Abstract
Three dimensional mass spectral imaging (3D MSI) is an exciting field that grants the ability to study a broad mass range of molecular species ranging from small molecules to large proteins by creating lateral and vertical distribution maps of select compounds. Although the general premise behind 3D MSI is simple, factors such as choice of ionization method, sample handling, software considerations and many others must be taken into account for the successful design of a 3D MSI experiment. This review provides a brief overview of ionization methods, sample preparation, software types and technological advancements driving 3D MSI research of a wide range of low- to high-mass analytes. Future perspectives in this field are also provided to conclude that the positive and promises ever-growing applications in the biomedical field with continuous developments of this powerful analytical tool.
Visualization of the distribution of specific molecules in cells, organs and organisms allows a better understanding of the molecular interactions and underlying mechanism of biological processes. A combinesd use of immunohistochemical techniques and microscopy allows for the indirect mapping of analytes with high spatial resolution but limited chemical information since most of the methods require a priori knowledge of the analytes. Recent advances to integrate the high sensitivity and chemical specificity of MS with its imaging capability give rise to mass spectral imaging (MSI). MSI is a new analytical tool that does not require a priori knowledge to investigate the spatial arrangement of multiple analytes in biological tissues with high specificity.
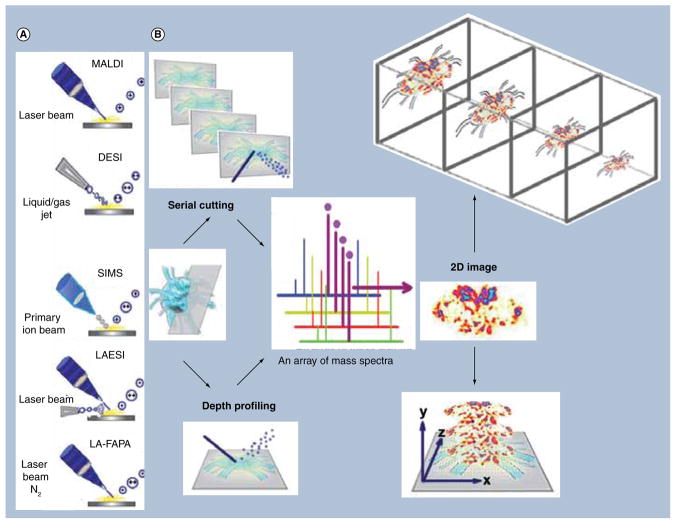
Mass spectral imaging is widely conducted in the microprobe mode. The analytes of interest are desorbed/ionized or volatized from a well-defined area (pixel) upon irradiation by laser, primary ion beam or gas/liquid jet, delivering a mass spectrum of selected mass range at each pixel. An array of mass spectra is acquired from a predefined 2D grid and subsequently processed into multiple cohesive images by selecting any individual peak of desorbed ions. MSI in microscope mode has also been reported by irradiating a much larger area of sample compared with the microprobe mode and measuring the relative intensity of analytes in the area by ion optics. Various ionization techniques have been employed for MSI (Figure 1A), such as MALDI, secondary ion MS (SIMS) and desorption ESI (DESI), which will be discussed in the following sections.
Figure 1. Overall workflow of 3D mass spectral imaging methodology showing (A) ionization method and (B) two types of tissue preparation strategies for mass spectral analysis, serial cutting and depth profiling.
Acquisition of 3D models takes place by collecting mass spectra for each pixel and processing this array into a representative 2D image. 2D images are then stacked in a 3D context to create a 3D model.
DESI: Desorption ESI; LAESI: Laser ablation ESI; LA-FAPA: Laser ablation coupled to a flowing atmospheric pressure afterglow; SIMS: Secondary ion MS.
In addition to planar 2D distribution, 3D scanning for a deeper view of analytes of interest from biological tissues can be provided by MSI. The additional spatial dimension enables the interrogation of analyte expression patterns in a tissue volume, delivering contextual information to 2D images and eliminating the possibility of neglecting small anatomical structures. 3D MSI methods can be divided into two major types, serial cutting and depth profiling (illustrated in Figure 1B), depending on the ionization method employed. Serial cutting generally involves slicing the specimen into thin serial sections at an appropriate interval and performing MSI on each consecutive section. 3D MSI of MALDI and DESI have been reported using this method [1,2]. Another method, depth profiling, is performed with ionization techniques such as SIMS, laser ablation ESI (LAESI) and laser ablation coupled to a flowing atmospheric pressure afterglow (LA-FAPA [3–5]). It is usually applied by conducting 2D spatial analysis while exposing a new, deeper surface of specimen to be scanned. A stack of 2D images representing a selected ion from the same tissue or section can be subsequently stitched together to reconstruct a 3D volumetric distribution of that ion. All of the 3D MSI studies to date are summarized in Table 1. The 3D distribution obtained from MSI can be further integrated and correlated with other imaging modalities, providing a more comprehensive and accurate molecular description of various biological processes.
Table 1.
Comparison of 3D mass spectral imaging methods and performance.
| Ionization technique | 3D MSI methods | Chemical species analyzed in 3D | Lateral resolution (μm) | Pros | Cons | Ref. |
|---|---|---|---|---|---|---|
| MALDI | Serial cutting | Peptides, proteins and lipids | 50–100 | Wide mass range | Matrix interference and vacuum method | [18,92,94] |
| SIMS | Depth profiling | Lipids, metabolites, elements and single cell | <1 | Minimal sample pretreatment and complementary to MALDI | Limited mass range, vacuum method and matrix interference | [34,37,47,116, 117,127–129,139] |
| DESI | Serial cutting | Lipids and metabolites | 180–200 | Minimal sample pretreatment, ambient method and high sensitivity | Limited mass range, sensitive to surface, ill-defined sampling area and analyte washing effect | [1,49,57] |
| LAESI | Depth profiling | Metabolites | 300 | Minimal sample pretreatment and ambient method | Limited mass range and water-rich target needed | [4,49] |
| LA-FAPA | Depth profiling | Pharmaceuticals | 30–50 | Minimal sample pretreatment | Limited mass range | [5] |
MSI: Mass spectral imaging; SIMS: Secondary ion MS; DESI: Desorption ESI; LAESI: Laser ablation ESI; LA-FAPA: Laser ablation flowing atmospheric pressure afterglow.
Methodological developments
Ionization techniques
Matrix-assisted laser desorption/ionization
Since its introduction in 1987 by Karas and Hillenkamp [6], MALDI has shown its revolutionary power by providing a wide range of intact, large biomolecules, in particular proteins, peptides and oligonucleotides, with a ‘soft’ and efficient ionization source. More recently, the boundary to analysis of low-molecular weight compounds has been removed by new matrices and innovative instrumentation, enabling studies of small-molecule drug compounds, metabolites and lipids [7–9]. In addition to the wide mass range (~1.5 million Da [10]) provided by coupling with a TOF analyzer, MALDI also delivers several other unique features for complex biological system analysis, such as a high tolerance for salts and buffers, relatively simple spectra with mostly singly charged ions and low femtomole to attomole sensitivity.
The routine MALDI analysis involves a simple mixing of analytes with a surplus of matrix, usually at a molar ratio of 1:103–105 [11], but an equimolar model has been introduced by Shroff et al. [12]. The matrix is of paramount importance in MALDI. The matrix extracts analytes from the complex sample and forms co-crystals as the solvent evaporates. The analyte-doped matrix absorbs the majority of laser energy, UV or infrared (IR), and explosively desorbs from the target upon laser irradiation above fluence threshold [13], carrying the incorporated analyte into the gas phase without analyte degradation. In the gas phase, the desorbed species, including analytes, have various proton/cation affinities, while the matrix and added acids function as a pool of protons. Thereafter, a number of reactions occur in the ablated plume due to proton, cation and electron transfer [14,15], causing the predominance of singly charged ions [16].
The ability to obtain detailed spatial distributions of individual molecules from a tissue sample is of tremendous value to the biological sciences. The first work bridging MALDI-MS with molecular imaging was reported by the Caprioli group and involved the imaging of peptides and proteins from fresh tissue sections or blots on a MALDI target coated with C18 beads [17]. The capability to display molecular localization based on individual m/z and provide additional sequence information utilizing postsource decay demonstrated the power of this essential tool for unraveling and understanding molecular complexity [17]. Since this study, interest in MALDI-based MSI technology has been growing exponentially.
In a typical 2D MSI experiment, a frozen tissue section is thaw-mounted onto a sample plate, uniformly deposited with matrix, dried in a vacuum desiccator and introduced into the vacuum inlet of a mass spectrometer [18]. During this process, matrix solvents extract analytes from the tissue, quench endogenous proteolytic enzyme activity and eventually evaporate, leaving the analyte-doped matrix crystals. Then, a laser beam is rastered across the entire tissue surface over predefined 2D grids [19]. A mass spectrum is acquired from ions desorbed and ionized from each irradiation surface spot and is recorded corresponding to each grid coordinate. From the intensity of a designated m/z ion detected in each spectrum, a 2D ion density map can be constructed, showing its relative abundance in specific regions. A deeper, volumetric view of analytes is subsequently achieved by serial cutting. A stack of 2D images from serially cut tissue sections with appropriate spacing is obtained and stitched together with 3D processing and visualization software to construct a 3D volumetric model (Figure 1).
Matrix-assisted laser desorption/ionization-MSI experiments are normally performed using either N2 (337 nm) or neodymium-doped yttrium aluminium garnet (355 nm) lasers. Typical pulse lengths are 3 ns or less, and the laser repetition rate is 200–1000 Hz or higher [20]. A limitation of MALDI-MSI is the relatively poor spatial resolution (typically 50–100 μm, but can be further reduced to 20 μm) [3] unable to satisfy the need for analyzing cellular features. Increasing the resolution into the low micrometer scale by focusing the laser beam has been accomplished [21,22], but the available amount of material per imaged spot is reduced quadratically with reduction of the spot diameter, challenging the sensitivity of current MS instrumentation. Recently, Rompp et al. coupled an atmospheric pressure (AP) source to a scanning microprobe MALDI mass spectrometer [23], which was previously reported by Spengler et al. [24]. The innovative set-up of AP scanning microprobe MALDI Fourier transform ion cyclotron resonance offered high lateral (10 μm) and mass spectral resolution with high sensitivity. An alternative approach is microscope MALDI-MSI, which employs a defocused UV laser to envelop a large matrix-coated area. Then, ion optics transfer and magnify the ion packet produced by the single laser shot, relying on a position-sensitive detector to record the resulting 2D images of ion distribution [25,26]. The high spatial resolution, increased speed and greater versatility of potential ion sources offered by the microscope approach appear to have potential. However, the method is complicated by technical constraints, such as the need for specialized ion optics, a fast detector and computing software to reconstruct ion images.
One characteristic that plagues MALDI for application in MSI is the requirement for sample preparation and the need to analyze specimens under vacuum, limiting the possibility to study live biological samples. An ionization source operated at AP, termed AP-MALDI, circumvents the aforementioned limitations. Although a UV laser is more commonly employed, an IR laser beam tuned to a vibrational band of water utilizes water as the matrix for desorption/ionization of ions, eliminating the use of exogenous matrix. Therefore, this method is promising for small-molecule imaging. To date, AP-IR-MALDI has been utilized to capture metabolic transport in plant vasculature [27] and metabolite distribution on plant skin [28]. However, it is significantly limited by the diffraction-limited spot size of the laser (250 μm, further reduced to 40 μm by oversampling) [28] and relatively poor signal to noise ratio.
