Abstract
Spirochetes causing Lyme borreliosis are obligate parasites that can only be found in a tick vector or a vertebrate host. The ability to survive in these two disparate environments requires up and downregulation of specific genes by regulatory circuits that remain largely obscure. In this work on the Lyme spirochete, B. burgdorferi, we show that a disruption of the hrpA gene, which encodes a putative RNA helicase, results in a complete loss in the ability of the spirochetes to infect mice by needle inoculation. Studies of protein expression in culture by 2D gels revealed a change in the expression of 33 proteins in hrpA clones relative to the wild-type parent. Quantitative characterization of protein expression by iTRAQ analysis revealed a total of 187 differentially regulated proteins in an hrpA background: 90 downregulated and 97 upregulated. Forty-two of the 90 downregulated and 65 of the 97 upregulated proteins are not regulated under any conditions by the previously reported regulators in B. burgdorferi (bosR, rrp2, rpoN, rpoS or rrp1). Downregulated and upregulated proteins also fell into distinct functional categories. We conclude that HrpA is part of a new and distinct global regulatory pathway in B. burgdorferi gene expression. Because an HrpA orthologue is present in many bacteria, its participation in global regulation in B. burgdorferi may have relevance in other bacterial species where its function remains obscure. We believe this to be the first report of a role for an RNA helicase in a global regulatory pathway in bacteria. This finding is particularly timely with the recent growth of the field of RNA regulation of gene expression and the ability of RNA helicases to modulate RNA structure and function.
Introduction
Lyme borreliosis is common in the northern hemisphere and is now the most frequent tick-borne disease in North America and Europe [1], [2]. The causative agents, Borrelia burgdorferi and related species, are obligate parasites that survive through a complex enzootic cycle involving a tick vector and a vertebrate host. Differential gene regulation in these two environments is an important feature for successful adaptation to both the tick vector and the infected animal (see [3], [4], [5] for recent reviews). About 150 genes appear to be differentially regulated in B. burgdorferi, depending upon environmental conditions and tick or host factors required for survival in these very different settings [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16].
There is much that remains unknown about the regulatory pathways in Borrelia species, which as in other organisms can function at the transcriptional and translational stages and at steps in between. Studies in B. burgdorferi are complicated by the need for growth in ticks and animals to accurately characterize global gene regulation. Nonetheless, several different regulatory molecules and pathways that play a role in differential gene expression have been identified, including the BosR regulator [5], [14], [15], [16], [17], [18], alternative σ factors RpoS and RpoN [7], [12], [19], [20] and the RpoN activator Rrp2 [21], [22], [23]. The two-component response regulatory system Rrp1-Hpk1 [24] as well as the DNA binding and bending protein Hbb [25], and DNA supercoiling [26], [27] also play a role in the modulation of gene expression in B. burgdorferi. RNA regulation has also been recently reported in B. burgdorferi; the RNA regulator DsrA has been shown to regulate the expression of rpoS and ospC [28] and the RNA chaperone Hfq appears to be involved in regulating the expression of pathogenicity factors [29].
RNA helicases are important enzymes present in virtually all living organisms. They unwind double stranded RNA in an energy-dependent manner and are involved in a wide variety of RNA metabolic functions [30], [31], [32], [33]. HrpA, a DEAH-box RNA helicase has been shown to be involved in processing of daa mRNA from a fimbrial operon in E. coli. The processing event results in a stable mRNA and upregulation of daa expression relative to other proteins encoded by the polycistronic transcript [34]. The HrpA protein also appears to be involved in physical interactions with a variety of ribosomal proteins in E. coli, either directly, or indirectly through RNA interaction [35], consistent with a possible regulatory role at the translational level. Other than these two reports, no other information on the function of HrpA in any bacterium exists in the current literature.
As part of our ongoing work on antigenic variation in B. burgdorferi, we generated a disruption of the hrpA (bb0827) gene, which encodes a putative RNA helicase ( Fig. 1 ), to see if loss of this function would have an effect upon antigenic switching at the vlsE locus. Surprisingly, the gene disruption resulted in a complete loss of infectivity and the modulation of the expression of about 180 B. burgdorferi proteins. Our findings suggest that HrpA is involved in a global regulatory pathway and may have relevance to regulation of virulence in other pathogens and to global regulatory mechanisms in bacteria in general.
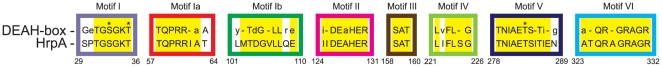
Figure 1. Schematic representation of conserved DEAH-box RNA helicase motifs in the B. burgdorferi HrpA protein.
The sequence and location of conserved motifs of the DEAH-box family aligned with B. burgdorferi HrpA are shown. Amino acids in the DEAH-box consensus that are conserved at least 80% are shown in capital letters, and small letters represent the amino acids with 50–70% conservation. For further details regarding the DEAH-box motifs see [31], [32]. * denotes either T or S at three positions in the consensus sequence. The regions highlighted in yellow are perfect matches between the B. burgdorferi HrpA protein and the consensus sequence and the numbers below the boxes represents the position of the conserved motifs in HrpA.
Results
Construction of hrpA and bb0826 gene disruptions in B. burgdorferi
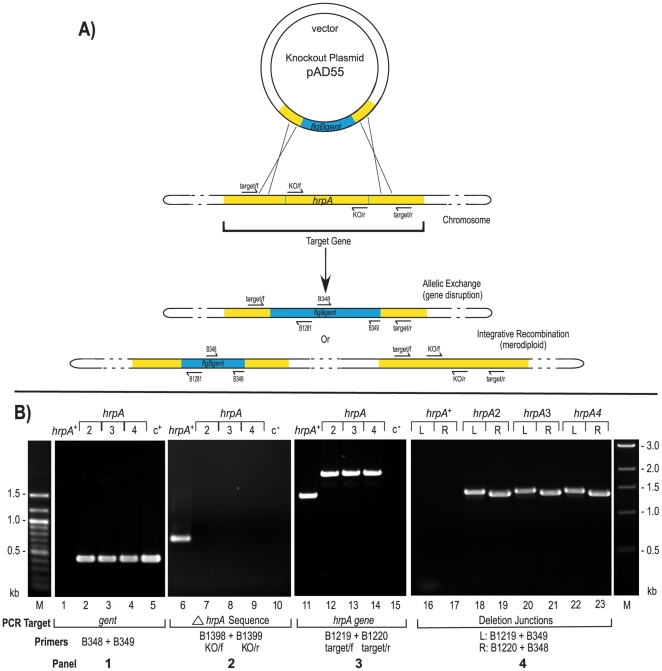
Disruption of the hrpA (bb0827) gene in B. burgdorferi was accomplished by allelic exchange [36]. A knockout plasmid ( Fig. 2A ) was constructed in E. coli, in which the central 500 bp of the hrpA gene was deleted and replaced with a gentamycin resistance cassette (aacC1) under the control of the B. burgdorferi flgB promoter. The orientation of the resistance cassette relative to the target genes are shown in Table 1 . A knockout plasmid was also constructed for bb0826, the gene downstream from hrpA, which contains an RNA binding motif and therefore, was considered as a protein that might function together with HrpA. The constructs were used to transform infectious B. burgdorferi B31 clone 5A4 [37]. The transformants recovered were screened by PCR as shown in Fig. 2B (results for bb0826 not shown). First, the presence of the gentamycin resistance cassette was verified (Panel 1, lanes 2–4), followed by confirmation that the central 500 bp of the target gene was no longer present (panel 2, lanes7–9). To confirm that the recovered mutants carried only the disrupted hrpA gene and were not merodiploids, the presence of only the disrupted gene carrying the gent cassette (2.1 kb) was verified (Panel 3, lanes 12–14) along with the absence of the wild-type gene (1.5 kb, lane 11). Finally, the correct insertion site was confirmed using PCR primers to uniquely amplify left and right side deletion junctions (Panel 4, lanes 18–23). In addition, the structural integrity of the gene disruptions and the presence of only a single disrupted gene were independently demonstrated by Southern hybridization using probes specific for the gentamycin cassette and the deleted portion of the hrpA or bb0826 gene (see Fig. S1 for the Southern blot of the hrpA gene disruption).
