Abstract
Dimethyl sulfoxide (DMSO) can effectively form a glassy matrix necessary for dynamic nuclear polarization (DNP) experiments. We tested the effects of 13C enrichment in DMSO on DNP of [1-13C]pyruvate doped with trityl radical OX063Me. We found that the polarization build-up time τ of pyruvate in 13C-labelled DMSO glassing solution is twice as fast as the unenriched DMSO while the NMR enhancement was unchanged. This indicates that 13C-13C spin diffusion is a limiting factor in the kinetics of dynamic nuclear polarization in this system, but it has a minimal effect on the absolute value of polarization achievable for the target.
1. Introduction
In vivo nuclear magnetic resonance (NMR) spectroscopy and MR imaging of nuclei other than proton is limited by the inherently low sensitivity that is a consequence of the minute differences in populations of the Boltzmann nuclear energy levels. Dynamic nuclear polarization (DNP) can produce higher nuclear polarization through the transfer of electron spin polarization from paramagnetic species (organic free radicals or paramagnetic metal complexes) to coupled nuclear spins by microwave irradiation of the sample in a frozen glass matrix at low temperatures (around 1 K) [1, 6]. DNP was originally used to produce polarized targets for neutron scattering experiments [1, 6], but it gained practical importance for chemical and biological applications when it was shown in ground breaking experiments that rapid dissolution and transfer of compounds hyperpolarized in the solid state can lead to massive increases in liquid state NMR sensitivity [2]. Hyperpolarized 13C-labeled metabolites, especially [1-13C]pyruvate, have successfully been used to study metabolism by in vivo NMR spectroscopy and imaging. Excellent results were achieved with signal enhancements of 10,000 being routinely realized [4, 8, 9, 10, 11, 13, 14, 15, 16, 18, 19, 20, 21, 22, 24, 25, 26]. Recent experiments at higher field have produced even larger enhancements and confirmed that the DNP mechanism in effect when the trityl radical is used with carbon-13 remains thermal mixing [17]. Pyruvic acid makes a good target for DNP due to two fundamental physical reasons. First, the neat liquid is dense in 13C label. This allows 13C spin diffusion to proceed rapidly following polarization of 13C spins close to the paramagnetic centers [3, 7, 28]. Second, neat pyruvic acid naturally forms a glass at low temperatures. Glass formation is also essential for spin diffusion to occur, resulting in homogeneous polarization throughout the sample. Samples that do not glass well must be suspended in the appropriate glassing matrix, usually composed of water and a small molecule that interrupts the formation of crystals at freezing temperatures; dimethyl sulfoxide (DMSO) and glycerol are two commonly used glassing agents. Unfortunately, these matrices usually give lower polarization with longer build up times, presumably due to dilution of 13C label in the target compound into the unlabelled matrix. More effective matrices for compounds that do not self-glass would increase the applicability of DNP in general.
Gunther and co-workers have demonstrated that glassing compounds containing methyl groups display unusual interference effects with the DNP process [29]. This was attributed to the Haupt effect, a mechanism by which methyl groups become polarized at low temperatures due to restricted methyl rotation [12]. New work from the same group used [2-13C]acetone to improve the nuclear spin diffusion in the sample [23]. With this observation in mind, we undertook a DNP study of sodium-[1-13C]pyruvate using 13C-DMSO as the glassing component in a 50/50 water/DMSO matrix. We have observed that an increase in the rate of nuclear polarization buildup but have not realized the gain in polarization reported earlier [29]. Our observations agree with the model of a spin diffusion barrier surrounding the paramagnetic center [3] which can be overcome by increasing the density of 13C in the sample.
2. Materials and methods
2.1. Sample preparation
Samples of [13C]DMSO were received as a gift from Sigma-Aldrich Isotec. [1-13C]Na-pyruvate (Sigma-Aldrich) was used without further purification. The trityl radical was purchased from Oxford Instruments Molecular Biotools (Tubney Woods, Abingdon, UK). Samples were made by mixing H2O with equal parts of DMSO either unlabeled or 13C-enriched. [1-13C]pyruvate was dissolved into the matrix to achieve a concentration of 0.8 M in the DNP target. Trityl radical was present at 15 mM concentration in all samples.