Secondary ion MS
Secondary ion MS is a technique that has long been established for surface chemical analysis and is also utilized for 3D imaging of biological systems. Ionization takes place by first bombarding a solid sample under a high vacuum with high energy ions from a ‘primary’ ion source [29]. Primary ion impact causes momentum transfer to the sample’s surface and atomic layers underneath. Factors such as ion mass, energy, target geometry and atomic/molecular structure influence primary ion collisions, and many publications, including a review by Fletcher et al., detail the theories regarding particle ejection from cluster bombardment well [29–32]. Following primary ion impact, energy returns to the sample surface, and locations where surface-binding energies are exceeded experience ejection, or ‘sputtering’ of atomic and molecular species. Ionic species ejected from this process are called ‘secondary’ ions, and the number of sputtered secondary ions per primary ion impact is termed the secondary ion yield. Most sputtered material is neutral for analytes under typical experimental conditions, and secondary ion yields of the majority of organic species are theoretically on the order of 10−4 to 10−6 [29,33,34]. Even with the relatively low ion yield, secondary ions can be drawn into a mass analyzer, typically a TOF analyzer, for surface chemistry analysis. Two imaging techniques with SIMS exist. The microprobe technique consists of focusing the primary ion beam to a small diameter at the surface and rastering the beam across the sample [29]. Mass spectra are acquired for each pixel to construct a spatially resolved chemical image. In the microscope/stigmatic imaging mode, the primary beam is unfocused across the entire region to be analyzed [29]. The mass spectrometer’s ion optics preserve the spatial localization of sputtered ions to create a magnified chemical image of the sample on a position-sensitive detector [29]. Furthermore, two areas of application using SIMS have been practiced since its inception. The first, dynamic SIMS, maps elemental species as a function of depth as high primary ion doses erode the sample during sputtering [35]. This analysis mode is generally used in the semiconductor field. The second method is static SIMS and utilizes a low primary ion dose to analyze the native chemistry of the surface since statistically a point should experience only one primary impact [35]. This method is typically used for analyzing molecular surface chemistry.
Although the erosive process of sputtering and surface analysis allows for depth profiling and consequently 3D imaging, the study of biological systems has presented many analytical difficulties. High-energy atomic primary ions such as Ga+ and In+ liquid-metal-ion guns (LMIGs) are capable of analyzing tissue sample surfaces with excellent lateral resolutions in the nanometer range [36]. However, these high-energy primary atomic ions inflict severe subsurface chemical damage, and ion impacts must be limited to ensure pristine material is impacted by each primary ion. Consequently, even when utilizing a TOF mass analyzer to maximize secondary ion collection, the number of ions collected per unit area limited the useful spatial resolution to approximately 1 μm [37]. In biological analyses, the minimal yield of bioorganic molecular ions above m/z 300 compounded sensitivity limitations.
Metal cluster and polyatomic ion sources provide significantly greater ion yields of higher mass molecular ions [38–42]. Aun+ and Bin+ (n = 1–7) cluster LMIGs have been coupled with TOF-SIMS instruments to obtain images of cholesterol, fatty acids, sulfatides, phosphatidylinositols and triglycerides from mouse brain tissue [43–46]. These ions significantly increase the secondary ion yield of molecular species in the m/z 400–3000 range, and spatial resolution for high ion yield molecular ions is below 1 μm [47]. Unfortunately, subsurface chemical damage still occurs because of cluster fragmentation into high-energy atomic ions upon primary ion impact, limiting the useful spatial resolution [37].
Molecular primary ions such as SF5+ and more commonly C60+ deliver large increases in mid-sized molecular and fragment ion yields as well as generating minimal subsurface chemical damage [33,37,40,48]. The absence in chemical damage from primary ion impact allows for molecular depth profiling and 3D imaging. Although the molecular ion beams are harder to focus than liquid-metal beams, 200-nm resolution is still attainable [47]. For maximum resolution, molecular depth profiling is combined with imaging etching in some cases. This technique requires etching away surface monolayers with a C60 source and acquiring images with a LMIG source because of its higher lateral resolution [34]. The use of C60+ ion beams allows for the possibility of SIMS to provide a complementary method to MALDI for obtaining molecular information in biological samples. As suggested by Jones et al., while SIMS will not likely be able to match MALDI’s capabilities in protein analysis, it can easily perform spatially resolved analyses of small molecules [47].
Although SIMS has many advantages in 3D imaging (e.g., no tissue sectioning and ability to detect small molecules), many disadvantages also exist. Sample handling of biological samples is complicated due to SIMS being a vacuum technique in comparison to other 3D MS techniques such as DESI and LAESI [47,49,50]. As with many MS techniques, the matrix effect is also a challenge in SIMS. The composition of an analyte’s immediate environment influences its ionization probability in SIMS, and knowledge of the matrix effect with respect to organic molecules is needed for correct interpretation of results [47].
Desorption ESI
One complicating factor in current 3D MSI is that the sample is generally introduced into a vacuum or a region coupled to a vacuum system. DESI is a simple, ambient sampling approach for MS analysis. Ionization occurs by channeling charged droplets and ions of solvent from an electrospray source onto the surface of interest [50]. The charged particles impact the surface to yield gaseous ions from chemical species originally present. Mass spectra are similar to normal ESI spectra, showing singly and multiply charged molecular ions [50]. Imaging is accomplished by mounting a DESI ion source onto a 3D manual translational moving stage with a 360° rotational stage to adjust the incident impact angle [51]. The translational moving stage allows for the DESI source to be rastered across the surface of interest, generating mass spectra at each ‘pixel.’ 3D imaging is accomplished by serially sectioning tissue, performing 2D MSI on each slice with DESI and then combining each 2D image to construct one 3D image [1].
Desorption ESI’s ambient nature and softness allow for the examination of chemical distributions on a variety of unmodified natural surfaces. The spatialization of endogenous chemicals such as inks, drugs of abuse, pharmaceutical molecules and biological molecules have been analyzed on many objects such as fingerprints, documents and tissue sections [52–57]. As DESI is carried out under ambient conditions, the only sample-pretreatment step is tissue sectioning. This avoids the possibility of exogenous compound contamination [57]. Spatial resolution in DESI imaging is generally reported to be approximately 180–200 μm, although values of approximately 40 μm have been reported [57,58].
One useful feature in DESI is reactive DESI. This method adds reagents to the spray solvent to harness solution-phase chemical reactions, increasing specificity of ionization for particular analytes such as cholesterol, anabolic steroids, cis-diols, phosphate esters and cyclic acetals [54,59–62]. Cholesterol imaging was accomplished at 200-μm resolution within 1 h using reactive DESI [54].
Although DESI has many unique advantages over vacuum MS methods, some disadvantages exist. With the exception of some hormones such as the adrenal, the low concentrations of hormones in biological tissue make their detection with DESI challenging [57,63]. Furthermore, efficient ionization of lipids with relatively low proton or cation affinities is difficult, causing their signal suppression by other lipids. As mentioned before, the spatial resolution of DESI is approximately 180–220 μm, which is comparable to that of LAESI but surpassed by SIMS for small-molecule analysis [47,57]. Solutions are currently being undertaken to increase this resolution, such as using the microscope rather than microprobe imaging mode and examining restricted regions at high spatial resolutions with fine needles that remove material to be examined in the open environment [26,56,64].
Laser ablation ESI
Laser ablation ESI is another ambient ionization method useful for MSI. This technique is designed primarily for biological samples containing water [49]. The sample is analyzed at AP, and a mid-IR laser beam at a wavelength of 2940 nm excites the -OH vibrations in the sample’s water molecules [65]. Phase explosion causes a rapid microscale ablation, and a mixture of molecules, clusters and particulate matter ejects from the surface. Ejected biomolecules coalesce with charged droplets formed from an electrospray source, and a portion of them is converted into gas-phase ions for introduction into a mass analyzer [65]. For MSI experiments, the sample holder is mounted on a motorized positioning stage and scans the target in the x–y plane while a manual translation stage positions the holder in the z-direction for signal optimization [66]. Once again, spectra are taken for each pixel on the sample surface. Depth profiling is also possible with LAESI as consecutive laser shots to the same point on a biological surface ablate deeper layers of tissue [67]. The coupling of LAESI depth profiling with its lateral imaging capabilities allows for 3D molecular imaging [4]. In LAESI 3D imaging, a three-axis translation stage is positioned to scan the sample in 2D. At each lateral grid point, a depth profile is constructed with multiple laser pulses and generated ions are recorded by the mass analyzer [4].
As LAESI is specifically tailored to analyze water-containing biological samples, it is well suited for studying the localization of analytes in tissue. It has been used to analyze metabolites in single cells, small metabolites and lipids in rat brain tissue, and inks and metabolites on leaf tissue [49,65–68]. Furthermore, a reactive LAESI technique has been utilized to improve collisional activated dissociation fragmentation of lipids for identification of fatty acyl chains that simply involves adding Li2SO4 to the electrospray solution in the LAESI source in order to produce lithiated ions from ablated lipids [69]. As with DESI, LAESI is an ambient technique free from matrix effects and contamination. Lateral resolution is similar to that reached by DESI at approximately 300 μm, while depth resolution is approximately 30–40 μm [4].
Benefits of LAESI are numerous, but several disadvantages to this technique are apparent. The first most obvious disadvantage is that a water-rich target is needed for the ablation process. As a result, significantly higher laser fluences are required to ablate tissues with lower mid-IR absorbances (e.g., dry skin, bone, nail and tooth) [49]. Variations of water content and tensile strength in a sample can also affect LAESI ion yield and changing imaging results [49]. As with DESI, spatial resolution is surpassed in small-molecule imaging by SIMS, but improvements in spatial resolution are thought to be possible [47].
Laser ablation-flowing AP afterglow
The development of plasma-based ambient desorption/ionization sources, such as flowing AP afterglow (FAPA), direct analysis in real time and plasma-assisted desorption/ionization, is an emerging field that aims to eliminate sample pre-treatment and separation steps. These sources make use of gaseous reagent ions or metastables directed to the ion source and heated by a grid electrode to become millimeric reactive protonated water clusters for analyte desorption/ionization. A wide variety of compounds can be ionized under the optimized operating conditions. Unfortunately, the large diameter of the reactive liquid jet hinders their use for MSI. To circumvent this situation, Hieftje et al. coupled FAPA to neodymium-doped yttrium aluminium garnet LA in an enclosed chamber [5]. The sample is ablated by focusing a 266-nm laser beam on a spot. The resulting aerosol travels along a stream of nitrogen to the afterglow region, undergoes desorption/ionization and is then detected by TOF-MS. Although slight fragmentation was observed for low molecular weight analytes in some cases, the degree of fragmentation was apparently independent of laser irradiance. To date, LA-FAPA imaging is still limited to small molecules (1000 Da), but the absence of matrix interference and sample preparation begets its opportunity in imaging drug compounds and their metabolites in a complex environment. Additionally, LA allows 3D imaging of analytes through depth profiling and makes this technique promising. Most recently, the first work using a new ambient air technique, named IR LA metastable-induced chemical ionization, was reported using a similar setup, but with an open-air ionization configuration [70]. Herein, an enlarged x–y sample stage enabled MSI of sizable tissue, and the possibility of deposition of ablated material in the ion source enclosure walls was eliminated. Unfortunately, no 3D imaging with this technique has been reported yet.