Figure 2. hrpA gene disruption and confirmation.
A) Gene disruption strategy. The infectious B. burgdorferi strain B31, clone 5A4 (B31-5A4) was transformed with a knockout plasmid carrying a 1 kb gentamicin cassette (blue) that replaced the central 500 bp of the hrpA gene (yellow) as described in Materials and Methods. The two possible outcomes of recombination events with the target gene are shown: allelic exchange would result in gene disruption while integrative recombination of the knockout plasmid would result in merodiploid formation. The position of PCR primers used for construct verification are shown by arrows on the schematic. B) PCR verification of the hrpA disruption. Each gene disruption was subjected to four PCR analyses. Panel 1) The presence of the gentamicin resistance cassette was confirmed as shown. The shuttle vector pBSV2G [38] served as the positive control (c+) for amplification of the gent cassette (lane 5). Panel 2) The portion of hrpA expected to be deleted in a gene disruption was not detected in hrpA2, 3 or 4 (lanes 7, 8 and 9). Lane 10 was a negative control (c−) that lacked DNA template. Panel 3) The size of the hrpA gene was compared in the three mutant strains. The expected 2.1 kb gene disruption products were observed (lanes 12, 13 and 14) in comparison to the 1.5 kb product from the wild-type hrpA gene (lane 11). Lane 15 was a negative control (c−) that lacked DNA template. Panel 4) Confirmation of the correct insertion site was performed using combinations of the target gene primers and primers internal to the gentamicin cassette to amplify the hrpA boundaries. The left boundary in the hrpA knockout clones displayed the expected 1.4 kb product (lanes 18, 20 and 22) and the right boundary showed the expected product of approximately 1.3 kb (lanes 19, 21 and 23). A 100 bp ladder on the left side of Fig. 1B is relevant to the two left panels, and a 1 kb ladder on the right side applies to the two right panels (M). The schematic in part A of the figure is modified from [13].
Table 1. Plasmids and strains used in this study.
| Gene target | Locus | Description | Plasmid | E. coli strains | gent polarity | B. burgdorferi mutant strains |
| hrpA | bb0827 | RNA helicase | pAD55 | GCE1567 | forward | hrpA2 (GCB1164), hrpA3 (GCB1165), hrpA4 GCB1166) |
| bb0826 | bb0826 | hypothetical prot | pPOH57-1 | GCE2149 | forward | bb0826-11 (GCB544) |
| bb0826 | bb0826 | hypothetical prot | pPOH57-2 | GCE2150 | reverse | bb0826-3 (GCB543) |
All genetic constructs were analyzed for plasmid content, which can affect infectivity. No plasmid loss was observed for GCB1164 (hrpA2) and GCB1165 (hrpA3). GCB1166 (hrpA4) was lacking cp32-3 and cp32-6; cp32-3 is not required for infectivity [37] and the effect of loss of cp32-6 has not been previously reported. GCB543 (bb0826-3) and GCB544 (bb0826-11) both contained a full plasmid complement. Finally, analysis of transcription of the downstream genes bb0825 and bb0826 in the hrpA mutant strains was performed by RT-PCR to determine whether the hrpA gene disruptions had a polar effect and displayed reduced expression of the downstream genes; no decrease in downstream gene expression was observed in any of the three hrpA mutant clones (see Fig. S2). All three clones displayed wild-type morphology and normal growth in BSK-II media (data not shown).
Effect of hrpA and bb0826 gene disruptions on C3H/HeN mouse infections
The mutant strains were each used to infect C3H/HeN mice using an inoculum of 1×103 spirochetes at two locations (see Materials and Methods). At seven days post-infection spirochetes were not recovered from the blood of any of the mice inoculated with the hrpA mutant clones, in contrast to the bb0826 mutants and the control group where all the cultures were positive for spirochetes ( Table 2 ). Similarly, ear cultures at day 21 were all negative for the mice inoculated with hrpA mutant spirochetes, but 100% positive for the bb0826 mutants and the wild-type spirochetes. Finally, at day 35 when all cultures from heart, bladder, ear and joint were positive for wild-type and bb0826 mutant B. burgdorferi, no positive cultures were recovered from the mice infected with the hrpA mutant clones. Mutation of the hrpA gene, therefore, appeared to obliterate spirochete infectivity. Attempts to complement the non-infectious phenotype by supplying the hrpA gene in trans on the shuttle vector pBSV2 [38] were unsuccessful (data not shown). Difficulty in complementing mutants in B. burgdorferi is not unusual and is elaborated upon in the Discussion.
Table 2. Effect of a mutation in the B. burgdorferi hrpA (bb0827) or bb0826 gene on infection of C3H/HeN mice.
| B. burgdorferi genotype | Strain | Total micea | Day 7 Bloodb | Day 7 Infection | Day 21 Ear | Day 21 Infection | Day 35c Heart | Day 35c Bladder | Day 35c Joint | Day 35c Ear | Total sitesd | Day 35 Infection |
| B31 5A4 (wt) | GCB933 | 18 | 18/18 | 100.0% | 18/18 | 100.0% | 4/4 | 4/4 | 4/4 | 4/4 | 16/16 | 100.0% |
| hrpA2 | GCB1164 | 3 | 0/3 | 0.0% | 0/3 | 0.0% | 0/3 | 0/3 | 0/3 | 0/3 | 0/12 | 0.0% |
| hrpA3 | GCB1165 | 3 | 0/3 | 0.0% | 0/3 | 0.0% | 0/3 | 0/3 | 0/3 | 0/3 | 0/12 | 0.0% |
| hrpA4 | GCB1166 | 3 | 0/3 | 0.0% | 0/3 | 0.0% | 0/3 | 0/3 | 0/3 | 0/3 | 0/12 | 0.0% |
| bb0826-3 | GCB543 | 2 | 2/2 | 100.0% | 2/2 | 100.0% | 2/2 | 2/2 | 2/2 | 2/2 | 8/8 | 100.0% |
| bb0826-11 | GCB544 | 2 | 2/2 | 100.0% | 2/2 | 100.0% | 2/2 | 2/2 | 2/2 | 2/2 | 8/8 | 100.0% |
There is a large number of mice in the control group because the hrpA mutants were initially assessed as part of a larger group of mutants.
Values listed correspond to number of positive cultures/number of sites tested.
Four mice infected with B. burgdorferi 5A4 were chosen as positive controls for organ harvests at day 35.
Number of positive tissue sites/number of sites tested.
