Abstract
Streptococcus mutans is the primary cariogen that produces several virulence factors that are modulated by a competence-stimulating peptide (CSP) signaling system. In this study, we sought to determine if proteases produced by early dental plaque colonizers such as Streptococcus gordonii interfere with the subsequent colonization of S. mutans BM71 on the existing streptococcal biofilms. We demonstrated that S. mutans BM71 colonized much less efficiently in vitro on streptococcal biofilms than on Actinomyces naeslundii biofilms. Several oral streptococci, relative to A. naeslundii, produced proteases that inactivated the S. mutans CSP. We further demonstrated that cell protein extracts from S. gordonii, but not from A. naeslundii, interfered with S. mutans BM71 colonization. In addition, S. mutans BM71 colonized more efficiently on the sgc protease knockout mutant of S. gordonii than on the parent biofilms. In conclusion, proteases of early colonizers can interfere with subsequent colonization by S. mutans in vitro.
Keywords: Streptococcus mutans, Streptococcus gordonii, Actinomyces naeslundii, biofilm, protease, quorum sensing
Introduction
Dental caries is among the most common chronic infectious diseases occurring in humans (NIDCR, CDC, 2002). Streptococcus mutans is the primary cariogen that produces several virulence factors. It is also implicated in some cases of infective endocarditis (Blanco-Carrion, 2004). S. mutans CSP (encoded by the comC gene) and a signal transduction system (a quorum-sensing system encoded by the comD and comE genes) mediate a variety of virulence characteristics, such as biofilm formation (Li et al., 2002b; Yoshida and Kuramitsu, 2002), bacteriocin production (Yonezawa and Kuramitsu, 2005), acid tolerance (Li et al., 2002a), antimicrobial sensitivity (Matsumoto-Nakano and Kuramitsu, 2006), and natural genetic transformation (Cvitkovitch, 2001).
Although S. mutans has been studied extensively relative to cariogenesis, there has been limited information relative to its interactions with other oral bacteria. Experiments carried out on laboratory animals have shown that selective suppression of S. mutans inhibits dental caries formation. For example, inoculation of low-virulence bacteria such as Streptococcus salivarius reduced the number of S. mutans in dental plaque and the subsequent levels of caries (Tanzer et al., 1985). An understanding of the interactions between S. mutans and other oral plaque bacteria as demonstrated in this study may provide insight into novel strategies for preventing oral streptococcal infections.
Mono-strain biofilm formation of S. mutans has been demonstrated to be a CSP-dependent quorum-sensing phenomenon (Li et al., 2002b; Yoshida and Kuramitsu, 2002). Elimination of CSP by knocking out the comC gene results in the formation of abnormal biofilms, either with altered architecture or with reduced biomass. However, dental plaque is composed of multiple bacteria that colonize in a sequential manner. We have demonstrated that one of these proteases, S. gordonii challisin (a serine protease encoded by the sgc gene), is able to inactivate S. mutans CSP and thereby to antagonize CSP-dependent bacteriocin production (Wang and Kuramitsu, 2005). Since biofilm formation is another property modulated by S. mutans CSP, we hypothesize that attenuation of S. mutans CSP will also affect S. mutans’ sequential establishment in multi-species biofilms.
Materials & Methods
Bacterial Strains and Media
S. mutans BM71 and its comC mutant (erythromycin-resistant), S. mutans BM71pTet [S. mutans BM71 containing a tetracycline-resistant plasmid (Xie et al., 2007)], S. mutans BM71 or GS5, S. gordonii Challis and its sgc knockout mutant (erythromycin-resistant) (Wang and Kuramitsu, 2005), S. gordonii 10558, Streptococcus mitis 10712, Streptococcus oralis KS32AR, Streptococcus sanguinis 10556, as well as A. naeslundii 12104 were maintained on Todd-Hewitt broth (THB) agar plates supplemented with erythromycin or tetracycline (10 µg/mL) where indicated. Bacteria were routinely cultured in THB. A group C streptococcal strain RP66 was used as an indicator for assays of S. mutans BM71 bacteriocin activity (Paul and Slade, 1975).