Instrumentation development
MALDI instrumentation development
The need to improve MSI performance has led to many recent instrumentation developments for 2D imaging that are readily applicable to 3D imaging. Bruker Daltonics has developed a proprietary kHz smartbeam-II™ MALDI laser integrated with a novel FlashDetector™ on their MALDI-TOF/TOF mass spectrometer, ultrafleXtreme™ [71]. The patented 1-kHz laser allows for focus diameters down to 10 μm for high resolution imaging with a resolving power of less than 40,000 across the peptide mass range and beyond. Furthermore, the 1000-Hz data acquisition rate for TOF and TOF/TOF operation allows for high-throughput sampling that can also be applied to high-speed 3D imaging [71]. Similarly, the Karger laboratory used a 2-kHz laser on a homebuilt MALDI-TOF instrument with an approximately 140-μm laser spot diameter and approximately 10,000 resolving power, analyzing 625 fractions of an 16O/18O differentially labeled tryptic digest from an Escherichia coli lysate [72]. Although this laser was not specifically used for MSI, its application could allow for high-throughput 3D imaging. Applied Biosystems has introduced a continuous laser rastering technique where, instead of using a conventional ‘spot-to-spot’ approach that utilizes a time-consuming ‘stop-and-go’ method for acquiring spectra, the laser rasters continuously in rows across the sample surface [73]. On the QSTAR® Elite system, the combination of oversampling and continuous rastering allowed for voxel resolutions of 50 × 50 μm without increasing acquisition time [72]. Furthermore, the Caprioli group has developed MALDI-TOF instruments that utilize the continuous scanning mode with a 5-kHz laser and show significant acquisition time improvements over conventional MALDI-TOF systems [74].
Deficiencies in the mass resolving power of TOF-MS limit its usefulness for MALDI-MSI since a loss of chemical and spatial information can occur. MALDI FT-ICR has been used to generate accurate mass measurements that provide molecular specificity for ion images of drugs and metabolites in tissue on the basis of elemental composition [75]. Structural confirmation of targeted compounds was made from accurate mass fragment ions generated in a quadrupole collision cell [75]. The ability of FT-ICR to perform tandem-MS has been utilized for the identification of peptides in the rat brain, but one downside of this technique is its low-throughput due to a lengthy acquisition time [76]. Our group has used a MALDI LTQ Orbitrap mass spectrometer for high mass resolution and high mass accuracy imaging of several neuropeptide families in a lobster brain [77]. The previously mentioned Fourier transform ion cyclotron resonance MSI experiment acquired data at a relatively slow rate (15 s/pixel) [76]. However, we acquired MALDI Orbitrap data at a rate of approximately 6.6 s/pixel, and the MALDI Orbitrap mass spectrometer can operate at even higher speeds (1.2–1.8 s/pixel) [77,78]. Ion mobility-based MSI is another technique aiming to address the low mass resolving problem in TOF-MS. In ion mobility-MS (IM-MS) imaging applications, the analyte is first ionized by MALDI and directed into the IM drift tube. The drift tube contains a neutral background drift gas that collides with ions traversing the drift cell [79]. Ions are separated according to their collisional cross-section (apparent surface area), allowing for a gas-phase separation dimension orthogonal to the MS dimension and identification of isobaric analytes extracted from tissue [79–81]. IM-MS imaging has been used to differentiate a nominally isobaric peptide and lipid deposited on a mouse liver thin tissue section, different isobaric tryptic peptides in rat brain tissue, phosphatidylcholine and cerebroside species from rat brain sections, and an anticancer drug, vinblastine, from an endogenous isobaric lipid [80,82–84]. Although no 3D MALDI-MSI studies have been performed with IM-MS at the present time, application of this technique seems likely.
SIMS instrumentation development
Although it has been previously mentioned that the advances in cluster-beam source technology have improved sensitivity, mass range and lateral resolution in biological SIMS, novel instrumentation has also improved upon SIMS technology. Cluster SIMS experiments have become routine for the characterization of complex organic samples, but commercial TOF-SIMS instruments are not equipped with MS/MS capabilities required for analysis. The Winograd laboratory used a high-performance hybrid quadrupole orthogonal design to explore the combination of cluster SIMS and MALDI [85]. The instrument is a C60 SIMS hybrid-quad-rupole orthogonal-TOF-MS capable of performing MS/MS. It has been applied to map the spatial distribution of the antibiotic gramicidin S under a copper grid with a lateral resolution of approximately 25–30 μm and is sufficiently sensitive to conduct MS/MS experiments during imaging [85]. Another major advantage of this instrument is that it uses a continuous (dc) primary ion beam rather than a pulsed beam, increasing incident ion beam current by as much as four orders of magnitude, while improving signal to noise ratio by several orders of magnitude [85].
The Vickerman group also designed an instrument that overcomes many drawbacks of TOF-SIMS (e.g., long analysis times and incompatibility of generating short pulses for high mass resolution with attaining high spatial resolution) by removing the pulsed primary ion beam [37]. Instead, a continuous supply of secondary ions is produced from a surface using a primary ion beam in dc mode. The complete mechanism behind this instrument is given in detail by Vickerman’s group, but the result is an imaging SIMS instrument with the mass detection and range of a TOF analyzer, the possibility of nanoscale imaging without sacrifice of mass resolution, mass accuracy of 5 ppm and an ultimate spot size of 200 nm delivered by the C603+ ion beam [37]. The instrumental concept has been tested through successful high mass resolution subcellular imaging in 2D and 3D, and MS/MS capability provides further structural elucidation.
Biological sample preparation
Sample preparation prior to imaging experiments is key to high-quality, reproducible and reliable results. In this section, we briefly discuss several crucial steps in sample-preparation protocol, ranging from tissue collection, storage, sectioning and pretreatment to matrix application in the case of MALDI and matrix-enhanced-SIMS (ME-SIMS). Additionally, the experimental requirements and supporting software for 3D imaging are reviewed.
General considerations for tissue collection & preparation
Tissue storage & preservation
Regardless of tissue type, integrity and morphology of the tissue should be maintained, while delocalization and degradation of bio-molecules must be avoided. The most common procedure immediately after tissue harvesting is to snap-freeze and store the tissue at −80°C until use. Another method involves loosely wrapping the tissue in aluminum foil and gently placing it into liquid nitrogen, ice-cold ethanol or isopropanol bath for 30–60 s [86]. The tissue stabilization step is recommended to minimize the sample aging effect during storage. Focusing microwave irradiation [87] and thermal transfer [88,89] can effectively denature the active proteolytic enzymes and prevent further proteome degradation. Formalin-fixed paraffin embedding is an alternative method to prevent postmortem degradation preferred by pathologists, but it is not highly compatible with direct tissue analysis by MSI [90]. Recently, ethanol-fixed paraffin-embedded tissue specimen was reported to generate high-quality MS images and be compatible with histological staining [91], promising an MSI-friendly substitute for formalin-fixed paraffin embedding.
Tissue sectioning & attachment
Typically in 2D imaging, a flat section approximately 10–20 μm thick sampled from the interior tissue is required for high-quality imaging. For ease of cryosectioning, tissues are usually embedded in a supporting media. Polymer-containing embedding media such as optimal cutting temperature, Tissue-tek and carboxy-methylcellulose often produce intense interfering background signals [86,92] and should be avoided. Ice is commonly used for imaging analytes in the low mass range since it does not contaminate MSI analysis [93]. An alternative, gelatin, has been shown to provide minimal interference peaks, while preserving the integrity of tissue sections [2,94]. Typically after cryo-sectioning, the tissue section is thaw-mounted to the MALDI target plate [86]. For histological purposes, metallic plates have been replaced by conductive indium-tin-oxide glass slides [95,96]. An alternative to the transferring/attaching approach involves the use of adhesive double-sided conductive tape to affix the tissue section to the sample plate [90]. This method has been increasingly utilized in recent years because it is amenable to the subsequent washing step.
Sample pretreatment
Washing the tissue sections prior to matrix application fixes the tissue and removes salts and lipids from tissue at the same time without delocalization of proteins and peptides [97], resulting in increased signal and ion yields. Several combinations of organic solvents such as ethanol, methanol, acetone, hexane and chloroform have been systematically explored for tissue washing [98,99]. However, the washing procedure could cause the migration of low molecular weight analytes, and it is typically not recommended for analysis of small molecules including metabolites, lipids and hydrophilic peptides.
Tissue section preparation
The following preparation steps of tissue-section analysis are tailored and grouped based on the MSI technique employed.
Tissue section preparation: MALDI
The paramount role of matrix type in MALDI-MSI necessitates the exercise of caution when selecting matrix and application methods. An appropriate matrix needs to be chosen to achieve the highest signal intensity and largest number of analyte signals with minimal interfering matrix peaks. Sinapinic acid is normally employed for large protein analysis, while α-cyano-4-hydroxycinnamic acid is most commonly applied to peptide and small protein analysis [86,100]. 2,5-dihydroxy benzoic acid (DHB) can also be used in small protein and peptide analysis [2,94]. Matrices such as 2,6-dihydroxycinnamic acid and DHB are typically used in lipid analysis [83,86,94]. Unfortunately, all of the aforementioned matrices produce clusters of adduct peaks in the low mass range, rendering metabolite and drug detection difficult. Recently, an acid–base model was proposed to circumvent the mass congestion by manipulating analyte–matrix interactions, and a class of highly basic matrices, including 9-aminoacridine and 8-bis(dimethyl-amino) naphthalene, demonstrated efficient detection of acidic metabolites in the negative mode with negligible matrix background [12,101,102]. This finding increases the possibility of developing more effective matrices, such as a binary matrix of acidic α-cyano-4-hydroxycinnamic acid and basic 9-aminoacridine reported to effectively suppress matrix cluster formation in both positive and negative modes [103]. Furthermore, ionic matrices have demonstrated improved spectral quality, crystallization and vacuum stability, and they have become increasingly utilized in the MSI field [104–107].
The desire to obtain high-resolution images of biomolecules at the cellular scale from biological tissues can be impeded by large analyte-doped matrix crystals. Gold nanoparticles serve as a potential substitute for conventional matrices in high-resolution imaging and have been successfully applied for mapping lipids in rat brain [83].
Although matrix selection is important, matrix application method selection is critical as well. In fact, a recent comparison of matrix coating methods for neuronal tissue preparation showed that while dry matrix coating favored the detection of lipids, regular matrix coating with aqueous solvent enabled extraction and imaging of neuropeptides from brain tissue section [94]. Matrix is normally deposited either as a homogeneous layer (spray coating) or as discrete spots (microspotting). For spray coating, pneumatic sprays (pneumatic sprayer, airbrush or TLC sprayer) and vibrational sprays (ImagePrep Device from Bruker Daltonics) can be used to apply a uniform matrix layer consisting of small droplets [2,94,95,108]. Manual sprayers usually produce good tissue coverage and a homogenous matrix layer with small crystals, but large variability and possible analyte migration hinders this device. An automatic device, such as ImagePrep, is believed to improve the reproducibility of manual spraying. A mist containing smaller droplets can be created with electrospray deposition for performing high-resolution imaging experiments [109]. Microspotters deposit pl-sized droplets of matrix according to a predefined array, and multiple rounds of spotting are usually required for enough matrix coverage. Different automatic spotting devices have been developed, such as the desktop inkjet printer, CHIP, produced by Shimadzu (equipped with inkjet-style piezonozzles), and the acoustic reagent multispotter [110–112]. Other solvent free methods such as sublimation and dry coating eliminate the chance of analyte diffusion induced by wet application and are deemed useful for lipid imaging. However, these methods yield relatively low sensitivities for MSI analyses of other analytes because of limited analyte–matrix interactions [113,114]. A histology-compatible preparation method enables simultaneous tissue fixation and matrix deposition with a high lateral resolution [115].
Tissue section preparation: SIMS
The advantage of SIMS over MALDI is that only drying is necessary after tissue attachment. For 3D cellular SIMS-MSI, the protocol has been successfully performed by simply plunging freezing cells in liquid propane, lyophilizing for dryness and freeze fracturing for histological corrections (optional). Small molecules, such as steroids, lipids and elements, have been mapped by 3D SIMS-MSI [35,116–118]. Surface modification approaches, such as metal-assisted SIMS and ME-SIMS, aid in secondary ion yields of large molecules but have only been applied to 2D MSI [105,109]. In metal-assisted SIMS, a very thin layer (~1 nm) of a metal (e.g., gold or silver) is deposited on the sample surface by a sputter coater. In ME-SIMS, electrospray deposition is commonly used for matrix application to ensure crystal size remains consistent with the high-resolution-driven technique.
