Abstract
Mitochondria are essential organelles required for a number of key cellular processes. As most mitochondrial proteins are nuclear encoded, their efficient translocation into the organelle is critical. Transport of proteins across the inner membrane is driven by a multicomponent, matrix-localized “import motor,” which is based on the activity of the molecular chaperone Hsp70 and a J-protein cochaperone. In Saccharomyces cerevisiae, two paralogous J-proteins, Pam18 and Mdj2, can form the import motor. Both contain transmembrane and matrix domains, with Pam18 having an additional intermembrane space (IMS) domain. Evolutionary analyses revealed that the origin of the IMS domain of S. cerevisiae Pam18 coincides with a gene duplication event that generated the PAM18/MDJ2 gene pair. The duplication event and origin of the Pam18 IMS domain occurred at the relatively ancient divergence of the fungal subphylum Saccharomycotina. The timing of the duplication event also corresponds with a number of additional functional changes related to mitochondrial function and respiration. Physiological and genetic studies revealed that the IMS domain of Pam18 is required for efficient growth under anaerobic conditions, even though it is dispensable when oxygen is present. Thus, the gene duplication was beneficial for growth capacity under particular environmental conditions as well as diversification of the import motor components.
Keywords: J-protein, mitochondria, protein translocation, Hsp70, gene duplication
Introduction
Translocation of proteins from the cytosol into the mitochondrial matrix is a critical process, as most mitochondrial proteins are encoded by nuclear DNA and synthesized on cytosolic ribosomes. Transfer across the impermeable inner mitochondrial membrane is achieved by the action of a highly conserved, multiprotein complex composed of two parts, a translocase through which the proteins pass and an import motor that drives their translocation into the organelle (Chacinska et al. 2009; Mokranjac and Neupert 2009). The driving force of the import motor is provided by an Hsp70 of the mitochondrial matrix (mtHsp70, known as Ssc1 in Saccharomyces cerevisiae). Ssc1 is tethered to the translocon and binds exposed hydrophobic amino acid segments as the unfolded polypeptide enters the matrix while ATP hydrolysis drives forward movement of the polypeptide.
This binding of Ssc1 to the translocating polypeptide requires the intrinsic ATPase activity of Ssc1, which is stimulated by a partner J-protein via its highly conserved J-domain. In S. cerevisiae, two J-protein partners of Ssc1, Pam18 and Mdj2, are found associated with the translocon (Mokranjac et al. 2005). Pam18 is the principal J-protein functioning with Ssc1 in protein import (D'Silva et al. 2003; Mokranjac et al. 2003; Truscott et al. 2003). An essential protein that spans the inner membrane, Pam18, has three domains (fig. 1A): an N-terminal 60-amino acid domain localized to the intermembrane space (IMS), an internal membrane-spanning domain, and a C-terminal J-domain, which extends into the matrix. Although the J-domain and membrane-spanning domain were found to be essential, no phenotypic effect was found upon deletion of the N-terminal IMS domain (Mokranjac et al. 2007; D'Silva et al. 2008). Mdj2, like Pam18, contains a single transmembrane-spanning domain and a C-terminal J-domain localized in the matrix but lacks an N-terminal IMS domain (fig. 1A). Although Pam18 is essential, the absence of Mdj2 has no known phenotype (Westermann and Neupert 1997). However, when overexpressed, Mdj2 allows growth of cells lacking Pam18 (Mokranjac et al. 2005). The presence of two J-proteins associated with the inner mitochondrial matrix translocase in S. cerevisiae is perhaps not surprising, as the progenitor of S. cerevisiae underwent a whole genome duplication (Wolfe and Shields 1997; Dietrich et al. 2004; Dujon et al. 2004; Kellis et al. 2004). Following genome duplication, two gene copies were maintained in at least one species in 20–26% of the cases analyzed (Byrne and Wolfe 2005), likely resulting in specialization, redundancy, or distribution of functions between the duplicate genes.
FIG. 1.
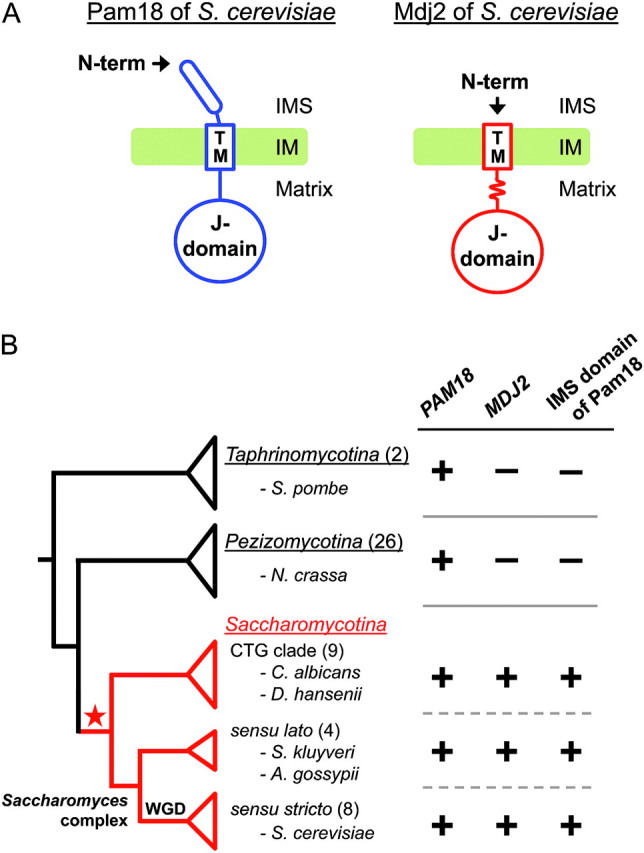
Domain structure and phylogenetic distribution of PAM18 and MDJ2. (A) Domain structures and topologies of Saccharomyces cerevisiae Pam18 (left) and Mdj2 (right) in the IMS, inner membrane, and matrix of the mitochondria. Pam18: IMS domain, amino acids 1–60; transmembrane (TM) domain, amino acids 66–85; and matrix domain, amino acids 86–168, which includes the J-domain (amino acids 101–168). Mdj2: TM domain, amino acids 1–24; matrix domain, amino acids 25–146, containing a J-domain (amino acids 89–138). The additional approximately 30 amino acids of Mdj2 compared with Pam18, between the TM domain and the J-domain, is indicated by a wavy line. All positions are with respect to the S. cerevisiae reference sequence. (B) The phylogenetic distributions of genes and domains are presented in a simplified cladogram of the fungal phylum Ascomycota, which consists of three subphyla Taphrinomycotina, Pezizomycotina, and Saccharomycotina. The species clades are represented as separate triangles. The Saccharomycotina species were separated into two major clades: the Saccharomyces complex and the CTG clade including Candida species (Scannell et al. 2007). The Saccharomyces complex, consisting primary of species from the genera Saccharomyces and Kluyveromyces, was further divided into the lineage that underwent whole genome duplication (WGD) and the non-WGD lineage. One or two representative species of each clade are named for clarity, with the total number of surveyed species indicated in parentheses. The branch leading to the Saccharomycotina is colored in red. The presence (+) and absence (−) of the orthologs/domains are indicated. The phylogenetic position of the duplication event of the ancestral PAM18 gene is marked by a star.
To better understand the function of J-proteins in mitochondrial import, we undertook evolutionary and molecular genetic analyses. We found that the PAM18/MDJ2 gene pair was not a product of the whole genome duplication, but rather resulted from a more ancient gene duplication corresponding with cladogenesis between the fungal subphyla Saccharomycotina and Pezizomycotina. Strikingly, the origin of the novel IMS domain of Pam18 coincides with the duplication event. In vivo analysis revealed that although IMS domain of Pam18 of S. cerevisiae, a facultative anaerobe, is dispensable in the presence of oxygen, it plays a critical role under anaerobic conditions. These results suggest that the gene duplication of the PAM18/MDJ2 gene pair may have been one of many key steps leading to functional diversification in respiration and mitochondrial function observed between the Saccharomycotina and Pezizomycotina clades.
Materials and Methods
Identification of PAM18 and MDJ2 Orthologs by Reciprocal BLAST Analysis
Using protein sequences of S. cerevisiae Pam18 and Mdj2 as queries, basic local alignment search tool (BLAST) searches (Altschul et al. 1990, 1997) were performed with National Center for Biotechnology Information server (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the Sanger Institute (http://www.sanger.ac.uk/DataSearch/blast.shtml). To assess gene presence and absence, BlastP, TBlastN, and BlastX were utilized. Unannotated, putative open reading frames (ORFs) were identified with ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Results from each BLAST search that were statistically significant were then used as query sequences in the BlastP search against the S. cerevisiae protein database to identify reciprocal-best-blast sequences as orthologs.