Sequential Biofilm Formation
To form initial biofilms, we inoculated overnight pure cultures of early colonizers—including S. gordonii Challis or 10558, the S. gordonii Challis sgc mutant, S. mitis, S. oralis, S. sanguinis, A. naeslundii or S. mutans BM71 or GS5 (as controls) in THB—into autoclaved ¼-strength THB supplemented with 0.01% mucin (Sigma Chemical Co., St. Louis, MO, USA) (BM, biofilm media) (Li et al., 2001) in 48-well polystyrene microtiter plates (107 cfu/mL). After a 24-hour incubation at 37°C in the presence of CO2 under static conditions, the planktonic cells were discarded, and biofilms were washed twice with PBS for further removal of the planktonic cells. For subsequent sequential biofilm formation, BM71pTet in the stationary phase (in THB with tetracycline) was diluted in fresh THB without tetracycline (1:2) and cultured at 37°C for 1 hr to bring the BM71pTet cells to the same growth conditions. The cultured BM71pTet were further diluted in fresh BM (1:1000), and 0.5 mL of BM71pTet was added to each well containing initial biofilms as described above and incubated at 37°C without CO2 under static conditions for another 4 hrs. The sequentially formed biofilms were washed twice with PBS for removal of the non-adherent bacteria. THB (0.5 mL/well) was added to the washed biofilms, and sonication was applied to each well 5 times by means of a Branson sonifier (output 3, cycle 30). The disrupted biofilm bacteria were further diluted in PBS (1:100), and 10 or 100 µL were plated on THB agar plates supplemented with tetracycline (10 µg/mL). For initial biofilm assessment, the subsequent addition of BM71 was omitted, and the disrupted initial biofilm bacteria were plated on THB agar plates. The plates were cultured at 37°C for 48 hrs under anaerobic conditions for subsequent colony counting.
Effects of Early Colonizers on the CSP of S. mutans
Supernatants from S. gordonii or A. naeslundii grown to the stationary phase in THB were filtered through 0.22-µm filters to eliminate cellular components. The supernatants were then neutralized with NaOH to pH 7.0-7.5. Exogenous synthetic S. mutans CSP [amino acid sequence: SGSLSTFFRLFNRSFTQALGK (Li et al., 2001), synthesized by Sigma-Genosys, The Woodlands, TX, USA; 2.5 µg/mL, final concentration] was incubated with the supernatants at 37°C for 2 hrs. CSP thus treated was added to S. mutans BM71 comC mutant cells (107 cfu) in 0.9 mL of THBY (THB supplemented with 3% yeast extract) to a final concentration of 0.25 µg/mL. After a 24-hour incubation at 37°C, the supernatants from the cultures were passed through 0.22-µm filters, and the bacteriocin level produced by the comC mutant was determined by the agar well assays as described below.
Agar Well Assays
The supernatant fluids containing bacteriocin from the S. mutans BM71 comC mutant cultures were obtained as described above. The supernatants were added into pre-cut wells in THB agar plates and incubated at 37°C for 24 hrs to facilitate the absorption of the supernatants into the agar surrounding the wells. The wells were then filled using THB with 1% low-melting-temperature agarose, and the plates were overlaid with the indicator strain RP66 (106 cfu) in 3 mL THB with 1% low-melting-temperature agarose. After 24 hrs of further incubation at 37°C under anaerobic conditions, the diameters of the inhibition zones surrounding the wells were measured.
Azocasein Assays
Proteolytic activities against azocasein from several early colonizers were measured according to a modified method (Sojar et al., 1993). Briefly, 50 mL overnight cultures of S. gordonii or A. naeslundii were centrifuged at 4000 × g for 10 min. After being washed once with PBS, the cell pellets were utilized in the azocasein assays. OD420nm measured in these assays indicated the levels of protease activity present.
Extraction of Cell Proteins from Early Colonizers
Cell proteins from S. gordonii or A. naeslundii were extracted according to a modified method (Wang et al., 2009). Briefly, 50 mL overnight cultures of S. gordonii or A. naeslundii were centrifuged at 4000 × g for 10 min. After being washed once with PBS, the cells were treated with Lysing Matrix B and FastPrep, followed by sonication. The cell extracts were then centrifuged at 12,000 × g for 10 min. The supernatants from the cell extracts were concentrated with PEG 8000, and final protein concentrations were adjusted to 100 mg/mL.
Statistical Analysis
We performed Student’s t test to determine significance. A difference was considered significant when a p value < 0.05 was obtained.
