stable. There is a growing body of evidence suggesting that TA systems are involved in programmed cell death, bacterial stringent response to amino acid starvation, and reversible bacteriostasis (persistence or dormancy), and that these and other roles influence biofilm formation (Kim et al., 2009; Makarova et al., 2009). T. denticola 35405 contains 33 predicted TA systems, and a high proportion of these (25/33) showed an increase in expression when the bacterium was grown as a monospecific biofilm (Mitchell et al., 2010). The large number of these systems in T. denticola and their increased levels of expression in mature biofilm suggest that not only may they play a role in biofilm persistence, but they may also represent a mechanism of resistance to various environmental assaults, including diverse antibiotics and other drugs (Lewis, 2000; Jayaraman, 2008).
Transposases
T. denticola possesses a large, unusual family of genes with significant similarity to transposases. Transposases are enzymes that "cut and paste" mobile genetic elements from one position to another within the genome. Twenty-five of the 35 putative transposase genes were significantly up-regulated in the biofilm relative to planktonic cells. This could lead to extensive chromosomal re-arrangement, resulting in the development of population-level diversity, or may represent a novel gene-regulatory mechanism (Mitchell et al., 2010). A functional lysogenic bacteriophage, φtd1, was discovered during T. denticola biofilm growth that may be capable of transferring virulence-related genes through horizontal gene transfer. The bacteriophage is 37,920 bp in length and has a GC content of ~37%, similar to that of T. denticola (Mitchell et al., 2010). Analysis of these data together indicates that there is a higher potential for genetic mobility in T. denticola when growing as a biofilm, and that these systems are important for biofilm persistence and therefore virulence of this bacterium.
Outer Sheath Proteins
Cell-surface components, especially proteins, act as the sensors and effectors of interactions with the host and other bacteria and are largely the targets of the host adaptive immune response. One of the unifying features of the Red Complex bacteria that are responsible for chronic periodontitis progression is their high levels of extracellular proteolytic activity that is mediated by cell-surface-located proteases. Since the outer membrane of T. denticola is significantly different from that of Gram-negative bacteria, it is usually referred to as the outer sheath.
Dentilisin
Dentilisin has been proposed to be a major T. denticola virulence factor, since it is an active cell-surface-located protease that cleaves at phenylalanyl/alanyl and prolyl/alanyl bonds (Uitto et al., 1988a,b; Grenier et al., 1990; Makinen et al., 1995; Ishihara et al., 1996; Beausejour et al., 1997). It contributes to disease progression by disrupting or modulating intercellular host signaling pathways and degrading host cell matrix proteins. Dentilisin potentially allows for penetration of epithelial cell layers by T. denticola by degradation of intercellular adhesion proteins (Chi et al., 2003) and modulates host cell immune responses by degradation of interleukin-1β (IL-1β), IL-6, tumor necrosis factor alpha (TNF-α), and monocyte chemoattractant protein 1 (Miyamoto et al., 2006; Okuda et al., 2007). The protease occurs as part of a complex of three proteins, the 72-kDa subtilisin-like protease (dentilisin, PrtP, chymotrypsin-like protease) and two auxiliary proteins, PrcA1 (~40 kDa) and PrcA2 (~30 kDa) (Lee et al., 2002; Ishihara et al., 2004). Recent work has shown that the dentilisin protease complex is encoded by an operon containing three genes, prcB, prcA, and prtP (Bian et al., 2005). The protein encoded by prcA is hydrolyzed to produce two proteins (PrcA1 and PrcA2) of various sizes, depending on the strain. prcB is expressed at a lower level than prtP and prcA, and although recombinant PrcB associates with PrtP, its role in complex formation and activity is currently unknown (Bian et al., 2005). Dentilisin may also play a role in processing of other cell-surface components, such as Msp, and is proposed to associate with this protein. As noted above, dentilisin is also proposed to interact with the fimbriae of P. gingivalis and participate in the co-aggregation of the bacteria. The dentilisin-PrcA complex has been compared with the major virulence factors of P. gingivalis, the RgpA-Kgp proteinase-adhesin complexes, since each complex adheres to and degrades fibrinogen (Bamford et al., 2007). It has been suggested that T. denticola and P. gingivalis synergistically hydrolyze fibrinogen, thereby promoting vascular disruption, bleeding, and inflammation and retarding tissue repair (Bamford et al., 2007).
Trypsin-Like Protease Activity
Trypsin-like protease activity has been reported from T. denticola (Ohta et al., 1986). The responsible enzyme, oligopeptidase B, is encoded by opdB (TDE2140), has similarity to prolyl oligopeptidases, and has been shown to cleave only C-terminal to Arg-residues (Fenno et al., 2001; Lee and Fenno, 2004). In addition to the 78-kDa OpdB described by Fenno et al. (2001), a second putative OpdB (TDE1195) of the same mass and sharing 45% amino acid sequence similarity (28% identity) was found by Veith et al. (2009). Although the activity of this putative protease is unknown, it is possible that the second OpdB may be a Lys-specific protease, which together would explain the trypsin-like proteolytic activity described for T. denticola. Per cell, T. denticola 35405 has approximately ten-fold less Arg-specific activity than P. gingivalis W50 when tested with the chromogenic substrate BApNA (unpublished).
Major Sheath Protein
Msp, a member of the Tpr (T. pallidum repeat) protein family, is the most abundant protein in the T. denticola outer membrane (or sheath) and is one of the most-studied T. denticola proteins. It is a β-barrel, integral outer sheath protein that acts as a porin and has surface-exposed loops that are able to bind to a variety of host proteins. Msp has been proposed to mediate colonization of host tissues and has cytopathic pore-forming activity against cultured epithelial cells (Fenno and McBride, 1998; Ellen, 2006).
Msp appears to be a ubiquitous outer sheath protein in oral treponemes, although there is considerable variation in the sequence among species and strains (Fenno et al., 1997; Lee et al., 2005). The T. denticola Msps can be divided into 3 domains, the highly conserved 203-residue N-terminal and 271-residue C-terminal domain, plus a central variable region of ~70 residues (Edwards et al., 2005). Differences in Msp may be a defining element of strain serotype, because the strain-specific Msp correlates with different serotypes (Capone et al., 2008). Edwards et al. showed that the variable region contains the dominant B-cell epitopes for animals immunized with T. denticola cells. A recombinant Msp protein and fragments expressed in E. coli demonstrated that, in addition to its known binding to laminin and immobilized fibronectin, Msp also binds keratin, collagen Type 1, fibrinogen, hyaluronic acid, and heparin. The major binding epitopes were located in the N-terminal domain of the protein, and this region of the protein was surface-exposed (Edwards et al., 2005).
Msp is one of the immunodominant T. denticola antigens recognized by human serum antibodies (Capone et al., 2008) and murine anti-T. denticola polyclonal antibodies (SF Lee et al., 2009; Veith et al., 2009). Numerous studies have shed light on the mechanisms by which Msp interaction leads to actin remodeling and reorganization in host cells, and how this is likely to impair neutrophil chemotaxis and phagocytic activity (Batista da Silva et al., 2004; Puthengady et al., 2006; Amin et al., 2007; Jobin et al., 2007; Magalhães et al., 2008). In contrast to the studies that showed induction of IL-8 expression from various cells following exposure to Msp, Brissette et al. (2008) showed that exposure to T. denticola did not induce IL-8 production by primary gingival epithelial cells (PGEC). IL-8 induces migration of neutrophils to sites of infection. Dysregulation of efficient neutrophil activation and migration is one means by which T. denticola could evade the host immune response and persist at a site. The T. lecithinolyticum Msp stimulates the up-regulation of signaling pathways in monocytes, inducing IFN-β expression and IFN-stimulated genes (SH Lee et al., 2009).