To determine whether an IMS domain was present in each orthologous protein, the amino acid sequence identified as a Pam18 or an Mdj2 orthologous protein was further analyzed to specify the location of a membrane-spanning segment. The putative transmembrane position across a protein sequence was located by comparing the results from three prediction methods of transmembrane segments: HMMTOP2 (Tusnady and Simon 1998, 2001), Split4 (Juretic et al. 2002), and TMpred (Hofman and Stoffel 1993). Then, the presence or absence of an IMS domain in an orthologous protein was comprehensively evaluated in view not only of its transmembrane position but also of sequence comparison with the other orthologous proteins. The full-length sequence data of Pam18 and Mdj2 orthologous proteins were aligned with the ClustalW program (Thompson et al. 1994) and corrected by manual inspection.
Sequence Data Set for Phylogenetic Reconstruction
The coding sequence data of PAM18 and MDJ2 for phylogenetic reconstruction were obtained from 21 fungi in the subphylum Saccharomycotina. Five sequences of the PAM18/MDJ2 orthologs from non-Saccharomycotina fungi were included as outgroups (subphylum Taphrinomycotina—Schizosaccharomyces pombe; subphylum Pezizomycotina—Aspergillus fumigatus, Coccidioides immitis, Gibberella zeae, and Neurospora crassa). To construct the phylogenetic trees from sequence data, the coding sequences were first translated to amino acid sequences, and the IMS domains were removed from PAM18 orthologs. Alignments were then generated using Dialign-TX, which has a segment-based approach that was reported to outperform other multiple alignments of locally-related sequences (Subramanian et al. 2008). After all gap positions were removed manually, sequence alignments of 96 amino acids in length were obtained, consisting mainly of highly conserved J-domains and moderately conserved transmembrane segments. The amino acid alignments were reverse translated into nucleotide sequences to obtain the codon alignments for the following tree construction.
Phylogenetic Tree Construction of PAM18 and MDJ2
Based on the gap-free multiple alignment described above, the optimal gene tree was derived using MrBayes ver. 3.1.2 (Ronquist and Huelsenbeck 2003) under a general time reversible model (GTR) with a proportion of invariant sites, assuming gamma distribution of rate variation across sites (GTR + I + G model). This model was estimated as the best fitting substitution model by MrModeltest version 2.3 (Nylander et al. 2004). Furthermore, the site-specific rate model was used to analyze the protein-coding sequences of the present study, allowing each codon position to evolve under different rates. Other model parameters (statefreq, revmat, shape, and pinvar) were also unlinked across each codon position. The Bayesian analyses were performed with two independent runs (each with four chains), sampling each 100 generations. The analyses were considered finished when the average standard deviation of split frequencies between two runs dropped below 0.01, which required 4,000,000 generations, and the first 25% of sampled trees were discarded as burn-in before estimation of posterior probabilities for branch support. The obtained phylogenetic trees were drawn using Tree Explorer of Molecular Evolutionary Genetics Analysis (MEGA) software 4.0 (Tamura et al. 2007). In the reduced phylogenetic tree of figure 2, the clades of orthologs from the Saccharomycotina species were further divided into two major clades: the Saccharomyces complex and the CTG clade, which is composed of a monophyletic group of taxa that translate the CTG codon into a serine rather that leucine (Kurtzman and Robnett 2003; Fitzpatrick et al. 2006; Moura et al. 2010).
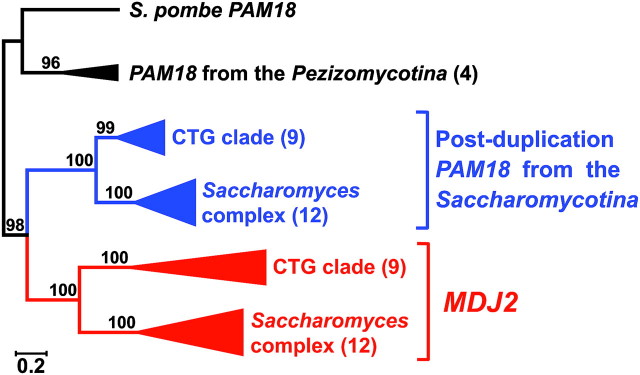
FIG. 2.
Phylogeny of PAM18 and MDJ2. The number along branches refers to the Bayesian posterior probability (X100) supporting the respective branch. The number of samples in each clade is given in parentheses. The orthologs from the Saccharomycotina species were divided into the CTG clade and the Saccharomyces complex. The scale bar represents 0.2 substitutions per site. The branches of each ortholog are colored: red, MDJ2; blue, post-duplication PAM18 from the Saccharomycotina fungi; and black, PAM18 from the non-Saccharomycotina fungi. For the complete phylogenetic tree, see supplementary figure S1, Supplementary Material online.
Calculation of the Substitution Nucleotide Rates
Nonsynonymous substitution rates (the number of substitutions per nonsynonymous site, dN) and synonymous substitution rates (the number of substitutions per synonymous site, dS) were determined for each branch of the nucleotide tree (fig. 2) by using maximum likelihood approach with codeml program in PAML package ver. 4.2b (Yang 2007). Codon frequencies were calculated from the average nucleotide frequencies at the three codon positions (codon frequency model F3 × 4). To avoid falling into suboptimal likelihood peaks, the program was run multiple times in each analysis of this study, using different initial parameters (initial dN/dS ratio or ω = 0.01 or 1, and initial transition to transversion rate ratio or κ = 0.4 or 4). Under the branch-specific model estimating different dN and dS values among a priori specified branches (Yang 1998; Yang and Nielsen 1998), three models of sequence evolution were considered: 1) one-ratio model (a single ω ratio, assuming that all branches have the same ratio of ω), 2) two-ratio model (a single ω ratio for PAM18 and a second ω ratio for MDJ2), and 3) three-ratio model (two separate ω ratios for PAM18 of the Saccharomycotina and of the non-Saccharomycotina and a single ω ratio for MDJ2). Statistical significance was determined by likelihood ratio tests (a statistical test of the goodness-of-fit between two models) assuming a chi-square distribution. The two-ratio model provided a significantly better fit to the data than the one-ratio model (P < 0.05) in the likelihood ratio test. On the other hand, the three-ratio model was not a better model than the two-ratio model statistically (P > 0.05). Therefore, the dS and dN parameters of branches in the result of the two-ratio model were adopted.
Codon Usage Bias Estimation
Assuming that optimal codons are significantly common among closely related Saccharomycetes species, the codon adaptation index values of their PAM18 and MDJ2 coding sequences were calculated with CodonW program (Peden 1999) (http://codonw.sourceforge.net/) using the reference set for the optimal codons of S. cerevisiae (Sharp and Cowe 1991).
Yeast Strains, Media, and Growth Conditions
In this study, strains of S. cerevisiae were derived from PJ53 (James et al. 1997), which is isogenic to W303: trp1-1/trp1-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 ade2-1/ade2-1 can1-100/can1-100 GAL2-Δ/GAL2-Δ met2-Δ1/met2-Δ1 lys2-Δ2/lys2-Δ2. Haploid strains were constructed by tetrad dissection of appropriate diploid strains. Yeast strains deleted for PAM18 (pam18::TRP1), TIM44 (tim44::LYS2), and SSC1 (ssc1::LEU2) have been described previously (Maarse et al. 1992; Gambill et al. 1993; D'Silva et al. 2003). Strains having deletions of PAM17 and TIM21 were constructed by replacing their coding regions with a hygromycin B resistance (hph) gene (Goldstein and McCusker 1999) yielding pam17::HPH and with a KanMX module gene yielding tim21::kanMX, respectively. To obtain single deletion mutant of MDJ2, the DNA between the BamHI and MfeI sites in MDJ2 (−47 to +465) was replaced with the TRP1 gene yielding mdj2::TRP1. For the double deletion mutant of PAM18 and MDJ2, pam18::TRP1 was further genetically modified by replacing MDJ2 with a KanMX module to generate pam18::TRP1 mdj2::kanMX.
Yeast cells were cultivated, where appropriate, in yeast extract/peptone/dextrose media, synthetic complete media (SC), synthetic dropout media, sporulation media, or media containing 5-fluoroorotic acid (Boeke et al. 1984; Sherman et al. 1986). As S. cerevisiae cells become auxotroph for sterols and unsaturated fatty acids under anaerobic conditions (Andreasen and Stier 1953, 1954), media were supplemented with 20 μg/ml Ergosterol (MP Biomedicals) and 5 mg/ml Tween 80 (Sigma-Aldrich Corporation) for anaerobic culture of the yeast. The same lipid-supplemented media were also used for the aerobic culture in the control experiments of cell growth. Anaerobic conditions for cell cultures were maintained using GasPak EZ Anaerobe Pouch System (Becton, Dickinson and Company).