Tissue section preparation: DESI, LAESI & LA-FAPA
Desorption ESI is an ambient ionization technique, featuring minimal sample preparation and soft ionization. DESI MSI experiments can be performed after thaw-mounting the tissue section (typically 4–10 μm thick) onto an indium-tin-oxide glass slide [51,119,120]. The DESI source’s pre-optimized geometry and other operation conditions ensure good and reproducible performance. Two other ambient sources, LAESI and LA-FAPA, also do not require involved sample preparation [5,27,28].
3D volume reconstruction & processing software
Three dimensional images can be constructed from a stack of 2D images and provide additional information approximately the distribution of molecules along the z-axis registered by MSI of the entire sample. As shown in Figure 1B, serial cutting and depth profiling are employed to perform 3D volumetric reconstruction. In serial cutting MSI, the whole sample is cut into serial sections that later undergo the identical processing method under the same conditions. After acquiring 2D MSI, post-acquisition processing software, such as BioMap [201], MATLAB, Origin, Lispix (a National Institute of Standards and Technology written software) and propriety programs for MSI systems (e.g., FlexImaging from Bruker Daltonics and TissueView from Applied Biosystems/MDS) can be used to tailor 2D images to enhance visualization of discrete anatomical structures. 3D modeling software tools, such as Amira, Image J [202] and 3D-Doctor (Able), can be employed to assemble the 2D images in a 3D context to construct the 3D model. The correction and registration of a 3D model can be accomplished by defining the physical dimensions of each 2D image in software and calibrating each of the 2D images using its pixel size and x, y coordinates with optical images since each spot corresponds to one pixel in MSI images [18]. Depth profiling involves using a desorbing agent such as a primary ion or laser beam to remove the superficial material from the sample’s surface before/while recording the imaging data of tissue surface. SIMS, LAESI and LA-FAPA have been employed to construct 3D images by depth profiling. The 3D images, similarly, are produced by assembling a stack of 2D images generated by each layer of the depth profile from the imaged sample. MATLAB, Origin and VoxBlast (VayTek, Inc. Fairfield, IA, USA) have been utilized in depth profiling MSI to plot the volumetric 3D cubes for individual peaks or groups of peaks.
3D MSI applications
3D single cell imaging
The ability to extract localized, chemical data from single cells without the use of tags is essential to understanding and monitoring cellular processes. MS imaging is a technique that can be applied to extract the native chemistry from single cells without using tags. Although advancements in MALDI-MSI such as microscope mode and oversampling allow for a lateral resolution of 4 and 25 μm, respectively, few 2D and no 3D single-cell imaging publications utilizing MALDI-MSI exist [121,122]. Neuropeptide distribution in cultured bag cell neurons has been analyzed with MALDI-MSI, and cholesterol population in astrocytes, surface metabolites of Arabidopsis thaliana and secondary metabolites of A. thaliana and Hypericum species have all been imaged at the single cell level with LDI-MSI [123–126]. Furthermore, LAESI has been applied to acquire metabolite profiles in single cells by pulsing a mid-IR laser through the etched tip of a GeO2-based glass fiber [68].
Secondary ion MS is the primary MSI technique for 3D imaging of single cells. In order to study the chemical composition of specialized subcellular regions hidden beneath the surface of mitotic cells, Chandra performed 3D SIMS on human glioblastoma T98G cells grown on silicon chips and cryogenically prepared with a sandwich freeze-fracture technique [127]. In this study, a dynamic SIMS primary ion beam was rastered across a 250-μm region to image 12C, 23Na, 24Mg, 39K and 40Ca [127]. Mass images were acquired at increasing depths, and the 3D 40Ca distribution in dividing cells was studied.
The Vickerman laboratory demonstrated the first use of a TOF-SIMS instrument with a C60 primary ion source to depth profile freeze-dried Xenopus laevis oocytes. Subsequent imaging of the revealed subsurfaces allowed for generation of 3D molecular images of cholesterol and lipids from m/z 540–570 and 800–1000 in positive ion mode. Negative ion mode yielded images of lipid fatty acid side chain distributions [35]. The X. laevis oocyte is a common cell model and is a simple model to use due to its large size (0.8–1.3 mm diameter) and resistance to osmotic changes [128]. However, 3D imaging with SIMS was logically found to be much more time consuming than 2D. Furthermore, changes in topography during analysis proved to be complicating, but proof-of-principle was still demonstrated for 3D imaging with C60+ ions [35].
As stated before, cluster ion LMIGs can be coupled with C60+ ion sources to combine the fine focus of an LMIG with the C60+ primary ion beam’s ability to etch away damaged surface. A study by Breitenstein et al. used this dual-beam setup to determine the chemical composition of the surface with Bi3+ primary ions while C60+ ions intermittently sputtered away damaged surface [116]. Using this setup, normal rat kidney cells grown on cover slips were analyzed by TOF-SIMS 3D microarea analysis. Lateral and vertical resolution in this experiment were found to be 350 and 100 nm, respectively, detecting intact molecules up to a m/z ratio of 800 [116]. Another publication by Nygren et al. analyzed the 3D distribution of phosphocholine, sodium and potassium ions in thyroid tumor cells using the Bi3+ and C60+ dual-beam setup [129]. The data obtained indicated that potassium ions are compartmentalized in thyroid tumor cells, and the lateral resolution in this study was found to be 300 nm.
A disadvantage of TOF-SIMS in imaging is very short (ns) primary ion beam pulses are required for high mass resolution, but these short pulses are incompatible with obtaining high spatial resolution images used in the 3D studies for single cell imaging above. The low duty cycle of the pulsed ion beam also means that imaging and depth-profiling experiments can take a long time to complete. According to a publication by the Vickerman group, a 3D SIMS imaging analysis with a primary ion beam dose density of 1 × 1013 ions cm−2 (static limit) and an accumulated ion dose of 1 × 1015 ions cm−2 from a 100 × 100 μm2 area would require over a month. Further specifications include using a focused ion beam providing 10 pA of target current using 50-ns primary ion pulses on an instrument running at 10 kHz [37]. As mentioned in the instrumentation developments section, the Vickerman group developed a TOF-SIMS instrument where the MS is decoupled from the sputtering event so that dc beams can be used without any loss in mass resolution, reducing the 3D imaging time to 30 min [37]. The instrument was used to construct 3D molecular images of lipids in benign prostatic hyperplasia cells, HeLa cells and human cheek cells. High mass resolution and high spatial imaging resolution were demonstrated in three dimensions [37].
Drug & pharmaceutical analysis
An important role for MSI is the localization of small molecular mass pharmaceutical compounds in tissue. Several techniques have been developed to image pharmaceutical compounds laterally in biological tissue with MALDI-MSI [7,130–133]. MALDI-MSI has also been applied to imaging drugs in whole body sections of animals dosed orally and intravenously and in rat lung sections through inhalation [93,134,135]. Another utility for MALDI-MSI is the direct analysis of pharmaceutical tablets to assess the homogeneity of active drug compounds [136]. DESI is an ambient MSI technique that has been used to localize clozapine and its N-desmethyl metabolite in histological sections of brain, lung, kidney and testis [120]. A recently introduced matrix-free desorption/ionization technique, nanostructure initiator MS has imaged clozapine and its N-desmethyl metabolite in rat brain tissue with greater sensitivity than MALDI as no matrix is necessary with nanostructure initiator MS [137].
Although the studies above highlight 2D MSI studies of pharmaceuticals in tissue, 3D TOF-SIMS has been applied to analyze pharmaceutical distributions in a coronary stent. The 3D spatial distribution of pharmaceutical molecules in a coronary stent coating was analyzed in order to visualize drug distribution according to elution time of the stent [138]. The TOF-SIMS instrument analyzed the 3D localization of sirolimus in poly(lactic-co-glycolic acid) matrix as a function of elution time with an Au+ LMIG coupled to a C60+ ion beam. The examined stent contained large portions of the surface and subsurface channels composed primarily of sirolimus, followed by a drug-depleted region, and then relatively homogeneous sirolimus dispersion in the polymer matrix. In order to determine pharmaceutical elution from the stent, 3D chemical distributions were analyzed with TOF-SIMS by characterizing stents at 0 h, 1 h and 1 day. Elution was found to occur on the drug-enriched surface region much quicker compared with the gradual elution in the subsurface regions, revealing that much of the drug (~55%) had eluted in the first day [138]. Besides SIMS, another depth profiling technique, LA-FAPA has also been used to image pharmaceuticals, caffeine and acetaminophen, and a substance in the protective coating layer from a tablet [5]. A great improvement in resolution, 20 μm laterally and 40 μm vertically, compared with previous FAPA configuration was achieved with high sensitivity (LOD of 5 fmol for caffeine) and at similar speed as MALDI-MSI [5].
Metabolites & lipids
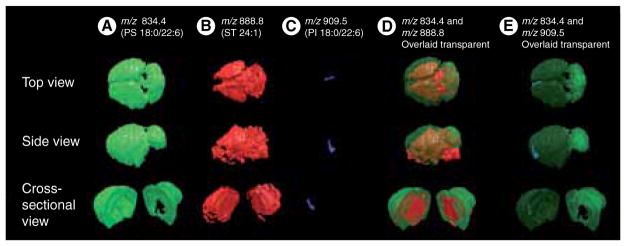
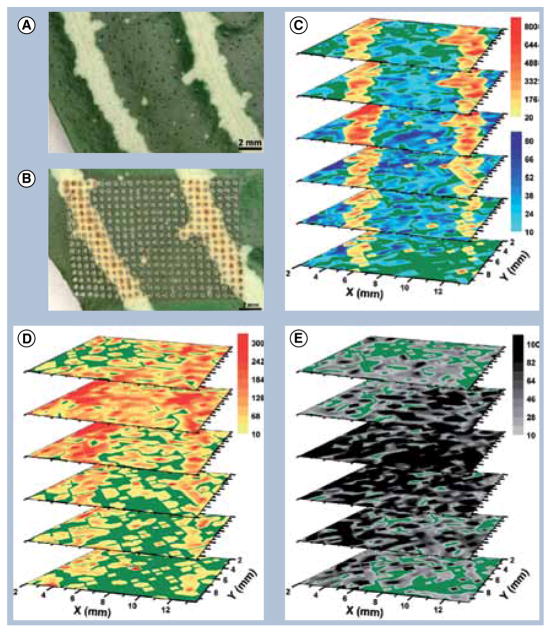
Metabolomics and lipidomics are two emerging fields involving the study of small-molecule metabolite profiles and the full complement of cellular lipids in biological systems. 3D MSI techniques have been employed in both areas by providing the 3D spatial distributions of these important molecules in biological tissues. Although many 3D experiments analyzing small molecules are performed with SIMS techniques, MALDI and DESI have imaged lipids in three dimensions, whereas LAESI has been used for the 3D MSI analysis of metabolites. In a 3D MALDI-MSI experiment performed by our laboratory, Cancer borealis brain tissue was used to investigate the different localization patterns of the two chemically similar phosphatidylcholine and sphingomyelin lipid classes [94]. While phosphatidylcholine was primarily distributed in the major lobes of the brain, sphingomyelin had higher abundance in the fibers, suggesting different functions in the nervous system [94]. DESI 3D MSI techniques have been employed to analyze deprotonated free fatty acids, phosphatidylserines, phosphatidylinositols and sulfatides in mouse brain tissue [1]. 2D ion images showing distributions of lipids phosphatidylserines 18:0/22:6, sulfatides 24:1 and phosphatidylinositols 18:0/22:6 were overlaid to construct the 3D model as shown in Figure 2, compromising image quality in favor of time by selecting 36 tissue sections out of 500 [1]. Nevertheless, this methodology provided the 3D distribution of lipids of interest. The Vertes group presented the first example of 3D MSI at AP through 3D LAESI imaging by combining lateral imaging with LAESI’s potential for molecular depth profiling [4]. LAESI 3D imaging mapped spatial variations in metabolites found in A. squarrosa plant leaves and assigned by accurate mass measurements, isotope distribution analysis, and collisional activated dissociation experiments coupled with plant metabolomic database searches [4]. Figure 3 shows the LAESI 3D images acquired from imaging the distribution of kaempferol/luteolin, chlorophyll a, acacetin and kaempferol (diacetyl coumarylrhamnoside).
