Abstract
Background. Hepatitis C virus (HCV) chronically infects >170 million persons worldwide and is a leading cause of cirrhosis and hepatocellular carcinoma. The identification of more effective and better-tolerated agents for treating HCV is a high priority. We have reported elsewhere the discovery of the anti-HCV compound ceestatin using a high-throughput screen of a small molecule library.
Methods. To identify host or viral protein targets in an unbiased fashion, we performed affinity chromatography, using tandem liquid chromatography/mass spectrometry to identify specific potential targets.
Results. Ceestatin binds to 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) synthase and irreversibly inhibits HMG-CoA synthase in a dose-dependent manner. Ceestatin’s anti-HCV effects are reversed by addition of HMG-CoA, mevalonic acid, or geranylgeraniol. Treatment with small interfering RNA against HMG-CoA synthase led to a substantial reduction in HCV replication, further validating HMG-CoA synthase as an enzyme essential for HCV replication.
Conclusions. Ceestatin therefore exerts its anti-HCV effects through inhibition of HMG-CoA synthase. It may prove useful as an antiviral agent, as a probe to study HCV replication, and as a cholesterol-lowering agent. The logical stepwise process employed to discover the mechanism of action of ceestatin can serve as a general experimental strategy to uncover the targets on which novel uncharacterized anti-HCV compounds act.
Hepatitis C virus (HCV) chronically infects >170 million persons worldwide. Left unchecked, HCV leads to the development of cirrhosis in as many as 30% of patients [1]. Because cirrhosis frequently leads to liver failure and hepatocellular carcinoma, HCV-related liver disease is now the leading indication for liver transplantation worldwide [1]. The best current therapy for chronic HCV infection, a combination of pegylated interferon (PEG-IFN) and ribavirin, produces sustained virologic response in only about half of treated individuals [1]. Furthermore, these agents are associated with a high rate of side effects, ultimately causing 20% of patients to discontinue therapy. The identification of more effective and better tolerated agents is therefore a high priority.
A major strategy in the search for anti-HCV agents has been the targeting of viral proteins. Indeed, a number of small molecule inhibitors of the HCV NS3-4A serine protease and NS5B RNA-dependent RNA polymerase are in clinical development and show great promise. Unfortunately, the strong selection pressure imposed by these agents, coupled with the error-prone nature of the viral RNA polymerase, has led to the rapid selection of drug-resistant HCV strains [2].
Another approach to control HCV without selecting resistant variants is to target host cofactors that support the viral life cycle. Like all viruses, hepatitis C is dependent on host proteins for viral entry, uncoating, replication, virion assembly, and egress. Targeting these cofactors is likely to impose a higher barrier to viral resistance and also offers the possibility of blocking the HCV life cycle at multiple, complementary steps. For example, 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitors have been shown to inhibit HCV replication [3–6]. In addition, small molecule inhibitors of cyclophilin B, which mediates the binding of RNA to the viral polymerase [7], inhibit HCV replication [8].
One strategy that we have successfully used to discover small molecule inhibitors of HCV replication is high-throughput screening of chemical libraries [3]. If the libraries screened contain novel compounds, then one may discover new antiviral compounds. Novel compounds, however, often are poorly characterized biologically, so the target on which they act is unknown. At present, there is no established consensus strategy for determining the mechanism of action of uncharacterized anti-HCV or other antiviral compounds.
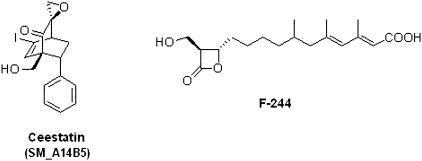
We have reported elsewhere the discovery of a novel class of epoxides, the SM library, capable of inhibiting HCV replication in vitro in both the OR6 full-length HCV replicon [9] and the cell culture infectious strain JFH1 [10] HCVcc [11]. Structure-activity relationship analysis of the SM library led to the design and synthesis of ceestatin (SM_A14B5) [11], the most potent member of the SM class of epoxides (Figure 1). It had an IC50 of 3.5 μmol/L in the OR6 full-length HCV replicon and 5 μmol/L in JFH1 HCVcc [11]. No cytotoxicity was observed at these concentrations [11]. In this study, we found that ceestatin exerts its antiviral effects against HCV through inhibition of the host cofactor HMG-CoA synthase.