Construction of Plasmids
As described previously (D'Silva et al. 2003), PAM18 was obtained by polymerase chain reaction (PCR)-amplifying genomic DNA from position −303 to +702 and cloned into pRS315 (CEN6 LEU2) or into pRS316 (CEN6 URA3) (Sikorski and Hieter 1989). Mutants of PAM18 were constructed by site-directed mutagenesis by using QuikChange protocol (Stratagene). The construction of the Pam18Δ1–60 protein made use of existing an AUG codon at amino acid position 61 within the sequence while the start codons were introduced to allow translation of the Pam18Δ1–25 and Pam18Δ1–43 proteins. The plasmid construction of pRS314 (CEN6 TRP1) carrying the wild-type or P419S-mutated SSC1 was described previously (Miao et al. 1997; Liu et al. 2001). The plasmid construction of pRS314 carrying the wild-type or R180A-mutated TIM44 was also described previously (Liu et al. 2003; Schiller et al. 2008). For overexpression of Pam18Δ1–60 (amino acids 61–168), the corresponding coding sequence was PCR amplified and cloned into the BamHI/XhoI sites of p415 (CEN6 LEU2) placing it under the control of a constitutive promoter derived from the gene encoding translation elongation factor 1α (TEF promoter) (Mumberg et al. 1995). For overexpression of Mdj2, the full-length coding sequence of MDJ2 was PCR amplified from genomic DNA and cloned into the BamHI/XhoI sites of p416 (CEN6 URA3) placing it under the control of a constitutive promoter derived from the gene encoding glyceraldehyde-3-phosphate dehydrogenase (GPD promoter).
Analysis of Protein Expression with Tricine–SDS-PAGE and Western Blotting
For the protein extraction from yeast cells for electrophoretic analysis, yeast cells were scraped off agar-plates using toothpicks. Then, whole cell extracts were prepared by treating cells with 0.1 M NaOH and subsequently boiling for 3 min in sodium dodecyl sulfate (SDS) sample buffer (Kushnirov 2000). To separate the extracted proteins, Tricine–SDS-PAGE was applied according to the procedures described previously (Schagger 2006). Mini-PROTEAN 3 Cell (Bio-Rad Laboratories) was used for this electrophoresis as vertical electrophoresis apparatus. Each lysate equal to 0.1 OD600 cells (optical density at 600 nm = 0.1) was loaded on the gel whose concentration of acrylamide was 18% (separating) and 4% (stacking). The electrophoresis was started with an initial voltage of 30 V. After the sample has completely entered the stacking gel, the voltage was increased up to 150 V. After the completion of electrophoresis, the proteins were transferred to a polyvinylidene fluoride membrane (0.2 μm pore size, Millipore Corporation) by electroblotting with a tank transfer system or Trans-Blot cell (Bio-Rad Laboratories). Immunoblot analyses with polyclonal antibodies against Pam18, Tim23, or Mdj2 were performed using a chemiluminescence reaction kit (Western Lightning Plus, PerkinElmer) according to the manufacturer's suggestion.
Analysis of Precursor Accumulation In Vivo
Yeast cells carrying either wild-type or mutant form of PAM18 were grown on SC solid media supplemented with Ergosterol and Tween 80 (SC + Erg/Tw) under anaerobic conditions at 30 °C for 2.25 days. After growth on solid media, cells were scraped from the plates, the cultured cells were subjected to alkali treatment, followed by 3 min boiling in SDS sample buffer. Total cell lysates were analyzed by Laemmli–SDS-PAGE (Laemmli 1970) and by immunoblot analysis using Hsp60-specific antibody.
Antibodies
Preparation of antibodies against Pam18, Tim23, and Hsp60 were described previously (D'Silva et al. 2003, 2008). Because the Pam18 antibody was prepared against the C-terminus of the protein (amino acids 80–168), it reacts equally well with full-length Pam18, Pam18Δ1−60 (amino acids 61–168), and the other truncated Pam18 variants (Pam18Δ1–25, amino acids 26–168; Pam18Δ1–43, amino acids 44–168). For production of antibody against Mdj2, 6xHis-tagged coding sequence of Mdj2 protein (amino acids 20–146) was cloned into pET-3a vector (Novagen). The protein expression from the plasmid construct was induced with isopropyl-β-d-thiogalactopyranoside in Escherichia coli C41 (DE3) strain (Miroux and Walker 1996). After purification, the His-tagged protein was injected into rabbits for Mdj2-specific polyclonal antibody production.
Miscellaneous
Yeast transformation with exogenetic DNA was conducted by the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (Gietz and Woods 2002). Transformed cells were selected on the appropriate selective media.
Results
The PAM18/MDJ2 Pair Arose by Duplication in a Common Ancestor of the Saccharomycotina
As a first step in elucidating the origin of the PAM18 and MDJ2 parologs, located on chromosomes XII and XIV, respectively, we determined their phylogenetic distribution in fungal genomes using BLAST analyses (supplementary table S1, Supplementary Material online). Within Saccharomycotina, all species exhibited the presence of both PAM18 and MDJ2. However, outside the subphylum, each species exhibited only a single ORF when queried using either PAM18 or MDJ2 sequence. Reciprocal BLAST matches of each single copy gene against the S. cerevisiae proteome were consistently more similar to PAM18 than to MDJ2. For each species in Saccharomycotina, two ORFs were identified that resulted in significant BLAST e-values when probed with either PAM18 or MDJ2. In each case, reciprocal best BLAST analysis consistently distinguished between PAM18 and MDJ2 putative orthologs.
Based on the above BLAST results, we concluded the duplication event that gave rise to the PAM18 and MDJ2 pair occurred prior to the whole genome duplication in a common ancestor of the Saccharomycotina clade (fig. 1B). However, it was also possible that the presence of only a single mitochondrial J-protein in non-Saccharomycotina fungal species was due to multiple losses of one member of the gene pair that originated from an earlier duplication event. Therefore, we determined whether a single copy J-protein was present in nonfungal eukaryotes via BLAST analysis in well-annotated genomes among plantae, metazoa, and protozoa. The BLAST search using the sequence of S. cerevisiae MDJ2 and PAM18 as a query provided one or more significant hits in each surveyed species. However, the reciprocal BLAST search against S. cerevisiae database revealed that they were all more closely related to PAM18 than to MDJ2 (supplementary table S1, Supplementary Material online), indicating homology to PAM18 outside the fungi.
Next, we analyzed the phylogenetic relationship of PAM18 and MDJ2 orthologs from Saccharomycotina fungi as well as selected single copy orthologs from non-Saccharomycotina fungi. Results of Bayesian reconstruction of the nucleotide-based phylogeny indicated that orthologs formed strongly supported monophyletic clades among taxa within the Saccharomycotina (fig. 2 and supplementary fig. S1, Supplementary Material online), consistent with a single gene duplication event. Congruent monophyletic patterns were also observed with maximum likelihood methods (data not shown). Taken together, our analyses indicate that duplication of the ancestral single copy PAM18/MDJ2 gene gave rise to the two J-proteins, Pam18 and the fungal-specific Mdj2, prior to the diversification of the Saccharomycotina clade.
MDJ2 Is Rapidly Evolving
Next, we compared rates of evolution of PAM18 and MDJ2. The nucleotide-based tree and the amino acid–based tree (supplementary fig. S2, Supplementary Material online) exhibited longer branches for the MDJ2 clade compared with PAM18. The analysis of codon evolution further indicated that both synonymous and nonsynonymous rates of evolution are higher for the MDJ2 gene than the PAM18 gene (fig. 3A). In addition, our analyses of average levels of codon bias (fig. 3B) and expression studies (Holstege et al. 1998) indicate that MDJ2 is expressed at lower levels than PAM18. The observed lower levels of expression, reduced codon bias, and rapid rate of evolution for MDJ2 relative to PAM18 are consistent with previous observations that genes with lower levels of expression tend to evolve more rapidly (Drummond et al. 2005; Drummond and Wilke 2009). This accelerated rate of evolution for MDJ2 also explains why nonfungal mitochondrial J-proteins have higher BLAST similarity scores for PAM18.
FIG. 3.