2.2. Solid-state polarization
Approximately 100 μL of sample was used for the microwave frequency sweeps and for the measurement of the polarization build up times. A small aliquot of the sample (20 μL) was used for the NMR enhancement measurements. DNP was carried out using a HyperSense (Oxford Instruments Molecular Biotools) polarizer operating at 3.35 T. Microwave frequency sweeps were obtained using 10 MHz steps across a window of 0.30 GHz. Each data point polarized for 3 minutes prior to measurement. Polarization build up curves were measured in the solid state using a nominal flip angle of 5 degrees with a 300-second delay between acquisitions. Solid state spectra were acquired using a Varian (Varian Inc., Palo Alto, CA) console and a 20 degree pulse, without proton decoupling.
2.3. Liquid-state enhancement
The dissolution liquid, with approximately 4 mL volume, was transferred directly into a 10 mm NMR tube inside a Varian 9.4 T magnet via a FEP tube line with a transfer time of 8 s. The spectrum of the hyperpolarized pyruvate was recorded with 2-degree pulse and no proton decoupling. The total acquisition time was 1 s and the acquired spectrum (32,000 data points) was zero-filled and a 10 Hz exponential line broadening was applied prior to Fourier transformation. Spectra of the thermally polarized samples were obtained with a single 90-degree excitation pulse and the same receiver gain. NMR signal enhancements were calculated by taking the ratio of the integrated NMR intensities of the hyperpolarized signal (scaled by the sine of 2-degrees) over the thermal signal.
3. Results and Discussion
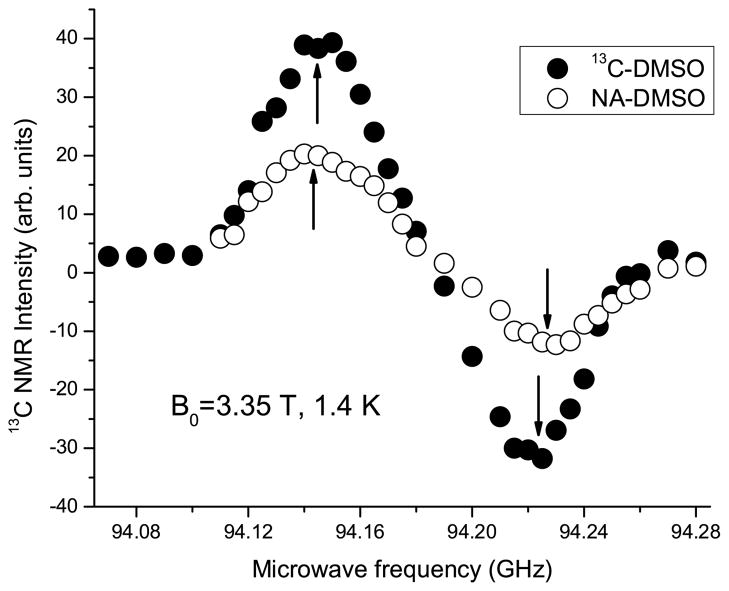
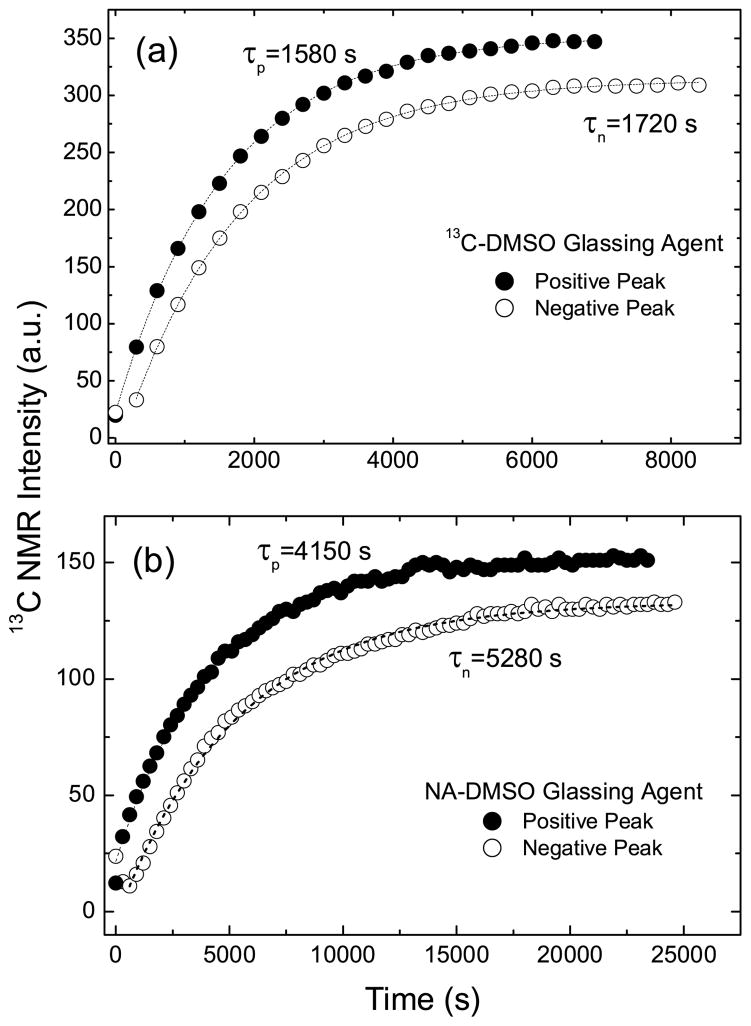
Figure 1 shows the microwave sweep spectra for samples of [1-13C]pyruvate dissolved in a matrix made with either natural abundance or 13C -labeled DMSO. The characteristic dispersive enhancement is observed for both samples. The apparent discrepancy in peak amplitudes between the samples is due to the lack of 13C label in the natural abundance sample. While the total amplitude of the signal in the solid state is small due to the short irradiation time, the curve sufficed to allow us to pick the optimal frequencies (94.15 and 94.24 GHz) for producing positive (ωe − ωn) and negative polarizations (ωe + ωn). Figure 2 shows the buildup curves for both natural abundance and 13C-enriched DMSO samples for irradiation at positive and negative polarization peaks. Previous results