Figure 1.
Chemical structures of ceestatin, previously designated as SM_A14B5, and F-244.
METHODS
HCV Polymerase Assays
This procedure was carried out as reported elsewhere by our laboratory (D. N. F.) [12].
HCV Helicase Assays
This procedure was carried out as reported elsewhere by our laboratory (D. N. F.) [13].
HCV Protease Assays
This procedure was performed as directed by the package directions of the SensoLyte 490 HCV Protease Assay Kit (AnaSpec).
HCV Genomic Sequence Analysis
This procedure was carried out as reported elsewhere by our laboratory (T. M. A.) [14]. (Please contact the authors for raw sequence data.)
Ceestatin Immobilization Procedure
Immobilization was performed as reported elsewhere by our laboratory (S. L. S.) [15].
Affinity Chromatography
Affinity chromatography was performed as described elsewhere by our laboratory (S. L. S.) [15].
Mass Spectrometry Sequence Analysis
Bands were excised from the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel for mass spectrometry sequence analysis. Sequence analysis was performed at the Harvard Microchemistry and Proteomics Analysis Facility by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry on a Thermo LTQ-Orbitrap mass spectrometer.
HMG-CoA Synthase Purification
This procedure was carried out as reported elsewhere in the literature [16–18].
Human Recombinant HMG-CoA Synthesis and Isolation
This procedure was carried out as reported elsewhere by our laboratory (H. M. M.) [19].
HMG-CoA Synthase Assay
This procedure was carried out as reported elsewhere in the literature [13–15] and by our laboratory (H. M. M.) [19].
Ceestatin Rescue Experiments
This procedure was carried out as reported elsewhere by our laboratory (R. T. C.) [3, 11], with modifications, as described below. OR6 cells stably harboring the full-length genotype 1 replicon ORN/C-5B/KE [9] were used (kind gift of Dr Nobuyuki Kato, Okayama University). This replicon was derived from the 1B-2 strain (strain HCV-O, genotype 1b), in which the Renilla luciferase gene is introduced as a fusion protein with neomycin to facilitate the monitoring of HCV replication. This construct contains a tissue culture adaptive mutation in the NS3 region. Cells were propagated in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum supplemented with 1% penicillin-streptomycin, and 500 μg/mL of Geneticin (Invitrogen). Cells were cultured in a 37°C, 5% CO2-humidified incubator for all experiments.
Cells were seeded into 96-well plates at a density of 2000 cells/well in 100 μL of medium. The cells were incubated for 24 hours at 37°C to obtain the optimal level of adherence. Solutions of compounds were added to wells to achieve the appropriate final concentrations. Mock solutions were used as a negative control, and PEG-IFN as a positive control. The plates were then incubated at 37°C with 5% CO2 for 72 hours before they were analyzed. Luminescent signal was generated using the Renilla luciferase assay kit (Promega) according to the manufacturer’s instructions. Signal was then detected using a LumiCount luminometer (Packard BioScience). Cell viability was assessed using CellTiter-Glo (Promega), following the manufacturer’s instructions. All experiments were performed in quadruplicate.
Huh-7.5.1 cells were propagated in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum supplemented with 1% penicillin-streptomycin. Cells were cultured in a 37°C, 5% CO2-humidified incubator for all experiments.
The cells were seeded at a density of 10000 cells/well in 500 μL of medium in 24-well plates and were allowed to attach overnight (∼24 hours) before removal of the medium and addition of 100 μL of JFH-1–infected medium. After the cells were incubated for 6 hours, the JFH-1–infected medium was removed and 500 μL of fresh medium was added. The cells were then incubated for another 48 hours before the addition of PEG-IFN and the test compounds at the appropriate concentrations (day 0), after which the plates were incubated for another 48 hours.
RNA was harvested using the RNeasy kit (Qiagen) according to the manufacturer’s directions. The RNA was converted to complementary DNA (cDNA) over a single polymerase chain reaction (PCR) cycle, using the GeneAmp RNA PCR kit (Applied Biosystems), according to the manufacturer’s instructions, and the JFH 1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers reported in the literature [10].