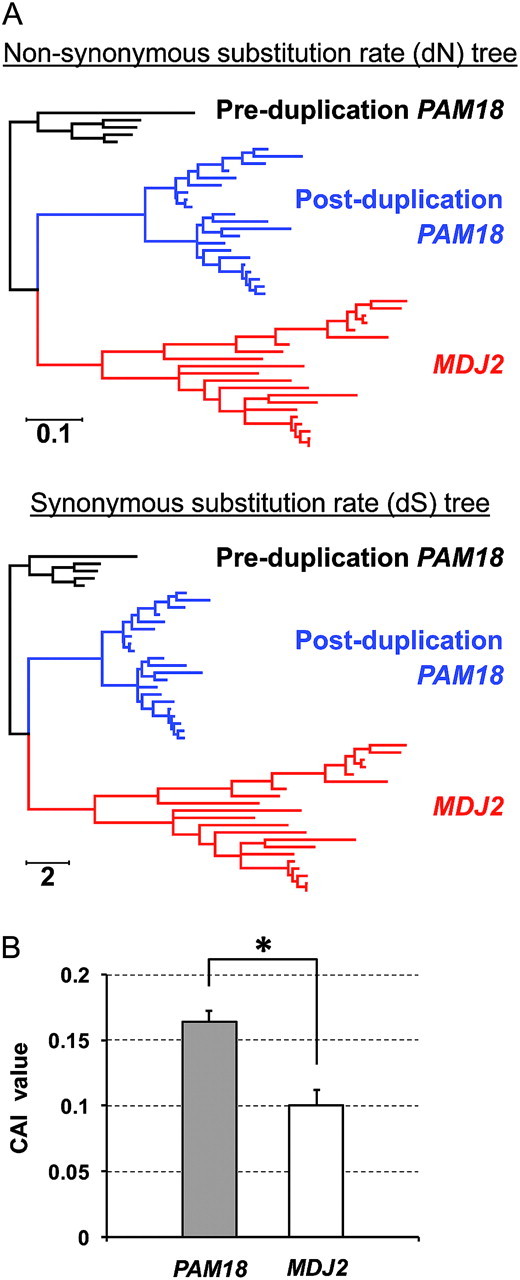
Analysis of codon usage in PAM18 and MDJ2. (A) Codon-based phylograms of PAM18 and MDJ2 orthologs. Shown is the nonsynonymous substitution rate tree (dN tree, top) or synonymous substitution rate tree (dS tree, bottom) whose branch lengths are proportional to nonsynonymous or synonymous substitution rates, respectively. Scale bars indicate substitution rates per site. Red, MDJ2; blue, post-duplication PAM18 from the Saccharomycotina fungi; and black, pre-duplication PAM18 from the non-Saccharomycotina fungi. (B) Codon adaptation index (CAI) values of PAM18 and MDJ2 of Saccharomyces species. The CAI values of the PAM18 and MDJ2 coding sequences from closely related Saccharomyces species (S. cerevisiae, S. paradoxus, S. mikatae, S. kudriavzevii, and S. bayanus) were calculated with CodonW program using the reference set of highly expressed genes in S. cerevisiae cells. Data are expressed as the mean and standard deviation of the CAI values from the five species (*P < 0.01, two-tailed t-test). The CAI calculation using the codon usage tables of each species genome (Nakamura et al. 2000) as the references also produced the same result in that the CAI values of MDJ2 are statistically lower than those of PAM18 among the Saccharomyces species (data not shown).
Acquisition of Fungal Pam18 IMS Domain Coincides with the Gene Duplication
Upon further examination of the PAM18/MDJ2 pair, we noted that all post-duplication Pam18 orthologous proteins have an extension N-terminal to the membrane-spanning region (figs. 1B and 4, supplementary table S1, Supplementary Material online). A qualitative comparison among these N-terminal extensions revealed a high degree of conservation, indicating ancient acquisition and subsequent negative selection for the maintenance of this domain. In addition, BLAST analyses using only the IMS domain of Pam18 as a query failed to return any significant matches among fungi, indicating that this novel domain is unlikely to have originated from domain shuffling of previously existing polypeptides.
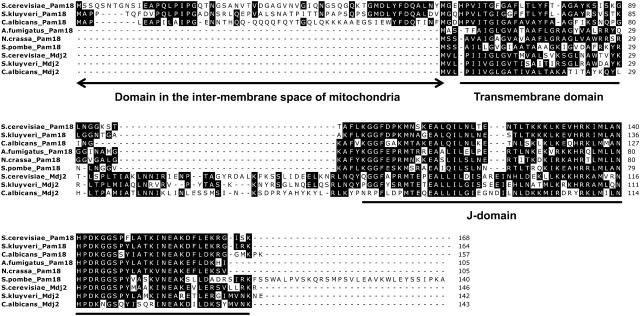
FIG. 4.
Amino acid sequence alignment of representative Pam18 and Mdj2 orthologous proteins. Sequences were aligned using the Dialign-TX program, followed by manual inspection. Similar residues in each column are shaded. These representative sequences of Pam18 and Mdj2 orthologous proteins were derived from three Saccharomycotina fungi (post-WGD yeast, Saccharomyces cerevisiae; pre-WGD yeast, S. kluyveri; and Candida species, C. albicans) and three non-Saccharomycotina fungi (Schizosaccharomyces pombe from the subphylum Taphrinomycotina; Aspergillus fumigatus and Neurospora crassa from the subphylum Pezizomycotina).
To investigate whether any nonfungal Pam18 orthologous proteins had an N-terminal extension sharing homology to fungal Pam18 IMS domains, we conducted a BLAST analysis of well-annotated genomes among plantae, metazoa, and protozoa (supplementary table S1, Supplementary Material online). Most nonfungal PAM18 orthologs lacked an N-terminal extension. An exception was a metazoan group consisting mostly of vertebrates. Each genome of the surveyed vertebrates encoded a pair of PAM18 homologs; in each case, one had a short N-terminal extension, approximately 35 amino acids in length. However, the sequence alignment of the fungal IMS domains (50–70 amino acids in length) and the nonfungal IMS domains showed no detectable homology (data not shown), suggesting that fungal and vertebrate IMS domains of Pam18 evolved independently from each other.
In addition to the novel domain found among all postduplication Pam18 orthologous proteins, we also noted an additional amino acid segment present in all post-duplication Mdj2 orthologous proteins, situated between the membrane-spanning domain and C-terminal J-domain (fig. 4B). Although the gain of this domain within Mdj2 is of interest, MDJ2 loss-of-function alleles have no known phenotype (Westermann and Neupert 1997); hence, we decided to focus on the novel IMS domain of Pam18.
The IMS Domain of Pam18 Is Required for Normal Growth and Protein Import in the Absence of Oxygen
The IMS domain of S. cerevisiae Pam18 has been reported to be unimportant for efficient protein import and thus vigorous cell growth (Mokranjac et al. 2007). However, the sequence conservation among post-duplication Pam18 IMS domains suggests that acquisition of the IMS domain could have important functional consequences. Therefore, we reassessed the ability of a Pam18 variant lacking the N-terminal 60 amino acids (Pam18Δ1–60) to grow under a variety of conditions. We tested growth on media containing different carbon sources at various temperatures. In addition, because previous comparative genomic studies have noted that differences in gene presence/absence between the Saccharomycotina and Pezizomycotina clades are related to mitochondrial and respiratory function, we also examined growth in anaerobic conditions. Even though no oxidative phosphorylation occurs in the absence of oxygen, other mitochondrial functions, such as Fe-S cluster biogenesis, are critical under these conditions (Groot et al. 1971; Trocha and Sprinson 1976; Lill and Muhlenhoff 2008). Consistent with previous reports (Mokranjac et al. 2007), we found no deleterious effect of the absence of the IMS domain of Pam18 under aerobic conditions (data not shown). In sharp contrast, under anaerobic conditions pam18Δ1−60 cells grew more slowly than cells expressing full-length Pam18 at all temperatures tested (fig. 5A and data not shown). This difference was especially apparent at low (18 °C) and at high (37 °C) temperatures.
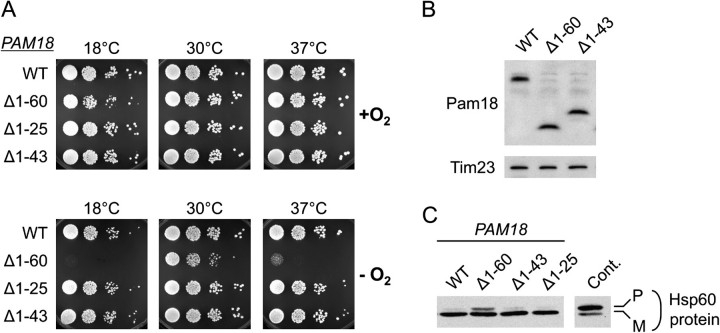
FIG. 5.