Tp92 is a 92-kDa T. pallidum subspecies pallidum surface-exposed antigen that induces a protective immune response (Cameron et al., 2000). Homologs of Tp92 have been identified in T. maltophilum, T. lecithinolyticum, T. socranskii, and T. denticola. Antisera raised to recombinant T. denticola Td92 have been used to show surface exposure of Td92 and inhibition of T. denticola binding to KB epithelial cells (Jun et al., 2008). All oral treponeme Tp92 homologs induced production of various cytokines, cyclooxygenase-2 (COX-2), and prostaglandin E2 from THP-1 cells and periodontal ligament cells, indicating a pro-inflammatory response.
Lipoproteins
Lipoproteins are the most abundant membrane-associated proteins found in spirochetes, and T. denticola 35405 is predicted to have 166 of them, the highest number for any of the sequenced spirochetes (Setubal et al., 2006). Lipoproteins are acylated at the N-terminus of the mature protein, and this anchors the protein to a membrane. Veith et al. (2009) detected 20 lipoproteins under a single set of growth conditions, including 11 uncharacterized putative lipoproteins, 2 hemin-binding proteins (HbpA, HbpB), 2 putative extracellular solute-binding lipoproteins, 4 putative oligopeptide/dipeptide ABC transporter peptide-binding proteins, and the dentilisin complex-associated polypeptide (PrcA). Fourteen lipoprotein genes had significant changes in expression when T. denticola was grown as a biofilm, including the outer membrane hemin-binding protein A that, along with 3 other putative lipoprotein encoding genes, was down-regulated in the biofilm (Mitchell et al., 2010). Analysis of these data indicates the importance of lipoproteins to the treponeme lifestyle.
OppA is a 70-kDa cell-surface, membrane-associated lipoprotein that has significant similarity to the solute binding protein of a highly conserved ATP-binding cassette-type transporter involved in peptide uptake and environmental signaling in a wide range of bacteria (Fenno et al., 2000). OppA can bind soluble host proteins such as plasminogen and fibrinogen, but not immobilized insoluble host proteins or epithelial cells. It has been proposed to act as an adhesin and to help decorate the surface of T. denticola with host proteins as a means of avoiding or delaying immune recognition. Group A streptococci and Streptococcus equi subsp. have been shown to be cleared more slowly from a host when coated with fibrinogen (Whitnack and Beachey, 1985; Poirier et al., 1989; Boschwitz and Timoney, 1994; Ellen, 2006; Bamford et al., 2007).
FhbB is a small (11.4 kDa) surface-exposed T. denticola lipoprotein that binds complement regulatory proteins of the factor H (FH) family. By using T. denticola dentilisin-deficient mutants, it was determined that after binding FH, it is cleaved by dentilisin to yield a FH subfragment of ~50 kDa. FH bound to dentilisin-deficient mutants was not cleaved and retained its ability to serve as a co-factor for factor I in the cleavage of C3b. The role of this unique binding and cleavage in T. denticola virulence is as yet unresolved, but it is likely to be linked to epithelial cell binding and invasion, subversion of the complement cascade, or tissue invasion (McDowell et al., 2007, 2009).
Outer Sheath Vesicles
Gram-negative bacteria have long been known to produce outer membrane vesicles (OMVs) (Devoe and Gilchrist, 1973; Nowotny et al., 1982; Grenier and Mayrand, 1987). Initially thought to be a result of random blebbing of the outer membrane, or sheath, producing small spherical vesicles of 50-100 nm in diameter, more recent studies have revealed that OMV formation is a highly regulated process which may increase the fitness of the bacterium in response to environmental cues (Wensink and Witholt, 1981; Kato et al., 2002; Wai et al., 2003). OMVs are considered potent virulence factors, since they possess adhesins, toxins, and proteolytic enzymes, can mediate bacterial aggregation and invasion, are cytotoxic, and can modulate the host immune response (Kuehn and Kesty, 2005). Besides affecting the host, OMVs may be important for securing a niche in the competitive environment of subgingival plaque by eliminating competitors via the delivery of proteases and toxins (Kadurugamuwa and Beveridge, 1996; Allan and Beveridge, 2003). OMVs also facilitate the remote delivery of labile signaling molecules and prevent their degradation by other micro-organisms (Z Li et al., 1996; Mashburn and Whiteley, 2005).
T. denticola outer sheath vesicles (OSVs) have been proposed as a long-range virulence factor that can penetrate tissues more readily than the bacterium itself (Cimasoni and McBride, 1987; Cockayne et al., 1989; Weinberg and Holt, 1991; Kuehn and Kesty, 2005). Application of T. denticola OSVs to Hep-2 epithelial cell monolayers disrupted the tight junctions, which might facilitate penetration into underlying tissues (Chi et al., 2003). However, the involvement of treponemal OSVs in disease remains to be properly explored. When these bacteria are grown in continuous co-culture with P. gingivalis, scanning electron microscopic analysis revealed not only their close association, but also the presence of OSV/OMVs on the surface of T. denticola (Fig. 2a). OSVs were also detected when T. denticola was grown as a monospecific biofilm (Fig. 2b). Recently, it was demonstrated that T. denticola lipooligosaccharide (LOS) and Msp induce macrophage tolerance to further stimulation (Nussbaum et al., 2009). Considering these components are found on T. denticola OSVs, initial exposure of macrophages to these OSVs might limit the clearance of the bacterium during subsequent encounters.
Motility
Motility and chemotaxis, like biofilm formation and interspecies co-operation, are not considered to be classic virulence factors of bacteria. However, in the context of T. denticola and its role in disease progression, it is likely that they are essential for the virulence of the bacterium. Of the sequenced treponemes, 5-6% of the genome is dedicated to motility and chemotaxis, implying its importance for survival and disease progression in the host.
Periplasmic Flagella
Unlike the exposed flagella of most motile bacterial species, the spirochetal flagella are located within the periplasmic space between the outer sheath and cytoplasmic membrane (Canale-Parola, 1978; Holt, 1978). In Treponema, up to 15 periplasmic flagella (PF) originate from each pole of the cell, entwine the cytoplasmic cylinder, and overlap at the cell center (Watson et al., 1951; Bergey and Holt, 1984). The organization of PF in the peptidoglycan-containing periplasm as well as other native cellular structures has recently been resolved by cryo-electron tomography (Izard et al., 2008, 2009). PF facilitate translocation in highly viscous environments which would usually slow or immobilize most externally flagellated bacteria (Fenno and McBride, 1998). PF are protected from the immobilization effects of flagella-specific antibodies produced by the host response to infection (Charon and Goldstein, 2002). Cytoplasmic filaments are cytoplasmic structures located directly beneath the PF. A CfpA-deficient mutant displayed reduced spreading ability compared with wild-type strains, implicating involvement of cytoplasmic filaments in motility (Izard et al., 2001; Vesey and Kuramitsu, 2004).