2D gel analysis of proteins from wild-type and hrpA mutant B. burgdorferi
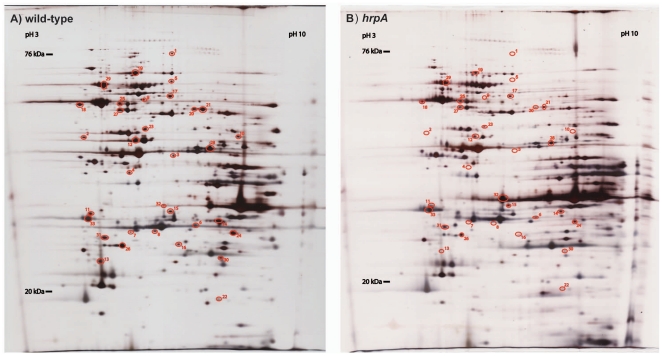
Because gene regulation by an RNA helicase is expected to occur primarily at the post-transcriptional level, we compared the protein content of wild-type B. burgdorferi B31 parent and their derivative hrpA mutant strains. Whole cell lysates were prepared from spirochetes grown to late log stage and were separated by two-dimensional gel electrophoresis. Silver staining methods were used to visualize proteins on the 2D gels. Approximately 600 protein spots were detected in the range of 10–80 kDa and the pH 3–10 area. The 2D gel profiles of the three hrpA mutant clones were indistinguishable from each other (data not shown). A comparison of the wild-type 2D gel profile compared to the hrpA3 mutant (GCB1165) is shown in Fig. 3 . In total, 33 protein spots were identified with changes in intensity between wild-type B. burgdorferi B31 and the hrpA mutant strains on silver stained analytical gels. Twenty of the proteins were identified by LC-MS/MS analysis of tryptic peptides coupled with a Mascot database search ( Table 3 ). For 13 spots, the identification was not successful due to low MS signals or Mascot scores that were below the threshold. Both identified and unidentified protein spots are listed in decreasing order based upon their estimated fold decrease or increase relative to wild-type ( Table 3 ). Out of the 20 identified spots, translation elongation factor Tu (EF-Tu, BB0476), and outer surface protein A (OspA, BBA15) were identified twice from different protein spots within one 2D gel, likely the result of protein modification. EF-Tu (spots 9 and 23) displayed the same migration in the isoelectric focusing dimension and ran with molecular masses of approximately 59 and 45 kDa, respectively. The 45 kDa spot corresponds to the full length protein and the higher mass band likely represents a modified form without any changes in pI. These two spots showed an estimated decrease of 8.27 and 2.62 fold in an hrpA background relative to wild-type. Interestingly, the OspA protein (spots 10 and 32) showed both a decrease of 6.81 (spot 10) and an increase of 8.01 fold (spot 32) in hrpA mutant strains. These two spots displayed vastly different migration in both dimensions, consistent with post-translational modification. Of the 18 proteins identified by 2D gels coupled with LC/MS-MS, 10 of these proteins were also identified by iTRAQ analysis (marked with asterisks in Table 3 ). The highest fold decrease (50.39) detected by 2D gels was from the large subunit of the putative phage terminase (BBM42). This protein is encoded by cp32-6, which was absent from one of the hrpA mutant clones used in this study, thereby contributing to an artificially high decrease in this protein relative to the wild-type strain. The average fold decrease from the two strains carrying cp32-6 was 34, still indicating a major change in expression.
Figure 3. Silver stained 2D gel images.
Whole cell protein extracts are shown for A) wild-type B. burgdorferi B31 clone 5A4 [37] and B) hrpA mutant strain GCB1165. Whole cell protein extracts were separated on non-linear pH 3–10 IPG strips in the first dimension and 12% SDS-PAGE gels in the second dimension. Differentially expressed protein spots are circled and numbered. Select proteins were identified by trypsin digestion followed by LC MS/MS analysis of spots from silver stained gels. Table S3 shows relative fold differences in expression.
Table 3. Changes in protein expression in hrpA mutant clones relative to wild-type B. burgdorferi as estimated by 2D gel electrophoresis.
| Spot | ORF | Protein Name | Decrease |
| 1 | BB_M42 | Phage terminase, large sub, pbsx fam | 50.39 |
| 2 | NA | NA | 41.31 |
| 3 | BB_N43 | Phage terminase, large sub, pbsx fam | 40.38 |
| 4 | BB_0831 | Xylose operon reg prot (XyIR-2) | 32.84 |
| 5* | BB_0366 | Vacuolar aminopeptidase I | 32.62 |
| 6 | NA | NA | 31.64 |
| 7 | NA | NA | 22.64 |
| 8 | NA | NA | 11.58 |
| 9* | BB_0476 | Translation elong factor Tu | 8.27 |
| 10 | BB_A15 | Outer surface protein A (OspA) | 6.81 |
| 11 | BB_J02.1 | Conserved hypothetical protein | 6.21 |
| 12 | NA | NA | 6.02 |
| 13 | NA | NA | 4.94 |
| 14 | BB_0658 | Phosphoglycerate mutase fam prot | 4.79 |
| 15* | BB_0215 | ABC trans peripl PO4 bind prot (PstS) | 4.24 |
| 16 | BB_0239 | Deoxyguan/deoxyadenosine kinase(Dck) | 4.11 |
| 17 | BB_0127 | Ribosomal protein S1 (RpsA) | 3.44 |
| 18 | NA | NA | 3.30 |
| 19* | BB_0540 | Translation elong factor G | 3.07 |
| 20 | NA | NA | 3.00 |
| 21* | BB_0020 | PF6F1P β sub | 2.84 |
| 22* | BB_0463 | Nucleoside- diphosph kinase (NDK) | 2.67 |
| 23* | BB_0476 | Translation elong factor Tu | 2.62 |
| 24* | BB_0375 | Pfs protein (Pfs-1) | 2.58 |
| 25* | BB_0513 | Phe-tRNA ligase alpha chain (PheRS) | 2.57 |
| 26 | BB_0559 | PTS system, gluce-spec IIA comp | 2.48 |
| 27 | NA | NA | 1.87 |
| 28 | NA | NA | 1.71 |
| 29 | NA | NA | 1.58 |
| 30 | NA | NA | 1.53 |
| 31 | NA | NA | 1.52 |
Changes in protein expression in hrpA mutants relative to wild-type B. burgdorferi as determined by 2D gel electrophoresis. Average fold increase or decrease was estimated as the mean value from two gels of the ratio between the spot volumes in wild-type Borrelia burgdorferi B31 and the hrpA mutant strains hrpA2 (GCB1164), hrpA3 (GCB1165) and hrpA4 (GCB1166). Spot volumes were estimated using REDFIN 2D gel analysis software.
*indicates the proteins identified with both 2D gel and iTRAQ analysis.
NA indicates spots that were not identified. The 20 most abundant spots with a changed expression pattern in the mutants were identified by LC MS/MS analysis as described in materials and methods.
iTRAQ proteome analysis for proteins differentially expressed in wild-type and hrpA mutant B. burgdorferi
Analysis by 2D gels is time consuming, only semi-quantitative, limited to analysis of single isoforms and requires gel spots that are resolvable as stained unique species. In contrast, iTRAQ analysis [39], [40] is expedient, allows simultaneous analysis of hundreds of proteins, is highly accurate, and captures data from all isoforms. To follow-up on the 2D gel results, quantitative iTRAQ analysis was performed on the hrpA mutant clones and the parental wild-type strain. For the iTRAQ proteome analysis, the goal was to create an experimental design that would allow control of experimental and technical variations as much as possible. Biological variation was controlled by pooling four independent cultures for each sample. In this way, protein from 32 cultures was analyzed in an 8-plex format. Protein extracts from the wild-type and each of the three mutant strains were analyzed in duplicate, where each of the eight samples was labeled with a different isobaric tag to allow simultaneous processing. Trypsinized proteins were analyzed and identified as described in Materials and Methods. Through the iTRAQ analysis, 370 proteins were identified (Table S2). Out of 370, 90 were significantly (P<0.05) downregulated in the hrpA mutant strains relative to the wild-type; 71 were encoded by the chromosome and 19 by plasmids (Table S3). An even greater number (97) of upregulated proteins were identified: 84 chromosomal and 13 plasmid-encoded (Table S4).