have shown that buildup rates are similar across the full microwave frequency range for the nitroxide based radicals[16]. The buildup rate time constant is shorter for irradiation at 94.15 GHz (ωe − ωn) in both cases. The solid state signal intensity is also lower at higher frequency for both samples, which can be attributed to the slightly asymmetric shape of the trityl ESR line[30].
Figure 1.
Microwave frequency sweep of [1-13C]pyruvate in 13C -labelled and naturally-abundant (NA) DMSO glassing matrices at 1.4 K and 3.35 T. The up and down arrows indicate the positive (ωe − ωn) and negative (ωe + ωn) polarization peaks, respectively.
Figure 2.
Polarization buildup curves of 100 uL samples containing 0.8 M [1-13C]pyruvate with 15 mM trityl in a) [13C]DMSO:water b) naturally abundant (NA) DMSO:water glassing matrices. The samples were irradiated at the positive and negative polarization peaks at 3.35 T and 1.4 K.
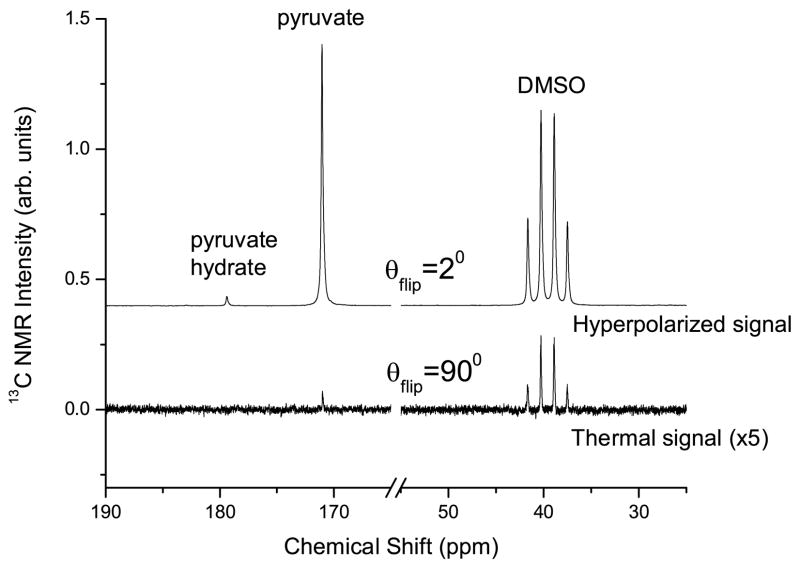
Table 1 gives the average 13C NMR enhancement and standard deviations for these experiments (N = 4). For liquid-state NMR enhancements, the 13C-labeled DMSO samples were polarized for 1.5 hrs while the NA-DMSO samples were polarized for 2.5 hrs prior to dissolution and transfer of the hyperpolarized sample to the 400 MHz liquid state NMR system. Figure 3 shows representative hyperpolarized and thermal NMR spectra used in the enhancement calculations. Our results show that the final polarization enhancement for the labelled versus unlabelled solvent samples are equivalent. Further dissolution experiments at intermediate time points confirmed that [1-13C]pyruvate polarization builds up faster for the [1-13C]DMSO samples, i.e., the faster solid state build up times are not due to the [13C]DMSO matrix alone but to the entire sample including the pyruvate. Increasing concentrations of 13C labeled target molecules, aside from the glassing matrix, was shown previously to shorten polarization times as well[5]. The solid state NMR signal of the thermally polarized sample was acquired, and a calculation using the Brillouin function at 3.35 T and 1.4 K assigned a polarization of 0.06 %. Following DNP to equilibrium, the signal increased by a factor of 150, corresponding to a polarization of 9.3 %. A similar calculation for the liquid state polarization, including a T1 correction for the 8 second transfer time, yielded a value of 8.8 % polarization for the pyruvate, agreeing well with the solid state calculation.