Chemotaxis
Chemotaxis allows motile bacteria to respond to environmental stimuli in a positive or negative manner. Genome sequencing and bioinformatic analyses predicted that T. denticola and T. pallidum possess a complete set of chemotaxis proteins required for signal perception, transduction, and adaptation. Environmental stimuli are detected through methyl-accepting chemotaxis proteins (MCPs) that traverse the inner membrane. These signals are transduced via the cytosolic chemotaxis proteins CheA, CheW,and CheY that modulate the direction of flagellar motor rotation. Adaptation in T. denticola is achieved via methylation/demethylation of the MCPs by the regulatory proteins, CheR and CheB, in a manner similar to that of well-studied organisms such as E. coli (Sim et al., 2005). Surprisingly high numbers of MCPs were identified in the T. denticola genome, suggesting chemotaxis responses toward a variety of compounds that might reflect the complexity of its ecological niche (Seshadri et al., 2004). Chemoattractants for T. denticola include glucose, serum, and albumin, which are possible indicators of damaged host tissues (Umemoto et al., 2001; Ruby et al., 2008). Allelic replacement mutagenesis of two MCP encoding genes, dmcA and dmcB, resulted in a decreased chemotactic response to serum and albumin (Kataoka et al., 1997; Li et al., 1999). As expected, a cheA knockout mutant, which lacks the central kinase of the chemotaxis pathway, failed to respond chemotactically to serum (Lux et al., 2002).
Epithelial Model Systems
The ability of T. denticola to penetrate and migrate through cultured tissue has been used as a model system to assess virulence (Lux et al., 2001). The immotile flgE flagella mutant (H Li et al., 1996) lost the ability to penetrate cultured layers formed by human gingival keratinocytes. The importance of a functional chemotaxis pathway for T. denticola virulence was underscored by the finding that a cheA mutant (Lux et al., 2002), as well as dmcA and dmcB mutants lacking specific MCPs, displayed significantly impaired tissue penetration despite retaining motility (Kataoka et al., 1997; Li et al., 1999). These studies implied that chemotaxis and motility may be significant for T. denticola, and presumably other oral treponemes, in the invasion of gingival tissues. Transcriptome analysis of T. pallidum showed significant up-regulation of genes involved in chemotaxis during infection (Smajs et al., 2005).
Tissue Penetration and Epithelial Cell Invasion
The role of bacterial tissue invasion and internalization by epithelial cells in the progression of periodontitis is still unclear. There appears to be little evidence of extensive tissue penetration by T. denticola in human chronic periodontitis; however, T. denticola has been detected between junctional epithelium cells, which are normally tightly associated (Saglie et al., 1982). Biopsy of tissue from canine periodontal lesions showed extensive tissue penetration by treponemes (Nordhoff et al., 2008b).
Invasion of, or internalization by, epithelial cells provides oral bacteria with a nutrient-rich environment that is partially protected from the host immune system. While not immediately contributing to disease, T. denticola may use the intracellular locale to persist safely and possibly replicate (Colombo et al., 2007; Johnson et al., 2008). During treatment of disease, intracellular bacteria are less likely to be physically removed by scaling and root planing and are more resistant to antibiotics. Furthermore, this intracellular population could constitute a reservoir of bacteria for the repopulation of treated subgingival sites in refractory periodontitis (Tribble and Lamont, 2010).
Lipooligosaccharides (Los)
LPS, also called endotoxin, forms the outer leaflet of the Gram-negative bacterial outer membrane. The treponemal outer sheath does not have a typical LPS, and the treponemes lack genes encoding the necessary enzymes for LPS synthesis (Fraser et al., 1998; Schultz et al., 1998; Norris and Weinstock, 2000; Schröder et al., 2000; Hashimoto et al., 2003a; Seshadri et al., 2004). The treponeme outer sheath (membrane) contains lipooligosaccharides (LOS), however, that have functional similarities to LPS. The T. denticola LOS have a diacylglycerol lipid anchor plus a core region consisting of hexose-hexosamine-hexose, but lack heptose, 3-deoxy-D-manno-2-octulosonic acid, and β-hydroxy fatty acids, which are core components of LPS. Strikingly, the presence of a glycerol-hexose unit and two glycerol-hexadecanoic acid fragments in the lipid anchor indicates a glycolipid membrane anchor typically found in the lipoteichoic acid of Gram-positive bacteria (Schultz et al., 1998).
Immunomodulation and Immuno-Evasion
The host innate immune system is the first line of defense against bacterial infection. Inhibition of components of the innate immune system is obviously of great advantage to a bacterium that has been shown to associate closely with epithelial cells in vivo. T. denticola can evade aspects of the innate immune defense by preventing efficient binding of antimicrobial peptides, such as β-defensins, that are produced by epithelial cells, and by inducing rapid efflux of some host defense peptides which enter the cytoplasm (Brissette and Lukehart, 2007). Recently, T. denticola has also been shown to suppress the production of β-defensin 3 by human gingival epithelial cells (Shin et al., 2010).
TLR-Mediated Responses
Individual Toll-like receptors (TLRs) on the surfaces of host cells recognize structurally conserved, pathogen-associated molecular patterns, such as LPS, flagellin, and peptidoglycan. TLR activation leads to induction of inflammation-related genes important for limiting infection. However, dysregulation of the inflammatory response can lead to over-expression and continued expression of inflammatory mediators that can result in the tissue destruction characteristic of periodontitis.
Bacterial LPS is sensed by host epithelial cells and macrophages via binding to TLR4 that triggers a series of intracellular signaling systems, leading ultimately to production of inflammatory mediators and migration of macrophages and neutrophils to a site of infection (Dauphinee and Karsan, 2006).T. denticola LOS stimulates macrophages via TLR4 and MyD88 (myeloid differentiation primary response gene 88) and induces macrophage tolerance to LPS. T. denticola LOS also stimulates fibroblasts, inducing them to produce a variety of inflammatory mediators, interleukin-6 (IL-6), IL-8, monocyte chemoattractant 1, nitric oxide, and prostaglandin E2, as well as matrix metalloproteinase 3 (MMP-3) (Tanabe et al., 2008). Several signaling proteins have been shown to be phosphorylated following LOS stimulation of fibroblasts, including Fos, NF-κB p50, and NF-κB p65, indicating an inflammatory response.
In addition to LOS, other T. denticola cellular components can stimulate an immune response. Whole T. denticola cells and Msp can activate macrophages in a TLR2-MyD88-dependent process, resulting in TNF-α production in a dose-dependent manner. These macrophages also became tolerant to stimulation by enterobacterial LPS (Nussbaum et al., 2009). The LPS tolerance induced by T. denticola components during polymicrobial infection or proliferation may be a general mechanism to evade bacterial clearance. In addition to induction of innate immune responses by TLR2 and TLR4 receptors, T. denticola cells also induce innate immune responses via a TLR2/TLR6 heterodimer with mitogen-activated protein kinases (MAPKs), extracellular regulated protein-serine kinases 1 and 2 (ERK1/2), and p38, playing major roles in the resulting pro- and anti-inflammatory cytokine production (Ruby et al., 2007).
T. denticola peptidoglycan can also stimulate factors of innate immunity in human macrophage-like cells, inducing inflammatory mediators including TNF-α, IL-1β, IL-6, IL-8, and the chemokine RANTES (regulated upon activation, normal T-cell expressed, and secreted) and matrix metalloproteinase 9 (MMP-9) (Tanabe et al., 2009).
Immunohistochemistry of gingival tissues taken from patients undergoing periodontal therapy revealed high levels of TLR2 and TLR5 receptors in comparison with healthy tissues and that gingival epithelial cells can be stimulated to produce IL-1β and TNF-α in response to stimulation by the TLR2 and TLR5 ligands HKLM and flagellin (from Salmonella typhimurium), respectively (Beklen et al., 2009). The authors suggested that these antigens would also be present in other periodontal pathogens, such as the flagellin of T. denticola, and these could also stimulate host epithelial cells via TLR5 and thus may promote inflammation.