The purity of the resulting JFH1 and GAPDH cDNA was validated by PCR using the GoTaq PCR kit (Promega) according to the manufacturer’s instructions, and the PCR products were separated on a 1% agarose gel. Quantitative real-time PCR was performed on the JFH1 and GAPDH cDNAs using the DyNAmo HS SYBR Green qPCR kit (New England Biolabs), according to the manufacturer’s instructions, and the BioRad IQ5 Multicolor RT PCR Detection System (BioRad). Data analysis was performed using the BioRad IQ5 Optical System Software (BioRad).
HCV Small Interfering RNA Inhibition Assays
This procedure was carried out as described elsewhere by our laboratory (R. T. C.) [20], with minor modifications, as described below. Knockdown of HMG-CoA synthase expression via small interfering RNA (siRNA) was performed in OR6 full-length replicon cells. All siRNAs were obtained from Thermo Scientific Dharmicon. Nontargeting siRNA, siRNA specifically targeting the 5’ HCV genome and HMG-CoA reductase, and IFN were used as controls. For each gene, the 4 individual siRNA duplexes were spotted into quadruplicate wells in 96-well plates to a final concentration of 50 nmol/L. To each well, diluted HiPerFect transfection reagent (Qiagen) was added and then 3000 OR6 cells were plated. Transfections were performed in duplicate 96-well plates. Renilla luciferase and CellTiterGlo (Promega) assays were performed 72 hours after transfection. Renilla luciferase activity was normalized to cellular adenosine triphosphate content, as determined by CellTiterGlo.
Infectious genotype 2a JFH1 HCV was prepared as described above. Huh7.5.1 and Huh7 cells were reverse-transfected in 96-well plates with siRNA duplexes under the same conditions as OR6 cells. siRNA-transfected cells were then infected with JFH1 virus at a multiplicity of infection of ∼.2. Total cellular and viral RNA was isolated after infection using RNeasy Mini columns (Qiagen) with on-column DNase digestion, reverse-transcribed by random priming with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), and then quantified by real-time PCR using the DyNAmo HS SYBR Green qPCR kit (Finnzyme). Efficiency-corrected relative quantification was used with GAPDH as an internal control.
RESULTS
Evaluation of Possible Viral Targets
We first sought to determine whether ceestatin demonstrated activity against the most important viral enzymatic targets. Ceestatin did not inhibit the activity of HCV polymerase, HCV helicase, or HCV protease at concentrations up to 100 μmol/L (data not shown).
In addition, prolonged (2-month) incubation of OR6 replicon cells with ceestatin at concentrations of up to 20 μmol/L did not lead to any diminution in the efficacy of ceestatin or any alterations in the sequence of the HCV genome (data not shown), suggesting that selection pressure was not being applied to other viral targets. These findings suggest that ceestatin inhibits HCV replication by acting on a host, rather than a viral, target.
Affinity Chromatography
Prior structure-activity relationship analysis of the SM library indicated that although ceestatin’s bridgehead primary alcohol substituent (Figure 1) was important in enhancing its activity relative to other compounds in the SM library, compounds in this library lacking bridgehead substituents also exhibit anti-HCV activity [11]. We could therefore use the bridgehead primary alcohol substituent as an attachment point to a solid support for affinity chromatography without affecting its ability to bind to its target.
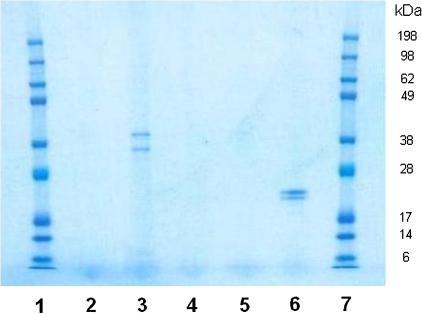
Affinity chromatography was carried out using methods developed by our laboratory [15]. Ceestatin was first immobilized on CarboxyLink coupling gel. A mock coupling gel was created by using ceestatin-free activated reagent solution in the immobilization procedure. Both compound-linked and mock coupling gel were treated with OR6 replicon cell lysate. In addition, compound-linked coupling gel was treated with cell lysate in the presence of ceestatin free in solution at a concentration of 100 μmol/L. The affinity-purified proteins were then separated by SDS-PAGE.