Results
S. mutans in vitro Colonization is More Efficient on Pre-existing A. naeslundii Compared with Biofilms of Other Early Colonizers
S. mutans BM71 in vitro colonization efficiencies on several pre-formed early-colonizer biofilms, including S. gordonii Challis or 10558, S. mitis, S. oralis, S. sanguinis, and A. naeslundii, were studied. Pre-formed S. mutans BM71 or GS5 biofilms, as well as blank 48-well polystyrene surfaces, served as internal controls. S. mutans BM71 colonization was significantly more efficient on the A. naeslundii biofilms and the control S. mutans biofilms than on those of S. gordonii Challis (Fig. 1). S. mutans BM71 colonization efficiencies on other early colonizers, such as S. gordonii 10558, S. mitis, S. oralis, and S. sanguinis, were comparable with that of S. gordonii Challis and significantly less relative to those on A. naeslundii and S. mutans (Appendix Fig.). S. mutans BM71 colonization on the blank polystyrene surfaces for 4 hrs was minimal (0.56 ± 0.03 × 105/well). The initial biofilms had comparable amounts of bacterial attachment (× 105): S. gordonii Challis, 13.5 ± 0.6; the S. gordonii Challis sgc knockout mutant, 12.9 ± 1.4; S. gordonii 10558, 13.1 ± 0.5; A. naeslundii, 6.9 ± 0.4; S. mitis, 6.5 ± 0.9; S. oralis, 9.4 ± 1.2; S. sanguinis, 12.6 ± 0.7; S. mutans BM71, 14.2 ± 1.3; and S. mutans GS5, 13.6 ± 1.1.
Figure 1.
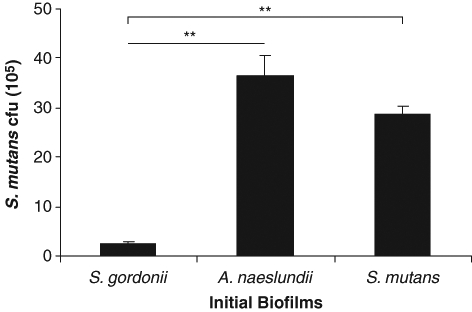
Colonization of S. mutans BM71 on pre-existing early-colonizer biofilms in vitro. Initial biofilms of S. gordonii Challis, A. naeslundii 12104, and S. mutans BM71 (as control) were formed in 48-well polystyrene microtiter plates. S. mutans BM71pTet was added on the washed initial biofilms and incubated for another 4 hrs. The sequentially formulated biofilms were disrupted by sonication after the planktonic cells were washed away. The TetR colonies of S. mutans BM71pTet were counted on THB agar plates supplemented with tetracycline and adjusted to cfu (105)/well. Data are the mean ± standard deviation of triplicate platings from 1 of 3 reproducible experiments. **p < 0.01.
S. gordonii, but not A. naeslundii, Inactivates S. mutans CSP
We have previously demonstrated that oral streptococci such as S. gordonii, S. mitis, S. oralis, and S. sanguis inactivated S. mutans CSP, which in turn interfered with bacteriocin production, a Com quorum-sensing-dependent phenomenon (Wang and Kuramitsu, 2005). Since S. mutans CSP also mediates S. mutans mono-biofilm formation, we carried out experiments to compare the ability of S. gordonii and A. naeslundii to inactivate S. mutans CSP. The supernatant fluids from A. naeslundii did not inactivate S. mutans CSP as did those from S. gordonii (Fig. 2). This difference may be one of the mechanisms that might allow S. mutans to colonize much more efficiently on A. naeslundii than on S. gordonii and other CSP-inactivating early colonizers.
Figure 2.
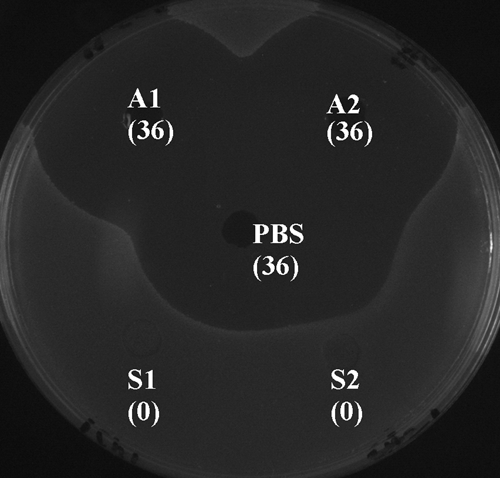
Determination of the effects of early colonizers on S. mutans CSP in agar well assays. Exogenous synthetic S. mutans CSP was incubated with early-colonizer culture supernatants for 2 hrs. CSP thus treated was used to stimulate bacteriocin production in S. mutans BM71 comC mutant cells. The bacteriocin level produced by the mutant cells was determined by agar well assays. A1 and A2, duplicates of A. naeslundii supernatants; PBS, a CSP positive control treated with PBS instead of culture supernatants; S1 and S2, duplicates of S. gordonii supernatants. The diameters of the inhibition zones surrounding the wells (mm) are shown in brackets.
We have previously demonstrated that S. gordonii did not degrade S. mutans bacteriocin in the well assays (Wang and Kuramitsu, 2005), which confirmed that the absence of S. mutans bacteriocin in our well assays was due to inactivation of S. mutans CSP by S. gordonii.