The fold decrease of the downregulated proteins was between 1.20 and 14.28 in an hrpA background. The downregulated proteins directly identified in this study were compared with genes previously reported to be regulated at the transcriptional level by microarray experiments [7], [12], [15], [21], [23], [24]. Out of 90 downregulated proteins identified in this study (Table S3), the RNA for four of them were reported as downregulated in an rrp2 mutant while nine were upregulated in an rrp2 background [21], [23]. In an rpoN mutant background four of the 90 were downregulated, 14 were upregulated and one was inconsistently reported as both down and up [7], [21]. In an rpoS mutant eight were downregulated and 10 were upregulated [7], [12], [21]. In an rrp1 mutant 17 were downregulated and one was upregulated [24]. Finally, in a bosR mutant [15], the transcripts of seven genes were upregulated and none were downregulated. In summary, of the 90 proteins downregulated in hrpA mutant clones, the transcription for 42 of them showed no detectable regulation by any of the known B. burgdorferi regulators noted above.
The fold increase of the upregulated proteins was between 1.2 and 11.77 in an hrpA background. The upregulated proteins directly identified in this study were compared with genes previously reported to be regulated by microarray experiments [7], [12], [15], [21], [23], [24]. Out of 97 upregulated proteins identified in this study (Table S4), five of them have been reported as upregulated in an rrp2 mutant while seven are downregulated in an rrp2 background [21], [23]. In an rpoN mutant background nine of the 97 were upregulated and 11 were downregulated [7], [21]. In an rpoS mutant, nine were upregulated and 11 were downregulated [7], [12], [21], [23], [24]. In an rrp1 mutant there were no upregulated proteins but eight were downregulated [24]. Finally, in a bosR mutant [15], the transcripts of three genes were upregulated and six were downregulated. In summary, of the 97 proteins upregulated in hrpA mutant clones, the transcription for 65 of them showed no detectable regulation by any of the known B. burgdorferi regulators noted above.
Discussion
RNA regulation is a burgeoning field in bacteria in general [41], [42], [43], [44], [45], [46], [47] and in the control of virulence in bacterial pathogens [48], [49]. RNA helicases are ubiquitous proteins that play a role in a wide variety of RNA metabolic functions including transcription, ribosome biogenesis, RNA unwinding, RNA-protein complex disruption and reorganization, RNA processing and RNA decay [30], [31], [32], [33]. Based upon sequence analysis, B. burgdorferi appears to have a single RNA helicase, the DEAH-box protein HrpA ( Fig. 1 ). In contrast, E. coli carries five DEAD-box helicases [50] as well as an HrpA orthologue [50]. HrpA from both E. coli [51] and B. burgdorferi (A. Salman-Dilgimen and G. Chaconas, unpublished) display ATPase activity in vitro. An in vitro helicase activity has not yet been reported for either protein, although the strong correspondence of the B. burgdorferi protein sequence to the eight motifs of ATP-dependent RNA helicases ( Fig. 1 ) strongly supports assignment as an RNA helicase. Further biochemical characterization of HrpA will be required to conclusively establish an RNA helicase function.
During our studies on the proteins involved in antigenic switching at the vlsE locus of B. burgdorferi [13] we found that disruption of the hrpA gene resulted in complete loss of infectivity of C3H/HeN mice by needle inoculation ( Table 2 ). This result was observed with three independent hrpA mutant clones, suggesting that the hrpA gene disruption was responsible for the phenotype, rather than some other defect introduced during genetic manipulation. To prove this point a wild-type hrpA gene with its native promoter (the topA promoter) was introduced into the mutant clones on an E. coli – B. burgdorferi shuttle vector, pBSV2 [38]. However, no restoration of infectivity was observed (data not shown). Genetic complementation in B. burgdorferi can sometimes be difficult to achieve. For example complementation of ruvA and ruvB using shuttle vectors could not be achieved in two independent laboratories [13], [52]. The reason for this remains unknown. We believe that the possibility of having a secondary mutation with an identical phenotype in infectivity and protein expression in all three of our clones is exceedingly low. We also showed that the hrpA gene disruption is not polar (Fig. S2) and that disruption of the downstream gene bb0826 resulted in a completely infectious phenotype. From our combined data we conclude that the hrpA gene is required for infectivity, at least by needle inoculation and that HrpA is, therefore, a virulence determinant [53].
As a preliminary step in investigating the loss of infectivity we analyzed the protein content of our three hrpA mutant clones grown in culture, relative to the wild-type parent and found indistinguishable patterns in the three mutant strains with an obvious difference in protein expression relative to the wild-type parent strain ( Fig. 3 and Table 3 ). To more thoroughly and quantitatively investigate the differences in protein expression in the wild-type and mutant strains we performed a proteome analysis using iTRAQ methodology [39], [40]. We identified 370 B. burgdorferi proteins, of which 187 showed significant changes in expression compared to the wild-type parent: 90 were downregulated (Table S3) relative to wild-type and 97 were upregulated (Table S4). When compared to the changes observed with mutants in other regulatory systems, 42 of the 90 downregulated proteins (Table S3) and 65 of the 97 upregulated proteins (Table S4) are not regulated under any conditions by the previously reported B. burgdorferi regulators bosR, rrp2, rpoN, rpoS or rrp1 [7], [12], [15], [21], [23], [24].
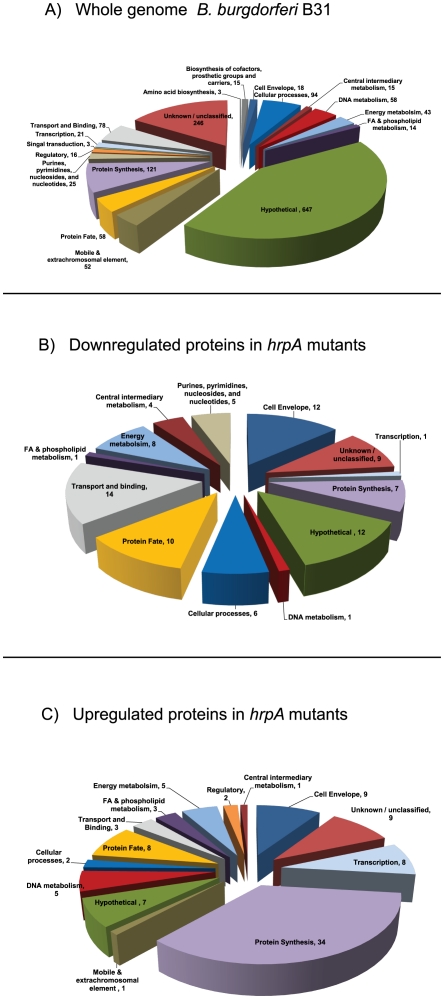
Functional categorization of the regulated proteins relative to the B. burgdorferi proteome ( Fig. 4 ) revealed that the most prominent downregulated categories were transport & binding, cell envelope, hypothetical proteins and protein fate. In contrast, the most prominent categories of upregulated proteins were protein synthesis, cell envelope, unclassified, transcription and protein fate, with protein synthesis representing 34% of the total. From the sum of the data discussed above we conclude that HrpA-mediated regulation is part of a new and distinct global regulatory pathway in B. burgdorferi gene expression. Since our iTRAQ analysis identified only a portion of the proteins in the total B. burgdorferi proteome, we have almost certainly identified only a subset of the HrpA-regulated B. burgdorferi proteins. Finally, it is noteworthy that HrpA itself has been reported to be slightly upregulated in an rpoN mutant [7].
Figure 4. Pie chart representations for functional categorization of B. burgdorferi B31 proteins.