Conclusion
Treponemes, including T. denticola, are able to suppress host responses to LPS, which may allow for the persistence of the bacterial consortia found associated with periodontal disease. Furthermore, T. denticola LOS has been shown to stimulate osteoclastogenesis and matrix metalloproteinase expression, which could exacerbate periodontal pathology (Choi et al., 2003). The characteristics of T. denticola that represent its major virulence factors in chronic periodontitis are: its motility and chemotaxis, which enable the bacterium to rapidly colonize new sites, penetrate deep periodontal pockets, and penetrate epithelial layers; its ability to interact synergistically with other periodontal pathogens on several levels; its ability to produce cytotoxic metabolites; and its ability to form biofilms and a range of cell-surface proteins to dysregulate the host defense to help protect the subgingival biofilm and cause host tissue destruction.
Acknowledgments
Research funds from the National Health and Medical Research Council of Australia (Oral Health Grant OH017, Project Grant Nos. 251758 and 400327), the Co-operative Research Centre for Oral Health Science, and the US National Institutes of Health (Grant No. 1R21 DEO1536-01) are gratefully acknowledged.
References
- Allan ND, Beveridge TJ. (2003). Gentamicin delivery to Burkholderia cepacia group IIIa strains via membrane vesicles from Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 47:2962-2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin M, Grove DA, Kapus A, Glogauer M, Ellen RP. (2007). An actin-stabilizing peptide conjugate deduced from the major outer sheath protein of the bacterium Treponema denticola. Cell Motil Cytoskeleton 64:662-674 [DOI] [PubMed] [Google Scholar]
- Ang CS, Veith PD, Dashper SG, Reynolds EC. (2008). Application of 16O/18O reverse proteolytic labeling to determine the effect of biofilm culture on the cell envelope proteome of Porphyromonas gingivalis W50. Proteomics 8:1645-1660 [DOI] [PubMed] [Google Scholar]
- Arnett HA, Viney JL. (2007). Considerations for the sensible use of rodent models of inflammatory disease in predicting efficacy of new biological therapeutics in the clinic. Adv Drug Deliv Rev 59:1084-1092 [DOI] [PubMed] [Google Scholar]
- Bamford CV, Fenno JC, Jenkinson HF, Dymock D. (2007). The chymotrypsin-like protease complex of Treponema denticola ATCC 35405 mediates fibrinogen adherence and degradation. Infect Immun 75:4364-4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartold PM, Gully NJ, Zilm PS, Rogers AH. (1991). Identification of components in Fusobacterium nucleatum chemostat-culture supernatants that are potent inhibitors of human gingival fibroblast proliferation. J Periodontal Res 26:314-322 [DOI] [PubMed] [Google Scholar]
- Batista da Silva A, Lee W, Bajenova E, McCulloch C, Ellen R. (2004). The major outer sheath protein of Treponema denticola inhibits the binding step of collagen phagocytosis in fibroblasts. Cell Microbiol 6:485-498 [DOI] [PubMed] [Google Scholar]
- Beauchamp RO, Jr, Bus JS, Popp JA, Boreiko CJ, Andjelkovich DA. (1984). A critical review of the literature on hydrogen sulfide toxicity. Crit Rev Toxicol 13:25-97 [DOI] [PubMed] [Google Scholar]
- Beausejour A, Deslauriers N, Grenier D. (1997). Activation of the interleukin-1beta precursor by Treponema denticola: a potential role in chronic inflammatory periodontal diseases. Infect Immun 65:3199-3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beklen A, Sorsa T, Konttinen YT. (2009). Toll-like receptors 2 and 5 in human gingival epithelial cells co-operate with T-cell cytokine interleukin-17. Oral Microbiol Immunol 34:38-42 [DOI] [PubMed] [Google Scholar]
- Bergey DH, Holt JG. (1984). Bergey’s manual of systematic bacteriology. Baltimore, MD: Lippincott, Williams & Wilkins [Google Scholar]
- Bian X, Wang H, Ning Y, Lee SY, Fenno JC. (2005). Mutagenesis of a novel gene in the prcA-prtP protease locus affects expression of Treponema denticola membrane complexes. Infect Immun 73:1252-1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond JP, Francklyn C. (2000). Proteobacterial histidine-biosynthetic pathways are paraphyletic. J Mol Evol 50:339-347 [DOI] [PubMed] [Google Scholar]
- Boschwitz JS, Timoney JF. (1994). Characterization of the antiphagocytic activity of equine fibrinogen for Streptococcus equi subsp. equi. Microb Pathog 17:121-129 [DOI] [PubMed] [Google Scholar]
- Brissette CA, Lukehart SA. (2007). Mechanisms of decreased susceptibility to β-defensins by Treponema denticola. Infect Immun 75:2307-2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette CA, Pham TT, Coats SR, Darveau RP, Lukehart SA. (2008). Treponema denticola does not induce production of common innate immune mediators from primary gingival epithelial cells. Oral Microbiol Immunol 23:474-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Zhang J, Hodgson JE. (1998). A bacterial antibiotic resistance gene with eukaryotic origins. Curr Biol 8:R365-R367 [DOI] [PubMed] [Google Scholar]
- Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. (2001). Universal trees based on large combined protein sequence data sets. Nat Genet 28:281-285 [DOI] [PubMed] [Google Scholar]
- Byrne SJ, Dashper SG, Darby IB, Adams GG, Hoffmann B, Reynolds EC. (2009). Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol Immunol 24:469-477 [DOI] [PubMed] [Google Scholar]
- Cameron CE, Lukehart SA, Castro C, Molini B, Godornes C, Van Voorhis WC. (2000). Opsonic potential, protective capacity, and sequence conservation of the Treponema pallidum subspecies pallidum Tp92. J Infect Dis 181:1401-1413 [DOI] [PubMed] [Google Scholar]
- Canale-Parola E. (1978). Motility and chemotaxis of spirochetes. Annu Rev Microbiol 32:69-99 [DOI] [PubMed] [Google Scholar]
- Capestany CA, Kuboniwa M, Jung IY, Park Y, Tribble GD, Lamont RJ. (2006). Role of the Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect Immun 74:3002-3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone R, Wang HT, Ning Y, Sweier DG, Lopatin DE, Fenno JC. (2008). Human serum antibodies recognize Treponema denticola Msp and PrtP protease complex proteins. Oral Microbiol Immunol 23:165-169 [DOI] [PubMed] [Google Scholar]
- Carlsson J. (1997). Bacterial metabolism in dental biofilms. Adv Dent Res 11:75-80 [DOI] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF. (2002). Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet 36:47-73 [DOI] [PubMed] [Google Scholar]
- Chen W, Kajiya M, Giro G, Ouhara K, Mackler HE, Mawardi H, et al. (2010). Bacteria-derived hydrogen sulfide promotes IL-8 production from epithelial cells. Biochem Biophys Res Commun 391:645-650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B, Qi M, Kuramitsu HK. (2003). Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res Microbiol 154:637-643 [DOI] [PubMed] [Google Scholar]
- Choi BK, Paster BJ, Dewhirst FE, Gobel UB. (1994). Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun 62:1889-1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BK, Nattermann H, Grund S, Haider W, Gobel UB. (1997). Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int J Syst Bacteriol 47:175-181 [DOI] [PubMed] [Google Scholar]
- Choi BK, Lee HJ, Kang JH, Jeong GJ, Min CK, Yoo YJ. (2003). Induction of osteoclastogenesis and matrix metalloproteinase expression by the lipooligosaccharide of Treponema denticola. Infect Immun 71:226-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L, Burgum A, Kolodrubetz D, Holt SC. (1995). The 46-kilodalton-hemolysin gene from Treponema denticola encodes a novel hemolysin homologous to aminotransferases. Infect Immun 63:4448-4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L, Ebersole JL, Holt SC. (1999). Hemoxidation and binding of the 46-kDa cystalysin of Treponema denticola leads to a cysteine-dependent hemolysis of human erythrocytes. Oral Microbiol Immunol 14:293-303 [DOI] [PubMed] [Google Scholar]