Table 1.
Liquid-state 13C NMR enhancement (N = 4) of [1-13C]pyruvate at 37°C immediately after dissolution of polarized samples in [13C]DMSO:water and NA-DMSO:water glassing matrices. The enhancement of self-glassing [1-13C]pyruvic acid is shown for comparison.
| Glassing Matrix | Polarization time | Positive Peak | Negative Peak |
|---|---|---|---|
| [13C]DMSO:water | 90 mins | 9120±1260 | 8800±1230 |
| NA-DMSO:water | 150 mins | 8760±860 | 7800±690 |
| [1-13C]pyruvic acid | 60 mins | 11650±300 | na |
Figure 3.
Top: Liquid-state 13C NMR spectrum of a [13C]DMSO sample in the hyperpolarized state, taken with a 2-degree flip angle, 8 seconds after dissolution. Total polarization time was 1.5 hours. Bottom: Thermal spectrum of the sample using a 90-degree flip angle.
The work of Saunders [29] showed that high polarizations levels were achieved for the CH3 groups of acetone and DMSO in 1:1:1 water/acetone/dmso mixtures. This observation led to DNP experiments with oxaloacetate which suggested that a glassy matrix containing abundant CH3 groups may be able to produce superior polarization levels when irradiating at ωe + ωn microwave frequencies for DNP. Further experiments showed an increase in polarization of ~5× when using [2-13C]acetone to polarize small molecules at natural abundance. These results led us to the experiments here that compared [13C]DMSO to DMSO in 50:50 water/DMSO mixtures.
For our system, the acetone portion of the Saunders et al. formulation was omitted for simplicity of sample preparation. The superior polarizations produced in the Saunders et al. report were not recapitulated in either the natural abundance or 13C-labelled DMSO samples (Table 1). However, as shown in Figure 2, the inclusion of [13C]DMSO in the glassing matrix significantly shortens the time needed to achieve maximum polarizations for the solid, frozen matrix and following dissolution (Table 1). The shortening of the build up time constant by increasing the concentration of 13C spins suggest that spin diffusion is the primary mechanism for propagation of the high polarization for the spins surrounding the paramagnetic centers into the bulk sample. The theory of spin diffusion, as first expounded by Bloembergen, detailed the return of nuclear spins to thermal equilibrium following saturation as mediated by paramagnetic centers[3]. This is the opposite of our case, as polarization proceeds outward from the paramagnetic center. Recently, a review of this specific phenomenon discussed qualitatively the expected impact of the spin diffusion barrier and various mechanisms by which the barrier can be traversed by high polarizations[27]. In order to see how spin diffusion might apply to our samples here, the density of the radical in the prepared samples versus the concentration of 13C nuclei must be considered. Since the samples are at 1.4 K, contributions to spin diffusion from motion in the matrix can be ignored. The 0.015 M concentration of radical corresponds to having paramagnetic centers that are 48 nanometers apart assuming equal distribution of the spins on a cubic matrix. Pure pyruvic acid is 14.4 M in concentration. If only the 1-position of pyruvic acid is labeled, then the average internuclear distance for 13C labels, assuming a uniform distribution in the sample, is 0.487 nanometers (or a 13C-13C dipolar coupling in the solid state of 66 Hz, neglecting angular factors and natural abundance carbons). This concentration of 13C spins amounts to ~960 spins/paramagnetic center. Calculation of this distance for the samples made with DMSO is more difficult, as the lattice of spins will be punctuated by the two methyl groups of DMSO that are only ~0.15 nanometers apart. If these two carbons are counted as a single point in the lattice, samples made with [13C]DMSO, water, and 0.8 M [1-13C]pyruvate give an inter-label distance of ~0.6 nanometers (~36 Hz homonuclear dipolar coupling) and a concentration of ~7.8M in 13C spins. If both carbons of DMSO are counted, then the concentration of 13C spins returns to a value similar to that of pyruvic acid, with the caveat that the labels are not spread evenly through the sample. If all the natural abundance carbons