Two unique bands were pulled down by ceestatin-linked coupling gel. These bands were not present when either mock coupling gel or ceestatin-linked coupling gel in the presence of 100 μmol/L ceestatin free in solution was used (Figure 2). These bands were discerned when the gel was stained with Novex Colloidal Blue (Invitrogen). Standard Coomassie Blue staining was not sensitive enough to detect any bands, whereas standard silver staining methods led to an overabundance of bands.
Figure 2.
Sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis gel of affinity chromatography experiments and controls, stained with Novex Colloidal Blue (Invitrogen). Lanes are as follows, left to right: sizing ladder (1), ceestatin-linked coupling gel with 100 μmol/L ceestatin (10 μL) (2), ceestatin-linked coupling gel (10 μL) (3), ceestatin-linked coupling gel with 100 μmol/L ceestatin (1 μL) (4), ceestatin-linked coupling gel (1 μL) (5), mock coupling gel (10 μL) (6), and sizing ladder (7). Note that 1 μL of SDS sample buffer solution provides an insufficient amount of protein to be visualized by this staining system (see lane 5 vs lane 3).
Mass Spectrometry Sequence Analysis
Both of the unique bands were excised from the gel and subjected to mass spectrometry sequence analysis. (Full sequence analysis can be obtained by contacting the authors.) Unique sequences corresponding to 53 potential proteins were found. As part of our analysis of the 53 potential polypeptides, our first triage step was to evaluate enzymes rather than structural proteins, because small molecules are more likely to modulate enzymatic activity than disrupt structural proteins. This reduced the number of potential target proteins to 13. Strikingly, we found that the only enzyme with specific sequences (TGVAPDVFAENM*K and ASSELFSQK) in both bands was HMG-CoA synthase. Because a small molecule inhibitor of this enzyme has been shown to inhibit HCV replication [5], we therefore sought to determine whether ceestatin acts as an inhibitor of HMG-CoA synthase.
HMG-CoA Synthase Assay
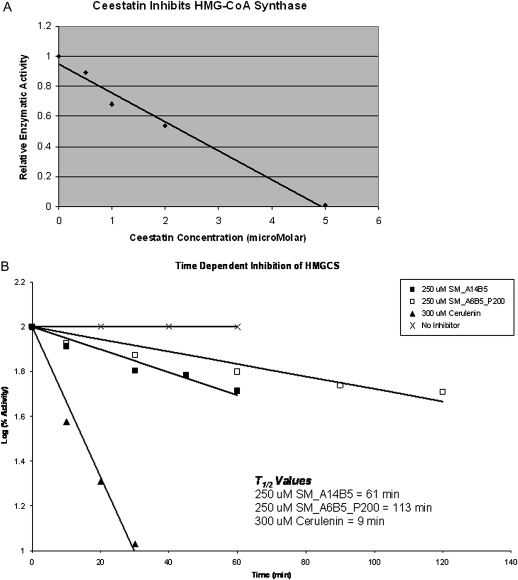
HMG-CoA synthase assays were performed as reported elsewhere in the literature [16–18]. HMG-CoA synthase was first tested against F-244 (Figure 1), a known small molecule inhibitor of the enzyme, at 1 μmol/L, the concentration at which F-244 exhibits its maximal inhibitory capability, according to other reports in the literature [16]. HMG-CoA synthase activity was then tested against ceestatin at concentrations of 100, 20, 10, 5, 2, 1, .5, .2, and 0 μmol/L. Ceestatin exhibited dose-dependent inhibition of the enzyme, with an IC50 of 2.3 ± .3 μmol/L and maximal inhibition first achieved at 5 μmol/L (Figure 3A). This maximum level of inhibition was equal in magnitude to that achieved by F-244 at its own maximal concentration of 1 μmol/L. Further kinetic studies were consistent with the role of ceestatin as an irreversible inhibitor of recombinant human HMG-CoA synthase (Figure 3B).
Figure 3.