- Chu L, Dong Z, Xu X, Cochran DL, Ebersole JL. (2002). Role of glutathione metabolism of Treponema denticola in bacterial growth and virulence expression. Infect Immun 70:1113-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L, Lai Y, Xu X, Eddy S, Yang S, Song L, et al. (2008). A 52-kDa leucyl aminopeptidase from Treponema denticola is a cysteinylglycinase that mediates the second step of glutathione metabolism. J Biol Chem 283:19351-19358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimasoni G, McBride BC. (1987). Adherence of Treponema denticola to modified hydroxyapatite. J Dent Res 66:1727-1729 [DOI] [PubMed] [Google Scholar]
- Cockayne A, Sanger R, Ivic A, Strugnell RA, MacDougall JH, Russell RR, et al. (1989). Antigenic and structural analysis of Treponema denticola. J Gen Microbiol 135:3209-3218 [DOI] [PubMed] [Google Scholar]
- Colombo A, da Silva C, Haffajee A, Colombo A. (2007). Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J Periodontal Res 42:236-243 [DOI] [PubMed] [Google Scholar]
- Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. (2009). Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol 80:1421-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashper SG, Ang CS, Veith PD, Mitchell HL, Lo AW, Seers CA, et al. (2009). Response of Porphyromonas gingivalis to heme limitation in continuous culture. J Bacteriol 191:1044-1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauphinee SM, Karsan A. (2006). Lipopolysaccharide signalling in endothelial cells. Lab Invest 86:9-22 [DOI] [PubMed] [Google Scholar]
- Demirkan I, Williams HF, Dhawi A, Carter SD, Winstanley C, Bruce KD, et al. (2006). Characterization of a spirochaete isolated from a case of bovine digital dermatitis. Appl Microbiol 101:948-955 [DOI] [PubMed] [Google Scholar]
- Devoe IW, Gilchrist JE. (1973). Release of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J Exp Med 138:1156-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. (2010). The human oral microbiome. J Bacteriol 192:5002-5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawi A, Hart CA, Demirkan I, Davies IH, Carter SD. (2005). Bovine digital dermatitis and severe virulent ovine foot rot: a common spirochaetal pathogenesis. Vet J 169:232-241 [DOI] [PubMed] [Google Scholar]
- Edwards AM, Jenkinson HF, Woodward MJ, Dymock D. (2005). Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect Immun 73:2891-2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftimiadi C, Valente S, Mangiante S, Mangiante PE, Niederman R. (1993). Short chain fatty acids produced by anaerobic bacteria inhibit adhesion and proliferation of periodontal ligament fibroblasts. Minerva Stomatol 42:481-485 [PubMed] [Google Scholar]
- Ellen RP. (2006). Virulence determinants of oral Treponemes. In: Pathogenic Treponema: molecular and cellular biology. Radolf JD, Lukehart SA, editors. Wymondham, Norfolk, UK: Caister Academic Press, pp. 357-386 [Google Scholar]
- Ellen R, Galimanas VB. (2005). Spirochetes at the forefront of periodontal infections. Periodontol 2000 38:13-32 [DOI] [PubMed] [Google Scholar]
- Evans NJ, Brown JM, Demirkan I, Murray RD, Birtles RJ, Hart CA, et al. (2009). Treponema pedis sp. nov., a spirochaete isolated from bovine digital dermatitits lesions. Int J Syst Evol Microbiol 59(Pt 5):987-991 [DOI] [PubMed] [Google Scholar]
- Fenno JC, McBride BC. (1998). Virulence factors of oral treponemes. Anaerobe 4:1-17 [DOI] [PubMed] [Google Scholar]
- Fenno JC, Wong GW, Hannam PM, Müller KH, Leung WK, McBride BC. (1997). Conservation of msp, the gene encoding the major outer membrane protein of oral Treponema spp. J Bacteriol 179:1082-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno JC, Tamura M, Hannam PM, Wong GW, Chan RA, McBride BC. (2000). Identification of a Treponema denticola OppA homologue that binds host proteins present in the subgingival environment. Infect Immun 68:1884-1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno JC, Lee SY, Bayer CH, Ning Y. (2001). The opdB locus encodes the trypsin-like peptidase activity of Treponema denticola. Infect Immun 69:6193-6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foschi F, Izard J, Sasaki H, Sambri V, Prati C, Muller R, et al. (2006). Treponema denticola in disseminating endodontic infections. J Dent Res 85:761-765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, et al. (1998). Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388 [DOI] [PubMed] [Google Scholar]
- Fukamachi H, Nakano Y, Okano S, Shibata Y, Abiko Y, Yamashita Y. (2005). High production of methyl mercaptan by L-methionine-alpha-deamino-gamma-mercaptomethane lyase from Treponema denticola. Biochem Biophys Res Commun 331:127-131 [DOI] [PubMed] [Google Scholar]
- Gemmell E, Bird PS, Carter CL, Drysdale KE, Seymour GJ. (2002). Effect of Fusobacterium nucleatum on the T and B cell responses to Porphyromonas gingivalis in a mouse model. Clin Exp Immunol 128:238-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier D. (1992). Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect Immun 60:5298-5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier D, Mayrand D. (1987). Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect Immun 55:111-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier D, Mayrand D. (2001). Cleavage of human immunoglobulin G by Treponema denticola. Anaerobe 7:1-4 [Google Scholar]
- Grenier D, Uitto VJ, McBride BC. (1990). Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect Immun 58:347-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffajee AD, Teles RP, Socransky SS. (2006). Association of Eubacterium nodatum and Treponema denticola with human periodontitis lesions. Oral Microbiol Immunol 21:269-282 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Asai Y, Jinno T, Adachi S, Kusumoto S, Ogawa T. (2003a). Structural elucidation of polysaccharide part of glycoconjugate from Treponema medium ATCC 700293. Eur J Cell Biol 270:2671-2679 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Ogawa S, Asai Y, Takai Y, Ogawa T. (2003b). Binding of Porphyromonas gingivalis fimbriae to Treponema denticola dentilisin. FEMS Microbiol Lett 226:267-271 [DOI] [PubMed] [Google Scholar]
- Henz SR, Huson DH, Auch AF, Nieselt-Struwe K, Schuster SC. (2005). Whole-genome prokaryotic phylogeny. Bioinformatics 21:2329-2335 [DOI] [PubMed] [Google Scholar]
- Hespell RB, Canale-Parola E. (1971). Amino acid and glucose fermentation by Treponema denticola. Arch Mikrobiol 78:234-251 [DOI] [PubMed] [Google Scholar]
- Hollmann R, Van der Hoeven HJ. (1999). Inability of intact cells of Treponema denticola to degrade human serum proteins IgA, IgG and albumin. J Clin Periodontol 26:477-479 [DOI] [PubMed] [Google Scholar]
- Holt SC. (1978). Anatomy and chemistry of spirochetes. Microbiol Rev 42:114-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. (2005). Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000 38:72-122 [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. (2006). The oral spirochetes: their ecology and role in the pathogenesis of periodontal disease. In: Pathogenic Treponema: molecular and cellular biology. Radolf JD, Lukehart SA, editors. Wymondham, Norfolk, UK: Caister Academic Press, pp. 323-356 [Google Scholar]
- Ibba M, Morgan S, Curnow AW, Pridmore DR, Vothknecht UC, Gardner W, et al. (1997). A euryarchaeal lysyl-tRNA synthetase: resemblance to class I synthetases. Science 278:1119-1122 [DOI] [PubMed] [Google Scholar]