S. gordonii Challis Exhibits Higher Protease Activity than A. naeslundii
To further investigate the mechanisms enhancing colonization of S. mutans on the A. naeslundii biofilms relative to those on S. gordonii, we compared the protease activities of S. gordonii and A. naeslundii. Utilizing azocasein assays to detect non-specific protease activities, we demonstrated that S. gordonii has significantly higher protease activity (0.39 ± 0.06 at OD420) than A. naeslundii (0.09 ± 0.06 at OD420) (p = 0.01). We also carried out zymography using ready-made casein gel and confirmed the results of the azocasein assays (data not shown). These results, along with the evidence that A. naeslundii did not inactivate S. mutans CSP, suggested that proteases from early plaque colonizers may play a role in subsequent S. mutans colonization.
Cell Extracts from S. gordonii, but not from A. naeslundii, Interfered with S. mutans Colonization
We next examined the formation of secondary S. mutans biofilms on A. naeslundii biofilms, with or without the cell extracts from S. gordonii or A. naeslundii. The cell extracts from S. gordonii, but not those from A. naeslundii, significantly interfered with the colonization of S. mutans BM71 on A. naeslundii (Fig. 3).
Figure 3.
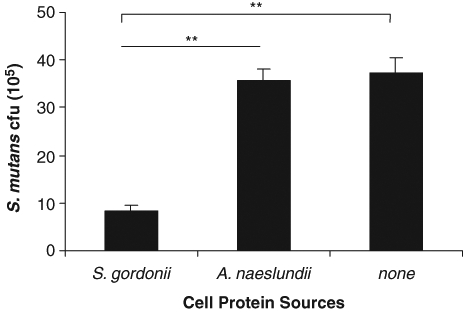
Effects of cell extracts from early plaque colonizers on the subsequent S. mutans BM71 colonization on A. naeslundii biofilms in vitro. Initial A. naeslundii biofilms were formed in 48-well plates. S. mutans BM71pTet, with or without cell extracts (at final concentrations of 1 mg/mL) from S. gordonii or A. naeslundii, was utilized to form the sequential biofilms on the A. naeslundii biofilms as described in MATERIALS & METHODS. S. mutans cfu represents the TetR colonies of S. mutans BM71pTet counted on THB agar plates supplemented with tetracycline, adjusted to cfu (105)/well. Data are the mean ± standard deviation of triplicate platings from 1 of 2 reproducible experiments. **p < 0.01.
Deletion of the sgc Gene Enhanced the Sequential S. mutans Colonization on S. gordonii Biofilms
To further confirm the role of proteases, we examined secondary S. mutans colonization on pre-existing biofilms of WT S. gordonii and its sgc knockout mutant in vitro. We have previously demonstrated that the sgc mutant produces reduced amounts of proteases and does not inactivate CSP (Wang and Kuramitsu, 2005). There was enhanced colonization of S. mutans BM71 on the sgc mutant biofilms, relative to that on the parental strain (Fig. 4).
Figure 4.
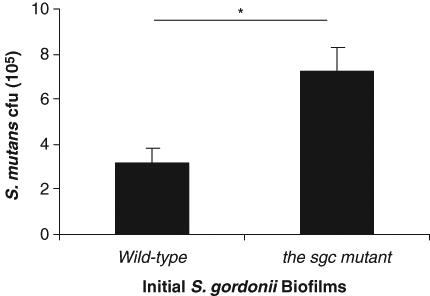
Deletion of the sgc gene in S. gordonii enhanced the subsequent in vitro colonization of S. mutans BM71. Initial biofilms of WT S. gordonii or its sgc knockout mutant were formed in 48-well plates. Sequential biofilms of S. mutans BM71pTet were formed as described in MATERIALS & METHODS. The TetR colonies of S. mutans BM71pTet were counted on THB agar plates supplemented with tetracycline and adjusted to cfu (105)/well. Data are the mean ± standard deviation of triplicate platings from 1 of 3 reproducible experiments. *p < 0.05.
Discussion
We have previously demonstrated that a S. gordonii protease, challisin, is able to inactivate S. mutans CSP and thereby to antagonize CSP-dependent bacteriocin production (Wang and Kuramitsu, 2005). In this study, we established a sequential duo-species biofilm model to test our hypothesis that proteases from early dental plaque colonizers also interfere with subsequent S. mutans colonization. S. mutans BM71 colonization on A. naeslundii biofilms was significantly more efficient than that on the biofilms of other early colonizers. One of the mechanisms causing this difference in S. mutans colonization might be due to lack of, or a reduced amount of, proteases from A. naeslundii biofilms, relative to streptococcal biofilms. Indeed, we demonstrated in this study that A. naeslundii cells exhibited much less protease activity relative to S. gordonii. This is the first evidence that proteases produced by an early colonizer can interfere with the subsequent colonization of S. mutans BM71.