Ceestatin inhibits 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) synthase. A, HMG-CoA synthase assay, demonstrating dose-dependent inhibition of enzymatic activity by ceestatin. The IC50 of ceestatin is ∼2 μmol/L. Maximum enzymatic inhibition is achieved by ceestatin at 5 μmol/L. B, Ceestatin (SM_A14B5) inhibits recombinant human HMG-CoA synthase in an irreversible fashion, with a t½ of ∼61 minutes. SM_A6B5_P200 is the parent compound from which ceestatin was derived [9]. It also demonstrates irreversible inhibition of HMG-CoA synthase. Cerulenin is a known powerful irreversible inhibitor of HMG-CoA synthase.
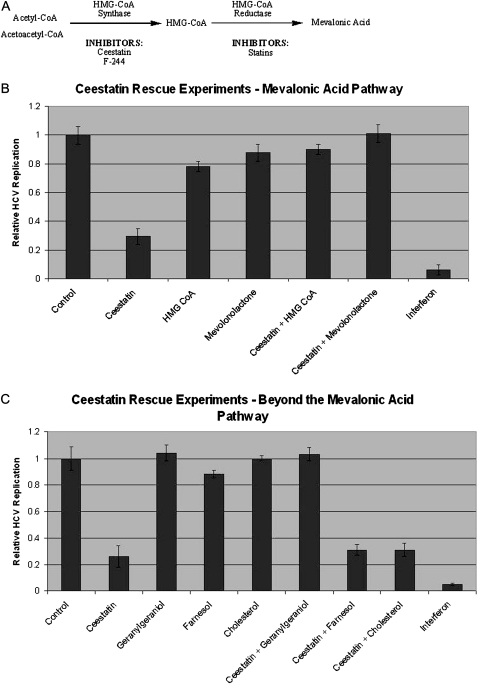
Ceestatin Rescue Experiments
As reported elsewhere, ceestatin at a concentration of 10 μmol/L inhibits HCV replication ∼70% [11]. We sought to determine whether restoring intermediates in the mevalonic acid pathway (Figure 4A) downstream of HMG-CoA synthase could reverse the anti-HCV effects of ceestatin and rescue HCV replication. Cotreatment with either ceestatin and mevalonic acid (supplied in its closed mevalonolactone form) or ceestatin and HMG-CoA, all at concentrations of 10 μmol/L, resulted in reversal of the anti-HCV effects of ceestatin (Figure 4B). We then sought to determine whether restoring intermediates downstream of the mevalonic acid pathway could also rescue HCV replication. Cotreatment with ceestatin and geranylgeraniol also reversed the anti-HCV effects of ceestatin, but cotreatment with either farnesol or cholesterol did not (Figure 4C). Treatment with HMG-CoA, mevalonolactone, or geranylgeraniol alone had little effect on HCV replication. No cytotoxicity was observed in any case, nor any significant differences in cell titer among the experimental conditions, as measured by CellTiterGlo.
Figure 4.
Ceestatin exerts its anti–hepatitis C virus (HCV) effects by inhibiting 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) synthase. A, The first portion of the mevalonate pathway. HMG-CoA synthase catalyzes the formation of HMG-CoA from acetyl-CoA and acetoacetyl-CoA. HMG-CoA reductase forms mevalonic acid from HMG-CoA. B, HCV replication assays, demonstrating the inhibition of HCV replication by ceestatin and the reversal of ceestatin’s antiviral effects by the addition of intermediates downstream of HMG-CoA synthase (HMG-CoA and mevalonolactone) in the mevalonic acid pathway. Ceestatin, HMG-CoA, and mevalonolactone were all tested at a concentration of 10 μmol/L. Although the level of HCV replication was normalized for cell titer, no cytotoxicity was observed in any case, nor any significant differences in cell titer among the experimental conditions, as measured by CellTiterGlo. C, HCV replication assays, demonstrating the inhibition of HCV replication by ceestatin and the reversal of ceestatin’s antiviral effects by the addition of geranylgeraniol, but not farnesol or cholesterol. Ceestatin, geranylgeraniol, farnesol, and cholesterol were all tested at concentrations of 10 μmol/L. Again, no cytotoxicity was observed in any case, nor any significant differences in cell titer among the experimental conditions, as measured by CellTiterGlo. Pegylated interferon-α-2b at a concentration of 1 ng/mL was used in these experiments. The data is reported for the OR-6 replicon cell line. Similar results were seen using JFH-1.