Figure 2. 3D models of mouse brain created with desorption ESI-mass spectral imaging.
3D constructions show the distributions of
(A) phosphatidylserines (PS) 18:0/22:6 in green, (B) sulfatides 24:1 in red, and (C) phosphatidylinositols 18:0/22:6 in blue. Overlaid distributions are shown for lipids (D) PS 18:0/22:6 and sulfatides 24:1, and (E) PS 18:0/22:6 and phosphatidylinositols 18:0/22:6.
Reprinted with permission from [1].
Figure 3. Metabolites mapped using laser ablation ESI 3D mass spectral imaging compared with tissue structure.
Optical images of Aphelandra squarrosa leaves (A) before and (B) after analysis. (C) stacked 2D laser ablation ESI-MS images of kaempferol/uteolin, (D) acacetin and (E) kaempferol-(diacetyl coumarylrhamnoside).
Reprinted with permission from [4].
The prospects of using SIMS for 3D biomolecular analysis of biological models including single biological cells improved substantially with the demonstration of C60+ primary ions. In a publication by the Winograd group, a TOF-SIMS instrument equipped with a C60+ ion source eroded membrane lipids with minimal chemical damage to the sample surface [139]. Furthermore, C60+ ions uncovered underlying biomolecules from lipid films with no significant loss of molecular profile data. As mentioned above, many SIMS 3D studies of lipids are performed at the single cell level because of its high lateral resolution and ability to ionize small molecules [35,37,116]. One 3D TOF-SIMS experiment analyzed the distribution of phosphocholine, sodium, and potassium ion in thyroid tumor cells [129]. The instrument utilized a dual beam, performing analysis with a Bi3+ LMIG after sequential sputtering with C60+ ions. A study by Jones et al. depth profiled rat brain tissue with a C60+ primary ion beam in order to perform 3D studies [117]. Results suggested that an ammonium formate wash be used to reduce salt levels within the tissue in order to better probe its chemistry. Depth profiling also showed that at room temperature under vacuum conditions, lipids migrate to the surface of the tissue. However, when the sample is in a frozen state, molecular redistribution is limited, shown by the constant distribution of the mobile molecule cholesterol [117].
Peptides & proteins
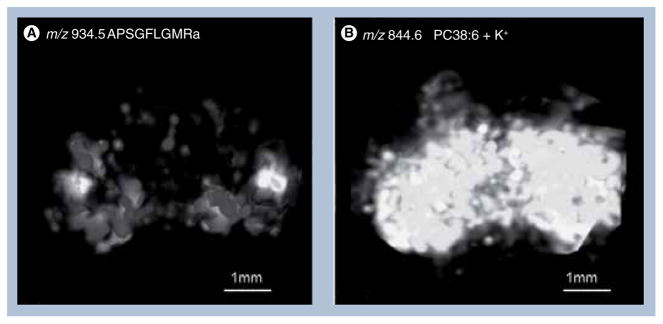
Matrix-assisted laser desorption/ionization has demonstrated its unparalleled ability in desorbing/ionizing relatively large biomolecules such as peptides and proteins without degradation. Therefore, it is the primary choice to satisfy the need of 3D imaging in the fields of peptidomics and proteomics. Using the common mouse brain model, Crecelius et al. reported the 3D visualization of myelin basic protein in the corpus callosum region and believed that reliable 3D mapping of any protein visible in ion images can be acquired by correlation with anatomical features provided by optical micrographs [92]. The alignment of MALDI-MSI with histological studies might lend insight into the underlying proteomic mechanism of disease [96], based on common observations that protein expression changes in specific regions can be correlated to certain diseases such as cancer or Alzheimer’s disease. A step-by-step protocol is well summarized by the Caprioli group, describing how to make 3D volume reconstructions out of 2D MALDI-MSI images [18]. In that review, substance P and PEP-19 in the rat ventral midbrain were mapped, demonstrating the feasibility of 3D peptide imaging. Crustacean brain has also been examined for the volumetric distribution of various neuropeptides as previously reported by our group [94]. By using seven sequential sections with an equal distance of 132 μm on the z-axis of the brain, over 20 neuropeptides belonging to eight families were mapped throughout 3D structures. A representative reconstructed 3D image of the neuropeptide C. borealis tachykinin-related peptide (CabTRP) 1a is shown in Figure 4. Moreover, an example of differentiating the 3D distribution of RFamide SMPSLRLRFa from other RFamide peptide isoforms demonstrated the advantage of 3D over 2D MSI: 3D imaging reveals more in-depth and detailed information regarding distributions of various peptides at different layers, which often would be missed in 2D imaging experiments.
Figure 4. 3D MSI of neuropeptides and lipids in crustacean brain.
(A) 3D reconstructed ion image of CabTRP 1a by MALDI mass spectral imaging prepared using regular matrix coating.
(B) 3D reconstructed ion image of lipid phosphatidylcholine 38:6 by MALDI mass spectral imaging prepared using dry matrix spraying technique for lipid detection.
Reprinted with permission from [2].
Topography corrections
The combination of different imaging techniques in neuroimaging with the goal of understanding brain functions in normal and diseased brain states is very powerful [140,141]. SIMS imaging has been coupled with various spectroscopy techniques for multi-modal imaging [92,142,143]. However, as previously mentioned, MALDI-MSI requires tissue to be sliced into sections, sometimes producing tissue tearing and deformation while creating difficulties in constructing a 3D visualization. A study by Caprioli’s laboratory helps develop a 3D visualization of proteomic data correlated to anatomical features in the brain [92]. Briefly, before MALDI-MSI analysis, a glass slide with a mouse brain section was thawed, and four black ink dots were attached to the slide framing the slice to use as a registration landmark. An optical image of this section and the surrounding landmarks was acquired. Next, MALDI matrix was sprayed uniformly over the entire mouse brain section, and mass spectra were acquired on a MALDI-TOF Voyager DE-STR mass spectrometer [92]. After MALDI-MSI imaging, tissue sections were Nissl stained after matrix removal for histological imaging. Data processing involved inter- and intra-section registration. Inter-section registration involved registering optical images to each other using a reference atlas. Intra-section registration mapped a MALDI-MSI image to its respective optical image based on landmarks visible in both images, including the black ink spots [92]. The 3D reconstruction of myelin basic protein in the corpus callosum of the mouse brain was carried out using this methodology. A registration error on the order of 30 μm and an additional error of 60 μm was caused by a height difference between the mouse brain section and the paper holding the toner spots [92]. Another study by the Caprioli group optimized the previous protocol to increase the number of molecular species analyzed [18]. Two sets of sections were collected, covering one set of sections with sinapinic acid for optimal protein detection and adjacent sections with DHB matrix for optimal low-mass species detection [18]. Once again, pixelated MSI images were exported and registered to the corresponding optical images. Approximately 1000 peaks could be observed with each dataset using this methodology. In yet another Caprioli study, 3D MALDI-MSI data was integrated with in vivo magnetic resonance imaging. 3D MALDI-MSI was extended to whole-animal tissue sections, and proteomic data was coupled with in vivo MRI [144]. Before MALDI-MSI analysis, high-resolution MRI of a tumor-laden mouse brain (longitudinal relaxation time [T1], transverse relaxation time [T2], and apparent diffusion coefficient) were produced. After MRI, each animal was sacrificed and prepared for blockface imaging acquisition. Tissue sections from the blockface volume were prepared for MALDI-MSI acquisition and analyzed on a MALDI TOF mass spectrometer (Autoflex II, Bruker Daltonics) [144]. The block-face volume was then co-registered to the magnetic resonance volume of the mouse’s head, and alignment of the 3D MALDI-MSI data with the blockface implicitly aligns the MSI data to the magnetic resonance volume. Results indicated a proof of principle for correlating postmortem proteomic data with in vivo anatomical imaging while also showing that proteomic features aligned well with corresponding regions in the MRI data [144].
Conclusion & future perspective
Mass spectral imaging is strongly poised to be a revolutionary tool to map thousands of molecules simultaneously by molecular weight ranging from proteins and peptides to metabolites and pharmaceutical compounds. 3D MSI improves upon this technique, enabling a detailed, volumetric visualization of analytes in the whole sample.
Since 3D MSI represents one of the newest additions to the MSI family, many developments and improvements involved in imaging acquisition speed, image resolution and data processing are occurring and promise ever-growing applications in biological sciences. The comparisons of 3D MS images and analyte profiles serve as a powerful discovery tool for pathologists to easily identify distinct ‘molecular signatures’ between healthy and diseased tissues without a priori knowledge. Furthermore, this unique 3D view can be cross-referenced to other target-specific imaging methods, such as immunohistochemistry. The addition of a third dimension to the previous 2D spatial mapping and imaging MSI methods increases the need to improve MSI’s throughput in the areas of sample preparation, data acquisition and MSI data file transfer, conversion and storage. Advanced computational tools and bioinformatics approaches will need to be developed to deal with the ever-increasing raw data file size from 3D-MSI experiments as well as to extract biologically relevant information from these large collections of MS spectra.
Three dimensional MSI is also of tremendous interest to the pharmaceutical industry by accurately assessing the distribution of a drug candidate, its metabolites and the endogenous molecules responsive to the drug based on specific mass information. Moreover, its correlation to physiological and structural information obtained from in vivo imaging techniques, such as MRI, has significant implications in functional proteomics. MSI had been viewed as an invasive imaging technique until ambient desorption and ionization methods including DESI and direct analysis in real time were developed. Ambient ion sources circumvent limitations imposed by vacuum operational conditions, and they are expected to induce the migration of the 3D MSI field from ex vivo to in vivo analyses of living systems, hopefully bringing 3D MSI to our everyday lives in the near future.
Executive summary.
Ionization techniques
MALDI-mass spectral imaging (MSI) allows for the mass spectral analysis and spatial mapping of compounds spanning a wide m/z range but suffers from matrix interference and its reliance on operating in vacuum.
Secondary ion MS (SIMS)-MSI has excellent lateral resolution in the sub-micrometre range but has a limited m/z range and its reliance on operating in vacuum.
Desorption ESI (DESI)-MSI is a soft, ambient ionization technique but also suffers from a limited m/z range and relatively poor spatial resolution as compared with MALDI-MSI.
Laser ablation ESI (LAESI)-MSI is an ambient ionization technique capable of depth profiling but requires a water-rich sample to analyze and has a limited m/z range.
LA-flowing atmospheric-pressure afterglow (FAPA) is an ambient ionization technique capable of depth profiling at relatively high resolution but has a limited m/z range.
Instrumentation developments
The development of high-frequency lasers and continuous rastering mode allow for high-throughput imaging.
Ion mobility MS imaging experiments add a dimension of separation useful for distinguishing between isobaric species.
SIMS developments include MS/MS instruments and dc beams to increase incident ion beam current and improve S/N.