A) The whole genome B.burgdorferi pie chart was created using data from The Comprehensive Microbial Resource database (http://cmr.jcvi.org/cgi-bin/CMR/shared/GetNumAndPercentGenesInARole.cgi). B) The pie chart showing proteins downregulated (total 90 proteins) in hrpA mutant clones was created using the data in Table S3. C) The pie chart showing proteins upregulated (total 97 proteins) in hrpA mutant clones was created using the data in Table S4.
Because an HrpA orthologue is present in many bacteria, its function in a global regulatory pathway in B. burgdorferi may have relevance in other bacterial species where its function remains obscure. The only known function of HrpA at this time is a single case where regulation of the expression of a fimbrial gene in E. coli has been reported [34]. Our work may, therefore, stimulate the investigation of possible global regulation of gene expression by HrpA in other bacteria. A role for an RNA helicase in global gene regulation in bacteria is both new and particularly timely with the recent growth of the field of RNA regulation of gene expression and the ability of RNA helicases to modulate RNA structure and function.
The reason why a complete loss of infectivity results from disruption of hrpA remains to be established. A notable point is that P66, which is required for infectivity, (Jenifer Coburn, personal communication) is downregulated five fold in the hrpA mutant and this may cause or contribute to the infectivity loss. In addition six of the 11 oligopeptide permeases (OppA-1, OppA-2, OppA-3, OppA-4, OppD and OppF) are all downregulated, which may compromise the ability of the spirochetes to survive in a mouse. Similarly, other transport systems were found to be downregulated in the hrpA strains (see Table S3) as were the protein chaperones GroEL and GroES. The inability to infect mice may result from a combined effect of the downregulation of these and other proteins. Alternatively, the upregulation of a variety proteins resulting from loss of hrpA may also have deleterious consequences for survival in the mouse. However, the experiments reported here have analyzed changes in protein expression during growth in culture. Therefore, we do not yet know the role of HrpA in the synthesis or stability of proteins in the tick vector or vertebrate host. Further studies will be required to assess the possibility of growing an hrpA mutant in ticks and in DMCs for such analyses. Growth of an hrpA mutant in ticks or DMCs would also aid further expression studies to be performed under different conditions to more thoroughly characterize the complete spectrum of HrpA regulated protein expression during the enzootic cycle.
In terms of the mechanisms by which HrpA might both upregulate and downregulate gene expression, it is too early to speculate. In the one case where HrpA has been shown to regulate fimbrial gene expression in E. coli, it is involved in the processing of a polycistronic mRNA [34]. HrpA may function in global regulation indirectly as a master regulator to turn on the expression of one or more activators or to turn off the expression of one or more repressors. We did not, however, uncover HrpA regulation of any of the known B. burgdorferi regulators such as BosR, Rrp2, RpoN, RpoS or Rrp1. Alternatively, HrpA might function directly in transcription or post-transcriptional events (see [30], [31], [32], [33]) for a large number of genes. The abundance of activities that can be modulated by RNA helicases, most of which are at the post-transcriptional level, leaves many possible mechanistic alternatives. Further studies on HrpA regulation in vivo as well as protein function in vitro will help to elucidate the mechanism of this fascinating protein, which may possibly hold a general role in global regulation of gene expression in bacteria.
Materials and Methods
Ethics statement
This study was carried out in accordance with the principles outlined in the most recent policies and Guide to the Care and Use of Experimental Animals by The Canadian Council on Animal Care. Our animal protocol (M08042) was approved by The Animal Care Committee of the University of Calgary.
Bacterial strains and culture conditions
E. coli DH5α was used for the construction and maintenance of the knockout plasmids. Infectious Borrelia burgdorferi B31 clone 5A4 [37] was used as the parental strain to generate the hrpA and bb0826 mutants. All B. burgdorferi clones were cultivated at 35°C (with a 1.5% CO2 environment) in BSK-II medium prepared in-house [54] and supplemented with 6% rabbit serum (Cedarlane Laboratories, Burlington, ON, Canada). To cultivate B. burgdorferi from mouse tissues, 1× Borrelia antibiotic cocktail (20 µg/ml phosphomycin, 50 µg/ml rifampicin and 2.5 µg/ml amphotericin B) was added to the culture media. Bacterial density was determined by dark-field microscopy using a Petroff-Hausser chamber.
Gene disruption in B. burgdorferi
Disruption of hrpA and bb0826 was performed as previously described [13]. Briefly, a 1.5 kb region containing the targeted gene was amplified from B. burgdorferi B31 clone 5A4 DNA [37] by PCR using target/f and target/r primers (see Table S1) and cloned into either pJET1.2/Blunt (Fermentas) for hrpA or pCR-BluntII-TOPO (Invitrogen) for bb0826. The central part of the target gene was removed by inverse PCR using outward-oriented primers containing a 5′ NheI restriction site and replaced by a gentamicin resistance cassette (aacC1) under the control of the flgB promoter. For the bb0826 knockout plasmid, the gentamicin resistance cassette was amplified from pBSV2g [55] using B415 and B416 primers containing 5′ NheI restriction site. For the disruption of the hrpA gene, the flgB promoter-driven gentamicin resistance cassette was fused to a T7 transcriptional terminator ( 5′ CTG CTA ACA AAG CCC GAA AGG AAG CTG AGT TGG CTG CTG CCA CCG CTG AGC AAT AAC TAG CA TAA CCC CTT GGG GCC TCT AAA CGG GTC TTG AGG GGT TTT TTG 3′ ) by overlap extension PCR. Resistance to gentamicin was used as the selectable marker. The orientation of the gentamicin resistance cassette relatively to the target gene was determined by PCR using either target/f or target/r primer, specific to the gene target, in combination with either B348 or B349 for the hrpA disruption or in combination with B348 or B1281 for the bb0826 knockout (See Figure 2 and Table 1 ). To generate hrpA and bb0826 gene disruptions, each knockout plasmid was used to transform infectious B. burgdorferi B31 clone 5A4 strain by electroporation. Gene disruption was confirmed by PCR and Southern blot hybridization as previously described [13], [56], [57] and as described in the Results section. Plasmid content was analyzed as previously described [13] and three isolates were chosen for further analysis. The three isolates were classified as independent clones based upon subsequent transformability and plasmid content.
RT-PCR analysis for the downstream genes of hrpA; bb0825 and bb0826
B. burgdorferi cultures were harvested by centrifugation when they reached the concentration of ∼1×108 cells/ml in 10 ml BSK-II medium prepared in-house. RNA was extracted using an Aurum Total RNA Mini Kit as per manufacturer's instructions. RNA concentrations were determined using a NanoDrop spectrophotometer and the integrity of the RNA was assessed by agarose gel electrophoresis. cDNA was generated for bb0825 and bb0826 by The RevertAid H Minus First Strand cDNA Synthesis Kit, Fermentas using gene specific primers (Table S1) according to manufacturer's instructions. For subsequent PCR reactions, 1 µl of cDNA was used as a template in 50 µl reactions with 10 pmoles of each primer. PCR reactions were run for 25, 30 and 35 cycles to ensure that saturation had not been reached.
Mouse infections
Three to four week old male C3H/HeN wild-type mice were obtained from Harlan Laboratories or Charles River Laboratories (St-Constant, QC). Mice were infected by intraperitoneal and dorsal subcutaneous injection of 100 µl containing 1×104 spirochetes/ml at each site. Infectivity and persistence of B. burgdorferi in mice was determined as previously described [13]. A 50 µl blood sample was collected from the saphenous vein seven days post-infection and cultivated as described above. At day 14 and 21, two ear punch biopsies were taken and cultivated for the presence of spirochetes. The heart, ear, bladder and joint were collected 35 days post-infection and cultivated. The presence of B. burgdorferi in culture was determined by dark-field microscopy.