HMG-CoA Synthase siRNA Knockdown
Small molecule inhibitors of HMG-CoA synthase may have off-target effects that could contribute to their ability to inhibit HCV replication. Because it has never before been demonstrated that specific inhibition of HMG-CoA synthase suffices to block HCV replication, we sought to confirm the specificity of this relationship.
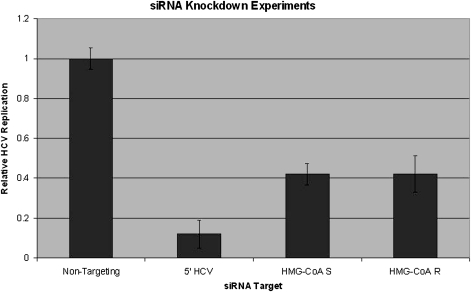
We therefore sought to determine whether knockdown of HMG-CoA synthase using RNA interference (RNAi) also produces an antiviral effect against HCV, using RNAi protocols developed by our group [20]. As a positive control, >90% knockdown of HCV replication relative to nontargeting siRNA was achieved by siRNA targeting the 5’-untranslated region of the HCV genome (Figure 5). Both siRNAs targeting HMG-CoA synthase and reductase achieved 60% knockdown of HCV replication relative to nontargeting siRNA, all at concentrations of 50 nmol/L (Figure 5). Quantitative PCR experiments demonstrated that expression is knocked down up to 10-fold by siRNA against HMG-CoA reductase and up to 25-fold by siRNA against HMG-CoA synthase, relative to nontargeting siRNA (data not shown).
Figure 5.
Inhibition of HMG-CoA synthase is sufficient to inhibit hepatitis C virus (HCV) replication. Small interfering RNA (siRNA) knockdown experiments demonstrate inhibition of HCV replication by siRNA targeting the 5′-untranslated region of the HCV genome, HMG-CoA synthase, and HMG-CoA reductase. Nontargeting siRNA was used as a negative control. Although the level of HCV replication is normalized for cell titer, no cytotoxicity was observed in any, case nor any significant differences in cell titer among the experimental conditions, as measured by CellTiterGlo. HMG-CoA R, HMG-CoA reductase; HMG-CoA S, HMG-CoA synthase.
DISCUSSION
Although much attention has been directed to the antiviral properties of the HMG-CoA reductase inhibitors, inhibition of the enzyme that immediately precedes it in the mevalonate pathway, HMG-CoA synthase, also inhibits HCV replication [5]. Other investigators have demonstrated that F-244 (L-659,699), a small molecule inhibitor of HMG-CoA synthase [21], inhibits HCV replication [5]. Herein, we report that ceestatin, a novel inhibitor of HCV replication identified in a compound screen [11], also inhibits HMG-CoA synthase. Furthermore, by restoring intermediates in the mevalonic acid pathway found downstream of HMG-CoA synthase, we were able to eliminate ceestatin’s anti-HCV effects and rescue HCV replication. Ceestatin, therefore, probably exerts its antiviral effects via HMG-CoA synthase inhibition.
One potential drawback of using small molecules as probes to examine biologic phenomena is that off-target effects may account for a portion of their activity. Although it is possible that ceestatin may act on other unknown targets, both our rescue and siRNA experiments strongly suggest that HMG-CoA synthase inhibition alone is sufficient to inhibit HCV replication. In fact, HMG-CoA synthase inhibition via siRNA knockdown inhibits HCV replication as much as the knockdown produced by siRNA against HMG-CoA reductase, indicating that both act as equivalent antiviral host targets.