Biological sample preparation
Matrix-based methods, MALDI, Meta-SIMS and ME-SIMS, involve coating matrix on sectioned tissue.
Matrix-free methods, SIMS, DESI, LAESI and LA-FAPA, require few preparative steps following tissue sectioning.
3D MSI applications
SIMS-MSI is currently the preferred method for single cell 3D imaging because of its superior lateral resolution compared with their methods.
Publications regarding 3D analysis of pharmaceuticals area are scarce and the area has great potential for expansion.
SIMS, DESI and LAESI are well suited for 3D lipid and metabolite analysis in a variety of tissue types.
MALDI-MSI are so far the best-recognized method for 3D visualization of peptides and proteins.
Topography corrections
A method correlating 3D proteomic data to anatomical features in the brain has been developed.
Optical images need to be registered to one another and then MALDI-MSI data can be registered to this optical image.
This methodology has also been used to integrate 3D MALDI-MSI data from whole-animal tissue sections with in vivo MRI data.
Key Terms
- Immunohistochemical
Process of localizing analytes (typically peptides or proteins known as antigens) in biological tissue through antibody-specific recognition
- Mass spectral imaging
Technique used in MS to visualize the spatial distribution of various compounds in a biological specimen by their molecular masses
- Microprobe mode
Utility of a highly focused laser or ion beam to construct spatially resolved images in mass spectral imaging
- Microscope mode
Utility of a defocused laser or ion beam coupled to a spatially resolving detector to conduct mass spectral imaging
- Voxel
Volumetric pixel
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure
Preparation of this manuscript was supported in part by National Science Foundation (CHE- 0957784), NIH through grant 1R01DK071801. Lingjun Li acknowledges an Alfred P Sloan Research Fellowship and a Vilas Associate Award. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1▪.Eberlin LS, Ifa DR, Wu C, Cooks RG. Three-dimensional vizualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angew Chem Int Ed. 2005;49(5):873–876. doi: 10.1002/anie.200906283. Good presentation of 3D model of mouse brain constructed by ambient mass spectra imaging (MSI) of lipids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li L. Imaging mass spectrometry of neuropeptides in decapod crustacean neuronal tissues. J Proteome Res. 2007;6(5):1782–1791. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd PJ, Schaaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J Mass Spectrom. 2001;36(4):355–369. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- 4▪.Nemes P, Barton AA, Vertes A. Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization-mass spectrometry. Anal Chem. 2009;81(16):6668–6675. doi: 10.1021/ac900745e. First publication verifying laser ablation ESI’s ability to image metabolites at ambient conditions in three dimensions. [DOI] [PubMed] [Google Scholar]
- 5.Shelley JT, Ray SJ, Hieftje GM. Laser ablation coupled to a flowing atmospheric pressure afterglow for ambient mass spectral imaging. Anal Chem. 2008;80(21):8308–8313. doi: 10.1021/ac801594u. [DOI] [PubMed] [Google Scholar]
- 6.Karas M, Bachmann D, Bahr U, Hillenkamp F. Matrix-assisted ultraviolet-laser desorption of nonvolatile compounds. Int J Mass Spectrom. 1987;78:53–68. [Google Scholar]
- 7.Reyzer ML, Hsieh YS, Ng K, Korfmacher WA, Caprioli RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 2003;38(10):1081–1092. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 8.Cohen LH, Gusev AI. Small molecule analysis by MALDI mass spectrometry. Anal Bioanal Chem. 2002;373(7):571–586. doi: 10.1007/s00216-002-1321-z. [DOI] [PubMed] [Google Scholar]
- 9.Svatos A. Mass spectrometric imaging of small molecules. Trends Biotechnol. 2010;28(8):425–434. doi: 10.1016/j.tibtech.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Schriemer DC, Li L. Detection of high molecular weight narrow polydisperse polymers up to 1.5 million daltons by MALDI mass spectrometry. Anal Chem. 1996;68(17):2721–2725. doi: 10.1021/ac960442m. [DOI] [PubMed] [Google Scholar]
- 11.Woods AS, Jackson SN. Brain tissue lipidomics: direct probing using matrix-assisted laser desorption/ionization mass spectrometry. AAPS J. 2006;8(2):E391–E395. doi: 10.1007/BF02854910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shroff R, Rulisek L, Doubsky J, Svatos A. Acid-base-driven matrix-assisted mass spectrometry for targeted metabolomics. Proc Natl Acad Sci USA. 2009;106(25):10092–10096. doi: 10.1073/pnas.0900914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhigilei LV, Garrison BJ. Microscopic mechanisms of laser ablation of organic solids in the thermal and stress confinement irradiation regimes. J Appl Phys. 2000;88(3):1281–1298. [Google Scholar]
- 14.Knochenmuss R, Zenobi R. MALDI ionization. The role of in-plume processes. Chem Rev. 2003;103(2):441–452. doi: 10.1021/cr0103773. [DOI] [PubMed] [Google Scholar]
- 15.Zenobi R, Knochenmuss R. Ion formation in MALDI mass spectrometry. Mass Spectrom Rev. 1998;17(5):337–366. [Google Scholar]
- 16.Karas M, Gluckmann M, Schafer J. Ionization in matrix-assisted laser desorption/ionization: singly charged molecular ions are the lucky survivors. J Mass Spectrom. 2000;35(1):1–12. doi: 10.1002/(SICI)1096-9888(200001)35:1<1::AID-JMS904>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 17.Caprioli R, Farmer T, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 18▪▪.Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Methods. 2008;5(1):101–108. doi: 10.1038/nmeth1145. Practical and comprehensive review of conducting 3D MALDI-MSI of peptides and proteins in tissues. [DOI] [PubMed] [Google Scholar]
- 19.Chaurand P, Schwartz SA, Caprioli RM. Imaging mass spectrometry: a new tool to investigate the spatial organization of peptides and proteins in mammalian tissue sections. Curr Opin Chem Biol. 2002;6(5):676–681. doi: 10.1016/s1367-5931(02)00370-8. [DOI] [PubMed] [Google Scholar]
- 20.Chughtai K, Heeren R. Mass spectrometric imaging for biomedical tissue analysis. Chem Rev. 2010;110(5):3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaurand P, Schriver KE, Caprioli RM. Instrument design and characterization for high resolution MALDI-MS imaging of tissue sections. J Mass Spectrom. 2007;42(4):476–489. doi: 10.1002/jms.1180. [DOI] [PubMed] [Google Scholar]
- 22.Koestler M, Kirsch D, Hester A, Leisner A, Guenther S, Spengler B. A high-resolution scanning microprobe matrix-assisted laser desorption/ionization ion source for imaging analysis on an ion trap/fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun Mass Spectrom. 2008;22(20):3275–3285. doi: 10.1002/rcm.3733. [DOI] [PubMed] [Google Scholar]
- 23.Rompp A, Guenther S, Schober Y, et al. Histology by mass spectrometry: label-free tissue characterization obtained from high-accuracy bioanalytical imaging. Angew Chem Int Ed. 2010;49(22):3834–3838. doi: 10.1002/anie.200905559. [DOI] [PubMed] [Google Scholar]
- 24.Spengler B, Hubert M. Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J Am Soc Mass Spectrom. 2002;13(6):735–748. doi: 10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- 25.Luxembourg SI, Mize TH, Mcdonnell LA, Heeren RMA. High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal Chem. 2004;76(18):5339–5344. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- 26.Klerk LA, Altelaar AFM, Froesch M, Mcdonnell LA, Heeren RMA. Fast and automated large-area imaging MALDI mass spectrometry in microprobe and microscope mode. Int J Mass Spectrom. 2009;285(1–2):19–25. [Google Scholar]
- 27.Li Y, Shrestha B, Vertes A. Atmospheric pressure infrared MALDI imaging mass spectrometry for plant metabolomics. Anal Chem. 2008;80(2):407–420. doi: 10.1021/ac701703f. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Shrestha B, Vertes A. Atmospheric pressure molecular imaging by infrared MALDI mass spectrometry. Anal Chem. 2007;79(2):523–532. doi: 10.1021/ac061577n. [DOI] [PubMed] [Google Scholar]
- 29.Fletcher JS, Lockyer NP, Vickerman JC. Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom Rev. 2010;30(1):142–174. doi: 10.1002/mas.20275. [DOI] [PubMed] [Google Scholar]
- 30.Sigmund P. Theory of sputtering. I Sputtering yield of amorphous and polycrystalline targets. Phys Rev. 1969;184(2):383–416. [Google Scholar]
- 31.Lehmann C, Sigmund P. On the mechanism of sputtering. Physica Stat Solidi (b) 1966;16(2):507–511. [Google Scholar]
- 32.Le Beyec Y. Cluster impacts at keV and MeV energies: secondary emission phenomena. Int J Mass Spectrom. 1998;174(1–3):101–117. [Google Scholar]
- 33.Cheng J, Wucher A, Winograd N. Molecular depth profiling with cluster ion beams. J Phys Chem B. 2006;110(16):8329–8336. doi: 10.1021/jp0573341. [DOI] [PubMed] [Google Scholar]
- 34.Cheng J, Kozole J, Hengstebeck R, Winograd N. Direct comparison of Au3+ and C60+ cluster projectiles in SIMS molecular depth profiling. J Am Soc Mass Spectrom. 2007;18(3):406–412. doi: 10.1016/j.jasms.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. TOF-SIMS 3D biomolecular imaging of xenopus laevis oocytes using buckminsterfullerene (C60) primary ions. Anal Chem. 2007;79(6):2199–2206. doi: 10.1021/ac061370u. [DOI] [PubMed] [Google Scholar]
- 36.Levisetti R, Hallegot P, Girod C, et al. Critical issues in the application of a gallium probe to high-resolution secondary ion imaging. Surf Sci. 1991;246(1–3):94–106. [Google Scholar]
- 37.Fletcher JS, Rabbani S, Henderson A, et al. A new dynamic in mass spectral imaging of single biological cells. Anal Chem. 2008;80(23):9058–9064. doi: 10.1021/ac8015278. [DOI] [PubMed] [Google Scholar]
- 38.Davies N, Weibel DE, Blenkinsopp P, Lockyer N, Hill R, Vickerman JC. Development and experimental application of a gold liquid metal ion source. Appl Surf Sci. 2003;203:223–227. [Google Scholar]
- 39.Wong SCC, Hill R, Blenkinsopp P, Lockyer NP, Weibel DE, Vickerman JC. Development of a C60+ ion gun for static SIMS and chemical imaging. Appl Surf Sci. 2003;203:219–222. [Google Scholar]
- 40.Weibel D, Wong S, Lockyer N, Blenkinsopp P, Hill R, Vickerman JC. A C60 primary ion beam system for time of flight secondary ion mass spectrometry: Its development and secondary ion yield characteristics. Anal Chem. 2003;75(7):1754–1764. doi: 10.1021/ac026338o. [DOI] [PubMed] [Google Scholar]
- 41.Kotter F, Benninghoven A. Secondary ion emission from polymer surfaces under Ar+, Xe+ and SF5+ ion bombardment. Appl Surf Sci. 1998;133(1–2):47–57. [Google Scholar]
- 42.Gillen G, Roberson S. Preliminary evaluation of an SF5+ polyatomic primary ion beam for analysis of organic thin films by secondary ion mass spectrometry. Rapid Commun Mass Spectrom. 1998;12(19):1303–1312. doi: 10.1002/(SICI)1097-0231(19981015)12:19<1303::AID-RCM330>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 43.Benguerba M, Brunelle A, Dellanegra S, et al. Impact of slow gold clusters on various solids – nonlinear effects in secondary ion emission. Nucl Instrum Methods Phys Res Sect B. 1991;62(1):8–22. [Google Scholar]