Importance of the IMS domain of Pam18 for growth and protein translocation under anaerobic conditions. Analysis of pam18-Δ cells carrying a centromeric plasmid (pRS315) with the indicated PAM18 alleles: full-length PAM18 (WT); pam18Δ1−60 (Δ1−60); pam18Δ1−25 (Δ1−25); and pam18Δ1−43 (Δ1−43). (A) Growth of cells expressing truncations of IMS domain of Pam18 under anaerobic conditions. Ten-fold serial dilutions of the cell suspensions were spotted on synthetic complete media supplemented with Ergosterol and Tween 80 (SC + Erg/Tw). After plating, cells were grown aerobically (+O2) or anaerobically (−O2) at the indicated temperatures. Culture periods: 5 days at 18 °C, 2 days (+O2) or 2.25 days (−O2) at 30 °C, 2.5 days (+O2), or 3 days (−O2) at 37 °C. (B) Expression levels of wild-type and truncated Pam18 proteins in anaerobically grown cells. After growth on SC + Erg/Tw media at 30 °C for 2.25 days under anaerobic conditions, extracts were prepared from cells that were scraped off agar-plates. The extracts were subjected to electrophoresis and analyzed by immunoblotting with an antibody against Pam18, which recognizes all Pam18 proteins with equal efficiency (see Materials and Methods). Tim23 protein was also detected as a loading control. (C) Accumulation of a mitochondrial precursor protein in vivo. Total protein extracts prepared from cells grown anaerobically on SC + Erg/Tw media at 30 °C for 2.25 days were subjected to immunoblot analysis using an antibody specific for Hsp60. Protein extract from a mutant known to be defective in Hsp60 import (pam18L150W) was used as a positive control. pam18L150W extract was made from cells grown aerobically in rich liquid media at 23 °C and heat-shocked at 37 °C for 7 h. Precursor (P) and mature (M) forms of Hsp60 are indicated.
To determine the region of the IMS domain critical for robust growth under anaerobic conditions, we created two smaller nested N-terminal truncations, lacking the first 25 (Pam18Δ1−25) or 43 amino acids (Pam18Δ1−43). We compared the growth of cells expressing these two variant alleles with cells expressing full-length Pam18 or the variant allele lacking the N-terminal 60 amino acids, Pam18Δ1−60. Both pam18Δ1−25 and pam18Δ1−43 cells grew similarly to cells expressing full-length Pam18 under anaerobic, as well as aerobic conditions (fig. 5A).
These results are consistent with amino acids 44–60, a conserved segment of the IMS domain of Pam18, being required for robust cell growth under anaerobic conditions. However, to ensure that the affects on growth were not due to differences in expression levels of the variants, we prepared cell extracts from anaerobically grown cells and examined the levels of the Pam18 proteins using antibodies specific for the matrix domain of Pam18 (fig. 5B). We found that full-length Pam18, Pam18Δ1−60, and Pam18Δ1−43 were all expressed at similar levels. Because only the cells expressing Pam18Δ1−60 exhibited an anaerobic growth defect, we conclude that the IMS domain plays an important function under these conditions, particularly at suboptimal growth temperatures.
As a component of the import motor, it has been established that Pam18 plays a critical role in the translocation of proteins into the mitochondrial matrix. We next wanted to determine if the growth defect of pam18Δ1−60 we observed in the absence of oxygen is consistent with a defect in protein translocation under this condition. Many mitochondrial proteins encoded by nuclear DNA, such as Hsp60, are synthesized as preproteins, having a mitochondrial targeting sequence that is cleaved by a processing protease upon entrance into the matrix. Thus, the accumulation of the precursor form of such proteins can be taken as a marker for defective protein import, as normally translocation is so rapid that the precursor form of such proteins is present at extremely low levels and nearly undetectable (Reid and Schatz 1982). Therefore, to assess the efficiency of protein import into the mitochondrial matrix, we monitored the accumulation of Hsp60 precursor by immunoblot analysis of cell extracts. As a control, we used a pam18 mutant that does not grow aerobically at 37 °C. After shift to the nonpermissive temperature, substantial accumulation of Hsp60 precursor was detected (fig. 5C). Precursor accumulation was also observed in extracts prepared from pam18Δ1−60 cells grown anaerobically at 30 °C, a temperature at which these cells grow more slowly than wild-type cells. However, negligible precursor was detected in pam18Δ1−25 and pam18Δ1−43 cells, as well as wild-type cells under these conditions. This precursor accumulation in pam18Δ1−60, but not the shorter truncation mutants, is consistent with an anaerobic growth defect of pam18Δ1−60 cells, being caused by inefficient import of proteins into mitochondria.
Other Mutants Defective in Protein Import Grow Equally Well in the Presence or Absence of Oxygen
The compromised growth of pam18Δ1–60 under anaerobic, but not aerobic, conditions raises the question as to whether particularly efficient function of the import apparatus of the inner membrane is required in the absence of oxygen. To address this question, we compared aerobic and anaerobic growth of several well-studied strains having mutations in or deletion of genes encoding several components of the import apparatus. All these mutant strains have been documented to have protein translocation defects. First, the effects of conditional mutations in two genes encoding essential components, Ssc1 and Tim44, the protein that tethers Ssc1 to the translocon, were tested. ssc1P419S (ssc1-2) cells are defective in protein import into the mitochondrial matrix due to a defective interaction with Tim44 (D'Silva et al. 2004). Consistent with previous results (Gambill et al. 1993; Voisine et al. 1999), ssc1P419S grew poorly at all temperatures and did not form colonies at 34 °C and above in the presence of oxygen. Growth of ssc1P419S under anaerobic conditions was very similar (fig. 6A). The second mutant tested, tim44R180A, is defective in regulation of the Ssc1–Tim44 interaction as well as in stable association with the translocon (Schiller et al. 2008). tim44R180A is temperature sensitive under aerobic conditions, unable to form colonies at 34 °C and above. The growth defect of tim44R180A was slightly more severe under anaerobic than aerobic conditions at all temperatures tested (fig. 6B). However, the difference between aerobic and anaerobic growth was not nearly as drastic as that found with pam18Δ1−60 (fig. 5A). The effects of the absence of two nonessential components of the translocation machinery, Pam17 and Tim21 (van der Laan et al. 2005; Hutu et al. 2008; Popov-Celeketic et al. 2008), which affect the general organization of the components of the translocation machinery, were also tested. pam17-Δ cells and tim21-Δ cells grew as well as wild-type cells under both anaerobic and aerobic conditions (fig. 6C). We conclude that defects in the translocation apparatus of the inner mitochondrial membrane do not generally cause enhanced growth defects under anaerobic conditions.
FIG. 6.
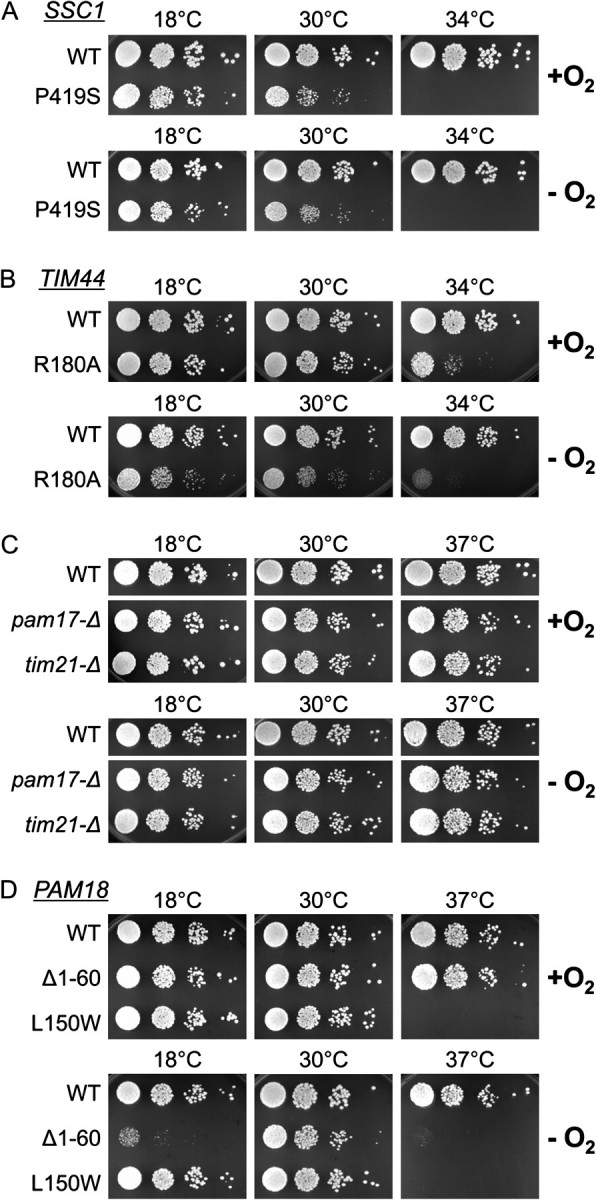
Similarity of aerobic and anaerobic growth of strains having defects in mitochondrial import caused by mutations in genes encoding various components of the translocation apparatus. Ten-fold serial dilutions of the indicated deletion mutants were spotted on SC + Erg/TW plates and grown in the presence (+O2) or absence (−O2) of oxygen at the indicated temperatures. In the case of the essential genes, SSC1 (A), TIM44 (B), or PAM18 (D), the deletion strains carried either the wild-type (WT) or mutant gene encoding the indicated amino acid alterations on the plasmid pRS315. In the case of the nonessential genes PAM17 and TIM21 (C), WT and complete deletion mutants were compared.