- Ikegami A, Honma K, Sharma A, Kuramitsu HK. (2004). Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infect Immun 72:4619-4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S, Onishi S, Kuramitsu HK, Sharma A. (2006). Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia”. Infect Immun 74:5023-5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Miura T, Kuramitsu HK, Okuda K. (1996). Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin). Infect Immun 64:5178-5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Kuramitsu HK, Miura T, Okuda K. (1998). Dentilisin activity affects the organization of the outer sheath of Treponema denticola. J Bacteriol 180:3837-3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Kuramitsu HK, Okuda K. (2004). A 43-kDa protein of Treponema denticola is essential for dentilisin activity. FEMS Microbiol Lett 232:181-188 [DOI] [PubMed] [Google Scholar]
- Izard J, Samsonoff WA, Limberger RJ. (2001). Cytoplasmic filament-deficient mutant of Treponema denticola has pleiotropic defects. J Bacteriol 183:1078-1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard J, Hsieh CE, Limberger RJ, Mannella CA, Marko M. (2008). Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J Struct Biol 163:10-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, et al. (2009). Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol 191:7566-7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman R. (2008). Bacterial persistence: some new insights into an old phenomenon. J Biosci 33:795-805 [DOI] [PubMed] [Google Scholar]
- Jobin MC, Virdee I, McCulloch CA, Ellen RP. (2007). Activation of MAPK in fibroblasts by Treponema denticola major outer sheath protein. Biochem Biophys Res Commun 356:213-218 [DOI] [PubMed] [Google Scholar]
- Johnson J, Chen R, Lenton P, Zhang G, Hinrichs J, Rudney J. (2008). Persistence of extracrevicular bacterial reservoirs after treatment of aggressive periodontitis. J Periodontol 79:2305-2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PW, Ng W, Tonzetich J. (1992). Modulation of human gingival fibroblast cell metabolism by methyl mercaptan. J Periodontal Res 27:476-483 [DOI] [PubMed] [Google Scholar]
- Jun HK, Kang YM, Lee HR, Lee SH, Choi BK. (2008). Highly conserved surface proteins of oral spirochetes as adhesins and potent inducers of proinflammatory and osteoclastogenic factors. Infect Immun 76:2428-2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. (1996). Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol 178:2767-2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CW, Lux R, Haake SK, Shi W. (2009). The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol 71:35-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka M, Li H, Arakawa S, Kuramitsu H. (1997). Characterization of a methyl-accepting chemotaxis protein gene, dmcA, from the oral spirochete Treponema denticola. Infect Immun 65:4011-4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Kowashi Y, Demuth DR. (2002). Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog 32:1-13 [DOI] [PubMed] [Google Scholar]
- Kesavalu L, Walker SG, Holt SC, Crawley RR, Ebersole JL. (1997). Virulence characteristics of oral treponemes in a murine model. Infect Immun 65:5096-5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavalu L, Holt SC, Ebersole JL. (1998). Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol Immunol 13:373-377 [DOI] [PubMed] [Google Scholar]
- Kesavalu L, Sathishkumar S, Bakthavatchalu V, Matthews C, Dawson D, Steffen M, et al. (2007). Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect Immun 75:1704-1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigure T, Saito A, Seida K, Yamada S, Ishihara K, Okuda K. (1995). Distribution of Porphyromonas gingivalis and Treponema denticola in human subgingival plaque at different periodontal pocket depths examined by immunohistochemical methods. J Periodontal Res 30:332-341 [DOI] [PubMed] [Google Scholar]
- Kim Y, Wang X, Ma Q, Zhang XS, Wood TK. (2009). Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol 191:1258-1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. (2009). Hydrogen sulfide: from brain to gut. Antioxid Redox Signal 12:1111-1123 [DOI] [PubMed] [Google Scholar]
- Klitgaard K, Boye M, Capion N, Jensen TK. (2008). Evidence of multiple Treponema phylotypes involved in bovine digital dermatitis as shown by 16S rRNA gene analysis and fluorescence in situ hybridization. J Clin Microbiol 46:3012-3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Parrish KD, Andersen RN, Greenberg EP. (1995). Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect Immun 63:4584-4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr (2002). Communication among oral bacteria. Microbiol Mol Biol Rev 66:486-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MJ, Kesty NC. (2005). Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev 19:2645-2655 [DOI] [PubMed] [Google Scholar]
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. (2007). Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev 71:653-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita-Ochiai T, Fukushima K, Ochiai K. (1995). Volatile fatty acids, metabolic by-products of periodontopathic bacteria, inhibit lymphocyte proliferation and cytokine production. J Dent Res 74:1367-1373 [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 62:1244-1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancero H, Niu J, Johnson PW. (1996). Exposure of periodontal ligament cells to methyl mercaptan reduces intracellular pH and inhibits cell migration. J Dent Res 75:1994-2002 [DOI] [PubMed] [Google Scholar]
- Lee SF, Andrian E, Rowland E, Marquez IC. (2009). Immune response and alveolar bone resorption in a mouse model of Treponema denticola infection. Infect Immun 77:694-698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kim KK, Choi BK. (2005). Upregulation of intercellular adhesion molecule 1 and proinflammatory cytokines by the major surface proteins of Treponema maltophilum and Treponema lecithinolyticum, the phylogenetic group IV oral spirochetes associated with periodontitis and endodontic infections. Infect Immun 73:268-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kim JS, Jun HK, Lee HR, Lee D, Choi BK. (2009). The major outer membrane protein of a periodontopathogen induces IFN-B and IFN-stimulated genes in monocytes via lipid raft and TANK-binding kinase 1/IFN regulatory factor 3. J Immunol 182:5823-5835 [DOI] [PubMed] [Google Scholar]
- Lee SY, Fenno JC. (2004). Expression of Treponema denticola oligopeptidase B in Escherichia coli. Curr Microbiol 48:379-382 [DOI] [PubMed] [Google Scholar]
- Lee SY, Bian XL, Wong GW, Hannam PM, McBride BC, Fenno JC. (2002). Cleavage of Treponema denticola PrcA polypeptide to yield protease complex-associated proteins Prca1 and Prca2 is dependent on PrtP. J Bacteriol 184:3864-3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. (2000). Programmed death in bacteria. Microbiol Mol Biol Rev 64:503-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruby J, Charon N, Kuramitsu H. (1996). Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol 178:3664-3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Arakawa S, Deng QD, Kuramitsu H. (1999). Characterization of a novel methyl-accepting chemotaxis gene, dmcB, from the oral spirochete Treponema denticola. Infect Immun 67:694-699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Clarke AJ, Beveridge TJ. (1996). A major autolysin of Pseudomonas aeruginosa: subcellular distribution, potential role in cell growth and division and secretion in surface membrane vesicles. J Bacteriol 178:2479-2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux R, Miller JN, Park NH, Shi W. (2001). Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect Immun 69:6276-6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux R, Sim JH, Tsai JP, Shi W. (2002). Construction and characterization of a cheA mutant of Treponema denticola. J Bacteriol 184:3130-3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhães MA, Sun CX, Glogauer M, Ellen RP. (2008). The major outer sheath protein of Treponema denticola selectively inhibits Rac1 activation in murine neutrophils. Cell Microbiol 10:344-354 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. (2009). Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen PL, Makinen KK. (1997). Gamma-glutamyltransferase from the outer cell envelope of Treponema denticola ATCC 35405. Infect Immun 65:685-691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen PL, Makinen KK, Syed SA. (1995). Role of the chymotrypsin-like membrane-associated proteinase from Treponema denticola ATCC 35405 in inactivation of bioactive peptides. Infect Immun 63:3567-3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn LM, Whiteley M. (2005). Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422-425 [DOI] [PubMed] [Google Scholar]