- 44.Touboul D, Halgand F, Brunelle A, et al. Tissue molecular ion imaging by gold cluster ion bombardment. Anal Chem. 2004;76(6):1550–1559. doi: 10.1021/ac035243z. [DOI] [PubMed] [Google Scholar]
- 45.Sjovall P, Lausmaa J, Johansson B. Mass spectrometric imaging of lipids in brain tissue. Anal Chem. 2004;76(15):4271–4278. doi: 10.1021/ac049389p. [DOI] [PubMed] [Google Scholar]
- 46.Touboul D, Kollmer F, Niehuis E, Brunelle A, Laprevote O. Improvement of biological time-of-flight-secondary ion mass spectrometry imaging with a bismuth cluster ion source. J Am Soc Mass Spectrom. 2005;16(10):1608–1618. doi: 10.1016/j.jasms.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Jones EA, Lockyer NP, Vickerman JC. Mass spectral analysis and imaging of tissue by TOF-SIMS – the role of buckminsterfullerene, C60+, primary ions. Int J Mass Spectrom. 2007;260(2–3):146–157. [Google Scholar]
- 48.Mahoney CM, Roberson SV, Gillen G. Depth profiling of 4-acetamindophenol-doped poly(lactic acid) films using cluster secondary ion mass spectrometry. Anal Chem. 2004;76(11):3199–3207. doi: 10.1021/ac035532n. [DOI] [PubMed] [Google Scholar]
- 49.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79(21):8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 50.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 51.Ifa DR, Wiseman JM, Song QY, Cooks RG. Development of capabilities for imaging mass spectrometry under ambient conditions with desorption electrospray ionization (DESI) Int J Mass Spectrom. 2007;259(1–3):8–15. [Google Scholar]
- 52.Ifa DR, Manicke NE, Dill AL, Cooks RG. Latent fingerprint chemical imaging by mass spectrometry. Science. 2008;321(5890):805. doi: 10.1126/science.1157199. [DOI] [PubMed] [Google Scholar]
- 53.Ifa DR, Gumaelius LM, Eberlin LS, Manicke NE, Cooks RG. Forensic analysis of inks by imaging desorption electrospray ionization (DESI) mass spectrometry. Analyst. 2007;132(5):461–467. doi: 10.1039/b700236j. [DOI] [PubMed] [Google Scholar]
- 54.Wu C, Ifa DR, Manicke NE, Cooks RG. Rapid, direct analysis of cholesterol by charge labeling in reactive desorption electrospray ionization. Anal Chem. 2009;81(18):7618–7624. doi: 10.1021/ac901003u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kertesz V, Van Berkel GJ, Vavrek M, Koeplinger KA, Schneider BB, Covey TR. Comparison of drug distribution images from whole-body thin tissue sections obtained using desorption electrospray ionization tandem mass spectrometry and autoradiography. Anal Chem. 2008;80(13):5168–5177. doi: 10.1021/ac800546a. [DOI] [PubMed] [Google Scholar]
- 56.Dill AL, Ifa DR, Manicke NE, et al. Lipid profiles of canine invasive transitional cell carcinoma of the urinary bladder and adjacent normal tissue by desorption electrospray ionization imaging mass spectrometry. Anal Chem. 2009;81(21):8758–8764. doi: 10.1021/ac901028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ifa DR, Wu C, Ouyang Z, Cooks RG. Desorption electrospray ionization and other ambient ionization methods: current progress and preview. Analyst. 2010;135(4):669–681. doi: 10.1039/b925257f. [DOI] [PubMed] [Google Scholar]
- 58.Kertesz V, Van Berkel GJ. Improved imaging resolution in desorption electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(17):2639–2644. doi: 10.1002/rcm.3662. [DOI] [PubMed] [Google Scholar]
- 59.Huang G, Chen H, Zhang X, Cooks RG, Ouyang Z. Rapid screening of anabolic steroids in urine by reactive desorption electrospray ionization. Anal Chem. 2007;79(21):8327–8332. doi: 10.1021/ac0711079. [DOI] [PubMed] [Google Scholar]
- 60.Chen H, Cotte-Rodriguez I, Cooks RG. Cis-diol functional group recognition by reactive desorption electrospray ionization (DESI) Chem Commun (Camb) 2006;(6):597–599. doi: 10.1039/b516448f. [DOI] [PubMed] [Google Scholar]
- 61.Song Y, Cooks RG. Reactive desorption electrospray ionization for selective detection of the hydrolysis products of phosphonate esters. J Mass Spectrom. 2007;42(8):1086–1092. doi: 10.1002/jms.1244. [DOI] [PubMed] [Google Scholar]
- 62.Augusti R, Chen H, Eberlin LS, Nefliu M, Cooks RG. Atmospheric pressure eberlin transacetalization reactions in the heterogeneous liquid/gas phase. Int J Mass Spectrom. 2006;253(3):281–287. [Google Scholar]
- 63.Wu C, Ifa DR, Manicke NE, Cooks RG. Molecular imaging of adrenal gland by desorption electrospray ionization mass spectrometry. Analyst. 2010;135(1):28–32. doi: 10.1039/b919816d. [DOI] [PubMed] [Google Scholar]
- 64.Piehowski PD, Davey AM, Kurczy ME, et al. Time-of-flight secondary ion mass spectrometry imaging of subcellular lipid heterogeneity: poisson counting and spatial resolution. Anal Chem. 2009;81(14):5593–5602. doi: 10.1021/ac901065s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nemes P, Woods AS, Vertes A. Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization-mass spectrometry. Anal Chem. 2010;82(3):982–988. doi: 10.1021/ac902245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vertes A, Nemes P, Shrestha B, Barton AA, Chen ZY, Li Y. Molecular imaging by mid-IR laser ablation mass spectrometry. Appl Phys A. 2008;93(4):885–891. [Google Scholar]
- 67.Nemes P, Barton AA, Li Y, Vertes A. Ambient molecular imaging and depth profiling of live tissue by infrared laser ablation electrospray ionization-mass spectrometry. Anal Chem. 2008;80(12):4575–4582. doi: 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]
- 68.Shrestha B, Vertes A. In situ metabolic profiling of single cells by laser ablation electrospray ionization-mass spectrometry. Anal Chem. 2009;81(20):8265–8271. doi: 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- 69.Shrestha B, Nemes P, Nazarian J, Hathout Y, Hoffman EP, Vertes A. Direct analysis of lipids and small metabolites in mouse brain tissue by AP IR-MALDI and reactive LAESI-mass spectrometry. Analyst. 2010;135(4):751–758. doi: 10.1039/b922854c. [DOI] [PubMed] [Google Scholar]
- 70.Galhena AS, Harris GA, Nyadong L, Murray KK, Fernandez FM. Small molecule ambient mass spectrometry imaging by infrared laser ablation metastable-induced chemical ionization. Anal Chem. 2010;82(6):2178–2181. doi: 10.1021/ac902905v. [DOI] [PubMed] [Google Scholar]
- 71.Schäfer R. Ultraflextreme: redefining MALDI-TOF-TOF-mass spectrometry performance. LC GC Eur. 2009;(Suppl S):26–27. [Google Scholar]
- 72.Moskovets E, Preisler J, Chen HS, Rejtar T, Andreev V, Karger BL. High-throughput axial MALDI-TOF MS using a 2-kHz repetition rate laser. Anal Chem. 2006;78(3):912–919. doi: 10.1021/ac051393t. [DOI] [PubMed] [Google Scholar]
- 73.Simmons DA. Improved MALDI-MS imaging performance using continuous laser rastering. AB Applied Biosystems, MDS Analytical Technologies; 2008. [Google Scholar]
- 74.Vestal C, Parker K, Hayden K, et al. Tissue imaging by 5 kHz high-performance MALDI-TOF. Presented at: American Society for Mass Spectromety Conference on Mass Spectrometry and Allied Topics; Philadelphia, PA, USA. 31 May–4 June 2009. [Google Scholar]
- 75.Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal Chem. 2008;80(14):5648–5653. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taban IM, Altelaar AF, Van Der Burgt YE, et al. Imaging of peptides in the rat brain using MALDI-FTICR mass spectrometry. J Am Soc Mass Spectrom. 2007;18(1):145–151. doi: 10.1016/j.jasms.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 77.Chen R, Jiang X, Conaway MC, et al. Mass spectral analysis of neuropeptide expression and distribution in the nervous system of the lobster homarus americanus. J Proteome Res. 2010;9(2):818–832. doi: 10.1021/pr900736t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verhaert PD, Pinkske M, Prieto Conaway MC. Tissue imaging of neuropeptides by MALDI Orbitrap MS. Presented at: American Society for Mass Spectromety Conference on Mass Spectrometry and Allied Topics; Philadelphia, PA, USA. 31 May–4 June 2009. [Google Scholar]
- 79.Fenn LS, Mclean JA. Biomolecular structural separations by ion mobility-mass spectrometry. Anal Bioanal Chem. 2008;391(3):905–909. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- 80.Mclean JA, Ridenour WB, Caprioli RM. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J Mass Spectrom. 2007;42(8):1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 81.Amstalden Van Hove ER, Smith DF, Heeren RM. A concise review of mass spectrometry imaging. J Chromatogr A. 2010;1217(25):3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 82.Stauber J, Macaleese L, Franck J, et al. On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J Am Soc Mass Spectrom. 2010;21(3):338–347. doi: 10.1016/j.jasms.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 83.Jackson SN, Ugarov M, Egan T, et al. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J Mass Spectrom. 2007;42(8):1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trim PJ, Henson CM, Avery JL, et al. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal Chem. 2008;80(22):8628–8634. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- 85.Carado A, Passarelli MK, Kozole J, Wingate JE, Winograd N, Loboda AV. C60 secondary ion mass spectrometry with a hybrid-quadrupole orthogonal time-of-flight mass spectrometer. Anal Chem. 2008;80(21):7921–7929. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38(7):699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 87.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4(9):1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 88.Skold K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andren PE. The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: stathmin 2–20 and peptides as sample quality indicators. Proteomics. 2007;7(24):4445–4456. doi: 10.1002/pmic.200700142. [DOI] [PubMed] [Google Scholar]
- 89.Svensson M, Boren M, Skold K, et al. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res. 2009;8(2):974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 90.Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J Proteome Res. 2007;6(4):1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 91.Chaurand P, Latham JC, Lane KB, et al. Imaging mass spectrometry of intact proteins from alcohol-preserved tissue specimens: Bypassing formalin fixation. J Proteome Res. 2008;7(8):3543–3555. doi: 10.1021/pr800286z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J Am Soc Mass Spectrom. 2005;16(7):1093–1099. doi: 10.1016/j.jasms.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 93.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI-mass spectrometry. Anal Chem. 2006;78(18):6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 94••.Chen R, Hui L, Sturm RM, Li L. Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J Am Soc Mass Spectrom. 2009;20(6):1068–1077. doi: 10.1016/j.jasms.2009.01.017. One of the first papers to report the utility of 3D MSI for volumetric visualization of both neuropeptides and lipids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI-mass spectrometry. J Proteome Res. 2006;5(11):2889–2900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 96.Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Integrating histology and imaging mass spectrometry. Anal Chem. 2004;76(4):1145–1155. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 97.Kaletas BK, Van Der Wiel IM, Stauber J, et al. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9(10):2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 98.Lemaire R, Wisztorski M, Desmons A, et al. MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal Chem. 2006;78(20):7145–7153. doi: 10.1021/ac060565z. [DOI] [PubMed] [Google Scholar]