of the DMSO are counted for the unlabeled DMSO sample, the internuclear 13C distance increases to 1.28 nanometers (~4 Hz homonuclear dipolar coupling) and the concentration of 13C spins is ~0.87 M, neglecting natural abundance contributions from the C2 and C3 positions of pyruvate. In this case, the number of of 13C’s/paramagnetic center falls to ~521. In all cases the 13C-13C internuclear distance is much shorter than the distance between the paramagnetic centers indicating that the spin diffusion regime should be an accurate description of the samples. Bloembergen described spin diffusion D in the following terms, D = Wa2, where W is a dipolar transition probability in sec−1 and a is the distance between the nuclear spins[3]. The final units of D are therefore in cm2/sec. Assuming a cubic lattice of spins which can participate in the spin diffusion, D is proportional to 1/a when all spins, not just the nearest neighbors are integrated over. If the ~60 minute τ of pyruvic acid is taken as the baseline, then the increased distance from point to point in the [13C]DMSO matrix (assuming the concentration of 7.8 M in 13C in the above calculation) as compared to the pyruvic acid should slow spin diffusion by a ratio of the internuclear distances, or 1.22. The ratio of τ’s is actually 1.5, which is in excellent agreement with the calculation. The difference is likely accounted for by the difference in chemical shifts between the DMSO carbons and the C1 of pyruvate. Any difference in energies for the spins acted upon by spin diffusion will lower the transition probability, W. Comparison of the internuclear distances for the labeled and unlabeled DMSO samples indicates that diffusion should slow by a factor of 2.14, when a factor of 1.67 was observed. This is still in decent agreement with the theoretical predictions. Therefore, it can be said that spin diffusion can reasonably account the the difference in polarization rates for the three types of samples.
The differences in polarizations can be explained in a qualitative manner. First, for the DMSO samples, even though the number of spins per paramagnetic center has increased when using the [13C]DMSO, the change is less than a factor of 2, and does not produce a noticeable change in the spin heat capacity of the sample. Therefore the samples produced with DMSO should give approximately the same final polarizations, which they do. However, these samples have lower polarizations than the pure pyruvic acid sample. Most likely this is due to the increased number of protons in the sample due to the DMSO and H2O. Substitution with D2O and [d6,13C]DMSO would likely improve the polarization levels to the level of the pyruvic acid preparation by lowering the heat capacity of the nuclear spin bath while maintaining the increased 13C density. Even though the protons have a Larmor frequency sufficient to push them primarily into the solid effect regime, some contact of the thermal reservoir can remain, as observed by de Boer for the narrow ESR line of BDPA[6]. Furthermore, inclusion of a Gd based relaxation agent in the preparation should improve the raw enhancement numbers as well. It is unclear why the results of Gunther and co-workers have not been reproduced here. Of note is their choice of target for DNP, namely compounds containing only 13C at natural abundance. In their case, perhaps the spin diffusion barrier is even more difficult to break through, and hence polarizations due to methyl rotation become important factors in determining the kinetics of the DNP process. This hypothesis agrees with the necessary condition for transport through the spin diffusion barrier; another energy reservoir that can match the energy differences for the spins within the scattering distance of the paramagnetic center to those outside it[27]. An extra benefit of including the [13C]DMSO is that it can serve as a useful internal polarization standard that is not subject to metabolism like the pyruvate. Due to the cost of trityl radical, these experiments were not duplicated with other target compounds like lactate or acetate, though we expect these results to be fully generalizable for 13C-labeled metabolites.