The mechanism by which inhibition of HMG-CoA synthase impairs HCV replication remains unknown. One possible hypothesis is based on observations noted in studies of the anti-HCV properties of the statins. HMG-CoA reductase inhibitors shut down cholesterol biosynthesis by preventing the formation of mevalonate from 3-hydroxy-3-methyl-glutaryl CoA. In addition to lowering intracellular levels of sterols, statins also reduce levels of isoprenoids, which are derived from mevalonate. Isoprenoids such as geranylgeranyl pyrophosphate serve as lipid attachments for a variety of intracellular signaling molecules. It has been reported that inhibition of geranylgeranylation, rather than the synthesis of cholesterol itself, is responsible for the inhibition of HCV RNA replication [3–6, 22]. In fact, the anti-HCV effect of the statins was reversed by addition of geranylgeraniol, but not by farnesol or cholesterol [3, 4, 6]. Because the HCV genome does not encode a geranylgeranylated protein, it is hypothesized that a host geranylgeranylated protein, such as FBL2 [22], must play an important role in HCV replication and that inhibition of the geranylgeranylation of this protein represents a potential strategy for blocking HCV replication. Thus, for statins to exert their anti-HCV effects, they must deplete mevalonate sufficiently to lower the cellular pools of geranylgeranyl pyrophosphates [3–6, 22]. Because HMG-CoA synthase immediately precedes HMG-CoA reductase in the mevalonate pathway, one can logically infer that the impairment of HCV replication resulting from its inhibition is mediated by the same mechanisms by which the statins exert their anti-HCV effects. Indeed, the anti-HCV effects of ceestatin were reversed by the addition of geranylgeraniol, but not by the addition of either farnesol or cholesterol, as has been observed previously with the statins (Figure 4B). These findings further underscore the importance of maintaining cellular pools of geranylgeraniol for HCV replication.
No general algorithm currently exists to identify the targets of novel uncharacterized anti-HCV compounds. We employed a systematic process to uncover the mechanism of action of ceestatin. Our paradigm, a stepwise logical method, begins with a broad assessment of activity against both specific viral enzymatic targets (through in vitro enzymatic assays) and other possible nonenzymatic viral targets (through sequence evolution analysis). Should the compound in question not act on a viral target, affinity chromatography would then be used to identify potential host targets, followed by the appropriate confirmatory biochemical assays for target validation. Following this paradigm, we first eliminated potential viral targets before embarking on the more technically challenging, but ultimately successful, search for the host target, HMG-CoA synthase.
In summary, we have found that ceestatin exerts its anti-HCV effects through inhibition of the host cofactor HMG-CoA synthase. Ceestatin may therefore prove to be useful not only as an antiviral agent, but also as a cholesterol-lowering agent. Furthermore, it also can be used as a small molecule probe to further define the relationship between HMG-CoA synthase, the mevalonate pathway, and HCV replication. Indeed, we have provided further evidence supporting the hypothesis that inhibition of the mevalonic acid pathway leads to depletion of cellular pools of geranylgeraniol necessary for HCV replication. Finally, the logical stepwise process employed to discover the mechanism of action of ceestatin can serve as a general experimental strategy to reveal the targets on which novel uncharacterized anti-HCV compounds act. Our discovery and characterization of the novel anti-HCV compound ceestatin strongly links basic research in chemical biology to an ongoing problem in the practice of medicine, thereby representing a paradigm of the power of translational medicine.
Funding
This work was supported by the National Institutes of Health (grants R01 AI069939 and K24 DK078772 to R. T. C. and K08 DK088951 to L. F. P.), the American Gastroenterological Association (L. F. P.), and the American Liver Foundation (L. F. P.). S. L. S. is a Howard Hughes Medical Institute Investigator.
Acknowledgments
Huh 7.5.1 cells were obtained from Dr Francis Chisari (Scripps Institute). OR6 cells were obtained from Drs N. Kato and M. Ikeda (Okayama University Graduate School of Medicine). A plasmid encoding the full-length JFH1 sequence was a gift of Dr T. Wakita (Tokyo Metropolitan Institute for Neuroscience). We would like to acknowledge the technical assistance provided by Dr William Lane and the Harvard Microchemistry Facility (Harvard University) for protein microsequencing. We would also like to thank Dr Andrew Tai for helpful discussions regarding the siRNA experiments (Massachusetts General Hospital).