- 99.Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J Am Soc Mass Spectrom. 2008;19(8):1069–1077. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Altelaar AFM, Luxembourg SL, Mcdonnell LA, Piersma SR, Heeren RMA. Imaging mass spectrometry at cellular length scales. Nat Protoc. 2007;2(5):1185–1196. doi: 10.1038/nprot.2007.117. [DOI] [PubMed] [Google Scholar]
- 101.Vermillion-Salsbury RL, Hercules DM. 9-aminoacridine as a matrix for negative mode matrix-assisted laser desorption/ionization. Rapid Commun Mass Spectrom. 2002;16(16):1575–1581. [Google Scholar]
- 102.Benabdellah F, Touboul D, Brunelle A, Laprevote O. In situ primary metabolites localization on a rat brain section by chemical mass spectrometry imaging. Anal Chem. 2009;81(13):5557–5560. doi: 10.1021/ac9005364. [DOI] [PubMed] [Google Scholar]
- 103.Zhong G, Lin H. A binary matrix for background suppression in MALDI-MS of small molecules. Anal Bioanal Chem. 2007;387(5):1939–1944. doi: 10.1007/s00216-006-1100-3. [DOI] [PubMed] [Google Scholar]
- 104.Armstrong DW, Zhang LK, He LF, Gross ML. Ionic liquids as matrixes for matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 2001;73(15):3679–3686. doi: 10.1021/ac010259f. [DOI] [PubMed] [Google Scholar]
- 105.Fitzgerald JJD, Kunnath P, Walker AV. Matrix-enhanced secondary ion mass spectrometry (ME SIMS) using room temperature ionic liquid matrices. Anal Chem. 2010;82(11):4413–4419. doi: 10.1021/ac100133c. [DOI] [PubMed] [Google Scholar]
- 106.Lemaire R, Tabet JC, Ducoroy P, Hendra JB, Salzet M, Fournier I. Solid ionic matrixes for direct tissue analysis and MALDI imaging. Anal Chem. 2006;78(3):809–819. doi: 10.1021/ac0514669. [DOI] [PubMed] [Google Scholar]
- 107.Tholey A, Zabet-Moghaddam M, Heinzle E. Quantification of peptides for the monitoring of protease-catalyzed reactions by matrix-assisted laser desorption/ionization mass spectrometry using ionic liquid matrixes. Anal Chem. 2006;78(1):291–297. doi: 10.1021/ac0514319. [DOI] [PubMed] [Google Scholar]
- 108.Schuerenberg M, Luebbert C, Deininger SO, Ketterlinus R, Suckau D. MALDI tissue imaging: mass spectrometric localization of biomarkers in tissue slices. Nat Methods. 2007;4(5):iii–iv. [Google Scholar]
- 109.Altelaar AFM, Klinkert I, Jalink K, et al. Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS- and MALDI-mass spectrometry. Anal Chem. 2006;78(3):734–742. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- 110.Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Anal Chem. 2006;78(3):827–834. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 111.Baluya DL, Garrett TJ, Yost RA. Automated MALDI matrix deposition method with inkjet printing for imaging mass spectrometry. Anal Chem. 2007;79(17):6862–6867. doi: 10.1021/ac070958d. [DOI] [PubMed] [Google Scholar]
- 112.Franck J, Arafah K, Barnes A, Wisztorski M, Salzet M, Fournier I. Improving tissue preparation for matrix-assisted laser desorption ionization mass spectrometry imaging. Part 1: using microspotting. Anal Chem. 2009;81(19):8193–8202. doi: 10.1021/ac901328p. [DOI] [PubMed] [Google Scholar]
- 113.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007;18(9):1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J Am Soc Mass Spectrom. 2008;19(6):882–886. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Agar NYR, Yang HW, Carroll RS, Black PM, Agar JN. Matrix solution fixation: histology-compatible tissue preparation for MALDI mass spectrometry imaging. Anal Chem. 2007;79(19):7416–7423. doi: 10.1021/ac071460e. [DOI] [PubMed] [Google Scholar]
- 116••.Breitenstein D, Rommel CE, Mollers R, Wegener J, Hagenhoff B. The chemical composition of animal cells and their intracellular compartments reconstructed from 3D mass spectrometry. Angew Chem Int Ed. 2007;46(28):5332–5335. doi: 10.1002/anie.200604468. Example of a dual beam setup in secondary ion mass spectrometry imaging single cells in three dimensions. [DOI] [PubMed] [Google Scholar]
- 117.Jones EA, Lockyer NP, Vickerman JC. Depth profiling brain tissue sections with a 40 keV C60+ primary ion beam. Anal Chem. 2008;80(6):2125–2132. doi: 10.1021/ac702127q. [DOI] [PubMed] [Google Scholar]
- 118.Fletcher JS, Henderson A, Biddulph GX, Vaidyanathan S, Lockyer NP, Vickerman JC. Uncovering new challenges in bio-analysis with TOF-SIMS. Appl Surf Sci. 2008;255(4):1264–1270. [Google Scholar]
- 119.Wiseman JM, Ifa DR, Venter A, Cooks RG. Ambient molecular imaging by desorption electrospray ionization-mass spectrometry. Nat Protoc. 2008;3(3):517–524. doi: 10.1038/nprot.2008.11. [DOI] [PubMed] [Google Scholar]
- 120.Wiseman JM, Ifa DR, Zhu YX, et al. Desorption electrospray ionization-mass spectrometry: imaging drugs and metabolites in tissues. Proc Natl Acad Sci USA. 2008;105(47):18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Luxembourg SL, Mize TH, McDonnell LAA, Heeren RM. High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal Chem. 2004;76(18):5339–5344. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- 122.Jurchen JC, Rubakhin SS, Sweedler JV. MALDI-MS imaging of features smaller than the size of the laser beam. J Am Soc Mass Spectrom. 2005;16(10):1654–1659. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 123.Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI-MS: distribution of neuropeptides within single neurons. Anal Chem. 2003;75(20):5374–5380. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- 124.Perdian DC, Cha S, Oh J, Sakaguchi DS, Yeung ES, Lee YJ. In situ probing of cholesterol in astrocytes at the single-cell level using laser desorption ionization mass spectrometric imaging with colloidal silver. Rapid Commun Mass Spectrom. 2010;24(8):1147–1154. doi: 10.1002/rcm.4491. [DOI] [PubMed] [Google Scholar]
- 125.Jun JH, Song ZH, Liu ZJ, Nikolau BJ, Yeung ES, Lee YJ. High-spatial and high-mass resolution imaging of surface metabolites of Arabidopsis thaliana by laser desorption-ionization mass spectrometry using colloidal silver. Anal Chem. 2010;82(8):3255–3265. doi: 10.1021/ac902990p. [DOI] [PubMed] [Google Scholar]
- 126.Holscher D, Shroff R, Knop K, et al. Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and hypericum species. Plant J. 2009;60(5):907–918. doi: 10.1111/j.1365-313X.2009.04012.x. [DOI] [PubMed] [Google Scholar]
- 127.Chandra S. 3D subcellular SIMS imaging in cryogenically prepared single cells. Appl Surf Sci. 2004;231:467–469. [Google Scholar]
- 128.Fletcher JS. Cellular imaging with secondary ion mass spectrometry. Analyst. 2009;134(11):2204–2215. doi: 10.1039/b913575h. [DOI] [PubMed] [Google Scholar]
- 129.Nygren H, Hagenhoff B, Malmberg P, Nilsson M, Richter K. Bioimaging TOF-SIMS: high resolution 3D imaging of single cells. Microsc Res Techniq. 2007;70:969–974. doi: 10.1002/jemt.20502. [DOI] [PubMed] [Google Scholar]
- 130.Reyzer ML, Caprioli RM. MALDI-MS-based imaging of small molecules and proteins in tissues. Curr Opin Chem Biol. 2007;11(1):29–35. doi: 10.1016/j.cbpa.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 131.Atkinson SJ, Loadman PM, Sutton C, Patterson LH, Clench MR. Examination of the distribution of the bioreductive drug AQ4N and its active metabolite AQ4 in solid tumours by imaging matrix-assisted laser desorption/ionisation-mass spectrometry. Rapid Commun Mass Spectrom. 2007;21(7):1271–1276. doi: 10.1002/rcm.2952. [DOI] [PubMed] [Google Scholar]
- 132.Signor L, Varesio E, Staack RF, Starke V, Richter WF, Hopfgartner G. Analysis of erlotinib and its metabolites in rat tissue sections by MALDI quadrupole time-of-flight-mass spectrometry. J Mass Spectrom. 2007;42(7):900–909. doi: 10.1002/jms.1225. [DOI] [PubMed] [Google Scholar]
- 133.Acquadro E, Cabella C, Ghiani S, Miragoli L, Bucci EM, Corpillo D. Matrix-assisted laser desorption ionization imaging-mass spectrometry detection of a magnetic resonance imaging contrast agent in mouse liver. Anal Chem. 2009;81(7):2779–2784. doi: 10.1021/ac900038y. [DOI] [PubMed] [Google Scholar]
- 134.Rohner TC, Staab D, Stoeckli M. MALDI mass spectrometric imaging of biological tissue sections. Mech Ageing Dev. 2005;126(1):177–185. doi: 10.1016/j.mad.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 135.Nilsson A, Fehniger TE, Gustavsson L, et al. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS ONE. 2010;5(7):e11411. doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Earnshaw CJ, Carolan VA, Richards DS, Clench MR. Direct analysis of pharmaceutical tablet formulations using matrix-assisted laser desorption/ionisation-mass spectrometry imaging. Rapid Commun Mass Spectrom. 2010;24(11):1665–1672. doi: 10.1002/rcm.4525. [DOI] [PubMed] [Google Scholar]
- 137.Yanes O, Woo HK, Northen TR, et al. Nanostructure initiator mass spectrometry: tissue imaging and direct biofluid analysis. Anal Chem. 2009;81(8):2969–2975. doi: 10.1021/ac802576q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fisher GL, Belu AM, Mahoney CM, Wormuth K, Sanada N. Three-dimensional time-of-flight-secondary ion mass-spectrometry imaging of a pharmaceutical in a coronary stent coating as a function of elution time. Anal Chem. 2009;81(24):9930–9940. doi: 10.1021/ac901587k. [DOI] [PubMed] [Google Scholar]
- 139.Kozole J, Szakal C, Kurczy M, Winograd N. Model multilayer structures for three-dimensional cell imaging. Appl Surf Sci. 2006;252(19):6789–6792. [Google Scholar]
- 140.Ashburner J, Csernansky JG, Davatzikos C, Fox NC, Frisoni GB, Thompson PM. Computer-assisted imaging to assess brain structure in healthy and diseased brains. Lancet Neurol. 2003;2(2):79–88. doi: 10.1016/s1474-4422(03)00304-1. [DOI] [PubMed] [Google Scholar]
- 141.Bjaalie JG. Localization in the brain: new solutions emerging. Nat Rev Neurosci. 2002;3(4):322–325. doi: 10.1038/nrn790. [DOI] [PubMed] [Google Scholar]
- 142.Le Naour F, Bralet MP, Debois D, et al. Chemical imaging on liver steatosis using synchrotron infrared and TOF-SIMS microspectroscopies. PLoS ONE. 2009;4(10):e7408. doi: 10.1371/journal.pone.0007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Petit VW, Refregiers M, Guettier C, et al. Multimodal spectroscopy combining time-of-flight-secondary ion mass spectrometry, synchrotron-FT-IR, and synchrotron-UV microspectroscopies on the same tissue section. Anal Chem. 2010;82(9):3963–3968. doi: 10.1021/ac100581y. [DOI] [PubMed] [Google Scholar]
- 144.Sinha TK, Khatib-Shahidi S, Yankeelov TE, et al. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat Methods. 2008;5(1):57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 201.MS Imaging. www.maldi-msi.org.
- 202.Image J. http://imagej.nih.gov/ij/download.html.