- McDowell JV, Frederick J, Stamm L, Marconi RT. (2007). Identification of the gene encoding the FhbB protein of Treponema denticola, a highly unique factor H-like protein 1 binding protein. Infect Immun 75:1050-1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Huang B, Fenno JC, Marconi RT. (2009). Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola. Infect Immun 77:1417-1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineoka T, Awano S, Rikimaru T, Kurata H, Yoshida A, Ansai T, et al. (2008). Site-specific development of periodontal disease is associated with increased levels of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia in subgingival plaque. J Periodontol 79:670-676 [DOI] [PubMed] [Google Scholar]
- Mitchell HL, Dashper SG, Catmull DV, Paolini RA, Cleal SM, Slakeski N, et al. (2010). Treponema denticola biofilm-induced expression of a bacteriophage, toxin-antitoxin systems and transposases. Microbiology 156(Pt 4):774-788 [DOI] [PubMed] [Google Scholar]
- Miyamoto M, Ishihara K, Okuda K. (2006). The Treponema denticola surface protease dentilisin degrades interleukin-1 beta (IL-1 beta), IL-6, and tumor necrosis factor alpha. Infect Immun 74:2462-2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molbak L, Klitgaard K, Jensen TK, Fossi M, Boye M. (2006). Identification of a novel, invasive, not-yet-cultivated Treponema sp. in the large intestine of pigs by PCR amplification of the 16S rRNA gene. J Clin Microbiol 44:4537-4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederman R, Zhang J, Kashket S. (1997). Short-chain carboxylic-acid-stimulated, PMN-mediated gingival inflammation. Crit Rev Oral Biol Med 8:269-290 [DOI] [PubMed] [Google Scholar]
- Nilius AM, Spencer SC, Simonson LG. (1993). Stimulation of in vitro growth of Treponema denticola by extracellular growth factors produced by Porphyromonas gingivalis. J Dent Res 72:1027-1031 [DOI] [PubMed] [Google Scholar]
- Nordhoff M, Rühe B, Kellermeier C, Moter A, Schmitz R, Brunnberg L, et al. (2008a). Association of Treponema spp. with canine periodontitis. Vet Microbiol 127(March):334-342 [DOI] [PubMed] [Google Scholar]
- Nordhoff M, Moter A, Schrank K, Wieler L. (2008b). High prevalence of treponemes in bovine digital dermatitis—a molecular epidemiology. Vet Microbiol 131(October):293-300 [DOI] [PubMed] [Google Scholar]
- Norris SJ, Weinstock GM. (2000). The genome sequence of Treponema pallidum, the syphilis spirochete: will clinicians benefit? Curr Opin Infect Dis 13:29-36 [DOI] [PubMed] [Google Scholar]
- Nowotny A, Behling UH, Hammond B, Lai CH, Listgarten M, Pham PH, et al. (1982). Release of toxic microvesicles by Actinobacillus actinomycetemcomitans. Infect Immun 37:151-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum G, Ben-Adi S, Genzler T, Sela M, Rosen G. (2009). Involvement of Toll-like receptors 2 and 4 in the innate immune response to Treponema denticola and its outer sheath components. Infect Immun 77:3939-3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien-Simpson NM, Paolini RA, Reynolds EC. (2000). RgpA-Kgp peptide-based immunogens provide protection against Porphyromonas gingivalis challenge in a murine lesion model. Infect Immun 68:4055-4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien-Simpson NM, Pathirana RD, Paolini RA, Chen YY, Veith PD, Tam V, et al. (2005). An immune response directed to proteinase and adhesin functional epitopes protects against Porphyromonas gingivalis-induced periodontal bone loss. J Immunol 175:3980-3989 [DOI] [PubMed] [Google Scholar]
- Ohta K, Makinen KK, Loesche WJ. (1986). Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect Immun 53:213-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, Kimizuka R, Miyamoto M, Kato T, Yamada S, Okuda K, et al. (2007). Treponema denticola induces interleukin-8 and macrophage chemoattractant protein 1 production in human umbilical vein epithelial cells. Microbes Infect 9:907-913 [DOI] [PubMed] [Google Scholar]
- Orth R, O’Brien-Simpson N, Dashper S, Walsh K, Reynolds E. (2010). An efficient method for enumerating oral spirochetes using flow cytometry. J Microbiol Methods 80:123-128 [DOI] [PubMed] [Google Scholar]
- Paster BJ, Dewhirst FE. (2000). Phylogenetic foundation of spirochetes. J Mol Microbiol Biotechnol 2:341-344 [PubMed] [Google Scholar]
- Paster BJ, Dewhirst FE. (2006). The phylogenetic diversity of the genus Treponema. In: Pathogenic Treponema: molecular and cellular biology. Radolf JD, Lukehart SA, editors. Wymondham, Norfolk, UK: Caister Academic Press, pp. 9-18 [Google Scholar]
- Pathirana RD, O’Brien-Simpson NM, Brammar GC, Slakeski N, Reynolds EC. (2007). Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect Immun 75:1436-1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier TP, Kehoe MA, Whitnack E, Dockter ME, Beachey EH. (1989). Fibrinogen binding and resistance to phagocytosis of Streptococcus sanguis expressing cloned M protein of Streptococcus pyogenes. Infect Immun 57:29-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle M, Bergsten C, Fernström LL, Höök H, Johansson KE. (2008). Isolation and characterization of Treponema phagedenis-like spirochetes from digital dermatitis lesions in Swedish cattle. Acta Vet Scand 50:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle M, Backhans A, Otman F, Sjölund M, Fellström C. (2009). Isolation of spirochetes of genus Treponema from pigs with ear necrosis. Vet Microbiol 139:279-283 [DOI] [PubMed] [Google Scholar]
- Puthengady Thomas B, Sun C, Bajenova E, Ellen RP, Glogauer M. (2006). Modulation of human neutrophil functions in vitro by Treponema denticola major outer sheath protein. Infect Immun 74:1954-1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radbruch A, Isaacs J. (2009). Animal models in infection and inflammation—chance and necessity. Eur J Immunol 39:1991-1993 [DOI] [PubMed] [Google Scholar]
- Reiffenstein RJ, Hulbert WC, Roth SH. (1992). Toxicology of hydrogen sulfide. Annu Rev Pharmacol Toxicol 32:109-134 [DOI] [PubMed] [Google Scholar]
- Robertson D, Smith AJ. (2009). The microbiology of the acute dental abscess. J Med Microbiol 58(Pt 2):155-162 [DOI] [PubMed] [Google Scholar]
- Rosen G, Genzler T, Sela MN. (2008). Coaggregation of Treponema denticola with Porphyromonas gingivalis and Fusobacterium nucleatum is mediated by the major outer sheath protein of Treponema denticola. FEMS Microbiol Lett 289:59-66 [DOI] [PubMed] [Google Scholar]
- Rother M, Bock A, Wyss C. (2001). Selenium-dependent growth of Treponema denticola: evidence for a clostridial-type glycine reductase. Arch Microbiol 177:113-116 [DOI] [PubMed] [Google Scholar]