4. Conclusion
It has been shown that inclusion of [13C]DMSO as part of the glassing matrix for a sample of sodium [1-13C]pyruvate increases the rate of the DNP process compared to samples made with natural abundance DMSO. However, in this case, it does not increase the total polarization of the pyruvate target. The influence of the spin diffusion barrier limiting the kinetics of the DNP process can be minimized by increasing the total amount of 13C in the sample.
Acknowledgments
The authors acknowledge the support from the National Institutes of Health grants number 1 R21 EB009147-01 and RR02584.
References
- 1.Abragam A, Goldman M. Principles of dynamic nuclear polarization. Rep Prog Phys. 1978;41:395. [Google Scholar]
- 2.Ardenkjaer-Larsen H, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase of signal-to-noise ratio of > 10, 000 in liquid-state NMR. Proc Nat Acad Sci. 2003;100:10158. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloembergen N. On the interaction of nuclear spins in a crystalline lattice. Physica. 1949;15:410. [Google Scholar]
- 4.Chen AP, Albers MJ, Cunningham CH, Kohler SJ, Yen YF, Hurd RE, Tropp J, Bok R, Pauly JM, Nelson SJ, Kurhanewicz J, Vigneron DB. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3 T-initial experience. Magn Reson Med. 2007;58:1099. doi: 10.1002/mrm.21256. [DOI] [PubMed] [Google Scholar]
- 5.Comment A, van den Brandt B, Uffmann F, Kurdzesau F, Jannin S, Konter JA, Haulte P, Wenckebach WTh, Greutter R, van der Klink JJ. Design and performance of a DNP prepolarizer coupled to a rodent MRI scanner. Conc Mag Res B: Mag Res Eng. 2007;31B:255. [Google Scholar]
- 6.de Boer W. Dynamic orientation of nuclei at low temperatures. J Low Temp Phys. 1976;22:185. [Google Scholar]
- 7.Dementyev AE, Cory DG, Ramanathan C. Rapid diffusion of dipolar order enhances dynamic nuclear polarization. Phys Rev B. 2008;77:024413. doi: 10.1103/PhysRevLett.100.127601. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher FA, Kettunen MI, Brindle KM. Biomedical applications of hyperpolarized 13C magnetic resonance imaging. Prog Nucl Magn Reson Spec. 2009;55:285. [Google Scholar]
- 9.Golman K, in’t Zandt R, Lerche MH, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 2006;66:10855. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- 10.Golman K, in’t Zandt R, Thaning M. Real-time metabolic imaging. Proc Natl Acad Sci. 2006;103:11270. doi: 10.1073/pnas.0601319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golman K, Petersson JS, Magnusson P, Johansson E, Akeson P, Chai C, Hansson G, Mansson S. Cardiac metabolism measured noninvasively by hyperpolarized 13C MRI. Magn Reson Med. 2008;59:1005. doi: 10.1002/mrm.21460. [DOI] [PubMed] [Google Scholar]
- 12.Haupt J. A new effect of dynamic nuclear polarization in a solid obtained by rapid change of temperature. Phys Lett. 1972;38A:389. [Google Scholar]
- 13.Hu S, Chen AP, Zierhut ML, Bok R, Yen YF, Schroeder MA, Hurd RE, Nelson SJ, Kurhanewicz J, Vigneron DB. In vivo carbon-13 dynamic MRS and MRI of normal and fasted rat liver with hyperpolarized 13C-pyruvate. Mol Imaging Biol. 2009;11:399. doi: 10.1007/s11307-009-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu S, Lustig M, Balakrishnan A, Larson PEZ, Bok R, Kurhanewicz J, Nelson SJ, Goga A, Pauly JM, Vigneron DB. 3D compressed sensing for highly accelerated hyperpolarized 13C MRSI with in vivo applications to transgenic mouse model of cancer. Magn Reson Med. 2010;63:312. doi: 10.1002/mrm.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamin Y, Gabellieri C, Smyth L, Reynolds S, Robinson SP, Springer CJ, Leach MO, Payne GS, Eykyn TR. Hyperpolarized 13C magnetic resonance detection of carboxypeptidase G2 activity. Magn Reson Med. 2009;62:1300. doi: 10.1002/mrm.22049. [DOI] [PubMed] [Google Scholar]