S. mutans BM71 exhibited enhanced colonization on the sgc mutant biofilms relative to that on the WT biofilms (Fig. 4). However, S. mutans colonization was enhanced only two-fold on the sgc knockout mutant, whereas there was 10-fold enhancement of S. mutans colonization on A. naeslundii biofilms, relative to that on those of S. gordonii. Therefore, although proteases of S. gordonii can play a role in interfering with subsequent S. mutans colonization, some other factor(s) may also be involved in the difference of S. mutans colonization on the biofilms of A. naeslundii and S. gordonii, which merits further investigation. In addition, other S. gordonii proteases besides challisin may interfere with subsequent S. mutans colonization.
Ultrasonic dispersion has been utilized in other studies (Olsen and Socransky, 1981; Li et al., 2001) to remove attached biofilms of S. mutans or disassociate chains formed by S. mutans in planktonic cultures before being plated on agar plates. In these studies, the integrity of the oral streptococcal cells after sonication was confirmed. We have verified the viability of planktonic S. mutans cells after sonication under our conditions. Higher colony counts were detected after sonication relative to counts before sonication (1.70 × 109/mL vs. 1.89 × 109/mL, respectively), probably due to dissociation of the chains formed by S. mutans. Oral Gram-positive bacteria exhibit thick cell walls that may protect them against the impact of sonication.
Bacteria interact with each other and with their hosts in natural environments. Since quorum sensing in bacteria modulates their virulence, it is reasonable to assume that other bacteria and the host may produce factors that could interfere with the quorum-sensing system of a bacterium for the purpose of either competition for their establishment in the environment or as a defense mechanism. We demonstrated here that the proteases from oral streptococci such as S. gordonii could interfere with secondary S. mutans colonization in duo-species biofilms. An understanding of interactions among different species of oral bacteria may provide valuable insight into the prevention of oral infections.
Supplementary Material
Acknowledgments
We thank M.M. Vickerman for helpful discussions. These studies were supported by grant DE017708 from the National Institute of Dental and Craniofacial Research.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Blanco-Carrion A. (2004). Bacterial endocarditis prophylaxis. Med Oral Patol Oral Cir Bucal 9(Suppl):44-51; 37-43 [PubMed] [Google Scholar]
- Cvitkovitch DG. (2001). Genetic competence and transformation in oral streptococci. Crit Rev Oral Biol Med 12:217-243 [DOI] [PubMed] [Google Scholar]
- Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. (2001). Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183:897-908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, et al. (2002a). A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 184:2699-2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lau PC, Tang N, Svensäter G, Ellen RP, Cvitkovitch DG. (2002b). Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol 184:6333-6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Nakano M, Kuramitsu HK. (2006). Role of bacteriocin immunity proteins in the antimicrobial sensitivity of Streptococcus mutans. J Bacteriol 188:8095-8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIDCR, CDC (2002). Oral Health US, 2002. NIDCR/CDC Dental, Oral and Craniofacial Data Resource Center. Available at: http://drc.hhs.gov/report.htm (URL accessed 10/07/2010).
- Olsen I, Socransky SS. (1981). Ultrasonic dispersion of pure cultures of plaque bacteria and plaque. Scand J Dent Res 89:307-312 [DOI] [PubMed] [Google Scholar]
- Paul D, Slade HD. (1975). Production and properties of an extracellular bacteriocin from Streptococcus mutans bacteriocidal for group A and other streptococci. Infect Immun 12:1375-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojar HT, Lee JY, Bedi GS, Genco RJ. (1993). Purification and characterization of a protease from Porphyromonas gingivalis capable of degrading salt-solubilized collagen. Infect Immun 61:2369-2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer JM, Kurasz AB, Clive J. (1985). Competitive displacement of mutans streptococci and inhibition of tooth decay by Streptococcus salivarius TOVE-R. Infect Immun 48:44-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BY, Kuramitsu HK. (2005). Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol 71:354-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BY, Wu J, Lamont RJ, Lin X, Xie H. (2009). Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol 47:3902-3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Lin X, Wang BY, Wu J, Lamont RJ. (2007). Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 153(Pt 10):3228-3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa H, Kuramitsu HK. (2005). Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob Agents Chemother 49:541-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Kuramitsu HK. (2002). Multiple Streptococcus mutans genes are involved in biofilm formation. Appl Environ Microbiol 68:6283-6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