References
- 1.Alter MJ. Epidemiology of hepatitis C. Hepatology. 2006;43:S207–20. [Google Scholar]
- 2.Reesink HW, Zeuzem S, Weegink CJ, et al. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology. 2006;131:997–1002. doi: 10.1053/j.gastro.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 3.Kim SS, Peng LF, Lin W, et al. A cell-based, high-throughput screen for small molecule regulators of hepatitis C virus replication. Gastroenterology. 2007;132:311–20. doi: 10.1053/j.gastro.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 4.Ye J, Wang C, Sumpter R, Brown MS, Goldstein JL, Gale M. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc Natl Acad Sci U S A. 2003;100:15865–70. doi: 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci U S A. 2005;102:2561–6. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeda M, Abe K, Yamada M, Dansako H, Naka K, Nobuyuki K. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology. 2006;44:117–25. doi: 10.1002/hep.21232. [DOI] [PubMed] [Google Scholar]
- 7.Watashi K, Ishii N, Hijikata M, et al. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111–22. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Flisiak R, Horban A, Gallay P, et al. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–26. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda M, Abe K, Dansako H, Nakamura T, Naka K, Kato N. Efficient replication of a full-length hepatits C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem Biophys Res Commun. 2005;329:1350–9. doi: 10.1016/j.bbrc.2005.02.138. [DOI] [PubMed] [Google Scholar]
- 10.Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–6. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 11.Peng LF, Kim SS, Matchacheep S, et al. Identification of novel epoxide inhibitors of HCV replication using a high-throughput screen. Antimicrob Agents Chemother. 2007;51:3756–9. doi: 10.1128/AAC.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heck JA, Lam AM, Narayanan N, Frick DN. Effects of mutagenic and chain terminating nucleotide analogs on enzymes isolated from various hepatitis C virus genotypes. Antimicrob Agents Chemother. 2008;52:1901–91. doi: 10.1128/AAC.01496-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belon CA, Frick DN. Monitoring helicase activity with molecular beacons. Biotechniques. 2008;45:433–40. doi: 10.2144/000112834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasprowicz V, Kang YH, Lucas M, et al. Hepatitis C virus (HCV) sequence variation induces an HCV-specific T-cell phenotype analogous to spontaneous resolution. J Virol. 2010;84:1656–63. doi: 10.1128/JVI.01499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Imber BS, Schreiber SL. Small-molecule reagents for cellular pull-down experiments. Bioconjug Chem. 2008;19:585–7. doi: 10.1021/bc700297j. [DOI] [PubMed] [Google Scholar]
- 16.Bell K, Saepudin E, Harrison P. Irreversible inhibition of 3-hydroxy-3-methylglutaryl-coenzyme A synthase from yeast by F-244 and (RS)-P-butyrolactone. Can J Chem. 1996;74:24–7. [Google Scholar]
- 17.Middleton B, Tubbs PK. 3-Hydroxy-3-methylglutaryl-CoA synthase from bakers' yeast. Biochem J. 1972;126:27–34. doi: 10.1042/bj1260027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rokosz LL, Boulton DA, Butkiewicz EA, et al. Human cytoplasmic 3-hydroxyl-3-methylglutaryl coenzyme A synthase: expression, purification, and characterization of recombinant wild-type and Cys129 mutant enzymes. Arch Biochem Biophys. 1994;312:1–13. doi: 10.1006/abbi.1994.1273. [DOI] [PubMed] [Google Scholar]
- 19.Skaff DA, Miziorko HM. A visible wavelength spectrophotometric assay suitable for high-throughput screening of 3-hydroxy-3-methylglutaryl-CoA synthase. Anal Biochem. 2010;396:96–102. doi: 10.1016/j.ab.2009.08.030. [DOI] [PubMed] [Google Scholar]
- 20.Tai AW, Benita Y, Peng LF, et al. A functional genomic screen identifies cofactors of hepatitis C virus replication. Cell Host and Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenspan MD, Yudkovitz JB, Lo CY, et al. Inhibition of hydroxymethylglutaryl-coenzyme A synthase by L-659,699. Proc Natl Acad Sci USA. 1987;84:7488–92. doi: 10.1073/pnas.84.21.7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Gale M, Keller BC, et al. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell. 2005;18:425–34. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]