- 16.Jannin S, Comment A, Kurdzesau F, Konter JA, Hautle P, van den Brandt B, van der Klink JJ. A 140 GHz prepolarizer for dissolution dynamic nuclear polarization. J Chem Phys. 2008;128:241102. doi: 10.1063/1.2951994. [DOI] [PubMed] [Google Scholar]
- 17.Johannesson H, Macholl S, Ardenkjaer-Larsen JH. Dynamic nuclear polarization of [1-13C]pyruvic acid at 4.6 T. J Magn Reson. 2009;197:167. doi: 10.1016/j.jmr.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 18.Keshari KR, Kurhanewicz J, Jeffries RE, Wilson DM, Dewar BJ, Van Criekinge M, Zierhut M, Vigneron DB, Macdonald JM. Hyperpolarized 13C spectroscopy and an NMR-compatible bioreactor system for the investigation of real-time cellular metabolism. Magn Reson Med. 2010;63:322. doi: 10.1002/mrm.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keshari KR, Wilson DM, Chen AP, Bok R, Larson PE, Hu S, Van Criekinge M, Macdonald JM, Vigneron DB, Kurhanewicz J. Hyperpolarized [2-13C]-fructose: a hemiketal DNP substrate for in vivo metabolic imaging. J Am Chem Soc. 2009;131:17591. doi: 10.1021/ja9049355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurdzesau F, van den Brandt B, Comment A, Hautle P, Jannin S, van der Klink JJ, Konter JA. Dynamic nuclear polarization of small labelled molecules in frozen water-alcohol solutions. J Phys D: Appl Phys. 2008;41:155506. doi: 10.1063/1.2951994. [DOI] [PubMed] [Google Scholar]
- 21.Kurhanewicz J, Bok R, Nelson SJ, Vigneron DB. Current and potential applications of clinical 13C MR spectroscopy. J Nucl Med. 2008;49:341. doi: 10.2967/jnumed.107.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson PE, Bok R, Kerr AB, Lustig M, Hu S, Chen AP, Nelson SJ, Pauly JM, Kurhanewicz J, Vigneron DB. Investigation of tumor hyperpolarized [1-13C]-pyruvate dynamic using time-resolved multiband RF excitation echo-planar MRI. Magn Reson Med. 2010;63:582. doi: 10.1002/mrm.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludwig C, Marin-Montesinos I, Saunders MG, Gunther UL. Optimizing the polarization matrix for ex situ dynamic nuclear polarization. J Am Chem Soc. 2010;132:2508. doi: 10.1021/ja909984w. [DOI] [PubMed] [Google Scholar]
- 24.Merritt ME, Harrison C, Storey CJ, Jeffrey FMH, Sherry AD, Malloy CR. Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proc Natl Acad Sci. 2007;104:19773. doi: 10.1073/pnas.0706235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merritt ME, Harrison C, Storey CJ, Sherry AD, Malloy CR. Inhibition of carbohydrate oxidation during the first minute of reperfusion after brief ischemia: NMR detection of 13CO2 and H13CO3. Magn Reson Med. 2008;60:1029. doi: 10.1002/mrm.21760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson SJ, Vigneron D, Kurhanewicz J, Chen A, Bok R, Hurd R. DNP-hyperpolarized 13C magnetic resonance metabolic imaging for cancer applications. Appl Magn Reson. 2008;34:533. doi: 10.1007/s00723-008-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramanathan C. Dynamic Nuclear Polarization and Spin Diffusion in Nonconducting Solids. Appl Magn Reson. 2008;34:409. [Google Scholar]
- 28.Schmugge TJ, Jeffies CD. High dynamic polarization of protons. Phys Rev. 1965;138:A1785. [Google Scholar]
- 29.Saunders MG, Ludwig C, Gunther UL. Optimizing the signal enhancement in cryogenic ex situ DNP-NMR spectroscopy. J Am Chem Soc. 2008;130:6914. doi: 10.1021/ja800971t. [DOI] [PubMed] [Google Scholar]
- 30.Wolber J, Ellner F, Fridlund B, Gram A, Johannesson A, Hansson G, Hansson LH, Lerche MH, Mansson S, Servin R, Thaning M, Golman K, Ardenkjaer-Larsen JH. Generating highly polarized nuclear spins in solution using dynamic nuclear polarization. Nucl Instr Meth in Phys Res A. 526:173. [Google Scholar]