Protein sample preparation
For 2D gel electrophoresis wild-type B. burgdorferi B31 clone 5A4 [37] and the hrpA mutant strains GCB1164 (hrpA2), GCB1165 (hrpA3) and GCB1166 (hrpA4) were grown to late log phase (∼1×108 cells/ml) in 100 ml BSK-II medium prepared in-house and harvested by centrifugation (8000× g, 15 min, 4°C). Cell pellets were washed twice with 50 ml of 50 mM Tris-HCl, pH 7.5 and centrifuged as above. The pellets were then suspended in lysis buffer containing 25 mM Tris-Base, 50 mM KCl, 3 mM EDTA, 3 mM, benzamidine, 2.1 µM leupeptine, 9 M Urea, 2% ampholyte 3–10, 1% TritonX-100, 70 mM DTT and protease inhibitor cocktail (according to the manufacturer's instructions, Sigma-Aldrich, Cat No. P8465). The cells were lysed by 5 cycles of freezing in liquid nitrogen and thawing at room temperature. The soluble whole cell protein extracts were collected by centrifugation at 50,000× g at room temperature for 45 minutes. The samples were then reduced by addition of tributylphosphine to 5 mM at room temperature with an incubation of one hour with occasional shaking. The samples were subsequently alkylated by addition of iodoacetamide to 15 mM with an incubation of 1.5 hours at room temperature in the dark.
For iTRAQ, samples were harvested and prepared as noted above, with some modifications. For iTRAQ, DTT was not included in the lysis buffer and the reduction and alkylation steps were excluded from the sample preparation method. Samples were then diluted with HPLC-grade H2O to a final concentration of 4 M urea and then precipitated overnight with ice-cold acetone at −20°C. Precipitated samples were collected by centrifugation at 8,000× g at 4°C for 20 minutes and the pellets were dried at room temparature for 30 minutes. Dried samples were submitted for analysis at the University of Victoria Genome BC Proteomics Center.
2D gel electrophoresis experiments and protein identification
Protein concentrations were determined using the Bradford protein assay [58] with BSA as a standard. For separation in the first dimension by isoelectric focusing (IPG gels, GE Healthcare Immobiline™ DryStrip, pH 3–10 NL, 18 cm) samples (150 µg for analytical and 250 µg for preparative gels) were applied on strips allowing an overnight rehydration in buffer containing 8 M urea, 2% (w/v) Triton X-100, 0.5% (v/v) ampholyte 3–10, 0.002% Bromophenol blue and 18 mM DTT. The strips were focused using the IPGphor system (Amersham) with a gradient voltage increase of 500 V for 1 hour, 1000 V for 8 hours, 8000 V for 3 hours and a constant 8000 V for 2.5 hours. The focused strips were stored at −80°C until the second dimensional run.
Prior to the second dimensional separation by 12% SDS PAGE, the strips were equilibrated sequentially in two different buffers. The strips were first incubated in equilibration buffer I containing 6 M urea, 75 mM Tris-HCl pH 8.8, 29% (v/v) glycerol, 2% SDS (w/v), 0.002% bromophenol blue and 64 mM DTT with a gentle shaking, for 15 minutes at room temperature. Following this, the strips were incubated in equilibration buffer II containing 6 M urea, 75 mM Tris-HCl pH 8.8, 29% (v/v) glycerol, 2% SDS (w/v), 0.002% bromophenol blue and 135 mM iodoacetamide for a further 15 minutes at room temperature with gentle shaking. Second dimensional runs were standard 12% SDS-PAGE gels (18 cm×20 cm). The runs were started at 90 V for one hour, followed by 160 V for one hour and 200 V for five hours.
Analytical 2D gels were silver stained [59], [60] (Heukeshoven method for analytical and Shevchenko method for preparative gels) and analytical gels were dried under vacuum in a gel dryer. Protein expression differences between wild-type B. burgdorferi B31 clone 5A4 and the hrpA mutant strains GCB1164, GCB1165 and GCB1166 were compared to determine differential expression using REDFIN 2D gel analysis software (Ludesi, Sweden). Protein abundance changes of 1.5 or greater for the average of the three mutant clones versus the wild-type were candidates for differentially expressed proteins and were circled and numbered in Fig. 3. Some of these spots were cut out from silver stained preparative gels for in-gel tryptic digestion and subsequent identification by LC- MS/MS at Southern Alberta Mass Spectrometry (SAMS) Centre for Proteomics, a Core Facility of the University of Calgary, Faculty of Medicine using a MASCOT (Matrix science Ltd., London United Kingdom, www.matrixscience.com) database search. For in-gel tryptic digestion, briefly, protein spots were cut out from preparative silver stained gels with a scalpel, minced into one mm3 pieces and transferred to pre-washed (60% Acetonitrile/0.1% TFA) microcentrifuge tubes. Spots were washed two times with HPLC-grade H2O for 10 minutes and then with freshly made destaining solution containing 30 mM K3Fe(CN)6 and 100 mM Na2S2O3, 2 times for eight minutes at room temperature. After destaining a second H2O wash was performed. Excessive washing and short periodic low speed vortexing were done when necessary till all the stain was removed. Following this, dehydration of the gel pieces was performed with an incubation in 100% acetonitrile for 10 minutes at room temperature. All the acetonitrile was then removed and opaque gel pieces were air-dried at room temperature and then rehydrated for 60 minutes at 4°C in 20–50 µl of trypsin working solution (50 mM ammonium bicarbonate, 10 mM CaCl2, 1% acetonitrile and 20 ng/µl trypsin, Princeton Separations, porcine, sequencing grade, modified). Samples were then incubated at 37°C overnight. The next day 1/5 volume 5% TFA was added to the samples followed by incubation at 60°C for 1 hour. Digested samples were then cleaned up using Zip Tips C18 (Millipore) and eluted with 10 µl elution solution (85% acetonitrile, 0.1% TFA). Eluted samples were stored at −20°C for further LC MS/MS analysis.
iTRAQ experimental design and analysis
The iTRAQ analysis was designed as an 8-plex experiment enabling two comparisons of wild-type B. burgdorferi to three separate hrpA mutant clones. To control for biological variation, 8 individual samples (B31-1, B31-2, 1164-1, 1164-2, 1165-1, 1165-2, 1166-1, and 1166-2) were prepared by pooling four independent cultures for each. Technical and experimental variations were controlled by having replicate samples (e.g. B31-1 and B31-2) digested separately and tagged with different isobaric tags in the 8-plex iTRAQ experiment.
Total protein extracts were submitted to the University of Victoria-Genome BC Proteomics Centre for iTRAQ analysis. Briefly, samples were reduced with TCEP (Tris[2-carboxyethyl] phosphine) and alkylated with MMTS (s-methyl thiomethanesulfonate). Proteins were then digested in solution with trypsin (Promega) and labeled with appropriate iTRAQ labels. Isobaric labels assigned to individual samples were as follows; B31-1/113, 1164-1/114, 1165-1/115, 1166-1/116, B31-2/117, 1164-2/118, 1165-2/119, 1166-2/121. The labeled peptides were combined and separated by strong cation exchange HPLC. Fractions were then analyzed by LC-MS/MS.
LC-MS/MS
LC-MS/MS analysis was performed using an integrated Famos autosampler, Switchos II switching pump, and UltiMate micro pump (LC Packings, Amsterdam) system with an Hybrid Quadrupole-TOF LC/MS/MS Mass Spectrometer (QStar Pulsar i) equipped with a nano-electrospray ionization source (Proxeon, Odense, Denmark) and fitted with a 10 µm fused-silica emitter tip (New Objective, Woburn, MA). Chromatographic separation was achieved on a 75 µm×15 cm C18 PepMap Nano LC column and a 300 µm×5 mm C18 PepMap guard column was in place before switching inline with the analytical column and the MS. The mobile phase (solvent A) consisted of water/acetonitrile (98∶2 (v/v)) with 0.05% formic acid for sample injection and equilibration on the guard column at a flow rate of 100 µl/min. A linear gradient was created by mixing solvent A with solvent B that consisted of acetonitrile/water (98∶2 (v/v)) with 0.05% formic acid at a reduced flow rate of 200 nl/min for high resolution chromatography and introduction to mass spectrometer.