Because the mutants described above did not have enhanced growth defects under anaerobic conditions, we next wanted to test whether the affect of the absence of the IMS domain of Pam18 reflected a general increase in demand for Pam18 function or a specific requirement for the IMS domain itself. We therefore tested another PAM18 mutation, pam18L150W, which causes a single amino acid alteration in the matrix domain (D'Silva et al. 2008). pam18L150W has a temperature-sensitive growth defect when grown aerobically (fig. 6D). However, it grew as well in the absence of oxygen as in the presence of oxygen. Thus, under the conditions tested, none of the mutants, with the exception of pam18Δ1–60, was more compromised in the absence of oxygen than in its presence. Thus, our results are consistent with the hypothesis that the IMS domain of Pam18 is particularly required under anaerobic conditions, which indicates that the gene duplication event and concomitant gain of the IMS domain generated some novel advantage under such conditions.
Mdj2 Is Not Required Under Anaerobic Conditions
In addition, we examined the anaerobic growth of cells lacking the nonessential Mdj2, the other descendant of the ancestral duplication, to determine if it plays an important role under anaerobic conditions as well. Consistent with previous reports (Westermann and Neupert 1997; Mokranjac et al. 2005), mdj2-Δ cells grew as well as wild-type cells under aerobic conditions. Nor was a growth defect was observed in the absence of oxygen (fig. 7A), indicating that Mdj2 is not specifically required under these conditions. To test for any synthetic genetic interaction between the IMS domain of Pam18 and Mdj2, we constructed a double mutant lacking both Mdj2 and the Pam18 IMS domain (pam18Δ1–60 mdj2-Δ). No significant growth difference between pam18Δ1-60 mdj2-Δ and pam18Δ1-60 cells were observed between aerobic and anaerobic conditions (fig. 7B).
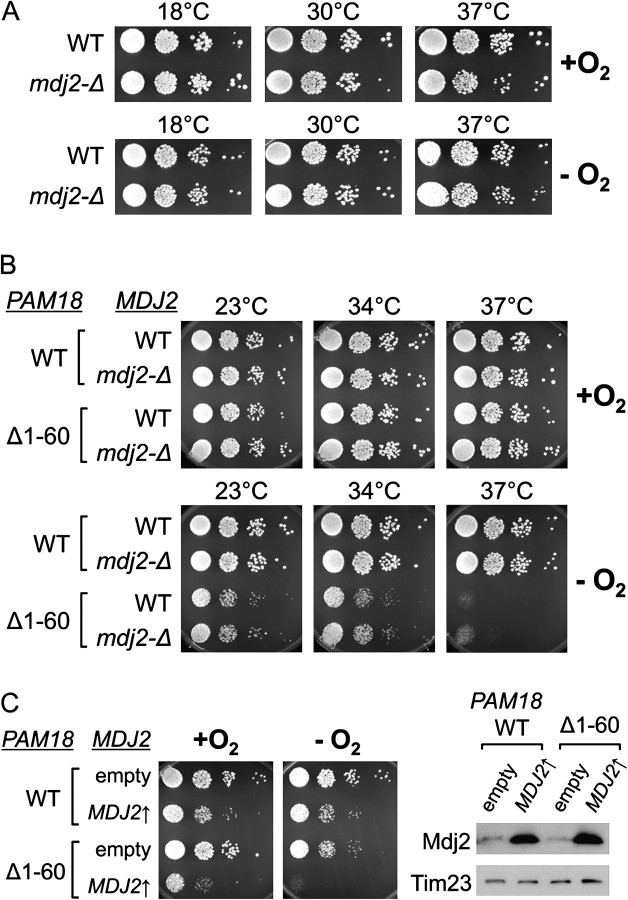
FIG. 7.
Mdj2 does not compensate for lack of the IMS domain of Pam18 in the absence of oxygen. (A) Cells lacking Mdj2 have no anaerobic growth defect. Ten-fold serial dilutions of wild-type (WT) and mdj2-Δ cells were spotted on SC + Erg/TW plates and grown in the presence (+O2) or absence (−O2) of oxygen at the indicated temperatures. (B) No synthetic genetic interaction between mdj2-Δ and pam18Δ1–60 mutations. The indicated combinations of mutants were grown in the presence (+) or absence (−) of oxygen as described in A. In the case of PAM18, the chromosomal copy was deleted, and the control WT Pam18 (WT) and Pam18Δ1−60 (Δ1−60) were expressed from the pRS315 plasmid. (C) Overexpression of Mdj2 does not rescue the anaerobic growth defect of pam18Δ1−60 cells. (Left) pam18-Δ cells expressing either full-length Pam18 (WT) or Pam18Δ1−60 (Δ1−60) from pRS315 as well as harboring either p416 having MDJ2 under the control of the GPD promoter (MDJ2↑) or p416 control vector (empty) were grown in the presence (+O2) or absence (−O2) of oxygen. SC-Ura-Leu dropout media with Ergosterol and Tween 80 supplement (SC-Ura-Leu + Erg/Tw) were used to ensure plasmid retention. Cells were grown at 34 °C aerobically for 2 days or anaerobically for 2.5 days. (Right) Whole cell lysates prepared from cells cultured on SC-Ura-Leu + Erg/Tw at 30 °C under anaerobic condition were used for the immunoblot analysis with antibodies against Mdj2 and Tim23, as the loading control.
To test whether Mdj2 could compensate for the lack of the Pam18 IMS domain, we then asked if overexpression of Mdj2 could restore the anaerobic specific defect of the pam18Δ1−60 cells. MDJ2 was placed under the control of the strong, constitutive GPD promoter, which resulted in overexpression of Mdj2 on the order of 10-fold (fig. 7C). Mdj2 was unable to compensate for the anaerobic function of the deleted IMS domain of Pam18, even when overexpressed. In fact, overexpression of Mdj2 was deleterious for cell growth of both wild-type and pam18Δ1−60 cells, under both aerobic and anaerobic conditions.
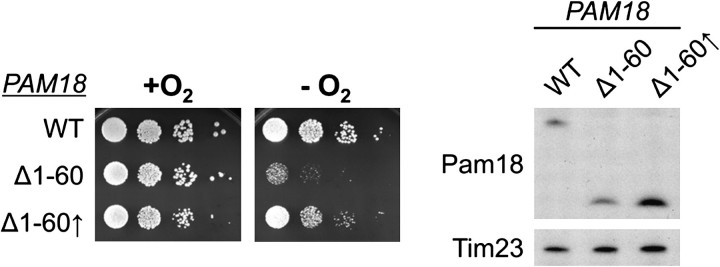
We also tested the affect of overexpression of Pam18Δ1−60. Contrary to the result with Mdj2, 3-fold overexpression of Pam18Δ1–60 partially suppressed the anaerobic growth defect caused by the lack of the IMS domain at 18 °C but only slightly improved growth at 37 °C (fig. 8, and data not shown). Together our results indicate that Mdj2 does not play a critical role under anaerobic conditions, whereas the IMS domain of Pam18 is important under this condition. However, the defect caused by the absence of the IMS domain can be partially overcome by overexpression of the variant lacking this domain.
FIG. 8.
Growth improvement by overexpressing Pam18Δ1−60 under anaerobic conditions. (Left) pam18-Δ cells carrying either full-length PAM18 (WT) or pam18Δ1–60 under the control of its endogenous promoter (Δ1−60) or the TEF promoter (Δ1–60↑) for overexpression. After spotting on SC + Erg/Tw media cells were grown at 18 °C for 5 days in the presence (+O2) or absence (−O2) of oxygen. (Right) Extracts of the indicated strains were prepared and examined by immunoblot analysis. The expression levels of the Pam18 proteins were assessed as described in figure 5B.