- Ruby J, Rehani K, Martin M. (2007). Treponema denticola activates mitogen-activated protein kinase signal pathways through Toll-like receptor 2. Infect Immun 75:5763-5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JD, Lux R, Shi W, Charon NW, Dasanayake A. (2008). Effect of glucose on Treponema denticola cell behavior. Oral Microbiol Immunol 23:234-238 [DOI] [PubMed] [Google Scholar]
- Saglie R, Newman MG, Carranza FA, Jr, Pattison GL. (1982). Bacterial invasion of gingiva in advanced periodontitis in humans. J Periodontol 53:217-222 [DOI] [PubMed] [Google Scholar]
- Sayers G, Marques PX, Evans NJ, O’Grady L, Doherty ML, Carter SD, et al. (2009). Identification of spirochetes associated with contagious ovine digital dermatitis. J Clin Microbiol 47:1199-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder N, Opitz B, Lamping N, Michelsen K, Zähringer U, Göbel U, et al. (2000). Involvement of lipopolysaccharide binding protein, CD14, and Toll-like receptors in the initiation of innate immune responses by Treponema glycolipids. J Immunol 165:2683-2693 [DOI] [PubMed] [Google Scholar]
- Schultz CP, Wolf V, Lange R, Mertens E, Weke J, Naumann D, et al. (1998). Evidence for a new type of outer membrane lipid in oral spirochete Treponema denticola: functioning permeation barrier without lipopolysaccharides. J Biol Chem 273:15661-15666 [DOI] [PubMed] [Google Scholar]
- Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, Dashper SG, et al. (2006). The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J Bacteriol 188:6376-6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela M. (2001). Role of Treponema denticola in periodontal diseases. Crit Rev Oral Biol Med 12:399-413 [DOI] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, et al. (2004). Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci USA 101:5646-5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setubal JC, Reis M, Matsunaga J, Haake DA. (2006). Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152(Pt 1):113-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Sojar HT, Glurich I, Honma K, Kuramitsu HK, Genco RJ. (1998). Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect Immun 66:5703-5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Inagaki S, Honma K, Sfintescu C, Baker PJ, Evans RT. (2005). Tannerella forsythia-induced alveolar bone loss in mice involves leucine-rich-repeat BspA protein. J Dent Res 84:462-467 [DOI] [PubMed] [Google Scholar]
- Shin JE, Kim YS, Oh JE, Min BM, Choi Y. (2010). Treponema denticola suppresses expression of human {beta}-defensin-3 in gingival epithelial cells through inhibition of the Toll-like receptor 2 axis. Infect Immun 78:672-679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim JH, Shi W, Lux R. (2005). Protein-protein interactions in the chemotaxis signalling pathway of Treponema denticola. Microbiology 151(Pt 6):1801-1807 [DOI] [PubMed] [Google Scholar]
- Simonson LG, McMahon KT, Childers DW, Morton HE. (1992). Bacterial synergy of Treponema denticola and Porphyromonas gingivalis in a multinational population. Oral Microbiol Immunol 7:111-112 [DOI] [PubMed] [Google Scholar]
- Smajs D, McKevitt M, Howell JK, Norris SJ, Cai WW, Palzkill T, et al. (2005). Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J Bacteriol 187:1866-1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. (1998). Microbial complexes in subgingival plaque. J Clin Periodontol 25:134-144 [DOI] [PubMed] [Google Scholar]
- Tanabe S, Bodet C, Grenier D. (2008). Treponema denticola lipooligosaccharide activates gingival fibroblasts and upregulates inflammatory mediator production. J Cell Physiol 216:727-731 [DOI] [PubMed] [Google Scholar]
- Tanabe SI, Bodet C, Grenier D. (2009). Treponema denticola peptidoglycan induces the production of inflammatory mediators and matrix metalloproteinase 9 in macrophage-like cells. J Periodontal Res 44:503-510 [DOI] [PubMed] [Google Scholar]
- Tonetti M, Eftimiadi C, Damiani G, Buffa P, Buffa D, Botta GA. (1987). Short chain fatty acids present in periodontal pockets may play a role in human periodontal diseases. J Periodontal Res 22:190-191 [DOI] [PubMed] [Google Scholar]
- Tribble GD, Lamont RJ. (2010). Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol 2000 52:68-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto VJ, Grenier D, Chan EC, McBride BC. (1988a). Isolation of a chymotrypsinlike enzyme from Treponema denticola. Infect Immun 56:2717-2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto VJ, Haapasalo M, Laakso T, Salo T. (1988b). Degradation of basement membrane collagen by proteases from some anaerobic oral micro-organisms. Oral Microbiol Immunol 3:97-102 [DOI] [PubMed] [Google Scholar]
- Umemoto T, Jinno T, Taiji Y, Ogawa T. (2001). Chemotaxis of oral treponemes toward sera and albumin of rabbit. Microbiol Immunol 45:571-577 [DOI] [PubMed] [Google Scholar]
- Veith PD, Dashper SG, O’Brien-Simpson NM, Paolini RA, Orth R, Walsh KA, et al. (2009). Major proteins and antigens of Treponema denticola. Biochim Biophys Acta 1794:1421-1432 [DOI] [PubMed] [Google Scholar]
- Vesey PM, Kuramitsu HK. (2004). Genetic analysis of Treponema denticola ATCC 35405 biofilm formation. Microbiology 150(Pt 7):2401-2407 [DOI] [PubMed] [Google Scholar]
- Wai SN, Lindmark B, Söderblom T, Takade A, Westermark M, Oscarsson J, et al. (2003). Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25-35 [DOI] [PubMed] [Google Scholar]
- Watson JH, Angulo JJ, Leon-Blanco F, Varela G, Wedderburn CC. (1951). Electron microscopic observations of flagellation in some species of the genus Treponema Schaudinn. J Bacteriol 61:455-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg A, Holt SC. (1991). Chemical and biological activities of a 64-kilodalton outer sheath protein from Treponema denticola strains. J Bacteriol 173:6935-6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink J, Witholt B. (1981). Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur J Biochem 116:331-335 [DOI] [PubMed] [Google Scholar]
- Whitnack E, Beachey EH. (1985). Inhibition of complement-mediated opsonization and phagocytosis of Streptococcus pyogenes by D fragments of fibrinogen and fibrin bound to cell surface M protein. J Exp Med 162:1983-1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Grishin NV, Tatusov RL, Koonin EV. (2001). Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol Biol 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C, Moter A, Choi BK, Dewhirst FE, Xue Y, Schüpbach P, et al. (2004). Treponema putidum sp. nov., a medium-sized proteolytic spirochaete isolated from lesions of human periodontitis and acute necrotizing ulcerative gingivitis. Int J Syst Evol Microbiol 54(Pt 4):1117-1122 [DOI] [PubMed] [Google Scholar]
- Yamada M, Ikegami A, Kuramitsu HK. (2005). Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol Lett 250:271-277 [DOI] [PubMed] [Google Scholar]
- Yano T, Yamagami R, Misumi K, Kubota C, Moe K, Hayashi T, et al. (2009). Genetic heterogeneity among strains of Treponema phagedenis-like spirochetes isolated from dairy cattle with papillomatous digital dermatitis in Japan. J Clin Microbiol 47:727-733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao ES, Lamont RJ, Leu SP, Weinberg A. (1996). Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol Immunol 11:35-41 [DOI] [PubMed] [Google Scholar]
- Yoneda M, Yoshikane T, Motooka N, Yamada K, Hisama K, Naito T, et al. (2005). Stimulation of growth of Porphyromonas gingivalis by cell extracts from Tannerella forsythia. J Periodontal Res 40:105-109 [DOI] [PubMed] [Google Scholar]
- Yoshimura M, Nakano Y, Yamashita Y, Oho T, Saito T, Koga T. (2000). Formation of methyl mercaptan from L-methionine by Porphyromonas gingivalis. Infect Immun 68:6912-6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmür R, et al. (2010). Oral biofilm architecture on natural teeth. PLoS One 5:e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]