Samples were brought up to 100 µl with 5% acetonitrile and 3% formic acid and transferred to autosampler vials (LC Packings, Amsterdam). 20 µl of sample were injected in 95% solvent A and allowed to equilibrate on the trapping column for 10 mins. Upon switching inline with the MS, a linear gradient from 95% to 40% solvent A was developed for 100 minutes and in the following 5 minutes the composition of the mobile phase was decreased to 20% solvent A before increasing to 95% solvent A for a 15 minute column re-equilibration prior to the next sample injection.
MS data was acquired automatically using Analyst QS 1.0 software Service Pack 8 (ABI MSD SCIEX, Concord, Canada). An information dependent acquisition method consisting of a 1 second TOFMS survey scan of mass range 400–1200 amu and two 2.5 second product ion scans of mass range 100–1500 amu. The two most intense peaks over 20 counts, with charge state 2–5 were selected for fragmentation and a 6 amu window was used to prevent the peaks from the same isotopic cluster from being fragmented again. Once an ion was selected for MS/MS fragmentation it was put on an exclude list for 180 seconds. Curtain gas was set at 23, nitrogen was used as the collision gas and the ionization tip voltage used was 2700 V.
All data files were searched for protein identification and relative abundance using Protein Pilot (V.3.0) and were searched against a custom B. burgdorferi B31 database. A positive protein identification required at least two matching peptide sequences with a minimum of a 95% confidence limit in ProtScore.
iTRAQ ratios were expressed as mutant/wild-type, using wild-type B31-1 and B31-2 samples (isobaric labels 113 and 117, respectively) as denominators in Protein Pilot software. Finally, the 12 iTRAQ ratios obtained (see Table S2) were used for subsequent data analysis. After transferring all data sets from Protein Pilot to Microsoft Excel, iTRAQ values were ranked according to the mean values calculated from 12 individual ratios. P values were calculated based upon the means of the 12 input ratios using a two-sided, one sample t-test with comparison against a theoretical value of 1.0 using GraphPad Instat (V.3.10). iTRAQ ratios ≥1.2 or ≤0.8 were considered as being differentially expressed if the P values were ≤0.05. Downregulated and upregulated proteins were grouped separately in Table S3 and Table S4, respectively. The total list of iTRAQ-identified proteins is shown in Table S2.
Supporting Information
Gene disruption and the absence of additional copies of the hrpA gene was confirmed by Southern hybridization. Genomic DNA was digested with HindIII and run on a 1.0% agarose gel with a 1 kb molecular weight ladder (M). Probes complementary to the gentamicin (gent) resistance cassette (left panel) and the portion of the hrpA gene deleted during gene disruption (right panel) were used for hybridization to duplicate blots. As expected, hybridization to the gent probe was not observed in the wild-type strain but was observed at the expected size (7.1 kb) for the three hrpA mutant strains. Conversely, hybridization to the deleted portion of hrpA was observed in the expected 6.5 kb fragment in the wild-type strain but not in the three hrpA mutant clones.
(EPS)
Region view and transcription patterns of hrpA , bb0825 and bb0826 . A) Schematic representation of hrpA and the two downstream genes on the B. burgdorferi chromosome. Arrows represent the direction of transcription and the numbers denote the coordinates on the chromosome. B) Ethidium bromide stained 1.4% agarose gel showing the products of RT-PCR reactions to assess the transcription patterns of genes bb0825 and bb0826 in the three hrpA mutant clones, along with a 100 bp molecular weight ladder (M). Panel 1) RT-PCR reactions for bb0825 in the wild-type parent strain (B. burgdorferi B31, clone 5A4) and in the mutants hrpA2, 3 and 4. The expected product size was 210 bp. Panel 2) RT-PCR reactions for bb0826 in the strains described in Panel 1. The expected product was 310 bp.
(EPS)
Primers used in this study.
(PDF)
Complete listing of iTRAQ results.
(PDF)
Downregulated B. burgdorferi proteins in hrpA mutant clones compared to wild-type based upon iTRAQ analysis.
(PDF)
Upregulated B. burgdorferi proteins in hrpA mutant clones compared to wild-type, based upon iTRAQ analysis.
(PDF)
Acknowledgments
We would like to thank Genevieve Chaconas for technical support, Morgan Khan (University of Calgary) for help with mass spectrometry, Derek Smith (University of Victoria) for help with the iTRAQ analysis and Tara Moriarty for helpful comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grant MOP 53086 from the Canadian Institutes of Health Research (http://www.cihr-irsc.gc.ca/e/193.html) to GC, who also holds a Canada Research Chair in the Molecular Biology of Lyme Borreliosis (http://www.chairs-chaires.gc.ca/home-accueil-eng.aspx) and a Scientist Award from the Alberta Heritage Foundation for Medical Research (http://www.chairs-chaires.gc.ca/home-accueil-eng.aspx). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stanek G, Fingerle V, Hunfeld KP, Jaulhac B, Kaiser R, et al. Lyme borreliosis: Clinical case definitions for diagnosis and management in Europe. Clin Microbiol Infect. 2010 doi: 10.1111/j.1469-0691.2010.03175.x. doi: 10.1111/j.1469-0691.2010.03175.x. [DOI] [PubMed] [Google Scholar]
- 2.Wormser GP, Dattwyler RJ, Shapiro ED, Halperin JJ, Steere AC, et al. The clinical assessment, treatment, and prevention of lyme disease, human granulocytic anaplasmosis, and babesiosis: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis. 2006;43:1089–1134. doi: 10.1086/508667. [DOI] [PubMed] [Google Scholar]
- 3.Skare JT, Carroll JA, X.F.Y, Samuels DS, Akins DR. Gene regulation, transcriptomics and proteomics. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Norfolk, UK: Caister Academic Press; 2010. pp. 67–101. [Google Scholar]
- 4.Pal U, Fikrig E. Tick interactions. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Norfolk, UK: Caister Academic Press; 2010. pp. 279–298. [Google Scholar]
- 5.Samuels DS, Radolf JD. Who is the BosR around here anyway? Mol Microbiol. 2009;74:1295–1299. doi: 10.1111/j.1365-2958.2009.06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, et al. Borrelia burgdorferi sigma54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci U S A. 2005;102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narasimhan S, Santiago F, Koski RA, Brei B, Anderson JF, et al. Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J Bacteriol. 2002;184:3122–3125. doi: 10.1128/JB.184.11.3122-3125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, et al. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci U S A. 2002;99:1562–1567. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokarz R, Anderton JM, Katona LI, Benach JL. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect Immun. 2004;72:5419–5432. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, et al. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dresser AR, Hardy P-O, Chaconas G. Investigation of the role of DNA replication, recombination and repair genes in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase. PLoS Pathogens. 2009;5:e1000680. doi: 10.1371/journal.ppat.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyde JA, Shaw DK, Smith R, 3rd, Trzeciakowski JP, Skare JT. The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both the oxidative stress response and pathogenic properties of the Lyme disease spirochete. Mol Microbiol. 2009;74:1344–1355. doi: 10.1111/j.1365-2958.2009.06951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, et al. BosR (bb0647) governs virulence expression in Borrelia burgdorferi. Mol Microbiol. 2009;74:1331–1343. doi: 10.1111/j.1365-2958.2009.06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouyang Z, Deka RK, Norgard MV. BosR (bb0647) Controls the RpoN-RpoS Regulatory Pathway and Virulence Expression in Borrelia burgdorferi by a Novel DNA-Binding Mechanism. PLoS Pathog. 2011;7:e1001272. doi: 10.1371/journal.ppat.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medrano MS, Ding Y, Wang XG, Lu P, Coburn J, et al. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J Bacteriol. 2007;189:2653–2659. doi: 10.1128/JB.01760-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyde JA, Shaw DK, Smith R, 3rd, Trzeciakowski JP, Skare JT. Characterization of a conditional bosR mutant in Borrelia burgdorferi. Infect Immun. 2010;78:265–274. doi: 10.1128/IAI.01018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elias AF, Bono JL, Carroll JA, Stewart P, Tilly K, et al. Altered stationary-phase response in a Borrelia burgdorferi rpoS mutant. J Bacteriol. 2000;182:2909–2918. doi: 10.1128/jb.182.10.2909-2918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, et al. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouyang Z, Blevins JS, Norgard MV. Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology. 2008;154:2641–2658. doi: 10.1099/mic.0.2008/019992-0. [DOI] [PubMed] [Google Scholar]