Discussion
The results from our evolutionary analyses indicated that duplication of an ancestral mitochondrial J-protein gene occurred prior to the divergence of the subphylum Saccharomycotina, where one duplicate gave rise to the rapidly evolving MDJ2 and the other give rise to PAM18, which subsequently gained a novel N-terminal IMS domain. Although a specific function of Mdj2 is yet to be identified, the work reported within demonstrated that the IMS domain of Pam18 is advantageous in the absence of oxygen.
Evolution of PAM18 and MDJ2 Gene Duplicates
Our analyses indicated that the emergence of the PAM18 and MDJ2 gene pair and the acquisition of the IMS domain of Pam18 corresponded to an ancient gene duplication event. BLAST analyses clearly revealed two paralogs in all Saccharomycotina, whereas all other fungi harbored only a single copy homolog. Phylogenetic reconstruction strongly supported two monophyletic clades of paralogs, each originating basal to the Saccharomycotina, again consistent with a single duplication event. In addition, both BLAST and phylogenetic methods elucidated orthologs of PAM18 and MDJ2 in each post-duplication species. Alignments of protein sequences among all homologs in fungi qualitatively demonstrated the origin of a novel protein domain within each paralog that coincides with the duplication event, the N-terminal IMS domain of Pam18, and an unstudied domain located between the internal membrane-spanning domain and the C-terminal J-domain of Mdj2, respectively.
Our analyses of protein and nucleotide trees, as well as the rate of change of both synonymous and nonsynonymous codons, indicated that the rates of evolution are higher for MDJ2 than PAM18. Previous studies have demonstrated that relatively lowly expressed genes tend to have reduced codon bias and relatively rapid rates of evolution in yeast (Drummond et al. 2005). Consistent with this hypothesis, MDJ2 exhibited reduced codon bias and mRNA levels under laboratory conditions have been reported to be on the order of 12-fold lower than those of PAM18 (Holstege et al. 1998). The relatively slower rate of evolution for more highly expressed genes is hypothesized to be due to stronger selection for translational efficiency and against translational errors for highly expressed genes in particular (Drummond and Wilke 2009). Although it is not clear whether lower levels of expression drive relaxed selection on proteins or rapid evolutionary change results in lower levels of gene expression, both of which are consistent with existing MDJ2 data, the correlations between expression levels, codon bias, and rates of evolution are particularly strong in yeast (Drummond et al. 2005; Drummond and Wilke 2009).
Despite the lack of phenotype of mdj2-Δ cells, there is no question that MDJ2 encodes a functional J-protein, as increasing its expression rescues the lethal phenotype caused by the absence of PAM18 (Mokranjac et al. 2005). However, its function in the wild-type cell remains unresolved. In our studies, we did not detect a growth defect of cells lacking Mdj2. An earlier study reported a genetic interaction between mdj2-Δ and the deletion of the gene encoding another mitochondrial J-protein, Mdj1, a soluble matrix J-protein involved in protein folding (Westermann and Neupert 1997). However, perhaps due to differences in strain background, we could not reproduce this reported slight temperature sensitivity of mdj1-Δ mdj2-Δ cells relative to mdj1-Δ cells (data not shown). But, this lack of an identifiable phenotype does not rule out the possibility that Mdj2 plays a significant role in the wild under conditions not typically used in the laboratory. Perhaps, it forms a low-abundance specialized translocation apparatus needed for efficient translocation of a subset of proteins under specific conditions. It is also plausible that only minor functional changes have evolved, and the observed increased rate of evolution is simply a by-product of a reduced level of gene expression and resultant relaxed negative selection pressures.
The mechanism by which one duplicate acquired the IMS domain after the PAM18/MDJ2 gene duplication event is a matter of conjecture. Two extreme possibilities come to mind. First, the IMS domain might have been acquired by domain shuffling from a preexisting gene (Babushok et al. 2007). Arguing against this idea, our BLAST analysis, using the amino acid sequence of S. cerevisiae Pam18 IMS domain as a query, revealed no region of significant similarity in other ORFs of the S. cerevisiae genome. Alternatively, a mutation(s) activating an in-frame AUG, or an ancestrally cryptic, start codon upstream from the original start codon of the ancestral gene might have been the instigating event for acquisition of the N-terminal extension forming the Pam18 IMS domain. One piece of evidence consistent with such a scenario is the conserved methionine residue near the beginning of the transmembrane domain, present in all but one of the fungal homologs analyzed. Regardless of the mechanism, the ability to unambiguously colocalize the duplication event and origination of the novel IMS domain, in its entirety, to such a short branch of the fungal tree, leads us to hypothesize that the duplication event itself was somehow key in the origin of the functionally novel IMS domain. Perhaps, the presence of the duplicate gene provisionally maintained essential mitochondrial import function while a specialized function of the novel domain evolved.
The requirement of the Pam18 IMS Domain under Anaerobic Conditions
Results reported here reveal the importance of the IMS domain of Pam18 for growth in the absence of oxygen. Mutations in genes encoding a variety of other components in the import apparatus had either little or no affect on growth in the absence, compared with the presence, of oxygen. In addition, another mutation in the region encoding the matrix domain of Pam18 grew as well anaerobically, as aerobically. Thus, we conclude that the IMS domain carries out a function particularly required under anaerobic conditions and does not simply reflect a need for a more efficient function of the import apparatus.
A clue to the possible function of IMS domain of Pam18 comes from studies with aerobically grown cells, as this domain is able to interact with Tim17, one of the two proteins that make up the core translocon through which the translocating polypeptides transits (Chacinska et al. 2005; D'Silva et al. 2008). Given what is known about the mitochondrial protein translocation system and the differences in structure between mitochondria of anaerobically and aerobically grown cells, we can speculate as to the possible reasons that the Pam18 IMS domain is required under these particular environmental conditions. First, it has been reported that under aerobic conditions, Pam18 associates, perhaps directly, with the respiratory chain by binding to Complex III/IV in the inner membrane (van der Laan et al. 2006; Wiedemann et al. 2007), leading to the idea that this interaction either promotes the assembly of the translocation complex or stabilizes Pam18's association with it (van der Laan et al. 2010). However, yeast cells do not possess a functional respiratory chain under anaerobic conditions (Groot et al. 1971; Rosenfeld et al. 2004). In addition, the lipid composition of the mitochondrial inner membrane is critical for maintaining the stability of the translocon complex and thus efficiency of the import process. For example, disruption of the synthesis of cardiolipin, a major component of the inner membrane, results in impairment of mitochondrial protein import (Jiang et al. 2000) and destabilization of the import apparatus of the inner membrane (Kutik et al. 2008; Tamura et al. 2009) as well as other multimeric protein complexes in the inner membrane (Pfeiffer et al. 2003). Mitochondria isolated from anaerobically grown cells have a lower percentage of cardiolipin in their mitochondrial membranes than do mitochondria from aerobically grown cells (Paltauf and Schatz 1969), thus potentially leaving multimeric complexes, such as the translocon, more prone to dissociation. These observations raise the intriguing possibility that the IMS domain of Pam18 may be critical in the absence of oxygen to stabilize the association of the import motor with the translocon or to precisely position it relative to other components of the motor itself.
Regardless of the reason for the requirement of IMS domain of Pam18 when oxygen is eliminated from the environment, it is intriguing that comparative genomic analyses have previously identified several other differences in mitochondrial related functions between Saccharomycotina and Pezizomycotina. For example, Saccharomycotina, unlike the Pezizomycotina, exhibit unusually rapidly evolving mitochondrial ribosomal proteins and excess novel gene clusters localized to the mitochondria (Arvas et al. 2007) as well as multiple differences in the presence and absence of various subunits of oxidative phosphorylation complexes I–V (Lavin et al. 2008). In addition, Saccharomycotina, but not the Pezizomycotina, have lost the ability of mitochondria to carry out fatty acid degradation via beta oxidation (Cornell et al. 2007; Shen and Burger 2009), whereas on the other hand, a regulatory mechanism causing rapid degradation of mRNA for proteins imported to the mitochondria via unique 3′ untranslated region binding sites is found only within the Saccharomycotina (Jiang et al. 2010). Ultimately these functional differences, along with the acquisition of the IMS domain of Pam18, may all play a role in the molecular diversification that underlies the overall differences in facultative anaerobic growth derived within the Saccharomycotina and potentially the fermentative capabilities of the more derived Saccharomyces.
Supplementary Materials
Supplementary table S1 and supplementary figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org).
Acknowledgments
We thank Jacek Kominek for thoughtful discussions and critical comments on this work. This work was supported by the National Institutes of Health Grants GM278709 (to E.A.C.) and Polish Ministry of Science and Higher Education Grant NN301419838 (to J.M.).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen AA, Stier TJ. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J Cell Physiol. 1953;41:23–36. doi: 10.1002/jcp.1030410103. [DOI] [PubMed] [Google Scholar]
- Andreasen AA, Stier TJ. Anaerobic nutrition of Saccharomyces cerevisiae. II. Unsaturated fatty acid requirement for growth in a defined medium. J Cell Physiol. 1954;43:271–281. doi: 10.1002/jcp.1030430303. [DOI] [PubMed] [Google Scholar]
- Arvas M, Kivioja T, Mitchell A, Saloheimo M, Ussery D, Penttila M, Oliver S. Comparison of protein coding gene contents of the fungal phyla Pezizomycotina and Saccharomycotina. BMC Genomics. 2007;8:325. doi: 10.1186/1471-2164-8-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok DV, Ostertag EM, Kazazian HH., Jr. Current topics in genome evolution: molecular mechanisms of new gene formation. Cell Mol Life Sci. 2007;64:542–554. doi: 10.1007/s00018-006-6453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Lind M, Frazier AE, et al. (12 co-authors) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Cornell MJ, Alam I, Soanes DM, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Talbot NJ, Oliver SG. Comparative genome analysis across a kingdom of eukaryotic organisms: specialization and diversification in the fungi. Genome Res. 2007;17:1809–1822. doi: 10.1101/gr.6531807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, et al. (14 co-authors) The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva P, Liu Q, Walter W, Craig EA. Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat Struct Mol Biol. 2004;11:1084–1091. doi: 10.1038/nsmb846. [DOI] [PubMed] [Google Scholar]
- D'Silva PR, Schilke B, Hayashi M, Craig EA. Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol Biol Cell. 2008;19:424–432. doi: 10.1091/mbc.E07-08-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva PD, Schilke B, Walter W, Andrew A, Craig EA. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc Natl Acad Sci U S A. 2003;100:13839–13844. doi: 10.1073/pnas.1936150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, et al. (67 co-authors) Genome Evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambill BD, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Groot GS, Kovac L, Schatz G. Promitochondria of anaerobically grown yeast. V. Energy transfer in the absence of an electron transfer chain. Proc Natl Acad Sci U S A. 1971;68:308–311. doi: 10.1073/pnas.68.2.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe Seyler. 1993;374:166. [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Hutu DP, Guiard B, Chacinska A, Becker D, Pfanner N, Rehling P, van der Laan M. Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol Biol Cell. 2008;19:2642–2649. doi: 10.1091/mbc.E07-12-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Pfund C, Craig EA. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem. 2000;275:22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- Jiang H, Guan W, Gu Z. Tinkering evolution of post-transcriptional RNA regulons: puf3p in fungi as an example. PLoS Genet. 2010;6:e1001030. doi: 10.1371/journal.pgen.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretic D, Zoranic L, Zucic D. Basic charge clusters and predictions of membrane protein topology. J Chem Inf Comput Sci. 2002;42:620–632. doi: 10.1021/ci010263s. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Kutik S, Rissler M, Guan XL, et al. (12 co-authors) The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol. 2008;183:1213–1221. doi: 10.1083/jcb.200806048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lavin JL, Oguiza JA, Ramirez L, Pisabarro AG. Comparative genomics of the oxidative phosphorylation system in fungi. Fungal Genet Biol. 2008;45:1248–1256. doi: 10.1016/j.fgb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- Liu Q, D'Silva P, Walter W, Marszalek J, Craig EA. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- Liu Q, Krzewska J, Liberek K, Craig EA. Mitochondrial Hsp70 Ssc1: role in protein folding. J Biol Chem. 2001;276:6112–6118. doi: 10.1074/jbc.M009519200. [DOI] [PubMed] [Google Scholar]
- Maarse AC, Blom J, Grivell LA, Meijer M. MPI1, an essential gene encoding a mitochondrial membrane protein, is possibly involved in protein import into yeast mitochondria. EMBO J. 1992;11:3619–3628. doi: 10.1002/j.1460-2075.1992.tb05446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Davis JE, Craig EA. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Berg A, Adam A, Neupert W, Hell K. Association of the Tim14.Tim16 subcomplex with the TIM23 translocase is crucial for function of the mitochondrial protein import motor. J Biol Chem. 2007;282:18037–18045. doi: 10.1074/jbc.M701895200. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Neupert W. Thirty years of protein translocation into mitochondria: unexpectedly complex and still puzzling. Biochim Biophys Acta. 2009;1793:33–41. doi: 10.1016/j.bbamcr.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Neupert W, Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Popov-Celeketic D, Berg A, Hell K, Neupert W. The import motor of the yeast mitochondrial TIM23 preprotein translocase contains two different J proteins, Tim14 and Mdj2. J Biol Chem. 2005;280:31608–31614. doi: 10.1074/jbc.M502397200. [DOI] [PubMed] [Google Scholar]
- Moura GR, Paredes JA, Santos MA. Development of the genetic code: insights from a fungal codon reassignment. FEBS Lett. 2010;584:334–341. doi: 10.1016/j.febslet.2009.11.066. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Syst Biol. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- Paltauf F, Schatz G. Promitochondria of anaerobically grown yeast. II. Lipid composition. Biochemistry. 1969;8:335–339. doi: 10.1021/bi00829a046. [DOI] [PubMed] [Google Scholar]
- Peden J. Analysis of codon usage [PhD thesis] Nottingham (UK): The University of Nottingham; 1999. [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- Popov-Celeketic D, Mapa K, Neupert W, Mokranjac D. Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 2008;27:1469–1480. doi: 10.1038/emboj.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid GA, Schatz G. Import of proteins into mitochondria. Extramitochondrial pools and post-translational import of mitochondrial protein precursors in vivo. J Biol Chem. 1982;257:13062–13067. [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosenfeld E, Schaeffer J, Beauvoit B, Salmon JM. Isolation and properties of promitochondria from anaerobic stationary-phase yeast cells. Antonie Van Leeuwenhoek. 2004;85:9–21. doi: 10.1023/B:ANTO.0000020268.55350.54. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Frank AC, Conant GC, Byrne KP, Woolfit M, Wolfe KH. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci U S A. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- Schiller D, Cheng YC, Liu Q, Walter W, Craig EA. Residues of Tim44 involved in both association with the translocon of the inner mitochondrial membrane and regulation of mitochondrial Hsp70 tethering. Mol Cell Biol. 2008;28:4424–4433. doi: 10.1128/MCB.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Cowe E. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991;7:657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- Shen YQ, Burger G. Plasticity of a key metabolic pathway in fungi. Funct Integr Genomics. 2009;9:145–151. doi: 10.1007/s10142-008-0095-6. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian AR, Kaufmann M, Morgenstern B. DIALIGN-TX: greedy and progressive approaches for segment-based multiple sequence alignment. Algorithms Mol Biol. 2008;3:6. doi: 10.1186/1748-7188-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Endo T, Iijima M, Sesaki H. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J Cell Biol. 2009;185:1029–1045. doi: 10.1083/jcb.200812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trocha PJ, Sprinson DB. Location and regulation of early enzymes of sterol biosynthesis in yeast. Arch Biochem Biophys. 1976;174:45–51. doi: 10.1016/0003-9861(76)90322-2. [DOI] [PubMed] [Google Scholar]
- Truscott KN, Voos W, Frazier AE, et al. (14 co-authors) A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J Cell Biol. 2003;163:707–713. doi: 10.1083/jcb.200308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Chacinska A, Lind M, Perschil I, Sickmann A, Meyer HE, Guiard B, Meisinger C, Pfanner N, Rehling P. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol Cell Biol. 2005;25:7449–7458. doi: 10.1128/MCB.25.17.7449-7458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Hutu DP, Rehling P. On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim Biophys Acta. 2010;1803:732–739. doi: 10.1016/j.bbamcr.2010.01.013. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr Biol. 2006;16:2271–2276. doi: 10.1016/j.cub.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Voisine C, Craig EA, Zufall N, von Ahsen O, Pfanner N, Voos W. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- Westermann B, Neupert W. Mdj2p, a novel DnaJ homolog in the mitochondrial inner membrane of the yeast Saccharomyces cerevisiae. J Mol Biol. 1997;272:477–483. doi: 10.1006/jmbi.1997.1267. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J Cell Biol. 2007;179:1115–1122. doi: 10.1083/jcb.200709087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007; 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 1998;46:409–418. doi: 10.1007/pl00006320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.