- 22.Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2003;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boardman BK, He M, Ouyang Z, Xu H, Pang X, et al. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun. 2008;76:3844–3853. doi: 10.1128/IAI.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers EA, Terekhova D, Zhang HM, Hovis KM, Schwartz I, et al. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol Microbiol. 2009;71:1551–1573. doi: 10.1111/j.1365-2958.2009.06621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medrano MS, Policastro PF, Schwan TG, Coburn J. Interaction of Borrelia burgdorferi Hbb with the p66 promoter. Nucleic Acids Res. 2010;38:414–427. doi: 10.1093/nar/gkp1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alverson J, Bundle SF, Sohaskey CD, Lybecker MC, Samuels DS. Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi. Mol Microbiol. 2003;48:1665–1677. doi: 10.1046/j.1365-2958.2003.03537.x. [DOI] [PubMed] [Google Scholar]
- 27.Beaurepaire C, Chaconas G. Topology-dependent transcription in linear and circular plasmids of the segmented genome of Borrelia burgdorferi. Mol Microbiol. 2007;63:443–453. doi: 10.1111/j.1365-2958.2006.05533.x. [DOI] [PubMed] [Google Scholar]
- 28.Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- 29.Lybecker MC, Abel CA, Feig AL, Samuels DS. Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2010;78:622–635. doi: 10.1111/j.1365-2958.2010.07374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2010;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 32.Linder P. Dead-box proteins: a family affair: active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 34.Koo JT, Choe J, Moseley SL. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol Microbiol. 2004;52:1813–1826. doi: 10.1111/j.1365-2958.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- 35.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 36.Rosa PA, Cabello F, Samuels DS. Genetic manipulation of B. burgdorferi. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Norfolk, UK: Caister Academic Press; 2010. pp. 189–219. [Google Scholar]
- 37.Purser JE, Norris SJ. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2000;97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart PE, Thalken R, Bono JL, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol. 2001;39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- 39.Brewis IA, Brennan P. Proteomics technologies for the global identification and quantification of proteins. Adv Protein Chem Struct Biol. 2010;80:1–44. doi: 10.1016/B978-0-12-381264-3.00001-1. [DOI] [PubMed] [Google Scholar]
- 40.Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct Genomic Proteomic. 2006;5:112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- 41.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caron MP, Lafontaine DA, Masse E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 2010;7:140–144. doi: 10.4161/rna.7.2.11056. [DOI] [PubMed] [Google Scholar]
- 43.Frohlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Jost D, Nowojewski A, Levine E. Small RNA biology is systems biology. BMB Rep. 2011;44:11–21. doi: 10.5483/BMBRep.2011.44.1.11. [DOI] [PubMed] [Google Scholar]
- 45.Lioliou E, Romilly C, Romby P, Fechter P. RNA-mediated regulation in bacteria: from natural to artificial systems. N Biotechnol. 2010;27:222–235. doi: 10.1016/j.nbt.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Liu JM, Camilli A. A broadening world of bacterial small RNAs. Curr Opin Microbiol. 2010;13:18–23. doi: 10.1016/j.mib.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, et al. RNAs: regulators of bacterial virulence. Nat Rev Microbiol. 2010;8:857–866. doi: 10.1038/nrmicro2457. [DOI] [PubMed] [Google Scholar]
- 49.Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8:116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 50.Jagessar KL, Jain C. Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. Rna. 2010;16:1386–1392. doi: 10.1261/rna.2015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jain C. Overexpression and purification of tagged Escherichia coli proteins using a chromosomal knock-in strategy. Protein Expr Purif. 2006;46:294–298. doi: 10.1016/j.pep.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 52.Lin T, Gao L, Edmondson DG, Jacobs MB, Philipp MT, et al. Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi. PLoS Pathogens. 2009;12:e1000679. doi: 10.1371/journal.ppat.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norris SJ, Coburn J, Leong JM, Hu LT, Hook M. Pathobiology of Lyme disease Borrelia. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Norwich, UK: Horizon Scientific Press; 2010. pp. 293–325. [Google Scholar]
- 54.Barbour AG. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 55.Elias AF, Bono JL, Kupko JJ, 3rd, Stewart PE, Krum JG, et al. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol. 2003;6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- 56.Margolis N, Samuels DS. Proteins binding to the promoter region of the operon encoding the major outer surface proteins OspA and OspB of Borrelia burgdorferi. Mol Biol Rep. 1995;21:159–164. doi: 10.1007/BF00997238. [DOI] [PubMed] [Google Scholar]
- 57.Bono JL, Elias AF, Kupko JJ, 3rd, Stevenson B, Tilly K, et al. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 59.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 60.Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988;9:28–32. doi: 10.1002/elps.1150090106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene disruption and the absence of additional copies of the hrpA gene was confirmed by Southern hybridization. Genomic DNA was digested with HindIII and run on a 1.0% agarose gel with a 1 kb molecular weight ladder (M). Probes complementary to the gentamicin (gent) resistance cassette (left panel) and the portion of the hrpA gene deleted during gene disruption (right panel) were used for hybridization to duplicate blots. As expected, hybridization to the gent probe was not observed in the wild-type strain but was observed at the expected size (7.1 kb) for the three hrpA mutant strains. Conversely, hybridization to the deleted portion of hrpA was observed in the expected 6.5 kb fragment in the wild-type strain but not in the three hrpA mutant clones.
(EPS)
Region view and transcription patterns of hrpA , bb0825 and bb0826 . A) Schematic representation of hrpA and the two downstream genes on the B. burgdorferi chromosome. Arrows represent the direction of transcription and the numbers denote the coordinates on the chromosome. B) Ethidium bromide stained 1.4% agarose gel showing the products of RT-PCR reactions to assess the transcription patterns of genes bb0825 and bb0826 in the three hrpA mutant clones, along with a 100 bp molecular weight ladder (M). Panel 1) RT-PCR reactions for bb0825 in the wild-type parent strain (B. burgdorferi B31, clone 5A4) and in the mutants hrpA2, 3 and 4. The expected product size was 210 bp. Panel 2) RT-PCR reactions for bb0826 in the strains described in Panel 1. The expected product was 310 bp.
(EPS)
Primers used in this study.
(PDF)
Complete listing of iTRAQ results.
(PDF)
Downregulated B. burgdorferi proteins in hrpA mutant clones compared to wild-type based upon iTRAQ analysis.
(PDF)
Upregulated B. burgdorferi proteins in hrpA mutant clones compared to wild-type, based upon iTRAQ analysis.
(PDF)